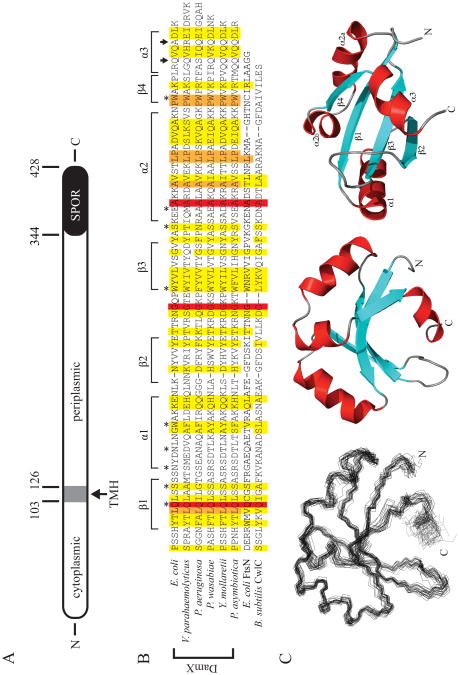
Figure 1.
The structure of DamX and its SPOR domain. (A) Cartoon of DamX from E. coli. DamX is predicted to comprise a 103 a.a. cytoplasmic domain, a 23 a.a. transmembrane helix (TMH), and a 302 a.a. periplasmic domain, of which the last 84 a.a. constitute the SPOR domain. (B) Sequence alignment of SPOR domains of several DamX's with the SPOR domains from CwlC and FtsN. The secondary structure elements determined for the SPOR domain of DamX from E. coli are shown above the alignment. Asterisks denote residues mutated in this study. Arrows indicate the site of C-terminal truncations. The alignment was obtained using CLUSTAL W (54) with the following sequences: E. coli DamX residues 344-428; Vibrio parahaemolyticus DamX residues 417-505; Pseudomonas aeruginosa DamX residues 464-551; Pectobacterium wasabaie WPP163 residues 251-336; Yersinia mollaretii DamX residues 249-333; Photorhabdus asymbiotica DamX residues 232-316; E. coli FtsN residues 244-319; and B. subtilis CwlC residues 182-255. (C) Solution structure of the SPOR domain (DamX residues 344-428). (Left) Side-on view of a backbone superposition of the 25 final structures. The average pairwise RSMD for residues is 0.795 Å. (Middle) Side-on and (Right) bottom-up views of a ribbon diagram of the domain, with β-strands in blue, α-helices in red, and loops in gray. Note that α2 is interrupted by a proline.