Abstract
The rhizobium-legume symbiosis is a model system for studying mutualistic interactions between bacteria and eukaryotes. Sinorhizobium sp. NGR234 is distinguished by its ability to form either indeterminate nodules or determinate nodules with diverse legumes. Here, we presented a high-resolution RNA-seq transcriptomic analysis of NGR234 bacteroids in indeterminate nodules of Leucaena leucocephala and determinate nodules of Vigna unguiculata. In contrast to exponentially growing free-living bacteria, non-growing bacteroids from both legumes recruited several common cellular functions such as cbb3 oxidase, thiamine biosynthesis, nitrate reduction pathway (NO-producing), succinate metabolism, PHB (poly-3-hydroxybutyrate) biosynthesis and phosphate/phosphonate transporters. However, different transcription profiles between bacteroids from two legumes were also uncovered for genes involved in the biosynthesis of exopolysaccharides, lipopolysaccharides, T3SS (type three secretion system) and effector proteins, cytochrome bd ubiquinol oxidase, PQQ (pyrroloquinoline quinone), cytochrome c550, pseudoazurin, biotin, phasins and glycolate oxidase, and in the metabolism of glutamate and phenylalanine. Noteworthy were the distinct expression patterns of genes encoding phasins, which are thought to be involved in regulating the surface/volume ratio of PHB granules. These patterns are in good agreement with the observed granule size difference between bacteroids from L. leucocephala and V. unguiculata.
Introduction
The rhizobium-legume symbiosis has been a model system for studying the mutualistic interactions for many years [1]. This system is characterized by its ability to form the symbiotic nodules, in which rhizobia differentiate into bacteroids to fix atmospheric nitrogen to the benefit of the legume while the host provides carbon sources to these microsymbionts. Symbiotic nodules could be simply categorized into either determinate or indeterminate nodules based on the activity of the nodule meristem. In the determinate nodules (such as those of Lotus japonicus, Glycine max and Phaseolus vulgaris), the meristem functions until the formation of the nodule primordium and produces synchronously developed infected cells, whereas the meristem persists in the indeterminate nodules (such as Medicago sativa, Pisum sativum and Vicia sativa) which are composed of different zones showing a clear developmental gradient: the apical meristem, the invasion zone, the interzone, the nitrogen-fixing zone, and the senescence zone [2], [3], [4]. Enlarged and nonreproductive bacteroids were thought to be a characteristic feature of indeterminate nodules of Inverse-Repeat Legume Clade (IRLC) including M. sativa, P. sativum and V. sativa, in contrast, morphologically unchanged and reproductive bacteroids were commonly found in determinate nodules of Milletioids plants including L. japonicus, G. max and P. vulgaris [5]. However, the latest study of the distribution of these two kinds of bacteroids in 40 legume species in the subfamily Papilionoideae [6] suggested that there was no clear correlation between nodule types and morphologies of bacteroids. Moreover, morphologically unchanged bacteroids might be the ancestral form to enlarged bacteroids [6].
High throughput transcriptomics and proteomics studies have revealed global gene expression profiles of enlarged bacteroids in indeterminate nodules [7], [8], [9], [10], [11] or morphologically unchanged bacteroids in determinate nodules [12], [13], [14], [15], [16]. Few of them compared host-specific bacteroid transcripts and/or proteins [11], [16]. Recently, exciting progresses have been made in identifying the determinants of the interactions between host cells and enlarged bacteroids in indeterminate nodules of Medicago and Pisum [17], [18], [19], [20], [21]. But little information was known for the persistence mechanisms of both types of bacteroids in determinate nodules and morphologically unchanged bacteroids in indeterminate nodules, partially due to different strategies used by diverse legume-rhizobium systems [2], [6], [22], [23]. Sinorhizobium sp. NGR234 is well known for its ability to nodulate legume hosts from as many as 112 genera forming either determinate or indeterminate nodules [24]. Thus NGR234 could serve as an excellent model for investigating the adaptation mechanisms of rhizobia to diverse conditions within different types of nodules. In this study, RNA-seq was used to investigate the transcriptomic differences between free-living NGR234 and NGR234 bacteroids in either determinate nodules of V. unguiculata or indeterminate nodules of L. leucocephala. Both common and distinct transcription patterns of NGR234 bacteroids in these two legumes were analyzed.
Materials and Methods
Growth Conditions for Bacterial Strain and Plants
The broad-host range strain Sinorhizobium sp. NGR234 [24] was grown in liquid TY (tryptone yeast extract) medium [25] at 28°C. Bacterial culture with OD600 = 0.6 (the optical density at 600 nm) was used for inoculating legume hosts V. unguiculata and L. leucocephala forming determinate and indeterminate nodules respectively with NGR234. Seeds of V. unguiculata were surface-sterilized by successive treatments with 95% ethanol for 30 sec and 0.2% (w/v) HgCl2 for 5 min, and were then washed for 10 times by using autoclaved deionized water. Seeds of L. leucocephala were first treated with sulfuric acid for 30 min, followed by washing six times in sterilized water. They were then surface-sterilized as described for seeds of V. unguiculata. All the surface-sterilized seeds were germinated on 0.6% agar–water plates in the dark at 28°C for 24–48 h. Germinated seeds were planted in vermiculite moisturized with low-N nutrient solution in Leonard jars [26] and were inoculated with 1 ml of bacterial culture with OD600 = 0.6 per plant. All the plants were grown at 24°C in a plant growth room with a daylight illumination period of 12 h. Nodules were harvested 21 DPI (days post inoculation) for V. unguiculata and 35 DPI for L. leucocephala when the acetylene reductase activity of nodules reached the peak level. Three biological replications were done. These nodules were either frozen in liquid nitrogen and stored at –80°C until RNA extraction, or used for sample preparations for electron and light microscopy.
Light and Electron Microscopy
Nodules were fixed in 2.5% glutaraldehyde in 0.05 M cacodylate buffer [27]. For light microscopy, fixed nodules were washed, dehydrated, embedded in Technovit 7100 (Kulzer Histo-Technik), according to the manufacturer’s instructions. Sections of 2 µm were cut on a Leica Ultracut C6i and stained with 1% toluidine blue for 40 sec. For electron microscopy, fixed nodules were washed with 0.1 M phosphate buffer and postfixed in 0.1 M phosphate buffer containing 1% (wt/vol) OsO4. The samples were then washed with 0.1 M phosphate buffer and dehydrated with increasing volumes of acetone (30%, 50%, 70%, 90%, and 100%). The samples were embedded in the SPURR epoxy. A Leica Ultracut C6i was used to obtain ultrathin sections (80 nm thick) of these nodule samples. The resulting sections were stained with uranyl acetate and lead citrate and finally observed in JEM-1230 transmission electron microscope.
Isolation and Purification of Bacteroids
A slightly modified approach described by Day et al. [28] was used for bacteroid isolation from frozen nodules of V. unguiculata and L. leucocephala. Briefly, 2–5 g nodules were extensively ground by using a pre-chilled pestle in 15 ml of extraction buffer (10 mM DTT, 300 mM sucrose, 10 mM phosphate buffer pH 7.0, 2 mM MgCl2 and 0.33 g PVP). In order to remove large particles of plant cell debris, the mixture was centrifuged at 400×g for 10min, 4°C. The supernatant was centrifuged at 12000×g for 10 min, 4°C. The resulting pellet containing bacteroids and small plant cell debris was resuspended in 10 ml extraction buffer. Then 10 ml 30% percoll, 5 ml 60% percoll and 5 ml 80% percoll were added into the same tube. After the centrifugation at 4000×g for 15 min, 4°C, the layer between 60%–80% percoll containing bacteroids was diluted into 20 ml using the extraction buffer. The resulting solution was transferred to the tube containing 5 mL 60% percoll and 5 mL 80% percoll and subject to another round of centrifugation at 4000×g for 15 min, 4°C. The cushion above 80% percoll containing bacteroids was resuspended in 20 mL 0.8% NaCl and centrifuged at 12000×g for 5 min. The pellet was collected for RNA extraction of bacteroids.
RNA Extraction
Bacteroids were ground by using a pestle in liquid nitrogen. They were then subjected to RNA extraction using QIAGEN RNeasy mini kit according to the manufacturer’s instructions. For free-living bacteria, RNA from a bacterial culture with OD600 = 0.5 was extracted by using the same QIAGEN RNeasy mini kit. RNA quality was assessed by using an Agilent 2100 Bioanalyzer. RNA integrity number (RIN, average ± SE) was 8.80±0.10, 9.87±0.13 and 9.93±0.07 for RNA sample of L. leucocephala bacteroids, V. unguiculata bacteroids and the free culture of NGR234, respectively, indicating the good quality of RNA samples in this study.
Library Construction and Strand-specific RNA Sequencing
Total RNA was sent to BGI-Shenzhen for further treatments, library construction and strand-specific RNA sequencing. Briefly, total RNA was treated with RNase-free DNase I for 30 min at 37°C to remove residual DNA. Total RNA was then treated with Ribo-Zero rRNA Removal Kit (Gram-Negative Bacteria) according to the manufacturer’s instructions to remove the ribosomal RNA before preparing RNA libraries for deep sequencing. 5 µg of total RNA was used as the starting material for treatment. The mRNA-enriched RNA was chemically fragmented to 150∼200 bp using divalent cations under elevated temperature. The cleaved RNA fragments were copied into first strand cDNA using reverse transcriptase and random primers. Non-incorporated nucleotides were removed and dTTP was substituted by dUTP during the synthesis of the second strand [29]. These cDNA then went through an end repair process, the addition of a single “A” base, and then ligation of adapters. The ligation products were then purified and subsequently digested with N-glycosylase (UNG; Applied Biosystems) to remove the second-strand cDNA. The products were then enriched by 15 cycles of PCR cycles with phusion polymerase to create the final cDNA library. Libraries were sequenced on an Illumina Hiseq 2000 platform.
Sequence Analyses
Clean reads were mapped to the reference genome of Sinorhizobium sp. NGR234 [30] using SOAP2 [31]. Mismatches no more than 5 bases were allowed in the alignment. To eliminate the influence of different gene length and sequencing discrepancy on the calculation of gene expression, the RPKM (reads per kilobase per million mapped reads) method was used to calculate gene expression level [32]. Genes with the ratio of RPKMs of the two samples above 2, Benjamini FDR (False Discovery Rate) ≤0.001 and the coverage value larger than 80% in the transcriptionally up-regulated condition were chosen as the differentially expressed genes (DEGs) between two samples. IGV [33] was used to visualize the expression patterns across the genome. KEGG pathway annotations for Sinorhizobium sp. NGR234 were retrieved from the KEGG database [34] and used in the pathway enrichment analysis of DEGs by using Gitools [35]. The Benjamini FDR corrected P value <0.05 (two-tailed) for Fisher exact test was used to define the enriched pathway.
qRT-PCR
To validate the results of RNA-seq, quantitative reverse transcription PCR (qRT-PCR) experiments were performed in triplicate for 13 genes having different expression profiles. Three biological replicates were analyzed. Single strand cDNA was synthesized by using the GoScript™ Reverse Transcription System kit (Promega). Quantitative PCR was performed by using 25 µL of Light Cycler 480 SYBR Green I Master (Roche) and a Light Cycler 480 real-time PCR system (Roche). The PCR procedures were as follows: 95°C for 2 min; 40 cycles of 95°C for 15 sec, 60°C for 1 min. PCR results were analyzed by relative quantification methods using the 16S rRNA gene (NGR_c26520) as the reference gene.
Results and Discussion
Distinct Characteristics of Nodules from V. unguiculata and L. leucocephala
Sinorhizobium sp. NGR234 formed spherical nodules on V. unguiculata and elongated nodules on L. leucocephala (Figure 1A and 1C). These elongated indeterminate nodules contain a typical meristem zone on the distal part of nodules (Figure 1D). However, bacteroids in determinate or indeterminate nodules of these two legume hosts show several distinct characteristics compared to those in the typical determinate (L. japonicus, G. max etc.) or indeterminate (M. truncatula, P. sativa etc.) nodules [2]. Bacteroids in nodules of V. unguiculata (Figure 1E) and L. leucocephala (Figure 1F) are <2.5 µm in length, suggesting that they all belong to the morphologically unchanged bacteroids [6]. Moreover, there is just one bacteroid surrounded by each peribacteroid membrane in nodules from these two legume hosts (Figure 1E and 1F). Notably, poly-3-hydroxybutyrate (PHB) granules in bacteroids of L. leucocephala nodules (Figure 1F) were larger than those in bacteroids of V. unguiculata (Figure 1E).
Figure 1. Nodule sections of Vigna unguiculata and Leucaena leucocephala.
(A–B) Nitrogen fixing nodules of V. unguiculata (21 days post inoculation, A) and L. leucocephala (35 days post inoculation, B). (C–D) Semithin sections of nodules from V. unguiculata (C) and L. leucocephala (D), the asterisk indicates the meristem zone of indeterminate nodules of L. leucocephala (D). (E–F) Ultrathin sections of nodules from V. unguiculata (E) and L. leucocephala (F), red arrowheads indicate the peribacteroid membrane; and PHB granules are seen as electron-transparent droplets in the cytoplasm of bacteroids.
Gene Expression Overview
RNA-Seq experiments produced 1.169 Gb sequences for the free-living bacteria, V. unguiculata and L. leucocephala bacteroids, respectively. As shown in Table 1, 97.92%, 87.23% and 56.83% of obtained reads in the three treatments were mapped to Sinorhizobium sp. NGR234 genome. These mapping results were consistent with a relatively higher level of the contamination by plant RNA in the RNA samples of L. leucocephala bacteroids than in V. unguiculata bacteroids (data not shown). However, around 80 Mb sequences from the L. leucocephala bacteroids’ sample were uniquely mapped to known CDS of Sinorhizobium sp. NGR234 genome (6.9 Mb), and this was among the largest RNA-seq data relative to the genome size of bacteria [36], [37], [38], [39].
Table 1. An overview of the RNA-Seq data.
| Free-living bacteria | V. unguiculata bacteroids | L. leucocephala bacteroids | ||||
| number | %* | number | %* | number | %* | |
| Total reads# | 12988910 | 12988910 | 12992456 | |||
| Reads mapped to genome | 12718213 | 11329803 | 7383865 | |||
| perfect match | 9760662 | 76.75% | 8638335 | 76.24% | 5642244 | 76.41% |
| < = 5 bp mismatch | 2957551 | 23.25% | 2691468 | 23.76% | 1741621 | 23.59% |
| reads mapped to rRNA | 1082203 | 8.51% | 2519658 | 22.24% | 2847571 | 38.56% |
| Reads mapped to gene | 7705821 | 3931113 | 1049762 | |||
| perfect match | 5920834 | 76.84% | 3022804 | 76.89% | 811630 | 77.32% |
| < = 5 bp mismatch | 1784987 | 23.16% | 908309 | 23.11% | 238132 | 22.68% |
| unique match | 7629003 | 99.00% | 3390818 | 86.26% | 886105 | 84.41% |
Note: #, 90-base reads; *, the percentage value relative to the reads mapped to genome or gene.
Transcriptomes of bacteroids from nodules of V. unguiculata (21 DPI) and L. leucocephala (35 DPI) were compared to that of free-living Sinorhizobium sp. NGR234 in exponential growth stage (Table S1). As shown in Table 2 and Table S2, among the 3143 DEGs between V. unguiculata bacteroids and free-living bacteria, 90.2% DEGs of pNGR234a, 66.4% DEGs of pNGR234b and 29.1% DEGs of the chromosome were up-regulated in V. unguiculata bacteroids. Similarly, 96.6% DEGs of pNGR234a, 84.1% DEGs of pNGR234b and 35.9% DEGs of the chromosome were up-regulated in L. leucocephala bacteroids compared to those of free-living bacteria (Table 2 and Table S2). These biased distribution patterns of DEGs in three replicons of Sinorhizobium sp. NGR234 (Figure 2) also suggested an active role of two plasmids in symbiotic adaptations to both determinate (V. unguiculata) and indeterminate (L. leucocephala) nodules. This is consistent with the view that rhizobial extrachromosomal elements are important in niche adaptations [40]. Moreover, V. unguiculata and L. leucocephala bacteroids shared a common subset of 2072 DEGs compared to free-living bacteria (Table S2), and 99.1% of these DEGs showed the same direction of regulation in both legumes hosts. Moreover, among these 2072 DEGs, 141/146 DEGs of pNGR234a, 391/523 DEGs of pNGR234b and 412/1403 DEGs of the chromosome were up-regulated in both V. unguiculata and L. leucocephala bacteroids. As shown in Table S3, the enrichment analysis revealed that up-regulated DEGs in bacteroids from both hosts were particularly enriched in the KEGG pathways for microbial metabolism in diverse environments (rhi01120), ABC transporters (rhi02010), nitrogen metabolism (rhi00910), fatty acid metabolism (rhi00071) and benzoate degradation (rhi00362). In contrast, down-regulated genes were enriched in the KEGG pathways for ribosomes (rhi03010), pyrimidine metabolism (rhi00240), flagellar assembly (rhi02040) and aminoacyl-tRNA biosynthesis (rhi00970). Despite these similar expression profiles, distinct transcriptional differences were also observed between bacteroids from V. unguiculata and L. leucocephala such as phenylalanine metabolism (rhi00360) (Table S2-S3 and see discussion below).
Table 2. Differentially expressed genes in three replicons of Sinorhizobium sp. NGR234.
| Comparisons | DEG | pNGR234a | pNGR234b | Chromosome |
| VB/F | 3143 | 166∶18* | 589∶298 | 603∶1469 |
| LB/F | 2780 | 201∶7 | 747∶141 | 604∶1080 |
| LB/VB | 1184 | 76∶28 | 428∶78 | 424∶150 |
Note: VB/F, V. unguiculata bacteroids vs free-living bacteria; LB/F, L. leucocephala bacteroids vs free-living bacteria; LB/VB, L. leucocephala bacteroids vs V. unguiculata bacteroids; DEG, differentially expressed genes in each comparison.
, in each replicon, the number of genes up-regulated: down-regulated in the corresponding condition.
Figure 2. Distributions of differentially expressed genes across replicons of Sinorhizobium sp. NGR234.
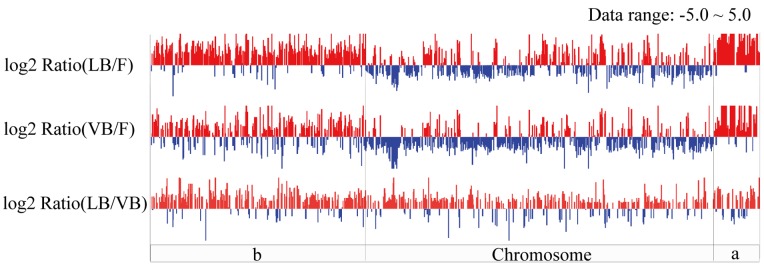
a, pNGR234a; b, pNGR234b; Chromosome, cNGR234; three replicons were separated by dashed lines. log2 Ratio(VB/F), the base 2 logarithm value for the ratio of expression level between bacteroids of Vigna unguiculata (VB) and free-living (F) Sinorhizobium sp. NGR234; LB/F, the ratio between L. leucocephala bacteroids and free-living bacteria; LB/VB, the ratio between L. leucocephala bacteroids and V. unguiculata bacteroids. The red bars above the horizontal line represent genes with the log2 Ratio values >1.0 whereas the blue bars below the horizontal line indicate genes with the log2 Ratio values<−1.0.
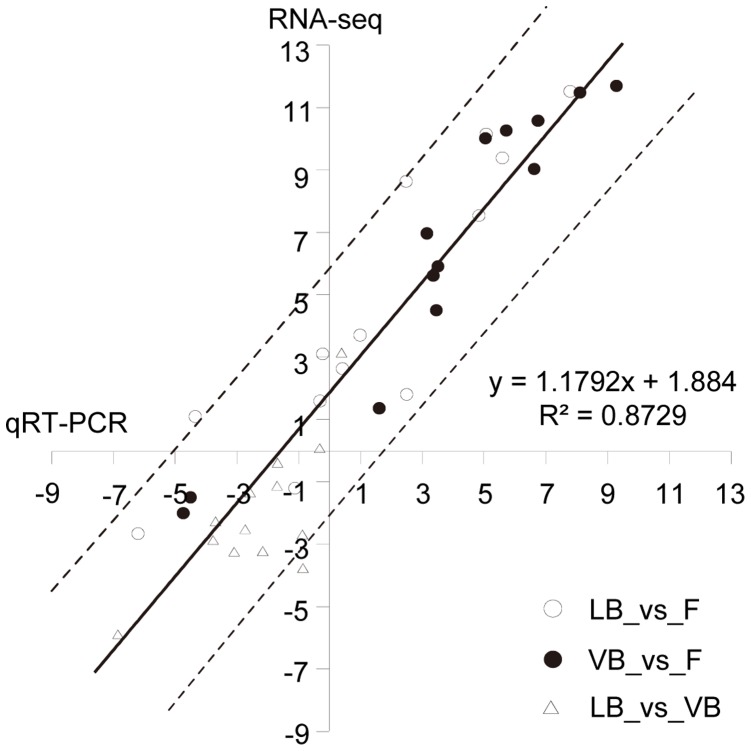
To validate the results of RNA-seq, we performed qRT-PCR on 13 genes with different expression profiles (Table S4). These include genes up-regulated or down-regulated in both L. leucocephala and V. unguiculata bacteroids with (or without) significant differences between expression levels in two hosts, and genes reversely regulated in bacteroids from L. leucocephala and V. unguiculata. As shown in Figure 3, the RNA-seq data agree well with the qRT-PCR data and Pearson correlation coefficient value was 0.934 (P<0.0001), despite the few differences that are often observed between qRT-PCR and microarrays results [10] [11] [41]. Similar results were obtained for qRT-PCR experiments with three independent biological replications (Table S4).
Figure 3. Validation of RNA-seq results by quantitative reverse transcription-polymerase chain reaction (qRT-PCR).
Expression of 13 genes was determined using qRT-PCR for the free-living Sinorhizobium sp. NGR234, L. leucocephala and V. unguiculata bacteroids. The expression ratios were calculated for L. leucocephala bacteroids vs. free-living (LB_vs_F), V. unguiculata bacteroids vs. free-living (VB_vs_F), and L. leucocephala bacteroids vs. V. unguiculata bacteroids (LB_vs_VB). The log2-transformed expression ratios from RNA-seq (vertical axis) and qRT-PCR (horizontal axis) are shown. The solid and dashed line(s) depicted the linear curve and the individual confidence intervals (95%). The linear equation and R square values are shown.
Growth Arrest of Bacteroids in Nodule Environments of V. unguiculata and L. leucocephala
Consistent with the non-growing state of nitrogen-fixing bacteroids, the Fisher exact test with Benjamini FDR corrected P values (two-tailed) revealed that the down-regulated genes in both L. leucocephala and V. unguiculata bacteroids (Table S3) were significantly (P<0.05) enriched in pathways for ribosomes (rhi03010), flagellar assembly (rhi02040), aminoacyl-tRNA biosynthesis (rhi00970) and pyrimidine metabolism (rhi00240) among others. Notably, all the flagellar assembly proteins except NGR_c28210 (the second copy of MotB) were down-regulated in bacteroids. This is in contrast to the induction of flagellar proteins in Bradyrhizobium japonicum by genistein application and the requirement of flagella in biofilm formation and competitive nodulation of S. meliloti [42], [43], [44], [45]. However, it was also reported that nonflagellated mutants of S. meliloti formed normal nitrogen-fixing nodules on alfalfa [46], suggesting that flagella are dispensable for function of bacteroids.
Growth arrest of bacteroids was also illustrated by the down-regulation of genes encoding DNA replication proteins (DNA polymerase I, DNA polymerase III subunits chi, DnaE, HolA and DnaQ, replicative DNA helicase, ribonuclease H), homologous recombination proteins (RecAR, Holliday junction DNA helicase RuvAB), chromosome partitioning proteins (ParABC), cell division proteins (FtsAIK1K2QZ1), peptidoglycan biosynthesis proteins (MurABCDEG, MraY, MviN, PbpB, D-alanyl-D-alanine carboxypeptidase NGR_c16080, D-alanine–D-alanine ligase NGR_c21040), core subunits of RNA polymerase (RpoABC), translation factors (initiation factors InfABC, elongation factors Ts, Tu, G and P, peptide chain release factors 1–3, ribosome recycling factor, release factor glutamine methyltransferase) and ATP synthase proteins (AtpABCDF1F2GH). A similar down-regulation of growth-associated processes was observed in bacteroids of S. meliloti, R. leguminosarum and Rhizobium etli [10], [11], [41].
ABC Transporters
Among the DEGs between bacteroids and the free-living form, up-regulated genes in bacteroids were particularly enriched in ABC transporters (Table S3 and Table 3). The strong up-regulation of transporters for phosphate (NGR_c01440-NGR_c01470, 7–18 folds, RPKM = 314–5089) in bacteroids is consistent with an earlier observation that a phosphate transporter mutant showed a deficiency in nitrogen-fixation [47]. Interestingly, transcriptions of genes coding phosphonate transporters (NGR_b13310-NGR_b13340, 58–436 folds, RPKM = 87–1576) and sn-glycerol 3-phosphate transporters (NGR_c36780-NGR_c36810, 50–428 folds, RPKM = 201–1771) were also strongly induced in bacteroids. These data suggest a phosphorus-limiting nodule environment for bacteroids. Under phosphorus-limiting conditions, S. meliloti could replace most of its phospholipids with membrane-forming lipids lacking phosphorus such as DGTS (diacylglyceryl-N,N,N-trimethylhomoserine) [48]. In NGR234 bacteroids from two test hosts, btaA and btaB (NGR_c21260- NGR_c21270, 47–123 folds) involved in the biosynthesis of DGTS were strongly up-regulated, implying that NGR234, regardless of test hosts, could form membrane lipids lacking phosphorus to adapt to the nodule environment. But available studies on S. meliloti suggested that phosphorus-free membrane lipids were not required for the symbiosis with alfalfa [49].
Table 3. Differentially expressed genes encoding ABC transporters.
| Substrate | Gene ID | RPKM_F | RPKM_LB | RPKM_VB | ||
| Up-regulated in bacteroids from both legume hosts | ||||||
| phosphate | NGR_c01440-NGR_c01470 | 42–323 | 314–4710 | 670–5089 | ||
| phosphonate | NGR_b13310-NGR_b13340 | 1–4 | 87–1576 | 116–1494 | ||
| sn-glycerol 3-phosphate | NGR_c36780-NGR_c36810 | 1–4 | 150–1711 | 267–1771 | ||
| aminoethane sulfonate | NGR_b17610-NGR_b17630 | 1–5 | 37–80 | 16–31 | ||
| aliphatic sulfonate | NGR_c25620-NGR_c25630, NGR_c25650 | 2–11 | 170–283 | 74–153 | ||
| osmoprotectants | NGR_b12990-NGR_b13020 | 5–24 | 31–312 | 19–185 | ||
| polar amino acids | NGR_b03480- NGR_b03510 | 1–26 | 13–157 | 14–439 | ||
| spermidine/putrescine | NGR_b12810-NGR_b12840 | 1–3 | 26–149 | 11–80 | ||
| iron (III) | NGR_c19530- NGR_c19550 | 1–6 | 56–377 | 56–369 | ||
| simple sugar | NGR_b03950, NGR_b03970-NGR_b03990 | 2–4 | 35–169 | 17–68 | ||
| multiple sugar | NGR_b02160-NGR_b02190; NGR_c06670-NGR_c06700; | 4–20 | 18–141 | 13–191 | ||
| NGR_b04010-NGR_b04030, NGR_b04050; | 7–69 | 30–221 | 26–515 | |||
| NGR_b20230-NGR_b20260 | 4–14 | 28–71 | 26–176 | |||
| Down-regulated in bacteroids from both legume hosts | ||||||
| alpha-glucoside | NGR_c03100-NGR_c03140 | 825–2951 | 35–305 | 12–256 | ||
| trehalose/maltose | NGR_b23110-NGR_b23140 | 62–364 | 16–37 | 1–12 | ||
| D-xylose | NGR_c24220-NGR_c24240 | 477–2200 | 20–28 | 6–15 | ||
| fructose | NGR_c01100-NGR_c01120 | 517–3865 | 42–436 | 52–949 | ||
| dipeptide | NGR_c03460-NGR_c03500 | 518–2642 | 56–1055 | 56–768 | ||
| lipoprotein-releasing | NGR_c10690-NGR_c10700 | 194–328 | 90–132 | 77–128 | ||
| lipopolysaccharide | NGR_c00180,NGR_c09020-NGR_c09030 | 165–342 | 66–87 | 44–78 | ||
| glycine betaine/proline | NGR_c27400- NGR_c27420 | 94–250 | 28–37 | 9–42 | ||
| multiple sugar | NGR_c33060- NGR_c33090; NGR_c30950-NGR_c30980 | 58–455 | 12–36 | 3–41 | ||
| simple sugar | NGR_c14510-NGR_c14540 | 97–854 | 24–356 | 19–296 | ||
| branched-chain amino acids | NGR_c25150, NGR_c25170-NGR_c25200 | 961–4353 | 105–1230 | 189–1296 | ||
| peptides/nickel | NGR_c24410-NGR_c24450 | 66–370 | 25–108 | 21–95 | ||
| iron complex | NGR_b02510-NGR_b02530; NGR_b11200-NGR_b11230 | 492–1936 | 31–38 | 69–260 | ||
Note: RPKM (reads per kilobase per million mapped reads); F, free-living bacteria; LB, L. leucocephala bacteroids; VB, V. unguiculata bacteroids.
The ssu genes (NGR_c25620- NGR_c25650, 10–133 folds) encoding a sulfonate monooxygenase (ssuD) and ABC transport proteins (ssuABC) for aliphatic sulfonate, and tauABC (NGR_b17610-NGR_b17630, 6–35 folds) encoding ABC transporters for aminoethane sulfonate were up-regulated in bacteroids. This is consistent with the utilization of aminoethane sulfonate as a sulfur source by S. meliloti and an increased expression of putative sulfonate monooxygenases in bacteroids of G. max and Sesbania rostrata [14] [50].
Transporter proteins of spermidine/putrescine (NGR_b12810-NGR_b12840) were specifically expressed in bacteroids (RPKM = 11–149 versus RPKM = 1–3 in the culture condition). This indicated bacterial uptake of these biogenic amines from plants [51].
Although transcription levels of certain transporter proteins for branched-chain amino acids (NGR_c25150, NGR_c25170-NGR_c25200, 3–11 folds) were down-regulated in bacteroids, their RPKM values were still at a relatively high level (105–1296). On the other hand, certain transporter proteins for polar amino acids (NGR_b03480- NGR_b03510) expressed higher in bacteroids (RPKM = 13–439) compared to the free-living form (RPKM = 1–26). Interestingly, two transporter proteins for branched-chain amino acids (NGR_c08790- NGR_c08800) were specifically up-regulated in V. unguiculata bacteroids (RPKM = 954–1030) compared to L. leucocephala bacteroids (RPKM = 39–48) and the free-living form (RPKM = 162–219).
Iron complex coding genes (NGR_b02510-NGR_b02530; NGR_b11200-NGR_b11230) were highly expressed in the free-living condition (RPKM = 322–1950 versus RPKM = 10–260 in bacteroids), whereas transcriptional levels of ferric ion transporter proteins (NGR_c19530- NGR_c19550, 35–122 folds) were specifically induced in bacteroids (RPKM = 56–377 versus RPKM = 1–6 in free-living condition). Certain ABC transporter proteins of osmoprotectants (NGR_b12990-NGR_b13020) were induced in bacteroids (RPKM = 19–312 versus RPKM = 5–24 in the free-living condition), while others (NGR_c27400- NGR_c27420) were repressed (RPKM = 9–42 versus RPKM = 94–250 in the free-living condition). Similar phenomena were observed for transporter proteins of simple and multiple sugars (Table 3). Moreover, a number of ABC transporters were significantly repressed in bacteroids such as transporter proteins of alpha-glucoside, trehalose/maltose, D-xylose, fructose, dipeptide, lipoprotein-releasing, lipopolysaccharide and peptides/nickel (Table 3). These expression patterns revealed a drastic transcriptional change of ABC transporters between bacteroids and free-living bacteria, suggesting the importance of compound exchange between two symbiotic partners. The observation of overrepresented DEGs in ABC transporters was also documented earlier for other rhizobia-legume symbioses [52].
Secretion Systems
A remarkable number of secretion systems were found in NGR234 [30]. Among the six Type I transporter genes, tolC (NGR_c13520) and prsDE (aprDE, NGR_b10690 and NGR_b10700) were down-regulated in bacteroids (2–3 folds), whereas NGR_c30050, NGR_c30060 and NGR_c30070 were up-regulated in bacteroids (5.5–16.3 folds). Although the prsD mutant of R. leguminosarum was defective in nitrogen fixation on peas [53], the role of the Type I secretion system in NGR234 remains elusive.
As to the Type II-linked protein secretion systems, the gsp cluster encoding general secretion pathway has restricted phyletic distribution among rhizobial species [30]. Our data provided evidence that this gsp cluster (NGR_c22980- NGR_c23100) was actively transcribed in the free-living condition (RPKM = 15–109) and showed a reduced expression level in bacteroids (RPKM = 0–34). The conserved twin arginine translocase (TAT) pathway is encoded by tatABC (NGR_c13710-NGR_c13730) which showed a constitutively high expression level (RPKM = 161–1194) in both free-living and bacteroids. The Sec-SRP (signal recognition particle) system includes inner-membrane proteins (SecD1D2EY, YajC and YidC), ATPase (SecA), SRP receptor (FtsY), targeting proteins (SecB and Ffh), and signal peptidase I. The expression levels of related coding genes (NGR_c26720 secA, NGR_c33550 secB, NGR_c02010 secD1, NGR_c13810 secD2, NGR_c11760 secE, NGR_c12100 secY, NGR_c13800 yajC, NGR_c00820 yidC, NGR_c32250 ffh, NGR_c08280 coding signal peptidase I) in the free-living condition were 2- to 10-fold of those in bacteroids.
NGR234 has two Type III clusters, and only T3SS-I locus (pNGR234a) was reported to modulate the nodulation of many legume hosts excluding L. leucocephala and V. unguiculata [30], [54]. However, it was reported that all of the T3SS-I locus genes were induced by flavonoids and that nolB, rhcJ, nolU and nolV were detected in mature nodules of V. unguiculata [55]. In this study, 13/20 and 19/20 T3SS-I locus genes (NGR_a00520- NGR_a00700, NGR_a00790) were up-regulated in V. unguiculata bacteroids (2- to 11-fold) and in L. leucocephala bacteroids (6- to 172-fold), respectively, compared to the free-living condition. In line with differential expression patterns of T3SS-I locus genes in two hosts, nopP (NGR_a00570), nopX (NGR_a00700) and nopL (NGR_a00770) encoding effector proteins were specifically up-regulated in L. leucocephala nodules (RPKM = 273–476 versus RPKM = 13–27 in V. unguiculata nodules). Considering the presence of bacteria from infection zone of L. leucocephala nodules, our finding is to a certain extent consistent with the view that the stimulation of T3SS coincides with development of the infection thread [56]. Although 8/22 and 22/22 T3SS-II locus genes (pNGR234b, NGR_b22800- NGR_b23010) were up-regulated in V. unguiculata bacteroids (2–356 folds) and in L. leucocephala bacteroids (2–132 folds) respectively, the deletion of seven T3SS-II locus genes (NGR_b22890-NGR_b22950) up-regulated in both V. unguiculata and L. leucocephala did not show any defects in symbiosis [30]. The Type IV cluster (NGR_b10250-NGR_b10360) was constitutively expressed, but at a very low level in both the free-living condition and nodules (RPKM = 1–40).
Surface Polysaccharides
An exo cluster of genes are involved in the synthesis of exopolysaccharides (EPS) in NGR234 [57]. In this study, exoKLAMONP (NGR_b18340- NGR_b18400, 2- to 8-fold), exoXU (NGR_b18280- NGR_b18290, 5- and 3- fold) and exoYFZ (NGR_b18270, NGR_b18260, NGR_b18240, 2- to 7-fold) were up-regulated in L. leucocephala bacteroids compared to the free-living form, whereas only exoY (NGR_b18270, 3-fold), exoX (NGR_b18280, 3-fold) and exoN (NGR_b18390, 3-fold) were up-regulated in V. unguiculata bacteroids. In line with these expression profiles, functional analyses have revealed that mutants of certain exo genes (exoY, exoF, exoQ, exoK, exoL, exoP) formed uninfected Fix- nodules on L. leucocephala but normal Fix+ nodules on V. unguiculata [57].
In the free-living culture, NGR234 synthesizes primarily rough lipopolysaccharides (LPS) and only trace amounts of smooth LPS [58]. A new smooth LPS species with a modified lipid A-Core and rhamnan O-antigen is induced by flavonoid and is present in bacteroids of V. unguiculata [58], [59], [60]. In line with these observations, putative O-antigen biosynthesis protein coding genes NGR_b11970 and NGR_b14100 involved in lipid A biosynthesis were up-regulated in bacteroids of both legumes (2- to 7-fold). Noteworthy the rgpF-rmlB gene cluster (NGR_a03500-NGR_a03580), necessary for the synthesis of rhamnan O-antigen [58], [59], [61], was expressed higher in bacteroids of L. leucocephala than in V. unguiculata bacteroids and in the free-living condition. It has been reported that this gene cluster is up-regulated in the following signaling pathways: NodD1→SyrM2→NodD2→FixF, and NodD1→TtsI→RmlB-WbgA [62], [63]. These regulation patterns led to the hypothesis that the genes absolutely required for rhamnan production are expressed after the bacteria have entered the plant but before they are released into cortical cells of the nodules [61]. Thus, the higher expression of rgpF-rmlB in L. leucocephala bacteroids might be due to the mixture of bacteroids with those bacterial cells in infection threads, a distinct characteristic for indeterminate nodules. In contrast to these up-regulated genes involved in the biosynthesis of lipid A and O-antigen in nodules from either L. leucocephala or both legumes, genes associated to the core region biosynthesis, lpsB and kdtA (NGR_c04250, NGR_c15710), the synthesis and modification of lipid A, lpxABD, acpXL (NGR_c18080, NGR_c13420, NGR_c13440, NGR_c13460, NGR_c18030), unusual sugar (NGR_c12790, NGR_c35510, NGR_c04250), and LPS ABC transporters (NGR_c00180, NGR_c09020-NGR_c09030) were down-regulated in bacteroids from both legumes (2- to 11-fold). This suggested a general down-regulation for LPS production in bacteroids. Similarly, lpxD (NGR_c13420 homolog) involved in lipid A synthesis was down-regulated in R. etli bacteroids [41]. However, mutants in either acpXL down-regulated in bacteroids or members of the rgpF-rmlB cluster could not form pink nodules with V. unguiculata but efficiently nodulate on L. leucocephala [61], [64].
These findings suggested that differences in nodule structures and associated characteristics between L. leucocephala and V. unguiculata may lead to the observed differential expression patterns of EPS and LPS biosynthesis genes. However, the symbiotic significance of these expression profiles, such as the observed expression of rgpF-rmlB cluster in L. leucocephala, may depend on their interactions with other cellular functions of bacteria and host responses.
Energy Metabolism
As expected, in microaerobic environment of nodules from both V. unguiculata and L. leucocephala, the nif genes directly involved in nitrogen fixation were strongly up-regulated. Symbiotic nitrogen fixation is a highly energy-demanding process [65]. In NGR234, there are two cluster of genes NGR_c22030-NGR_c22190 and NGR_c10480-NGR_c10630 encoding NADH dehydrogenase, and the former cluster was up-regulated in bacteroids while the latter was down-regulated. However, expression levels of the former genes (RPKM = 10–99) were quite low compared to the latter genes (RPKM = 150–707) in bacteroids. F-type H+-transporting ATPase coding genes (NGR_c31100-NGR_c31140 and NGR_c04470-NGR_c04500) were all down-regulated (2- to 6-fold). Both NGR_c25510-NGR_c25550 and NGR_c05230-NGR_c05300 encode cytochrome c oxidases, the former was up-regulated (11- to 41-fold, RPKM = 27–408) while the latter down-regulated (5- to 27-fold, RPKM = 15–249) in bacteroids. In NGR234, there are two clusters of genes encoding cbb3 oxidases, locus-I (NGR_c17970-NGR_c17990) and locus-II (NGR_c25780-NGR_c25810). Both loci were up-regulated in bacteroids, but showing a huge difference in expression levels: RPKM = 2453–3677 for locus-I and RPKM = 26–81 for locus-II. Interestingly, NGR_c01900 and NGR_c01910 encoding subunits of cytochrome bd ubiquinol oxidase were up-regulated in L. leucocephala (3- and 5-fold, respectively) but down-regulated in V. unguiculata (3- and 10-fold, respectively). However, it should be noted that expression levels of NGR_c01900 and NGR_c01910 in L. leucocephala were lower (RPKM = 49–51) than cbb3 oxidase locus-I (RPKM = 2453–3677). Although it was reported that Azorhizobium caulinodans uses both cytochrome cbb3 and bd as terminal oxidases for symbiotic nitrogen fixation [66], cbb3 oxidase could be the major terminal oxidase for symbiotic nitrogen fixation in bacteroids of both L. leucocephala and V. unguiculata. It has been reported that the constitutive expression of the thiamine biosynthetic pathway caused the production of cbb3 oxidase in the free-living condition and an increased capacity in nitrogen fixation during symbiosis [67], [68]. In this study we presented additional evidence that thiCOGE (NGR_b02900-NGR_b02930) and thiD (NGR_b18410) were up-regulated (2- to 13-fold) in bacteroids from nodules of both L. leucocephala (RPKM = 26 - 46) and V. unguiculata (RPKM = 44 -202), with a higher expression level in V. unguiculata bacteroids. This finding further supports the view that the thiamine biosynthesis genes are commonly expressed extrachromosomal genes in association with plants [40].
NGR_b03260- NGR_b03300 involved in pyrroloquinoline quinone (PQQ) biosynthesis were specifically activated in L. leucocephala bacteroids (RPKM = 169–580 versus RPKM = 4–19 in V. unguiculata bacteroids and the free-living form). PQQ has been found as a redox cofactor for membrane-bound dehydrogenases [69]. The presence of PQQ-dependent glucose dehydrogenase was also reported in rhizobia such as Rhizobium tropici and S. meliloti etc. [70]. The PQQ-linked glucose dehydrogenase has also been demonstrated as a requirement by S. meliloti for optimal nodulation efficiency and competitiveness on alfalfa roots [71]. Close to these pqq genes in NGR234 genome, NGR_b03250 encoding a periplasmic alcohol dehydrogenase with ferricytochrome c as the acceptor was specifically up-regulated in L. leucocephala bacteroids (RPKM = 1367 versus RPKM = 20 in V. unguiculata bacteroids and RPKM = 29 in the free culture). Moreover, NGR_b03210-NGR_b03240 within the same loci showed a similar expression trend as NGR_b03250- NGR_b03300, suggesting their potential role in adapting to the nodule environment of L. leucocephala. Proteins encoded by NGR_b03210-NGR_b03240 include a pseudoazurin, a signal transduction histidine kinase, a FIST containing signal transduction protein [72] and a LuxR family response transcriptional regulator. It would be interesting to study whether these signal transduction systems could regulate the expression of PQQ genes. It has also been shown that PQQ could work as an antioxidant protecting bacteria from oxidative damage, or as a nutrient to support bacterial growth [69]. However, the role of PQQ in rhizobium-legume symbiotic interactions remains largely unknown.
A cluster of genes (NGR_c14380-NGR_c14420) were expressed higher in V. unguiculata bacteroids (RPKM = 546–2522) than in L. leucocephala bacteroids (RPKM = 56–305) and the free-living form (RPKM = 9–20). These include genes encoding glutamine amidotransferase-like protein GlxB, putative glutamate synthase subunits GlxCD and glutamine synthetase GlnT. This is in contrast to the relatively low transcription level of glt genes (RPKM = 47–277) encoding glutamine synthetase, and NGR_c32060 (RPKM = 107–113) encoding glutamine amidotransferase in bacteroids from both hosts. Thus, in addition to the up-regulated glutamine synthetase GlnII (NGR_b20670, RPKM = 1807–3916 in V. unguiculata and L. leucocephala bacteroids versus RPKM = 281 in the free culture), NGR234 actively recruited another set of genes in glutamate metabolism in V. unguiculata nodules.
Role of Nitric Oxide
Recently, nitric oxide (NO) has been detected at different steps of the symbiosis between legumes and rhizobia [73], [74], [75]. Modulation of NO levels was demonstrated to be involved in the establishment and persistence of the symbiosis [73], [76], [77]. In this study, the up-regulation of NapABC (nitrate reductase, NGR_c10020-NGR_c10040, 10- to 15-fold) and NirK (nitrite reductase, NGR_c09950, 37- to 56-fold), and the down-regulation of NorC (nitric oxide reductase, NGR_c09850, 2- to 3-fold) suggested that bacteroids contributed to the NO pool within nitrogen-fixing nodules from both L. leucocephala and V. unguiculata. In line with this finding, in M. truncatula nodules formed by napA or nirK mutant of S. meliloti, the production of NO was decreased by about 35% compared with that of the wild-type control [78]. It was reported that cytochrome c550 is required for the succinate-dependent nitrite reduction and might be involved in electron transfer to the copper-containing nitrite reductase of B. japonicum [79]. In this study, NGR_b03130 encoding cytochrome c550 was specifically up-regulated in L. leucocephala nodules (RPKM = 186 versus RPKM = 5–6 in V. unguiculata nodules and in the free-living condition). In line with this, NGR_b03210, encoding a pseudoazurin which was demonstrated to be an electron donor to the copper-containing nitrite reductase in other denitrifying bacteria [80], was also strongly up-regulated in L. leucocephala nodules (RPKM = 449) compared to V. unguiculata nodules (RPKM = 20) and the free-living condition (RPKM = 11). Although cytochrome c550 is not required by B. japonicum for nitrogen-fixation in determinate nodules of G. max [81], potential roles of cytochrome c550 and/or pseudoazurin in indeterminate nodules such as L. leucocephala nodules are still unknown.
Carbon Metabolism
In bacteroids of nodules from both L. leucocephala and V. unguiculata, strongly up-regulated genes compared to free-living bacteria also included those encoding the C4-dicarboxylate transporter, DctA (NGR_b21870, 331- and 460-fold, respectively), the phosphoenolpyruvate carboxykinase, PckA (NGR_c33940, 182- and 219-fold, respectively), and a fructose-bisphosphate aldolase (NGR_c28100, 89- and 71-fold, respectively), the acetate kinase AckA (NGR_b13640, 118- and 182-fold, respectively) and the phosphate acetyltransferase Pta (NGR_b13630, 166- and 228-fold, respectively). These data are consistent with the requirement for succinate metabolism and acetyl-CoA [82], [83], suggesting that both L. leucocephala and V. unguiculata provide succinate to bacteroids as a carbon source.
In the comparison between L. leucocephala bacteroids and V. unguiculata bacteroids, DEGs were enriched in the phenylalanine metabolism pathway (rhi00360, Table S3). paa genes (NGR_c26090-NGR_c26200) were strongly up-regulated in V. unguiculata bacteroids (RPKM = 78–1195) compared to L. leucocephala bacteroids (RPKM = 12–157) and the free-living culture (RPKM = 7–73). Moreover, NGR_b02860, encoding a malonyl-CoA decarboxylase which catalyses the reaction of malonyl-CoA to acetyl-CoA and CO2, was also specifically up-regulated in V. unguiculata bacteroids. This may lead to a higher yield of acetyl-CoA in V. unguiculata bacteroids. NGR_c30010 (4-hydroxyphenylpyruvate dioxygenase), NGR_c29980 (homogentisate 1,2-dioxygenase) and NGR_c29960 (fumarylacetoacetate hydrolase) were expressed higher in V. unguiculata bacteroids (RPKM = 358–3133) than in L. leucocephala bacteroids (RPKM = 60–303) and the free-living form (RPKM = 75–360). Enzymes encoded by these genes are involved in the production of acetoacetate and fumarate. Therefore, the citrate cycle seems to be more active in V. unguiculata bacteroids than in L. leucocephala bacteroids.
A considerable number of DEGs encoding enzymes that participate in glyoxylate and dicarboxylate metabolism were also found (Table S3), suggesting a potential role of C2 metabolism in symbiosis. Recently, it was reported that this glyoxylate cycle of R. leguminosarum was more strongly induced in the rhizosphere of pea, the compatible legume host of this bacterium, than in that of alfalfa and sugar beet [82]. Moreover, the coincident expressions of glyoxylate cycle genes and a subset of nif genes in the chemoautotrophic culture of B. japonicum [84] imply potential coordination in transcriptions of these genes. However, S. meliloti mutants of two principle genes, aceA (encoding isocitrate lyase) and glcB (encoding malate synthase), in the glyoxylate cycle were not impaired in nodulation and nitrogen fixation on alfalfa [85]. NGR_c03920- NGR_c03950 expressed higher in V. unguiculata bacteroids (RPKM = 254–401) than in L. leucocephala bacteroids (RPKM = 55–105) and the free-living condition (RPKM = 24–40). They encode glycolate oxidase which catalyses the oxidation from glycolate and O2 to glyoxylate and H2O2. The glcD (NGR_c03920 homolog) mutant of A. caulinodans formed Fix- nodules on S. rostrata [86]. On the other hand, it has been reported, in S. meliloti-Medicago symbiosis, an optimal level of H2O2 is required in the normal progression of infection threads and the efficient release of bacteria into nodule cells [87], [88]. In this study, NGR_b11000 encoding catalase C (peroxidase) was up-regulated in both V. unguiculata bacteroids (RPKM = 19) and L. leucocephala bacteroids (RPKM = 48) compared to the free-living form (RPKM = 4). However, the symbiotic roles of glycolate oxidase and products from its activation in bacteroids remain elusive. Despite the unclear picture of the symbiotic role of glyoxylate cycle, there were large amounts of acetate and fatty acids in the nodules [89]. On the other hand, as shown in Table 3S, the down-regulation of fatty acid synthesis genes and the up-regulation of fatty acid metabolism genes in bacteroids might be related to the presence of fatty acids in nodules.
As shown in Figure 1E and 1F, PHB granules were observed in bacteroids of nodules from both L. leucocephala and V. unguiculata. In line with this observation, the acetyl-CoA acetyltransferase coding gene, phbA (NGR_c32720, 3-fold) and the poly-beta-hydroxybutyrate polymerase coding gene phbC1 (NGR_c34290, 4- and 7-fold, respectively) were up-regulated in both legume hosts while phbC2 (NGR_c14000) and phbB (NGR_c32710) were constitutively expressed. Noteworthy, phbZ encoding a polyhydroxybutyrate depolymerase (NGR_b03370) was up-regulated by 19-fold and 9-fold in L. leucocephala and V. unguiculata, respectively. bdhA2 encoding a 3-hydroxybutyrate dehydrogenase (NGR_c23850) was constitutively expressed. Therefore, PHB level is subject to strict modulation in bacteroids. It has been reported that PHB was not accumulated in the free-living culture of R. etli when biotin was added [90]. The transcription of bdhA in free-living S. meliloti was higher in the presence of added biotin [91]. These earlier reports suggested biotin-induced PHB degradation in the free-living condition. However, bioABDF involved in biotin synthesis were all up-regulated in L. leucocephala (3- to 9-fold) but down-regulated in V. unguiculata nodules (3-fold), indicating a potential complex regulation on PHB level in bacteroids.
Notably, PHB granules in L. leucocephala bacteroids were larger than those in V. unguiculata bacteroids (Figure 1E and 1F). Phasins encoded by phaP genes could bind to PHB granules and promote PHB synthesis by regulating the surface/volume ratio of PHB granules or by interacting with the polyhydroxyalkanoate synthase [92]. It was also reported that phaP1 and phaP2 in S. meliloti contributed to PHB accumulation in bacteroids and symbiotic nitrogen fixation [93]. NGR234 has three phaP homologs, phaP1 (NGR_c03360), phaP2 (NGR_c13240), phaP3 (NGR_a00900). As shown in Table 4, phaP3 seemed to be specifically and strongly expressed in bacteroids of both legume hosts while phaP1 and phaP2 were up-regulated in L. leucocephala bacteroids (1.8-fold) and phaP1 was down-regulated in V. unguiculata bacteroids. Therefore, these expression patterns of phaP homologs agree well with the difference in PHB granule size observed in L. leucocephala and V. unguiculata bacteroids.
Table 4. Expression patterns of phaP genes encoding phasins.
| phaP3 (408 bp) | phaP1(447 bp) | phaP2 (354 bp) | ||
| Unique reads | (F) | 2 | 8114 | 12203 |
| (LB) | 7063 | 1693 | 2592 | |
| (VB) | 21683 | 1271 | 6715 | |
| Coverage | (F) | 30.4% | 99.8% | 99.7% |
| (LB) | 99.8% | 99.8% | 99.7% | |
| (VB) | 99.8% | 99.8% | 99.4% | |
| RPKM | (F) | 0.64 | 2379.38 | 4518.51 |
| (LB) | 19536.37 | 4274.29 | 8263.17 | |
| (VB) | 15673.09 | 838.56 | 5594.2 | |
| Ratio | (LB/F) | 30404.78 | 1.8 | 1.83 |
| (VB/F) | 24392.3 | 0.35 | 1.24 | |
| (LB/VB) | 1.25 | 5.1 | 1.48 | |
| P-value | (LB/F) | 0 | 7.38E-94 | 2.42E-150 |
| (VB/F) | 0 | 0 | 6.1051E-44 | |
| (LB/VB) | 4.19E-56 | 0.00E+00 | 3.30E-60 | |
| FDR | (LB/F) | 0 | 9.25E-93 | 4.48E-149 |
| (VB/F) | 0 | 0 | 2.20E-43 | |
| (LB/VB) | 1.59E-54 | 0 | 1.35E-58 | |
Note: VB/F, V. unguiculata bacteroids vs free-living bacteria; LB/F, L. leucocephala bacteroids vs free-living bacteria; LB/VB, L. leucocephala bacteroids vs V. unguiculata bacteroids; RPKM (reads per kilobase per million mapped reads); FDR (false discovery rate).
Conclusions
Although Sinorhizobium sp. NGR234 is well known for its broad-host range and the ability of forming either determinate or indeterminate nodules on corresponding legumes [24], genome-level transcriptomic adaptions to determinate or indeterminate nodules of different legumes have not been investigated for this strain before. By using RNA-seq, we conducted a high-resolution transcriptomic analysis for NGR234 bacteroids in indeterminate nodules of L. leucocephala and in determinate nodules of V. unguiculata. Both common and distinct transcription patterns were uncovered for NGR234 bacteroids in these two non-model legumes. These shed new light on the mysterious mechanisms of rhizobial adaptations to diverse legume hosts.
Supporting Information
(XLS)
(XLS)
(XLS)
(XLS)
Acknowledgments
We thank Dr. Christian Staehelin for providing Sinorhizobium sp. NGR234, and Dr. Peter Mergaert and Dr. Esperanza Martinez-Romero for their helpful comments on the manuscript. We also thank Dr. Luz B. Gilbert for language revision.
Funding Statement
This study was funded by National Natural Science Foundation of China (31200002), National Natural Science Foundation of China (31170002), and Innovative Project of State Key Laboratory of Agrobiotechnology (SKLAB) Grant (2012SKLAB01-9). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Oldroyd GE, Murray JD, Poole PS, Downie JA (2011) The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45: 119–144. [DOI] [PubMed] [Google Scholar]
- 2. Kereszt A, Mergaert P, Kondorosi E (2011) Bacteroid development in legume nodules: evolution of mutual benefit or of sacrificial victims? Mol Plant Microbe Interact 24: 1300–1309. [DOI] [PubMed] [Google Scholar]
- 3. Vasse J, de Billy F, Camut S, Truchet G (1990) Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J Bacteriol 172: 4295–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Szczyglowski K, Shaw RS, Wopereis J, Copeland S, Hamburger D, et al. (1998) Nodule organogenesis and symbiotic mutants of the model legume Lotus japonicus . Mol Plant Microbe In 11: 684–697. [Google Scholar]
- 5. Mergaert P, Uchiumi T, Alunni B, Evanno G, Cheron A, et al. (2006) Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc Natl Acad Sci U S A 103: 5230–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oono R, Schmitt I, Sprent JI, Denison RF (2010) Multiple evolutionary origins of legume traits leading to extreme rhizobial differentiation. New Phytol 187: 508–520. [DOI] [PubMed] [Google Scholar]
- 7. Ampe F, Kiss E, Sabourdy F, Batut J (2003) Transcriptome analysis of Sinorhizobium meliloti during symbiosis. Genome Biol 4: R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Becker A, Berges H, Krol E, Bruand C, Ruberg S, et al. (2004) Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol Plant Microbe Interact 17: 292–303. [DOI] [PubMed] [Google Scholar]
- 9. Djordjevic MA (2004) Sinorhizobium meliloti metabolism in the root nodule: A proteomic perspective. Proteomics 4: 1859–1872. [DOI] [PubMed] [Google Scholar]
- 10. Capela D, Filipe C, Bobik C, Batut J, Bruand C (2006) Sinorhizobium meliloti differentiation during symbiosis with alfalfa: a transcriptomic dissection. Mol Plant Microbe Interact 19: 363–372. [DOI] [PubMed] [Google Scholar]
- 11. Karunakaran R, Ramachandran VK, Seaman JC, East AK, Mouhsine B, et al. (2009) Transcriptomic analysis of Rhizobium leguminosarum biovar viciae in symbiosis with host plants Pisum sativum and Vicia cracca . J Bacteriol 191: 4002–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pessi G, Ahrens CH, Rehrauer H, Lindemann A, Hauser F, et al. (2007) Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol Plant Microbe Interact 20: 1353–1363. [DOI] [PubMed] [Google Scholar]
- 13. Shimoda Y, Shinpo S, Kohara M, Nakamura Y, Tabata S, et al. (2008) A large scale analysis of protein-protein interactions in the nitrogen-fixing bacterium Mesorhizobium loti . DNA Research 15: 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Delmotte N, Ahrens CH, Knief C, Qeli E, Koch M, et al. (2010) An integrated proteomics and transcriptomics reference data set provides new insights into the Bradyrhizobium japonicum bacteroid metabolism in soybean root nodules. Proteomics 10: 1391–1400. [DOI] [PubMed] [Google Scholar]
- 15. Nomura M, Arunothayanan H, Van Dao T, Le HT-P, Kaneko T, et al. (2010) Differential protein profiles of Bradyrhizobium japonicum USDA110 bacteroid during soybean nodule development. Soil Science & Plant Nutrition 56: 579–590. [Google Scholar]
- 16. Koch M, Delmotte N, Rehrauer H, Vorholt JA, Pessi G, et al. (2010) Rhizobial adaptation to hosts, a new facet in the legume root-nodule symbiosis. Mol Plant Microbe Interact 23: 784–790. [DOI] [PubMed] [Google Scholar]
- 17. Haag AF, Baloban M, Sani M, Kerscher B, Pierre O, et al. (2011) Protection of Sinorhizobium against host cysteine-rich antimicrobial peptides is critical for symbiosis. Plos Biology 9: e1001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Van de Velde W, Zehirov G, Szatmari A, Debreczeny M, Ishihara H, et al. (2010) Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327: 1122–1126. [DOI] [PubMed] [Google Scholar]
- 19. Mulley G, White JP, Karunakaran R, Prell J, Bourdes A, et al. (2011) Mutation of GOGAT prevents pea bacteroid formation and N2 fixation by globally downregulating transport of organic nitrogen sources. Mol Microbiol 80: 149–167. [DOI] [PubMed] [Google Scholar]
- 20. Prell J, White JP, Bourdes A, Bunnewell S, Bongaerts RJ, et al. (2009) Legumes regulate Rhizobium bacteroid development and persistence by the supply of branched-chain amino acids. Proc Natl Acad Sci U S A 106: 12477–12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lodwig EM, Hosie AH, Bourdes A, Findlay K, Allaway D, et al. (2003) Amino-acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis. Nature 422: 722–726. [DOI] [PubMed] [Google Scholar]
- 22. Masson-Boivin C, Giraud E, Perret X, Batut J (2009) Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends Microbiol 17: 458–466. [DOI] [PubMed] [Google Scholar]
- 23. Tian CF, Zhou YJ, Zhang YM, Li QQ, Zhang YZ, et al. (2012) Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations. Proc Natl Acad Sci U S A 109: 8629–8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pueppke SG, Broughton WJ (1999) Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol Plant Microbe Interact 12: 293–318. [DOI] [PubMed] [Google Scholar]
- 25. Beringer JE (1974) R factor transfer in Rhizobium leguminosarum . J Gen Microbiol 84: 188–198. [DOI] [PubMed] [Google Scholar]
- 26.Vincent JM (1970) A manual for the practical study of root nodule bacteria. Oxford: Blackwell.
- 27. Van de Velde W, Guerra JC, De Keyser A, De Rycke R, Rombauts S, et al. (2006) Aging in legume symbiosis. A molecular view on nodule senescence in Medicago truncatula . Plant Physiol 141: 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Day DA, Price GD, Udvardi MK (1989) Membrane interface of the Bradyrhizobium japonicum- Glycine max symbiosis: peribacteroid units from soyabean nodules. Austra J Plant Physiol 16: 69–84. [Google Scholar]
- 29. Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, et al. (2009) Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Research 37: e123–e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schmeisser C, Liesegang H, Krysciak D, Bakkou N, Le Quere A, et al. (2009) Rhizobium sp. strain NGR234 possesses a remarkable number of secretion systems. Appl Environ Microbiol 75: 4035–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li R, Yu C, Li Y, Lam TW, Yiu SM, et al. (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967. [DOI] [PubMed] [Google Scholar]
- 32. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- 33.Thorvaldsdottir H, Robinson JT, Mesirov JP (2012) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform (doi:10.1093/bib/bbs017). [DOI] [PMC free article] [PubMed]
- 34. Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32: D277–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Perez-Llamas C, Lopez-Bigas N (2011) Gitools: analysis and visualisation of genomic data using interactive heat-maps. PLoS ONE 6: e19541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Perkins TT, Kingsley RA, Fookes MC, Gardner PP, James KD, et al. (2009) A strand-specific RNA-seq analysis of the transcriptome of the Typhoid Bacillus Salmonella Typhi. Plos Genetics 5: e1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yoder-Himes DR, Chain PS, Zhu Y, Wurtzel O, Rubin EM, et al. (2009) Mapping the Burkholderia cenocepacia niche response via high-throughput sequencing. Proc Natl Acad Sci U S A 106: 3976–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oliver HF, Orsi RH, Ponnala L, Keich U, Wang W, et al. (2009) Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics 10: 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Westermann AJ, Gorski SA, Vogel J (2012) Dual RNA-seq of pathogen and host. Nat Rev Microbiol 10: 618–630. [DOI] [PubMed] [Google Scholar]
- 40. López-Guerrero MG, Ormeño-Orrillo E, Acosta JL, Mendoza-Vargas A, Rogel MA, et al. (2012) Rhizobial extrachromosomal replicon variability, stability and expression in natural niches. Plasmid 68: 149–158. [DOI] [PubMed] [Google Scholar]
- 41. Vercruysse M, Fauvart M, Beullens S, Braeken K, Cloots L, et al. (2011) A comparative transcriptome analysis of Rhizobium etli bacteroids: specific gene expression during symbiotic nongrowth. Mol Plant Microbe Interact 24: 1553–1561. [DOI] [PubMed] [Google Scholar]
- 42. da Silva Batista JS, Hungria M (2012) Proteomics reveals differential expression of proteins related to a variety of metabolic pathways by genistein-induced Bradyrhizobium japonicum strains. J Proteomics 75: 1211–1219. [DOI] [PubMed] [Google Scholar]
- 43. Lang K, Lindemann A, Hauser F, Gottfert M (2008) The genistein stimulon of Bradyrhizobium japonicum . Mol Genet Genomics 279: 203–211. [DOI] [PubMed] [Google Scholar]
- 44. Ames P, Bergman K (1981) Competitive advantage provided by bacterial motility in the formation of nodules by Rhizobium meliloti . J Bacteriol 148: 728–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fujishige NA, Kapadia NN, De Hoff PL, Hirsch AM (2006) Investigations of Rhizobium biofilm formation. FEMS Microbiol Ecol 56: 195–206. [DOI] [PubMed] [Google Scholar]
- 46. Finan TM, Gough C, Truchet G (1995) Similarity between the Rhizobium meliloti fliP gene and pathogenicity-associated genes from animal and plant pathogens. Gene 152: 65–67. [DOI] [PubMed] [Google Scholar]
- 47. Bardin S, Dan S, Osteras M, Finan TM (1996) A phosphate transport system is required for symbiotic nitrogen fixation by Rhizobium meliloti . J Bacteriol 178: 4540–4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Geiger O, Rohrs V, Weissenmayer B, Finan TM, Thomas-Oates JE (1999) The regulator gene phoB mediates phosphate stress-controlled synthesis of the membrane lipid diacylglyceryl-N,N,N-trimethylhomoserine in Rhizobium (Sinorhizobium) meliloti . Mol Microbiol 32: 63–73. [DOI] [PubMed] [Google Scholar]
- 49. Lopez-Lara IM, Gao JL, Soto MJ, Solares-Perez A, Weissenmayer B, et al. (2005) Phosphorus-free membrane lipids of Sinorhizobium meliloti are not required for the symbiosis with alfalfa but contribute to increased cell yields under phosphorus-limiting conditions of growth. Mol Plant Microbe Interact 18: 973–982. [DOI] [PubMed] [Google Scholar]
- 50. Tsukada S, Aono T, Akiba N, Lee KB, Liu CT, et al. (2009) Comparative genome-wide transcriptional profiling of Azorhizobium caulinodans ORS571 grown under free-living and symbiotic conditions. Appl Environ Microbiol 75: 5037–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fujihara S (2009) Biogenic amines in rhizobia and legume root nodules. Microbes Environ 24: 1–13. [DOI] [PubMed] [Google Scholar]
- 52. Delmotte N, Ahrens CH, Knief C, Qeli E, Koch M, et al. (2010) An integrated proteomics and transcriptomics reference data set provides new insights into the Bradyrhizobium japonicum bacteroid metabolism in soybean root nodules. Proteomics 10: 1391–1400. [DOI] [PubMed] [Google Scholar]
- 53. Finnie C, Hartley NM, Findlay KC, JA D (1997) The Rhizobium leguminosarum prsDE genes are required for secretion of several proteins, some of which influence nodulation, symbiotic nitrogen fixation and exopolysaccharide modification. Mol Microbiol 25: 135–146. [DOI] [PubMed] [Google Scholar]
- 54. Viprey V, Del Greco A, Golinowski W, Broughton WJ, Perret X (1998) Symbiotic implications of type III protein secretion machinery in Rhizobium . Mol Microbiol 28: 1381–1389. [DOI] [PubMed] [Google Scholar]
- 55. Perret X, Freiberg C, Rosenthal A, Broughton WJ, Fellay R (1999) High-resolution transcriptional analysis of the symbiotic plasmid of Rhizobium sp. NGR234. Mole Microbiol 32: 415–425. [DOI] [PubMed] [Google Scholar]
- 56. Deakin WJ, Broughton WJ (2009) Symbiotic use of pathogenic strategies: rhizobial protein secretion systems. Nat Rev Microbiol 7: 312–320. [DOI] [PubMed] [Google Scholar]
- 57. Staehelin C, Forsberg LS, D'Haeze W, Gao MY, Carlson RW, et al. (2006) Exo-oligosaccharides of Rhizobium sp. strain NGR234 are required for symbiosis with various legumes. J Bacteriol 188: 6168–6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Reuhs BL, Relic B, Forsberg LS, Marie C, Ojanen-Reuhs T, et al. (2005) Structural characterization of a flavonoid-inducible Pseudomonas aeruginosa A-band-like O antigen of Rhizobium sp. strain NGR234, required for the formation of nitrogen-fixing nodules. J Bacteriol 187: 6479–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ardissone S, Noel KD, Klement M, Broughton WJ, Deakin WJ (2011) Synthesis of the flavonoid-induced lipopolysaccharide of Rhizobium Sp. strain NGR234 requires rhamnosyl transferases encoded by genes rgpF and wbgA. Mol Plant Microbe Interact 24: 1513–1521. [DOI] [PubMed] [Google Scholar]
- 60. Fraysse N, Jabbouri S, Treilhou M, Couderc F, Poinsot V (2002) Symbiotic conditions induce structural modifications of Sinorhizobium sp NGR234 surface polysaccharides. Glycobiology 12: 741–748. [DOI] [PubMed] [Google Scholar]
- 61. Broughton WJ, Hanin M, Relic B, Kopcinska J, Golinowski W, et al. (2006) Flavonoid-inducible modifications to rhamnan O antigens are necessary for Rhizobium sp. strain NGR234-legume symbioses. J Bacteriol 188: 3654–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Marie C, Deakin WJ, Ojanen-Reuhs T, Diallo E, Reuhs B, et al. (2004) TtsI, a key regulator of Rhimbium species NGR234 is required for type III-dependent protein secretion and synthesis of rhamnose-rich polysaccharides. Mol Plant Microbe Interact 17: 958–966. [DOI] [PubMed] [Google Scholar]
- 63. Kobayashi H, Graven YN, Broughton WJ, Perret X (2004) Flavonoids induce temporal shifts in gene-expression of nod-box controlled loci in Rhizobium sp. NGR234. Mole Microbiol 51: 335–347. [DOI] [PubMed] [Google Scholar]
- 64. Ardissone S, Kobayashi H, Kambara K, Rummel C, Noel KD, et al. (2011) Role of BacA in lipopolysaccharide synthesis, peptide transport, and nodulation by Rhizobium sp. strain NGR234. J Bacteriol 193: 2218–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dixon R, Kahn D (2004) Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol 2: 621–631. [DOI] [PubMed] [Google Scholar]
- 66. Kaminski PA, Kitts CL, Zimmerman Z, Ludwig RA (1996) Azorhizobium caulinodans uses both cytochrome bd (quinol) and cytochrome cbb(3) (cytochrome c) terminal oxidases for symbiotic N-2 fixation. J Bacteriol 178: 5989–5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Soberon M, Membrillo-Hernandez J, Aguilar GR, Sanchez F (1990) Isolation of Rhizobium phaseoli Tn5-induced mutants with altered expression of cytochrome terminal oxidases o and aa3. J Bacteriol 172: 1676–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. MirandaRios J, Morera C, Taboada H, Davalos A, Encarnacion S, et al. (1997) Expression of thiamin biosynthetic genes (thiCOGE) and production of symbiotic terminal oxidase cbb(3), in Rhizobium etli . J Bacteriol 179: 6887–6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Misra HS, Rajpurohit YS, Khairnar NP (2012) Pyrroloquinoline-quinone and its versatile roles in biological processes. J Biosci 37: 313–325. [DOI] [PubMed] [Google Scholar]
- 70. Bernardelli CE, Luna MF, Galar ML, Boiardi JL (2001) Periplasmic PQQ-dependent glucose oxidation in free-living and symbiotic rhizobia. Curr Microbiol 42: 310–315. [DOI] [PubMed] [Google Scholar]
- 71. Bernardelli CE, Luna MF, Galar ML, Boiardi JL (2008) Symbiotic phenotype of a membrane-bound glucose dehydrogenase mutant of Sinorhizobium meliloti . Plant Soil 313: 217–225. [Google Scholar]
- 72. Borziak K, Zhulin IB (2007) FIST: a sensory domain for diverse signal transduction pathways in prokaryotes and ubiquitin signaling in eukaryotes. Bioinformatics 23: 2518–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nagata M, Murakami E, Shimoda Y, Shimoda-Sasakura F, Kucho K, et al. (2008) Expression of a class 1 hemoglobin gene and production of nitric oxide in response to symbiotic and pathogenic bacteria in Lotus japonicus . Mol Plant Microbe Interact 21: 1175–1183. [DOI] [PubMed] [Google Scholar]
- 74. Meakin GE, Bueno E, Jepson B, Bedmar EJ, Richardson DJ, et al. (2007) The contribution of bacteroidal nitrate and nitrite reduction to the formation of nitrosylleghaemoglobin complexes in soybean root nodules. Microbiology 153: 411–419. [DOI] [PubMed] [Google Scholar]
- 75. Baudouin E, Pieuchot L, Engler G, Pauly N, Puppo A (2006) Nitric oxide is formed in Medicago truncatula-Sinorhizobium meliloti functional nodules. Mol Plant Microbe Interact 19: 970–975. [DOI] [PubMed] [Google Scholar]
- 76. Cam Y, Pierre O, Boncompagni E, Herouart D, Meilhoc E, et al. (2012) Nitric oxide (NO): a key player in the senescence of Medicago truncatula root nodules. New Phytol 196: 548–560. [DOI] [PubMed] [Google Scholar]
- 77. Pii Y, Crimi M, Cremonese G, Spena A, Pandolfini T (2007) Auxin and nitric oxide control indeterminate nodule formation. BMC Plant Biol 7: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Horchani F, Prevot M, Boscari A, Evangelisti E, Meilhoc E, et al. (2010) Both plant and bacterial nitrate reductases contribute to nitric oxide production in Medicago truncatula nitrogen-fixing nodules. Plant Physiol 155: 1023–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bueno E, Bedmar EJ, Richardson DJ, Delgado MJ (2008) Role of Bradyrhizobium japonicum cytochrome c550 in nitrite and nitrate respiration. FEMS Microbiol Lett 279: 188–194. [DOI] [PubMed] [Google Scholar]
- 80. Kukimoto M, Nishiyama M, Ohnuki T, Turley S, Adman ET, et al. (1995) Identification of interaction site of pseudoazurin with its redox partner, copper-containing nitrite reductase from Alcaligenes faecalis S-6. Protein Eng 8: 153–158. [DOI] [PubMed] [Google Scholar]
- 81. Bott M, Thony-Meyer L, Loferer H, Rossbach S, Tully RE, et al. (1995) Bradyrhizobium japonicum cytochrome c550 is required for nitrate respiration but not for symbiotic nitrogen fixation. J Bacteriol 177: 2214–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ramachandran VK, East AK, Karunakaran R, Downie JA, Poole PS (2011) Adaptation of Rhizobium leguminosarum to pea, alfalfa and sugar beet rhizospheres investigated by comparative transcriptomics. Genome Biol 12: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Barnett MJ, Toman CJ, Fisher RF, Long SR (2004) A dual-genome symbiosis chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc Natl Acad Sci U S A 101: 16636–16641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Franck WL, Chang WS, Qiu J, Sugawara M, Sadowsky MJ, et al. (2008) Whole-genome transcriptional profiling of Bradyrhizobium japonicum during chemoautotrophic growth. J Bacteriol 190: 6697–6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ramirez-Trujillo JA, Encarnacion S, Salazar E, de los Santos AG, Dunn MF, et al. (2007) Functional characterization of the Sinorhizobium meliloti acetate metabolism genes aceA, SMc00767, and glcB . J Bacteriol 189: 5875–5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Suzuki S, Aono T, Lee KB, Suzuki T, Liu CT, et al. (2007) Rhizobial factors required for stem nodule maturation and maintenance in Sesbania rostrata-Azorhizobium caulinodans ORS571 symbiosis. Appl Environ Microbiol 73: 6650–6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Jamet A, Mandon K, Puppo A, Herouart D (2007) H2O2 is required for optimal establishment of the Medicago sativa/Sinorhizobium meliloti symbiosis. J Bacteriol 189: 8741–8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Jamet A, Sigaud S, Van de Sype G, Puppo A, Herouart D (2003) Expression of the bacterial catalase genes during Sinorhizobium meliloti-Medicago sativa symbiosis and their crucial role during the infection process. Mol Plant Microbe Interact 16: 217–225. [DOI] [PubMed] [Google Scholar]
- 89. Johnson GV, Evans HJ, Ching T (1966) Enzymes of the glyoxylate cycle in rhizobia and nodules of legumes. Plant Physiol 41: 1330–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Encarnacion S, Dunn M, Willms K, Mora J (1995) Fermentative and aerobic metabolism in Rhizobium etli . J Bacteriol 177: 3058–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hofmann K, Heinz EB, Charles TC, Hoppert M, Liebl W, et al. (2000) Sinorhizobium meliloti strain 1021 bioS and bdhA gene transcriptions are both affected by biotin available in defined medium. FEMS Microbiol Lett 182: 41–44. [DOI] [PubMed] [Google Scholar]
- 92. York GM, Stubbe J, Sinskey AJ (2001) New insight into the role of the PhaP phasin of Ralstonia eutropha in promoting synthesis of polyhydroxybutyrate. J Bacteriol 183: 2394–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wang C, Sheng X, Equi RC, Trainer MA, Charles TC, et al. (2007) Influence of the poly-3-hydroxybutyrate (PHB) granule-associated proteins (PhaP1 and PhaP2) on PHB accumulation and symbiotic nitrogen fixation in Sinorhizobium meliloti Rm1021. J Bacteriol 189: 9050–9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLS)
(XLS)
(XLS)