Abstract
To understand the mechanisms leading to trastuzumab resistance in HER2-overexpressing breast tumors we created trastuzumab insensitive cell lines (SKBR3/100-8 and BT474/100-2). The cell lines maintain HER2 receptor overexpression, and show increase in EGFR. Upon trastuzumab treatment, SKBR3/100-8 and BT474/100-2 cell lines displayed increased growth rate and invasiveness. The trastuzumab resistance in SKBR3/100-8 and BT474/100-2 was accompanied with activation of the Wnt/β-catenin signaling pathway. Further investigation found that Wnt3 overexpression played a key role toward the development of trastuzumab resistance. The expression of Wnt3 in trastuzumab resistant cells increased nuclear expression of β-catenin and transactivated expression of EGFR. The increased Wnt3 in the trastuzumab resistant cells also promoted a parental EMT-like transition (epithelial to mesenchymal transition), increased N-cadherin, Twist, SLUG and decreased E-cadherin. Knockdown of Wnt3 by siRNA restored cytoplasmic expression of β-catenin, and decreased EGFR expression in trastuzumab resistant cells. Furthermore the EMT markers were decreased, E-cadherin was increased and the cell invasiveness was inhibited in response to the Wnt3 down-regulation. Conversely, SKBR3 cells which had been stably transfected with full-length Wnt3 exhibited EMT-like transition. The Wnt3 transfectants, SKBR3/Wnt3-7 and SKBR3/Wnt3-9, showed a significant decrease in E-cadherin and increase in N-cadherin, Twist and SLUG. The cells were less sensitive to trastuzumab compared to parental SKBR3 and vector transfected cells. In summary, our data suggests that Wnt3 overexpression activates Wnt/β-catenin signaling pathway that leads to transactivation of EGFR and promotes EMT-like transition. This could be an important mechanism leading to trastuzumab resistance in HER2 overexpressing breast cancer cells.
Keywords: Wnt3, β-catenin, EMT, trastuzumab, Breast Cancer
Introduction
Trastuzumab treatment has improved the overall survival rate of patients with HER2-overexpressing breast cancer. However, some patients with HER2-overexpressing breast cancer do not respond to trastuzumab therapy, as a single agent or in combination with chemotherapy, and the mechanisms underlying the resistance phenotype are not well understood. Some studies have suggested increase in HER2 receptor homo- and hetero-dimerization with other receptors of the ErbB family, such as epidermal growth factor receptor (EGFR), HER3, and HER4 (1, 2). These interactions activate intracellular signaling via the mitogen-activated protein kinase (MAPK), or phosphatidylinositol 3-kinase (PI3K) pathways (3, 4). We have previously shown that activation of PI3K/Akt pathway inhibited the transcription factor FOXO1A, resulting in nuclear export of p27kip1 and reduced the inhibitory properties of trastuzumab (5). Breast cancer patients with HER2-overexpressing tumors have increased active Akt (pAkt) in their tumors (6). The activation of PI3K/Akt and loss of PTEN may also result in accumulation of β-catenin, which suggests a crosstalk between the PI3K and Wnt signaling pathways (7-11).
The goal of this study is to understand the mechanisms leading to trastuzumab resistance in HER2 overexpressing breast tumors and which pathway specific genes may contribute to the resistance.
Materials and Methods
Cell lines and cell cultures
The human breast cancer cell lines SKBR3 (ATCC: HTB-30) and BT474 (ATCC: HTB-20) were obtained from the American Type Culture Collection. Unless otherwise stated, monolayer cultures of SKBR3 and BT474 cells were maintained in DMEM/F12 medium with 10% fetal bovine serum. The cell lines overexpressed the HER2/c-erb-2 (HER2) gene product. The trastuzumab resistant clones, SKBR3/100-8 and BT474/100-2 were generated from SKBR3 and BT474 cells respectively. In order to select trastuzumab resistant clones, SKBR3 and BT474 cells were plated in 24 well plates at low density and maintained in growth medium containing 10μg/ml, 50μg/ml and 100μg/ml of trastuzumab. The SKBR3/100-8 and BT474/100-2 clones were maintained in growth medium containing 100μg/ml of trastuzumab for over 2 years and 1 year, respectively. Both SKBR3/100-8 and BT474/100-2 were repeatedly confirmed as insensitive to trastuzumab. The SKBR3/Wnt3-7 and SKBR3/Wnt3-9 were generated by stable transfection of full length Wnt genes into SKBR3 cells, as well as clonal selection.
Microarray analysis
Total RNA was isolated from SKBR3 and SKBR3/100-8 cultured cells by using RNeasy micro kit (#74004, QIAGEN). The quality of RNA was determined by separation of the RNA via capillary electrophoresis using the Agilent 2100 Bioanalyzer. Whole Human Genome 4X44K (Cat#: G4112F, Agilent) expression array was used to compare the gene profiles between SKBR3 and SKBR3/100-8. Microarray slides were read using Agilent Scanner. Agilent Feature Extraction software version 9.13 was used to calculate the gene expression values. Differences of p<0.01 and ≥2-fold expression were considered as significant. The Wnt/β-catenin pathway regulated gene profiles in the cell lines were examined using Human Wnt/β-catenin Regulated cDNA plate array (Cat# AP-0171, Signosis; Sunnyvale, CA) according to manufacturer’s instructions.
siRNA knock-down genes
Wnt3 siRNA, a pool of 3 target-specific 19-25 nt siRNA (sc-41106, Santa Cruz Biotechnology), was used to knock-down the Wnt3 gene. The EGFR siRNA (sc-29301, Santa Cruz Biotechnology) targeting specific 20-25 nt siRNA and the ErbB-3 siRNA (sc-35327, Santa Cruz Biotechnology) targeting specific 19-25 nt siRNA were used to knock-down EGFR and HER3 gene expression, respectively.
siRNA-A negative sequence (sc-37007, Santa Cruz Biotechnology) was used in parallel for each knock-down experiment served as control. Lipofectamine™ 2000 transfection reagent (Cat#: 11668-019, Invitrogene) was used for transfection following the manufacturer’s instructions. Gene expression after siRNA knockdown was determined by PCR or quantitative reverse transcription-PCR (RT-Q-PCR), with specific primers (supplemental Table 1).
Overexpressing Wnt3 gene
Overexpressing Wnt3 was done by stable transfection of a full length Wnt3 gene (RG21115, Origene, Rockville, MD) into the cell; the empty vector (PS100010, Origene, Rockville, MD) was also transfected into SKBR3 cells as a control. LipofectamineTM PLUS reagent (Invitrogen) was used for transfection following the manufacturer’s instruction. The single clones from the Wnt3 transfected cells were selected by 400μg/ml G418 and confirmed by RT-Q-PCR and Western blot analysis.
Boyden Chamber Invasion assay
The invasive assay was done in 24-well cell culture chambers using inserts with 8-μm pore membranes precoated with Matrigel (28μg/insert; Sigma, Saint Louis, MO). Cell suspensions (2 × 105/mL) were placed in the upper wells and fibroblast-conditioned medium was filled in the lower wells. The cells were cultured for 24 hrs and then fixed by 0.5 ml of 0.5% Glutaraldehyde in 1×PBS and stained by 0.5 ml of 0.5% Toluidine Blue. The numbers of invaded cells were counted with 20X objective of microscope from 3 files per membrane and then normalized with total numbers of cells. Each experiment was performed twice and each condition was duplicated at each time.
Chromatin immunoprecipitation-Real-time PCR assay (ChIP-qPCR)
The chromatin-protein complex was prepared from SKBR3 and SKBR3/100-8 cells by immunoprecipitation of the chromatin with β-catenin antibody using Magna-ChIP assay kit (Cat# 17-10085, Millipore) following the manufacture’s instruction and then qPCR was performed. The primer sequences were designed to cover the LEF/TCF binding region of the promoter of EGFR (711-727): Lt: 5′-GCCTGGTCCCTCCTCCTC-3′ and Rt: 5′-GCTCTCCCGATCAATACTGG-3′. Fold enrichment in the β-catenin precipitated samples was calculated relative to the mock samples (precipitated with normal IgG). The data was also calculated as % input to ensure consistent results.
Immunoblotting analysis
The NE-PER Nuclear and Cytoplasmic Extraction reagents (Cat#: 78833, Thermo Scientific) were used to extract the nuclear and cytoplasmic protein following the manufacturer’s instructions. Immunoblotting analysis was performed with antibodies specific to HER2, E-cadherin, β-catenin, and Histone from cell signaling, EGFR, Wnt3, MMP-7, VEGF, α-Tubulin and β-actin from Santa Cruz Biotechnology.
Statistical analysis
The statistical significances of mean values among different cell lines were determined by one-way ANOVA first, then by Student t test, and the fold changes were analyzed by χ2 test. P-value < 0.05 was considered statistically significant.
Results
Characterization of trastuzumab resistant cell lines, SKBR3/100-8 and BT474/100-2
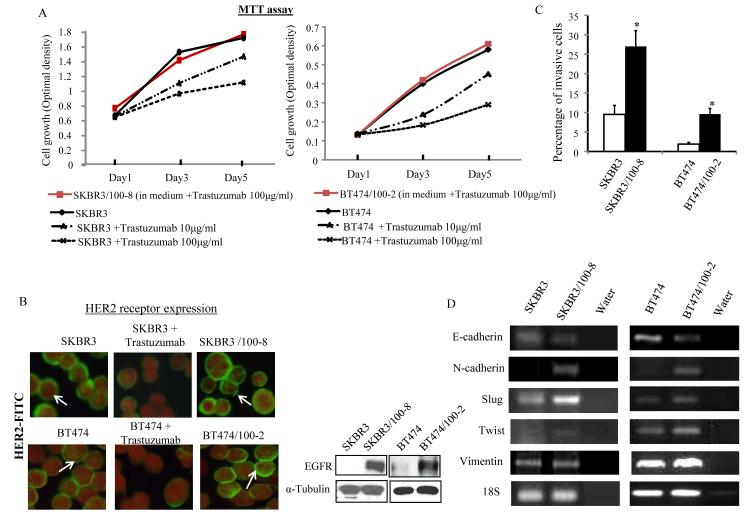
Data from MTT assay showed that the growth of SKBR3 and BT474 parental cells was significantly inhibited by fresh treatment with 10μg/ml and 100μg/ml of trastuzumab, while the growth of SKBR3/100-8 and BT474/100-2 cells which were maintained in media containing trastuzumab 100μg/ml were not inhibited in 5 days (Figure 1A). The number of cells overexpressing HER2 receptors was reduced in SKBR3 and BT474 cells upon fresh treatment with trastuzumab (10μg/ml) for 72 hrs. However, SKBR3/100-8 and BT474/100-2 cells maintained in growth media containing 100μg/ml trastuzumab for a long period of time, did not exhibit any significant difference in the overall number of HER2 receptor overexpression compared to the untreated parental cells (Figure 1B left). There was no change in phosphorylated HER2 receptors between the trastuzumab resistant and sensitive cells (supplemental Figure 1). Both SKBR3/100-8 and BT474/100-2 showed increased EGFR compared to parental cell lines (Figure 1B right). The HER2/HER3 dimerization was increased in SKBR3/100-8 (supplemental Figure 1), but no changes were found in BT474/100-2 (data not shown). Both SKBR3/100-8 and BT474/100-2 cells displayed significant enhanced invasive capacity (Figure 1C).
Figure 1. Characterization of trastuzumab resistant clones.
A, SKBR3/100-8 and BT474/100-2 were maintained in growth medium containing 100 μg/ml of trastuzumab. SKBR3 and BT474 were treated with trastuzumab at 0, 10μg/ml and 100μg/ml and MTT assay was performed at indicated days. B. SKBR3 and BT474 were treated with or without trastuzumab (10μg/ml) for 3 days. HER2 receptors were assessed by immunofluorescence analysis with FITC-labeled anti-HER2 antibody (green) and the cell nuclei were labeled by propidium iodide (red). The arrows indicate positive HER2 staining (right). The EGFR protein expression in indicated cell lines were measured by Western blot analysis and α-Tubulin was used as loading control (left). C, The cell invasiveness in the indicated cell lines were measured by Boyden Chamber Invasion assay as described in Method. The invasive cells (mean+SD from 5 different areas) were counted. *p<0.05 compared to untreated cells. D, mRNA levels of the indicated genes in SKBR3, SKBR3/100-8, BT474, and BT474/100-2 were analyzed by RT-PCR and “water” was used as vehicle control.
Compared to parental SKBR3 and BT474 cells, the SKBR3/100-8 and BT474/100-2 cells had decreased E-cadherin and increased mesenchymal markers, N-cadherin, SLUG, and Twist (Figure 1D). Vimentin expression increased in BT474/100-2 compared to BT474, but there was no difference between SKBR3/100-8 and SKBR3 cells (Figure 1D).
Deregulated Wnt pathway genes in the resistant clone, SKBR3/100-8 analyzed by Agilent chip microarray
Using the Agilent chip array (4X44K), we have identified over 3,000 genes. Many of these genes either increased or decreased in expression by 2 or more fold (p<0.01) in SKBR3/100-8 compared to parental cells, SKBR3.
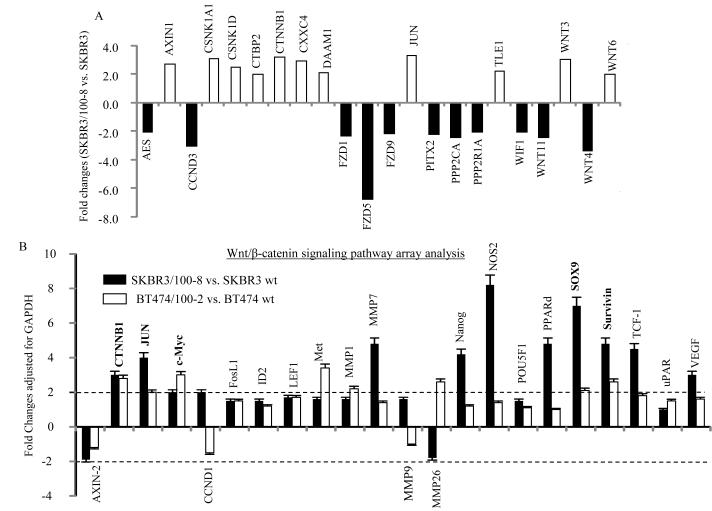
The differentially expressed molecules included 42 transcriptional factors (TFs) and 14 TFs were analyzed by ingenuity pathway analysis (IPA) as either activated (regulation z-score≥2) or inhibited (regulation z-scorein≤2) in SKBR3/100-8 compared to SKBR3 based on regulation z-score (Data not shown). Each TF is networking with a number of molecules (between 5 to 319 molecules). The differentially expressed genes in the profile were associated with over 200 signaling pathways and composed of 25 networks. Supplemental Figure 2 demonstrates breast cancer related signaling pathways that were regulated in the profile revealed by IPA. Approximately 95% of Wnt/β-catenin signaling genes were regulated (supplemental Figure 2). Compared to SKBR3 cells, 22 genes in the Wnt pathway were significantly deregulated in SKBR3/100-8. Eleven genes were up-regulated by 2 or more fold and 11 genes were down-regulated by 2 or more fold in SKBR3/100-8 cells compared to SKBR3 cells (Figure 2A). There were 19 Wnt ligands and 10 FZD receptors in the Agilent chip array. Among the 19 Wnt ligands, Wnt3 and Wnt6 were up-regulated by 3 and 2.1 fold (p<0.001) respectively, while Wnt4 and Wnt11 were down-regulated significantly in SKBR3/100-8 cells compared to SKBR3 cells (Table 1). Out of the 10 FZD receptors, FZD5, FZD1 and FZD9 were significantly down-regulated more than 2 fold in SKBR3/100-8 cells (Table 1). Except Wnt6 and FZD1 other Wnt ligands and FZDs were all confirmed having been regulated significantly in SKBR3/100-8 cells by RT-Q-PCR (supplemental Figure 3A). The Wnt3 and Wnt6 were also significantly up-regulated in BT474/100-2 compared to BT474 (supplemental Figure 3B).
Figure 2. Activation of Wnt signaling pathway in trastuzumab resistant cell lines.
A, Bar graph representing significantly up-regulated (open bars) or down-regulated (darkc bars) mRNA of Wnt pathway genes in SKBR3/100-8 compared to SKBR3 analyzed by microarray analysis. B, Bar graph of regulated Wnt/β-catenin pathway signaling in SKBR3/100-8 (dark bars) and BT474/100-2 (open bars) compared to SKBR3 and BT474 analyzed by specific Wnt/β-catenin mRNA array. The relative level of each indicated gene was adjusted with GAPDH and each bar indicates mean fold change and SD from four determinations.
Table 1.
Wnt ligands and Frizzed receptors expression in SKBR3/100-8 vs. SKBR3 analyzed by gene array analysis
| Name of genes | SKBR3/100-8 vs. SKBR3 (Fold changes) |
p-Value |
|---|---|---|
| Wnt1 | 1.1 | 0.5 |
| Wnt2 | 1.0 | 0.8 |
| Wnt2B | 1.1 | 0.3 |
| Wnt3 | 3.0 | <0.001 |
| Wnt3A | 1.0 | 0.8 |
| Wnt4 | −2.8 | <0.001 |
| Wnt5A | 1.7 | <0.001 |
| Wnt5B | −1.3 | 0.03 |
| Wnt6 | 2.1 | <0.001 |
| Wnt7A | −1.2 | 0.1 |
| Wnt7B | −1.5 | 0.005 |
| Wnt8A | −1.1 | 0.4 |
| Wnt8B | −1.0 | 0.7 |
| Wnt9A | −1.5 | 0.005 |
| Wnt9B | −1.0 | 0.7 |
| Wnt10A | −1.2 | 0.1 |
| Wnt10B | −1.0 | 0.6 |
| Wnt11 | −2.4 | <0.001 |
| Wnt16 | 1.1 | 0.4 |
| FZD1 | −2.3 | <0.001 |
| FZD2 | −1.4 | 0.03 |
| FZD3 | 1.1 | 0.5 |
| FZD4 | −1.9 | <0.001 |
| FZD5 | −6.7 | <0.001 |
| FZD6 | −1.0 | 0.8 |
| FZD7 | −1.2 | 0.2 |
| FZD8 | −1.0 | 0.8 |
| FZD9 | −2.2 | <0.001 |
| FZD10 | −1.0 | 0.8 |
Activation of Wnt/β-catenin signaling pathway promotes EMT phenotype and transcriptionally regulates EGFR in trastuzumab resistant cell lines
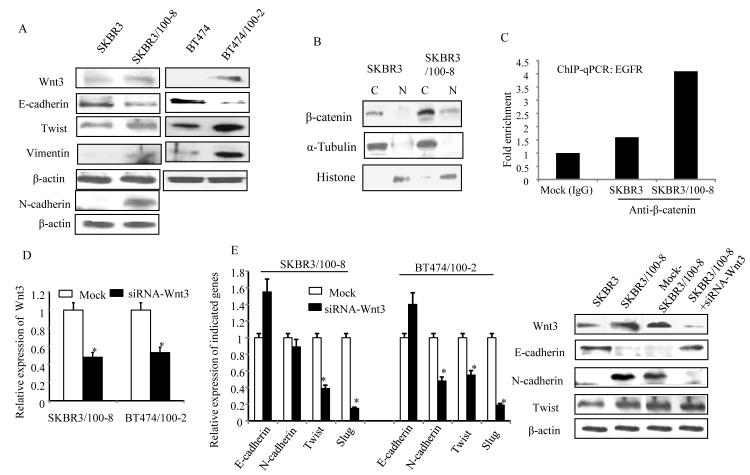
To further verify the activation of the Wnt/β-catenin signaling pathway in trastuzumab resistant cells a human Wnt/β-catenin regulated cDNA plate array was used. The array consisted of 22 Wnt/β-catenin pathway regulated genes; most genes in the array were up-regulated in the resistant clones, SKBR3/100-8 and BT474/100-2 cells (Figure 2B). Twelve genes were up-regulated >2 fold in SKBR3/100-8 compared to SKBR3, and eight genes were up-regulated ≥2 fold in BT474/100-2 compared to BT474. The β-catenin was up-regulated ~3 fold in both SKBR3/100-8 and BT474/100-2 clones. The c-Jun, c-Myc, Sox9 and survivin were also up-regulated ≥2 fold in both SKBR3/100-8 and BT474/100-2. The transcription factors, Nanog and TCF-1 were up-regulated 4 to 5 fold; in SKBR3/100-8 compared to SKBR3 (Figure 2B). The matrix metalloprotease family gene, MMP7 was up-regulated 5 fold in SKBR3/100-8 cells, and MMP26 was up-regulated 2.8 fold in BT474/100-2 cells. The VEGF (Vascular endothelial growth factor), NOS2, CCND1 and PPARd were also increased more than 2 fold in SKBR3/100-8 cells. Significant increases in the expression levels of several interesting genes in SKBR3/100-8 cells, including β-catenin, TCF-1, Nanog, c-Jun, c-Myc, PPARD, and Survivin, were further confirmed by RT-Q-PCR (supplemental Figure 4A). The protein levels of β-catenin, MMP7, and VEGF were confirmed and all increased in SKBR3/100-8 cells (supplemental Figure 4B). The up-regulation of Wnt3 in trastuzumab resistant cell lines, SKBR3/100-8 and BT474/100-2 was also verified by western blot analysis (Figure 3A). Figure 3A showed that protein level of Ecadherin was decreased and Twist was increased in both SKBR3/100-8 and BT474/100-2 cells. Even though the mRNA level of Vimentin was similar between SKBR3 and SKBR3/100-8 (Figure 1D), however, we found increased protein level of Vimentin in SKBR3/100-8 compared to SKBR3. The SKBR3/100-8 cells also had increased N-cadherin protein expression (Figure 3). The protein level of N-cadherin was undetectable in BT474 and BT474/100-2 cells. These results confirmed the PCR data of decreased E-cadherin and increased EMT markers in the trastuzumab resistant cell lines, as shown in Figure 1D. The decrease in E-cadherin and increase in Ncadherin were generally seen across most of SKBR3/100-8 cells, compared to SKBR3 (supplemental Figure 5A and 5B). The results implied that activation of Wnt signaling might promote EMT or partial EMT-like transition in the trastuzumab resistant cells. We further examined the localization of β-catenin protein expression in SKBR3 and SKBR3/100-8 cells. Data in Figure 3B showed that β-catenin was mainly present in the cytoplasm of SKBR3 cells. However, β-catenin was localized in both cytoplasm and nucleus of SKBR3/100-8 cells. Considering there may be heterogeneity within the cell line, immunofluorescence analysis was performed to examine the β-catenin expression and localization in SKBR3/100-8 and SKBR3 cells. The data in Supplemental Figure 5C confirmed the cell membrane and cytoplasmic expression of β-catenin in SKBR3. However, the β-catenin was observed in cell nuclei in most of the SKBR3/100-8 cells. The β-catenin translocates to cell nucleus could bind to TCF/LEF forming a β-catenin-TCF/LEF complex. Due to its transactivating ability, the β-catenin-TCF/LEF complex could bind to the promoter of genes and increase the gene expression that could be EGFR and/or HER2 in the trastuzumab resistant cell lines. To test our hypothesis ChIP-qPCR assay was performed. Figure 3C showed enriched β-catenin binding to the promoter of EGFR through the LEF/TCF binding region in SKBR3/100-8 compared to that in SKBR3. The data indicates that activation of Wnt/β-catenin signaling leads to transcriptional upregulation of EGFR in trastuzumab resistant cells. However, there was no difference in β-catenin binding to HER2 promoter between SKBR3 and SKBR3/100-8 cells (data not shown). It implies other mechanisms regulating the consistent overexpressing HER2 in SKBR3/100-8 cells.
Figure 3. Wnt3 promotes EMT and nuclear translocation of β-catenin in trastuzumab resistant cell lines.
A, The mRNA levels of the indicated genes were analyzed by RT-PCR. B, nuclear or cytoplasm expressions of β-catenin were determined by Western blot analysis. The α-Tubulin and Histone were used for confirmation of cytoplasmic and nuclear protein extraction respectively. C, ChiP-qPCR assay was performed as described in method. The bars indicated the fold enrichment of β-catenin binding to promoter of EGFR in SKBR3 and SKBR3/100-8 cells compared to Mock cells (immunoprecipitated with IgG). D and E (left). The cells were treated with siRNA-Wnt3 or negative sequences (Mock) for 72 hrs and RT-PCR was performed. The bars indicate relative expression of Wnt3 (D) and the indicated genes (E, left) (mean±SD from 3 determinations) and *P<0.05 compared to Mock cells. E. (right) cells were treated with siRNA-Wnt3 or negative sequence (Mock) for 72 hrs and the indicated protein expressions were analyzed by Western blot analysis. The β-actin was used as loading control.
Wnt3 knock-down restores the cytoplasmic expression of β-catenin, inhibits EMT-like transition and reduces cell invasiveness
siRNA knockdown of Wnt3 increased E-cadherin and decreased SLUG and Twist in both SKBR3/100-8 and BT474/100-2 cells compared to SKBR3 and BT474 cells (Figure 3D and 3E, left). The mRNA level of N-cadherin was decreased significantly in BT474/100-2 cells, but was not in SKBR3/100-8 cells (Figure 3E, left). The protein level of N-cadherin, however, was down regulated significantly in SKBR3/100-8 cells treated with siRNA-Wnt3 (Figure 3 right). Conversely the Twist protein level was not changed in SKBR3/100-8 after knockdown of Wnt3 (Figure 3E, right). The E-cadherin protein expression was up-regulated in SKBR3/100-8 after Wnt3 knockdown which was consistent with its mRNA level.
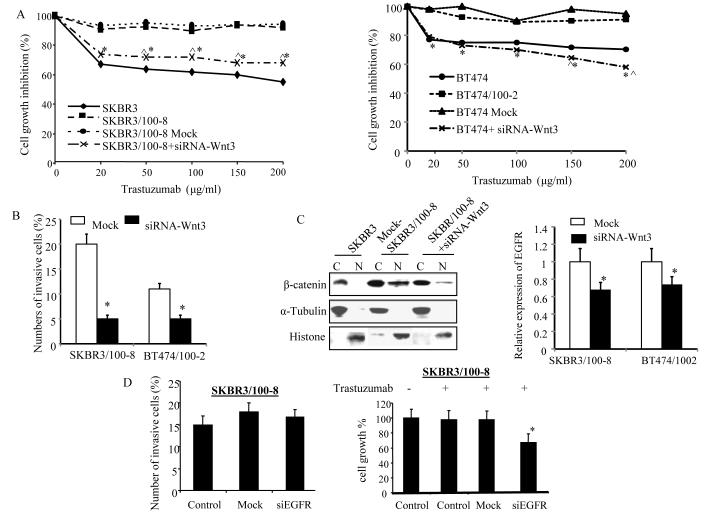
Next, we examined if Wnt3 knockdown could sensitize cells in response to trastuzumab. The SKBR3/100-8 and BT474/100-2 cells were treated with siRNA-Wnt3 or negative control sequence (siRNA-A) for 48 hrs followed by fresh treatment with 20μg/ml to 200μg/ml of trastuzumab for 72hrs. The data in Figure 4A demonstrates that 20μg/ml to 200μg/ml trastuzumab inhibited 35% to 45% cell growth in SKBR3 cells, but only ~8% in SKBR3/100-8 cells or SKBR3/100-8 cells treated with siRNA-A (Mock). In contrast, SKBR3/100-8 cells treated with Wnt3 siRNA, showed ~ 27 to 30% inhibitions with 20μg/ml to 200μg/ml trastuzumab. The cell growth inhibition was 25% to 30% in BT474 cells treated with 20μg/ml to 200μg/ml trastuzumab and a maximum of 10% inhibition of cell growth in BT474/100-2 and BT474/100-2 treated with siRNA-A (Figure 4A, right). While the cell growth was inhibited from 27% to 38% by 20μg/ml to 200μg/ml trastuzumab in BT474/100-2 cells treated with siRNA-Wnt3. The cell invasion was also significantly inhibited in SKBR3/100-8 and BT474/100-2 cells treated with siRNA-Wnt3 compared with the cells untreated with siRNA-Wnt3 or treated with siRNA-A (Figure 4B).
Figure 4. Knocking-down Wnt3 increases the cells’ response to trastuzumab and reduces cells’ invasiveness.
A, SKBR3/100-8 and BT474/100-2 were treated with siRNA-Wnt3 for 48 hrs and then treated with trastuzumab at 0, 20μg/ml, 50μg/ml, 100μg/ml, 150μg/ml and 200μg/ml for 3 days. The cell growth inhibitions by trastuzumab were determined by MTT assay. * p<0.05 compared to cells treated with sRNA negative sequence (Mock) and each data point is the mean of 6 determinations. B, Cells were treated with either siRNA-Wnt3 or negative sequence (Mock) for 48 hrs and invasive assay was performed as described in the Method. The bars indicate percentage of the invasive cells (mean+SD from 5 different areas) and * p<0.05. C, Cells were either treated with siRNA-Wnt3 or negative sequence (Mock) for 48hrs. The nuclear and cytoplasmic protein was extracted and β-catenin expression was analyzed by Western blot with antibody specific against β-catenin (left). α-Tubulin and Histone were used for confirmation of cytoplasmic and nuclear protein extraction, respectively. The relative expression of EGFR in the cells was determined by RT-Q-PCR (right). The bars indicate mean±SD from three determinations and *p<0.05. D, SKBR3/100-8 was treated with either siRNA-EGFR or negative sequence (Mock) for 48 hrs and invasive assay was performed as described in the Method (left). The bars indicate percentage of the invasive cells (mean+SD from 5 different areas). The cells were treated with or without trastuzumab (10μg/ml) for 3 days after knockdown EGFR. MTT assay was performed (right) and the bars indicate percentage of cells growth (mean±SD) from 6 determinations. *p<0.05 compared to control and Mock cells.
The down-regulation of Wnt3 by siRNA also resulted in decreased nuclear expression of β-catenin protein in SKBR3/100-8 cells and down-regulation of mRNA level of EGFR by 30% and 26% in SKBR3/100-8 and BT474/100-2, respectively (Figure 4C). In contrast, the mRNA level of Wnt3 was not changed after knockdown of EGFR (supplemental Figure 6).
Next, we pre-treated the SKBR3/100-8 with siRNA-EGFR, followed by treatment with 10μg/ml of trastuzumab. We found that down regulation of EGFR by siRNA-EGFR had no effect on cell invasiveness of SKBR3/100-8 cells, however, increased their response to trastuzumab induced inhibition of cell growth significantly (Figure 4D). The data suggests that the increase in EGFR in SKBR3/100-8 and BT474/100-2 leads to resistance to trastuzumab induced inhibition of cell growth, but may not be responsible for cell invasiveness directly. Since the SKBR3/100-8 showed increase in HER2/HER3 dimerization, similar experiments were performed on SKBR3/100-8 cells pre-treated with siRNA-HER3. An apparent reduction in invasion by HER3 knockdown did not achieve significance (supplemental Figure 7A). However, the cell growth inhibition by trastuzumab was increased significantly after HER3 knockdown (supplemental Figure 7B).
Thus, the data suggested that Wnt3 acted as a key mediator in the localization of β-catenin and controls EMT-like transition resulting in increase in cell invasion, proliferation, transactivation of EGFR. These events contribute to the increase in resistance to trastuzumab in HER2-overexpressing breast cancer cells.
Overexpressing Wnt3 promotes EMT and reduces sensitivity to trastuzumab in SKBR3 wild type cells
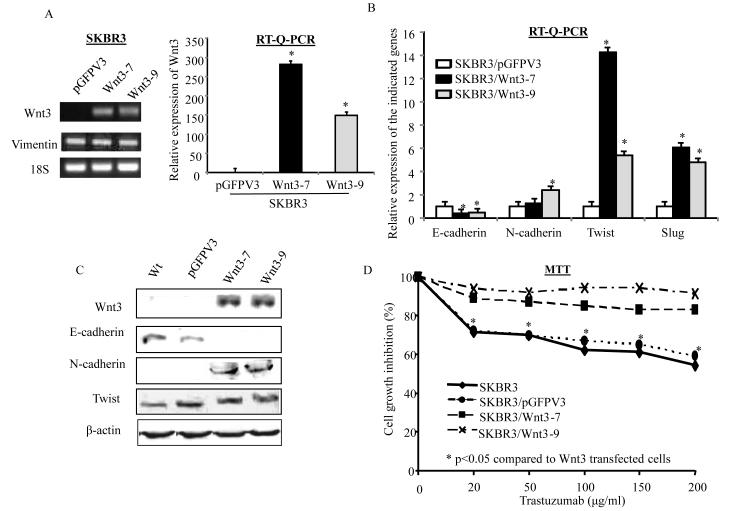
To ascertain the role of Wnt3 in EMT-like transition and trastuzumab resistance, a full length Wnt3 gene was stably transfected into parental or wild type SKBR3 cells. Figure 5A shows that the Wnt3 transfectants, SKBR3/Wnt3-7 and SKBR3/Wnt3-9 clones, had significant increase in the mRNA levels of Wnt3 compared to the vector transfectant, SKBR3/pGFPV3 cells. The mRNA level of E-cadherin was down-regulated and N-cadherin was up-regulated in SKBR3/Wnt3-7 and SKBR3/Wnt3-9 compared with SKBR3/pGFPV3 (Figure 5B). SKBR3/Wnt3-7 and SKBR3/Wnt3-9 also had significant increase in Twist and SLUG (Figure 5B), but no changes in Vimentin (Figure 5A). Data in Figure 5C confirmed that the protein level of E-cadherin was down-regulated and N-cadherin was up-regulated following the up-regulation of Wnt3 protein in both SKBR3/Wnt3-7 and SKBR3/Wnt3-9 cells. The Twist protein level, however, was not changed in these two Wnt3 transfectants (Figure 5C). The increased Wnt3 expression reduced the growth –inhibiting effect of trastuzumab. Figure 5D shows that after 3 days of trastuzumab (20μg/ml to 200μg/ml) treatment, cell growth was inhibited by 32% to 42% in the parental line, SKBR3, and 30% to 40% in SKBR3/pGFP3. However, cells transfected with full length Wnt3 showed significantly decreased sensitivity to trastuzumab. The cell growth inhibition by 20μg/ml to 200μg/ml of trastuzumab was less than 10% in SKBR3/Wnt3-7 and SKBR3/Wnt3-9 (Figure 5D). This data confirmed the role of Wnt3 in EMT-like transition on the cells that developed resistance to trastuzumab.
Figure 5. Overexpressing Wnt3 in SKBR3 cells promotes EMT and reduces sensitivity of trastuzumab.
A, PCR (left) and Q-PCR (right) analyses confirmed the overexpressing Wnt3 in the full length Wnt3 transfectants, Wnt3-7 and Wnt3-9 compared with empty vector transfected SKBR3 cells (pGFPV3). Vimentin expression in the indicated cells was determined by PCR (left). B, Relative expression of the indicated genes in SKBR3/pGFPV3 (open bar), SKBR3/Wnt3-7 (dark bar) and SKBR3/Wnt3-9 (gray bar) were determined by RT-Q-PCR. Each bar was the mean of 3 determinations with SD. *p<0.05 compared to SKBR3/pGFPV3. C, Western blot analysis was used to determine the protein levels of the indicated EMT marks in SKBR3 cells transfected with empty vector (pGFPV3), full length Wnt3 (Wnt3-7 and Wnt3-9) or without transfection (wt). D, MTT assay was performed in SKBR3, SKBR3/pGHPV3, SKBR3/Wnt3-7 and SKBR3/Wnt3-9 cells treated with the indicated doses of trastuzumab for 3 days. Each data point is the mean of 6 determinations. *p<0.05 compared to SKBR3/Wnt3-7 and SKBR3/Wnt3-9.
Discussion
The resistance to therapy is one of the biggest challenges in breast cancer treatment. Despite the success of trastuzumab as a monotherapy agent, a significant portion of HER2-positive breast cancers respond poorly to the treatment (12, 13). The underlying mechanisms responsible for the resistance are still not clear.
The EGFR/HER2 cross-talk has been suggested as one of the mechanisms of the development of trastuzumab resistance in HER2-overexpressing breast cancer cells (14, 15). Additionally, Wnt1 can transcriptionally regulate EGFR in HER2-overexpressing breast cancer cells (16).
We performed microarray analysis to identify the specific signaling pathway that mediated genes involved in HER2-overexpressing breast cancer cells’ resistance to trastuzumab. Twenty-two genes that are part of the Wnt pathway were either up or down-regulated two or more fold in SKBR3/100-8 cells compared to parental SKBR3 cells. In addition, a series of Wnt/β-catenin signaling molecules including β-catenin, c-Myc, c-Jun, Sox9, and survivin were up-regulated in both SKBR3/100-8 and BT474/100-2 cells.
Deregulation of the Wnt signaling pathway has been associated with the development and progression of several types of human cancers including colorectal cancer (CRC), prostate cancer, breast cancer and melanoma (11, 17-21). In CRC, the activation of the Wnt/β-catenin pathway is mainly due to mutation or lost function of the APC gene or protein in a ligand independent manner (11). The activation of the Wnt pathway in breast cancer, however, is more likely due to co-expression of Wnt ligands and FZD receptors (11, 22-25). The Wnt ligands, Wnt1, Wnt3a, Wnt4, Wnt5a, and Wnt7b have been reported to mediate cell proliferation and migration through canonical or noncanonical Wnt pathways in breast cancer (11, 16, 24, 25, 26). Data from our current study showed that Wnt3 was up-regulated upon the cells acquiring trastuzumab resistance in both SKBR3/100-8 and BT474/100-2 clones. The increased Wnt3 activated Wnt/β-catenin signaling pathway and nuclear translocation of β-catenin. The β-catenin in nucleus interacted with TCF/LEF and transactived expression of EGFR in trastuzumab resistant cells. It may also transactivate SLUG and induces EMT-like transition.
Converging evidence suggest that the canonical Wnt signaling pathway, through translocation of β-catenin, plays an important role in regulating EMT in different cancers including breast cancer (18, 27, 28). The EMT-like transition in cancer cells could promote tumor invasion and metastases, as well as mediate drug resistance (18, 27, 28, 29). Data from our current study concurred with those published observations.
The trastuzumab resistant clone, SKBR3/100-8, also showed increased pAkt level compared to the parental cells, SKBR3 (data not shown). The activation of pAkt followed by inactivation of GSK3β in SKBR3 cells has been shown in our previous study (5). The inactivation of GSK3β may also be partially responsible for the regulation of β-catenin without involving Wnt3. To confirm the specific role of Wnt3 in the development of trastuzumab resistance, the Wnt3 in SKBR3/100-8 and BT474/100-2 cells was knocked down by siRNA. These Wnt3 knockdown cells showed up-regulation of E-cadherin and down-regulation of EMT markers, N-cadherin, twist and SLUG. Furthermore, the cell invasiveness was reduced and the growth-inhibitory effects of trastuzumab were restored.
The Wnt3 knockdown also restored cytoplasmic expression of β-catenin and decreased EGFR expression. Down regulation of EGFR sensitized the cells’ response to trastuzumab induced growth inhibition. However, the nuclear translocation of β-catenin by Wnt3 did not increase the β-catenin binding to HER2 promoter at the TCF/LEF binding region (data not shown). Trastuzumab resistant cells continue to overexpress HER2 even in the presence of high concentration of trastuzumab at 100μ/ml of trastuzumab. Mechanism underlying this observation requires further investigations. Our current data confirm that expressing Wnt3 active Wnt/β-catenin signaling pathway, up-regulates EGFR and further leads to reduction of the growth-inhibiting effects of trastuzumab in HER2 overexpressing breast cancer cells. Hence, combination of trastuzumab and Lapatinib targeting both HER2 and EGFR could overcome the trastuzumab resistance.
In contrast to the knockdown strategy, a full length Wnt3 gene was stably transfected into a relatively sensitive to trastuzumab parental line, SKBR3 cells. The stably transfected Wnt3 clones tended to undergo EMT-like transition with down-regulation of E-cadherin and up-regulation of N-cadherin protein. The mRNA level of Twist and SLUG were significantly increased again. The Wnt3 transfectants were less responsive to trastuzumab-induced growth inhibition.
It has been suggested that Wnt3 has a key role in human breast, rectal, lung, and gastric cancers through activation of the canonical Wnt/β-catenin-TCF signaling pathway. The EMT-like phenotype has been observed in the development of resistance to Tamoxifen in ER-positive breast cancer cell line, MCF-7, and has been accompanied by a change in β-catenin phosphorylation (30). Our data from the current study provides an insight into the mechanism of HER2-overexpressing breast cancer cells that may become resistant to trastuzumab. This could be due to redundant expression of Wnt3.
Currently, Wnt signaling targeting includes small molecule inhibitors of the β-catenin/TCF signaling activity, as well as antibodies targeting Wnt1 and Wnt2 (31). Data from our current study suggests that specifically targeting Wnt3 could overcome trastuzumab resistance and benefit breast cancer patients with HER2-overexpressing tumors.
In conclusion, we have shown in the current study that the development of trastuzumab resistance in HER2 overexpressing breast cancer cells is accompanied by partial EMT-like transition. Expression of Wnt3 activation of the Wnt/β-catenin signaling pathway is one of the underlying mechanisms for the development of resistance to trastuzumab.
Supplementary Material
Acknowledgements
This work was supported in part by grants from NIH/National Cancer Institute 1U54CA14393-01; U56 CA101599-01; CA15083-25S3; R25DK067015-01; and Department of Defense Breast Cancer Research Program grant BC043180 to J.V. Vadgama; NIMHD U54MD007598 CDU-AXIS pilot project to Y. Wu.
Footnotes
Disclosure of conflicts of interest: No conflicts of interests were disclosed.
References
- 1.Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–60. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 2.Nahta R, Hung MC, Esteva FJ. The HER-2-targeting antibodies trastuzumab and pertuzumab synergistically inhibit the survival of breast cancer cells. Cancer Res. 2004;64:2343–6. doi: 10.1158/0008-5472.can-03-3856. [DOI] [PubMed] [Google Scholar]
- 3.Chan CT, Metz MZ, Kane SE. Differential sensitivities of trastuzumab (Herceptin)-resistant human breast cancer cells to phosphoinositide-3 kinase (PI-3K) and epidermal growth factor receptor (EGFR) kinase inhibitors. Breast Cancer Res treat. 2005;91:187–201. doi: 10.1007/s10549-004-7715-1. [DOI] [PubMed] [Google Scholar]
- 4.Weigelt B, Lo AT, Park CC, Gray JW, Bissell MJ. HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on 3D microenvironment. Breast Cancer Res treat. 2010;122:35–43. doi: 10.1007/s10549-009-0502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Y, Shang X, Sarkissyan M, Slamon D, Vadgama JV. FOXO1 is a target for HER2-overexpressing breast tumors. Cancer Res. 2010;70:5475–85. doi: 10.1158/0008-5472.CAN-10-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Y, Mohamed H, Chillar R, Clayton S, Slamon D, Vadgama JV. Clinical significance of Akt and HER2/neu overexpression in African-American and Latina women with breast cancer. Breast Cancer Res. 2008;10:R3. doi: 10.1186/bcr1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desbois-Mouthon C, Cadoret A, Blivet-Van Eggelpoël MJ, Bertrand F, Cherqui G, Perret C, et al. Insulin and IGF-1 stimulate the beta-catenin pathway through two signalling cascades involving GSK-3beta inhibition and Ras activation. Oncogene. 2001;20:252–9. doi: 10.1038/sj.onc.1204064. [DOI] [PubMed] [Google Scholar]
- 8.Petrocelli T, Slingerland JM. PTEN deficiency: a role in mammary carcinogenesis. Breast Cancer Res. 2001;3:356–60. doi: 10.1186/bcr322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Persad S, Troussard AA, McPhee TR, Mulholland DJ, Dedhar S. Tumor suppressor PTEN inhibits nuclear accumulation of beta-catenin and T cell/lymphoid enhancer factor 1-mediated transcriptional activation. J Cell Biol. 2001;153:1161–74. doi: 10.1083/jcb.153.6.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hashimoto M, Sagara Y, Langford D, Everall IP, Mallory M, Everson A, Digicaylioglu M, Masliah E. Fibroblast growth factor 1 regulates signaling via the glycogen synthase kinase-3beta pathway. Implications for neuroprotection. J Biol Chem. 2002;277:32985–91. doi: 10.1074/jbc.M202803200. [DOI] [PubMed] [Google Scholar]
- 11.Howe LR, Brown AMC. Wnt signaling and breast cancer (Review) Cancer Biol and Ther. 2004;3:36–41. doi: 10.4161/cbt.3.1.561. [DOI] [PubMed] [Google Scholar]
- 12.Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-over-expressing metastatic breast cancer. J Clin Oncol. 2002;2:719–26. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- 13.Nahta R, Yuan LX, Zhang B, Kobayashi R, Esteva FJ. Insulin-like growth factor-I receptor/human epidermal growth factor receptor 2 heterodimerization contributes to trastuzumab resistance of breast cancer cells. Cancer Res. 2005;65:11118–28. doi: 10.1158/0008-5472.CAN-04-3841. [DOI] [PubMed] [Google Scholar]
- 14.Ritter CA, Perez-Torres M, Rinehart C, Guix M, Dugger T, Engelmen JA, et al. Human breast cancer cells selected for resistance to trastuzumab in vivo overexpress epidermal growth factor receptor and ErbB ligands and remain dependent on the ErbB receptor network. Clin Cancer Res. 2007;13:4909–19. doi: 10.1158/1078-0432.CCR-07-0701. [DOI] [PubMed] [Google Scholar]
- 15.Diermeier S, Horváth G, Knuechel-Clarke R, Hofstaedter F, Szöllösi J, Brockhoff G. Epidermal growth factor receptor coexpression modulates susceptibility to Herceptin in HER2/neu overexpressing breast cancer cells via specific erbB-receptor interaction and activation. Exp Cell Res. 2005;304:604–19. doi: 10.1016/j.yexcr.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Schlange T, Matsuda Y, Lienhard S, Huber A, Hynes NE. Autocrine Wnt signaling contributes to breast cancer cell proliferation via the canonical Wnt pathway and EGFR transactivation. Breast Cancer Res. 2007;9:R63. doi: 10.1186/bcr1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurayoshi M, Oue N, Yamamoto H, Kishida M, Inoue A, Asahara T, et al. Expression of Wnt-5 is correlated with aggressiveness of gastric cancer by stimulating cell migration and invasion. Cancer Res. 2006;66:10439–48. doi: 10.1158/0008-5472.CAN-06-2359. [DOI] [PubMed] [Google Scholar]
- 18.Gupta S, Iljin K, Sara H, Mpindi JP, Mirtti T, Vainio P, et al. FZD4 as a mediator of ERG oncogene-induced Wnt signaling and epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2010;70:6735–45. doi: 10.1158/0008-5472.CAN-10-0244. [DOI] [PubMed] [Google Scholar]
- 19.Kobune M, Chiba H, Kato J, Kato K, Nakamura K, Kawano Y, et al. Wnt3/RhoA/ROCK signaling pathway is involved in adhesion-mediated drug resistance of multiple myeloma in an autocrine mechanism. Mol Cancer Ther. 2007;6:1774–84. doi: 10.1158/1535-7163.MCT-06-0684. [DOI] [PubMed] [Google Scholar]
- 20.Lalli E, Wakil AE. The Wnt/beta-catenin pathway in adrenocortical development and cancer. Mol Cell. Endocrinol. 2010;332:32–7. doi: 10.1016/j.mce.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 21.Kuorelahti A, Rulli S, Huhtaniemi I, Poutanen M. Human chorionic gonadotropin (hCG) up-regulates Wnt5 and Wnt7b in the mammary gland, and hCGβ transgenic female mice present with mammary gland tumors exhibiting characteristics of the Wnt/β-catenin pathway activation. Endocrinology. 2007;148:3694–3703. doi: 10.1210/en.2007-0249. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi-Yanaga F, Kahn M. Targeting Wnt signaling: Can we safely eradicate cancer stem cells? Clin Cancer Res. 2010;16:3153–62. doi: 10.1158/1078-0432.CCR-09-2943. [DOI] [PubMed] [Google Scholar]
- 23.Ayyanan A, Civenni G, Ciarloni L, Morel C, Mueller N, Lefort K, et al. Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notchdependent mechanism. PNAS. 2006;103:3799–804. doi: 10.1073/pnas.0600065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Givenni G, Holbro T, Hynes NE. Wnt1 and Wnt5a induce cyclin D1 expression through ErbB1 transactivation in HC11 mammary epithelial cells. EMBO. 2003;4:166–71. doi: 10.1038/sj.embor.embor735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan L, Coletta LD, Powell LK, Shen J, Thames H, Aldaz MC, et al. Activation of the canonical Wnt/β-catenin pathway in ATF3-induced mammary tumors. PLoS ONE. 2011;6:e16515. doi: 10.1371/journal.pone.0016515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benhaj K, Akcali KC, Ozturk M. Redundant expression of canonical Wnt ligands in human breast cancer cell lines. Oncol Rep. 2006;15:701–7. [PubMed] [Google Scholar]
- 27.Yook JI, Li XY, Ota I, Hu C, Kim HS, Kim NH, et al. A Wnt-Axin2-GSK3β cascade regulates Snail1 activity in breast cancer cells. Nat Cell Biol. 2006;8:1398–406. doi: 10.1038/ncb1508. [DOI] [PubMed] [Google Scholar]
- 28.Yee DS, Tang Y, Li X, Liu Z, Guo Y, Ghaffar S, et al. The Wnt inhibitory factor 1 restoration in prostate cancer cells was associated with reduced tumor growth, decreased capacity of cell migration and invasion and a reversal of epithelial to mesenchymal transition. Mol Cancer. 2010;9:162. doi: 10.1186/1476-4598-9-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asiedu MK, Ingle JN, Behrens MD, Radisky DC, Knutson KL. TGFβ/TNFα-mediated epithelial-mesenchymal transition generates breast cancer stem cells with claudin-low phenotype. Cancer Res. 2011;71:4707–19. doi: 10.1158/0008-5472.CAN-10-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiscox S, Jiang WG, Obermeier K, Taylor K, Morgen L, Burmi R, et al. Tamoxifen resistance in MCF7 cells promotes EMT-like behavior and involves modulation of β-catenin phosphorylation. Int J Cancer. 2006;118:290–301. doi: 10.1002/ijc.21355. [DOI] [PubMed] [Google Scholar]
- 31.Barker N, Clevers H. Mining the Wnt pathway for cancer therapeutics. Nat Rev Drug Discov. 2006;5:997–1014. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.