Abstract
Developmental programs are driven by transcription factors that coordinate precise patterns of gene expression. While recent publications have described the importance of coordinated action of transcriptional activators at multiple cis-regulatory modules or enhancers, the contribution of sequence-specific repressors to overall regulation and robustness of gene expression has been difficult to ascertain. The Ets transcriptional repressor Yan functions as part of a conserved network downstream of receptor tyrosine kinase (RTK) signaling in Drosophila. This network displays switch-like responsiveness to RTK signaling, with the transition from a high-Yan to a low-Yan state induced by mitogen-activated protein kinase (MAPK)-mediated phosphorylation and inactivation of Yan. The ability of Yan to self-associate through a conserved sterile α motif (SAM) is essential for Yan’s repressive ability, and has been suggested to allow spreading of Yan repressive complexes along chromatin. Such a mechanism has the potential to confer both signal responsiveness and robustness to the Yan network. To explore this spreading model, we compared the genome-wide chromatin binding profiles of wild-type vs. monomeric Yan. Consistent with the starting prediction, we found that wild type chromatin occupancy at genes encoding crucial developmental regulators and core signaling pathway components occurs as clusters of peaks that “spread” over multiple kilobases. However monomeric Yan, which fails to rescue a yan null mutation and displays significantly impaired repressive ability, exhibits a broadly similar occupancy profile to that of wild-type Yan, with multi-kilobase binding at developmentally important genes. This unexpected result suggests that SAM-mediated self-association does not mediate Yan recruitment to DNA or chromatin spreading, and raises the questions of why developmentally important genes require extensive Yan chromatin occupancy and how SAM-mediated polymerization might contribute to active repressive mechanisms in this context. In this Extra View article we discuss potential mechanisms by which Yan self-association and extended chromatin occupancy may contribute to robust regulation of gene expression.
Keywords: Drosophila, transcription, repressor, eye development, ChIP
Embryonic cells must faithfully execute specific developmental programs in the face of biological noise arising from stochasticity and environmental variation. For example, pattern formation in the early Drosophila embryo is driven by gene regulatory networks that are robust to both intrinsic and extrinsic noise.1-6 The inability to maintain robustness to noise will lead to aberrant patterning, and potentially, catastrophic failure of development. In contrast, other biological networks, for example those mediating the response to stress, rely on rapid regulatory changes at the expense of transcriptional precision.7-9 Since gene expression is controlled at multiple levels through regulation of transcription, translation and protein stability, it is likely that there are built-in mechanisms favoring either robustness or stochasticity at each of these steps.
As an example of transcription-level regulation, which is the topic of this article, recent studies of dorso-ventral embryonic patterning have revealed the role of auxiliary, or “shadow” enhancers, in conferring robustness. These auxiliary enhancers are intrinsically capable of driving gene expression in a pattern similar to that of the primary enhancer.10-13 Deletion of a shadow enhancer does not alter expression or fitness under optimal conditions, but compromises the ability of the system to buffer against noise. For example, a shadow enhancer identified downstream of the snail gene drives expression in a pattern broadly overlapping that of a previously characterized primary enhancer.11 Removal of either enhancer has no discernible phenotype in otherwise optimal conditions, but results in more variable snail expression upon either raising embryos at elevated temperature or reducing the genetic dose of dorsal, a critical activator of snail. Thus, regulation through these two semi-redundant elements buffers snail expression against both environmental and genetic perturbation. Although our understanding of auxiliary enhancers is presently limited to these few examples,10-13 their initial characterization underscores the importance of regulatory precision to developmental programs and suggests complex interactions between multiple cis-regulatory regions could provide diverse and widespread mechanisms for buffering gene expression. Here we present a speculative discussion of how regulation by the Drosophila ETS family transcriptional repressor Yan may confer robustness both at the level of individual gene expression and across developmental signaling networks.
Yan functions as part of a conserved network downstream of receptor tyrosine kinase (RTK) signaling to regulate gene expression programs that direct the differentiation of a variety of tissues and organs.14-22 The network displays switch-like behavior, transitioning from a high Yan to a low Yan state in response to RTK activation.23,24 In the initial “inactive” state, high Yan levels repress target gene expression to hold cells in an uncommitted progenitor state. Rapid degradation of Yan following RTK signaling switches the network to a low Yan “active” state in which expression of previously repressed genes can be turned on by the activator Pointed (Pnt) to drive specific cell fate transitions. The fidelity with which this system operates is perhaps best illustrated in the context of recruitment of photoreceptor fates in each of the ~800 ommatidia of a compound eye, a process that relies on the Yan-Pnt switch, and occurs with > 99% accuracy.25 To achieve this level of precision, we hypothesize that the Yan network must include mechanisms to buffer gene expression against noise that might otherwise induce inappropriate switching between states and consequent cell fate specification defects.
Over a decade ago, a potential mechanism was proposed to account for both robustness and signaling responsiveness of the Yan-Pnt switch, based on the ability of Yan to form helical polymers through homotypic sterile α motif (SAM) interactions.26-28 According to this model, Yan would be recruited to cis-regulatory enhancers with high-affinity GGA(A/T) ETS consensus binding sequences.29,30 Polymerization of Yan through its SAM domain would allow repressive complexes to spread along chromatin to occupy flanking regions that carry lower affinity binding sites. Functionally, spreading of Yan polymeric repression complexes would stabilize the inactive state of the network to prevent inappropriate induction of target gene expression in response to intrinsic fluctuations in MAPK signaling. By regulating the extent of polymerization across a particular locus, a cell might be able to set different thresholds of RTK signaling responsiveness at distinct target genes.
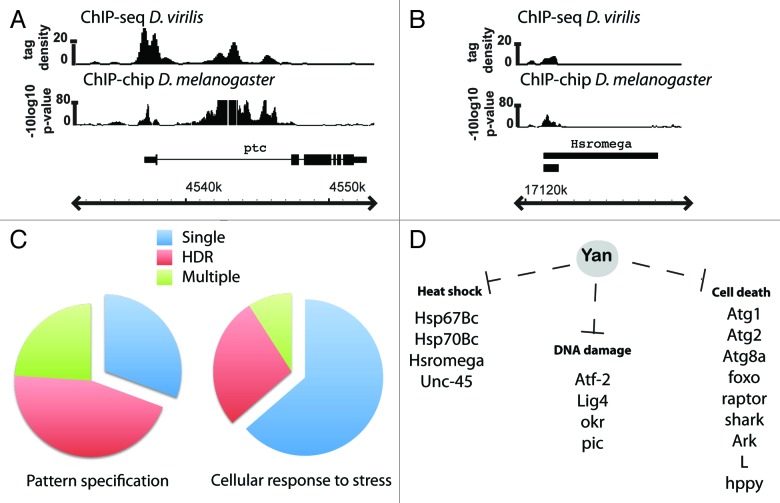
To explore these ideas, we examined occupancy patterns of endogenous wild-type Yan in stage 11 embryos from both D. melanogaster and D. Virilis, two species that diverged > 60 million years ago.31 Suggestive of Yan spreading along chromatin, we found that at ~25% of its putative target genes, Yan binding occurs as clusters of densely packed peaks spanning multiple kilobases. We refer to these as high-density regions (HDRs). The high degree of conservation of HDR-type Yan bound regions between D. melanogaster and D. virilis suggests they are important to gene regulation (Fig. 1A). Further, genes requiring conserved and complex regulation are themselves likely to be critical for development. Consistent with this prediction, gene ontology (GO) analysis revealed that Yan HDR binding occurs primarily at developmentally important genes, with significant enrichment of GO terms associated with signal transduction pathways and tissue specific networks. For example, genes associated with the GO term “Pattern specification process” are predominantly associated with HDRs (Fig. 1C). Thus the Yan HDR patterns map exactly to the set of genes that one predicts would require buffering mechanisms to stabilize and coordinate their expression, a possibility that we speculate on further below.
Figure 1. The complexity of Yan chromatin association patterns correlates with the predicted requirement for either stable or variable target gene expression. (A and B) Examples of conserved HDR (A) and single peak occupancy (B) patterns of wild-type Yan in D. melanogaster and D. virilis at the patched and Hsromega loci. ChIP-seq data are shown as smoothed tag density with a scale of number of reads per million. Gene structures shown below the ChIP patterns depict the plus strand with genomic coordinates indicated below. (C) Developmentally important genes and signaling factors, for example those associated with the GO term ‘Pattern specification process’, are predominantly associated with high-density Yan binding or multiple peaks, while genes associated with the GO term ‘cellular response to stress’ are predominantly associated with single isolated peaks. Yan binding at 271 “Pattern specification process” genes and 110 “Cellular response to stress” genes is summarized in the pie charts as percentage of genes bound with a single peak (blue), multiple peaks (green) or associated with an HDR (red). D) Yan putative single-peak targets include genes involved in cell death pathways and genes upregulated in response to either heat-shock or DNA-damage.
A further prediction of our hypothesis is that genes bound by single Yan peaks should be those affiliated with stochastic processes that require rapid, but not precise, regulation. To address this, we analyzed Yan binding patterns at genes associated with the GO term “cellular response to stress.” Of the 110 genes bound by Yan, 70 display a single-peak binding profile (Fig. 1B and C). These include factors implicated in the response to heat-shock, alcohol and DNA damage32-41 (Fig. 1D). Since these genes must be upregulated quickly in response to cellular stress, we predict that stochasticity will be favored and that they will have greater variability in expression under stable conditions than developmentally important genes. While 30 of the genes within this GO term category are associated with Yan HDRs, they include known regulators of development such as Notch, E(spl) and brinker.42
To address whether the Yan HDR signature reflects the proposed polymerization and spreading mechanism, we analyzed the chromatin occupancy patterns of Yan monomers.31 Previous work identified specific missense mutations within the SAM domain that block polymerization both in vitro and in Drosophila cells.26-28 Functionally, Yan monomers retain DNA binding capability, but do not repress transcription.26,43 To exclude potential artifacts associated with cDNA overexpression, we recombineered the V105R SAM domain missense mutation into the yan genomic locus and then crossed this transgene, which we refer to as monomeric Yan, into a yan null background. Careful controls confirmed the absence of maternally provided wild-type Yan in stage 11 yan null embryos, ensuring that all ChIP signal in this experiment derived from the YanV105R monomer.
In contrast to our starting prediction, we observed similar genome-wide occupancy patterns for wild-type and monomeric Yan, including prevalent HDR type binding. This suggests that SAM-mediated polymerization is not the primary determinant of Yan chromatin occupancy and argues against the polymerization-spreading model. Although Yan monomers can be recruited across multi-kb stretches of DNA, this appears insufficient to support transcriptional repression as the YanV105R transgene failed to rescue the yan null. Thus, the majority of yan;YanV105R animals die with the “anterior open” cuticular phenotype characteristic of yan null embryos.44 Based on these results, we propose that SAM-mediated polymerization is essential for active Yan-mediated repression, but not via a spreading mechanism. As analogous SAM-mediated polymerization-spreading models have been proposed to provide a mechanism of long-range repression for several other transcriptional regulators, most notably the polycomb repressor proteins,28,45-48 our unexpected results with Yan emphasize the importance of explicitly testing this widely accepted model.
If Yan's HDR binding profile does not reflect a polymerization-mediated mechanism of spreading along chromatin, then what might be the functional significance? Both the high degree of conservation of these patterns at specific genes between D. melanogaster and D. virilis and the prevalence of Yan HDR binding across key developmental regulators and signaling pathway genes, suggest contributions to gene expression regulation. Our broad hypothesis is that these patterns reflect a mechanism for maintaining the robustness of gene expression regulation in dynamic systems of elaborate spatial and temporal complexity.
The prevailing model of the Yan network is that Yan and Pnt compete for binding to ETS motifs to either repress or activate target gene expression, respectively. Genetic and biochemical analyses of known Yan/Pnt targets such as argos, eve, mae and prospero, are consistent with such a model.14,23,26,49-51 As part of the validation process for our Yan ChIP data, we expanded this set by showing that 16/18 Yan bound regions cloned from both HDR and single peak target genes can be activated by Pnt and repressed by Yan in cultured cell reporter assays.31 For genes regulated by standard Yan-Pnt switch like behavior, Yan HDR binding might stabilize the inactive state and prevent the switch from being flipped before a critical RTK signaling threshold was reached, thereby limiting transcriptional noise. In contrast, expression of genes at which Yan occupancy is limited to an individual binding site or element might be more prone to variation due to intrinsic fluctuation in MAPK activation or environmental stress. As currently there are no available ChIP data for Pnt, we do not yet know whether switch-like Yan-Pnt competition occurs across an entire HDR or at only a small subset of Yan-bound elements. Further, some Yan-bound genes may not use the Yan-Pnt switch at all, but may still rely on extensive Yan occupancy to buffer their expression. Below we present a series of models to consider how Yan might confer such regulation. Although the ideas are entirely speculative at this point, we hope they may provide an interesting framework for future investigations.
Given that we used chromatin derived from whole embryos for the Yan ChIP analysis,31 the HDR patterns may simply reflect the composite of enormous regulatory diversity across different cell types rather than regulatory complexity within individual cells. In other words, the spatial and temporal complexity in expression typical of developmental regulators and signaling factors might require extraordinary cell-to-cell heterogeneity in Yan occupancy at discrete cis-regulatory enhancers. While it will take single cell analysis to rule out this possibility, such a model predicts that we would observe very different ChIP profiles at different developmental stages. However, we found extensive overlap between Yan HDR patterns at stage 5–7 and stage 11, two developmental time points with diverse cell populations and gene expression profiles. A second prediction is that the majority of transcription factors that participate in developmental regulation should have similarly complex chromatin occupancy profiles. However meta-analysis of chromatin occupancy patterns of modEncode transcription factors revealed that most display a much lower extent of HDR type binding than we observed for Yan. Indeed, even for the three factors Kr, Ubx and Dll with the most prominent HDR patterns, the extent of high-density binding, in terms of both the average length of the region occupied and the extent to which signaling pathway components were bound, was less than for Yan. Finally, recent work from several other labs has confirmed that regulatory complexity can occur within single cells or tissues.52,53 Thus, although we expect that a modest fraction of Yan's occupancy patterns reflect binding to tissue-specific enhancers, for the purpose of the remaining discussion we accept as a reasonable assumption that the full complexity of the Yan HDR profile at a given locus could influence its expression in an individual cell.
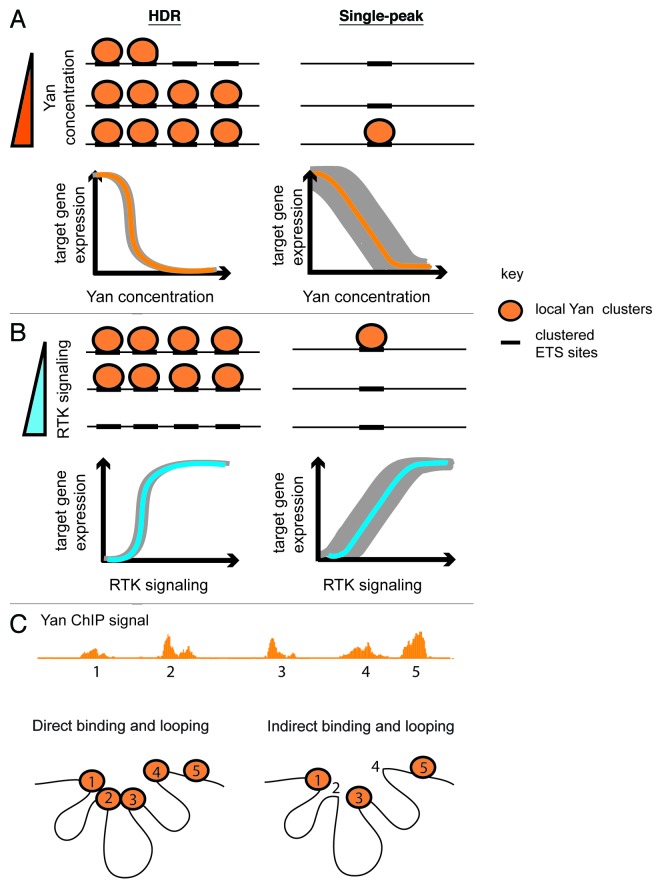
Considering that transcription factor occupancy and regulation of gene expression is determined by a complex combination of protein concentration, DNA-binding affinity, chromatin structure and the dynamics of transcriptional complex assembly/disassembly, what types of regulatory contributions to gene expression could be conferred by extensive occupancy of the Yan repressor across a locus? One possibility is that the primary role of Yan HDR-type binding is to maintain a high local Yan concentration to increase the probability of assembling active repressive complexes at the critical cis-regulatory elements. In this scenario, the ability to recruit or stabilize active repressive complexes from the neutral repository of HDR-bound Yan would require SAM-mediated self-association (Fig. 2A). HDR binding could both maintain the stability of the inactive state of the Yan-Pnt switch under optimal conditions and also provide a buffering mechanism against conditions that limit Yan availability. A testable prediction is that in a yan heterozygote, the expression of HDR category target genes should be essentially identical to wild type whereas expression of target genes bound by single Yan peaks should increase and become more variable.
Figure 2. Robustness through Yan HDR binding. Orange circles depict Yan while the cis-regulatory elements to which it binds are drawn as black rectangles on the DNA (black lines). As such elements generally contain clusters of ETS binding sites, each orange circle may represent multiple Yan molecules. (A) Binding of Yan across cis-regulatory elements comprising an HDR would maintain a locally high Yan concentration even if Yan levels in the cell are low. This would stabilize target gene repression over a wide range of Yan concentrations, and ensure a sharp switch-like off/on response when Yan concentration crossed the critical threshold. In contrast, Yan binding at a single element would be more prone to dissociation, with stable Yan occupancy only achieved at high Yan concentration. Such fluctuations in Yan occupancy would result in variable target gene expression at low Yan concentration (gray shading). (B) Extensive Yan binding across an HDR might protect against premature dissociation of Yan polymers in response to sub-threshold RTK signaling or intrinsic fluctuations in MAPK activation. Sustained and/or strong pathway activation would be required for cooperative removal of Yan from all elements and to activate gene expression. In contrast, Yan repressive complexes assembled at only a single element would be prone to dissociation in response to intrinsic MAPK fluctuation, leading to more variable gene expression (depicted by gray shading). (C) The 3D chromatin environment could contribute to the establishment and/or function of multi-kb HDR patterns. To illustrate this, a Yan high-density ChIP signature at an arbitrary locus is shown. The 5 ChIP peaks that define the HDR are labeled as 1–5. In the first scenario (left-most diagram, Direct binding and looping), all ChIP peaks in the sample HDR are directly bound by Yan. SAM-mediated interactions could either directly influence or indirectly exploit the 3D environment to assemble repressive complexes between elements 1–4 that interact in nonlinear space. Alternatively (right-most diagram, Indirect binding and looping), Yan occupancy across an HDR could involve direct binding of Yan to only a subset of sites (1,3 and 5 for example), with the ChIP signal at sites 2 and 4 coming about as an indirect consequence of the 3D environment that places those chromatin regions in close proximity to regions 1 and 3. Such indirect binding could be stabilized by Yan self-association and/or additional protein-protein interactions.
Alternatively, it is possible that Yan bound peaks across an HDR define discrete cis-regulatory elements that contribute to repression in an additive manner. These elements could include both bona fide shadow enhancers and elements essential for repression that lack autonomous regulatory capacity outside the HDR environment. The end result of such combinatorial regulation would be to set different thresholds of sensitivity to RTK signaling, with perhaps a graded response to different levels of MAPK activation depending on the extent of Yan binding. Such a system might not only confer differential sensitivity to levels of pathway activation, but might also be able to distinguish between the duration of a MAPK signal. Thus disassembly of Yan complexes across an HDR might require prolonged signaling whereas single-peak genes might respond to a short burst of RTK signal. Either scenario would effectively stabilize the expression of genes with extensive Yan occupancy signals against random fluctuations in MAPK signaling (Fig. 2B).
The importance of three-dimensional chromatin conformation to regulation of gene expression suggests another potential mechanism by which Yan occupancy across HDRs might stabilize repressive complexes across a locus (Fig. 2C). In this model, Yan polymers might directly promote or stabilize chromatin conformations that restrict access of other transcription factors and/or the basal transcriptional machinery. If true, then essential chromatin contacts that occur in wild-type animals between Yan-bound regions should be destabilized in the YanV105R monomeric background. Alternatively, Yan might not directly influence chromatin conformation or contacts, but might exploit the 3D environment to assemble repressive complexes that bridge linearly noncontiguous regulatory elements. Such 3D contacts could explain in part the multi-kb Yan HDR patterns we observe in our ChIP data sets. Thus, recruitment of Yan across an HDR could involve a combination of direct binding to clusters of ETS motifs in a cis-regulatory enhancer and indirect interactions with complexes brought into close proximity through the 3D chromatin environment. If correct, then if tested in isolation, only a subset of Yan-bound elements within an HDR should be sufficient to recruit Yan. Further, targeted genomic deletions of directly bound regions should disrupt long-range cooperative interactions and destabilize Yan occupancy across the entire HDR. In all scenarios, Yan repressive complexes within the complex 3D topology of an HDR would be protected from MAPK access, thus conferring robustness to intrinsic fluctuations in MAPK activation.
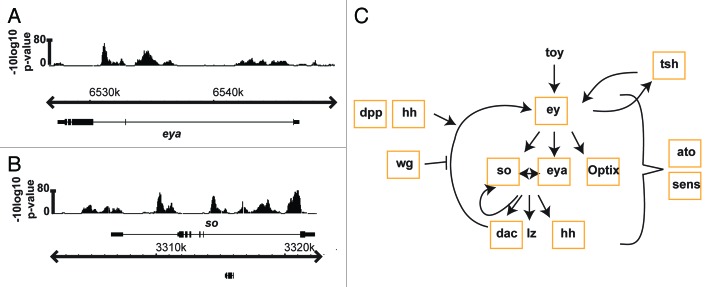
As chromatin conformation capture techniques have provided evidence of chromatin loops bringing otherwise distant genes into close proximity,54,55 Yan-mediated regulatory interactions could exist across multiple genes in a 3D environment. Very speculatively, this might coordinate expression levels across entire pathways or networks. A prime example of extensive Yan HDR occupancy across a group of functionally interconnected genes is seen in the retinal determination (RD) gene network. The RD network is comprised of a conserved group of transcription factors that collaborate with other signaling pathways and tissue-specific networks to direct many aspects of eye development.56,57 Two core RD genes, eyes absent (eya) and sine oculis (so) were recently identified as Yan targets 58 (Fig. 3A and B) and our ChIP data suggest an even broader involvement of Yan-mediated regulation within the RD network, with binding to eyeless (ey), eya, so, dachshund (dac), optix, teashirt (tsh), homothorax, nemo and distal antenna related (danr) (Fig. 3C).31 Given the extensive feedback interactions that occur within this and most other signaling networks, inappropriate fluctuations in gene expression might be quickly amplified, compromising output. Noise buffering mechanisms would be critical to prevent such amplification. Further, transcriptional responses to signaling inputs may require coordination across targets to ensure appropriate expression of key network nodes. Thus perhaps the prevalence of Yan HDR occupancy across the RD and other signaling networks provides both an extensive buffering mechanism and a novel level of pathway integration.
Figure 3. Yan occupancy patterns reveal the potential for extensive regulation of the retinal determination network. (A and B) Yan ChIP-chip patterns at the eya and so loci. Gene structures are shown below ChIP-chip patterns. Gene names are shown above or below the genome coordinates to depict + or – strands, respectively. (C) Genes within the retinal determination network bound by Yan are outlined with orange boxes.
In conclusion, we speculate that the complex Yan occupancy patterns identified in our ChIP study may reflect a mechanism for buffering expression of important developmental regulators against genetic and environmental noise. The particularly striking signatures observed across components of multiple signaling networks, including the RD network and the Notch, Wingless and EGFR pathways, make these appealing contexts for future exploration of this hypothesis.
Acknowledgments
We thank Jackie Gavin-Smyth, Matthew Slattery, Jean-François Boisclair Lachance, Trevor Davis, Charlene Hoi and Matthew Hope for critical reading of the manuscript, and members of the Rebay, Fehon, White and Carthew labs for helpful discussions of ideas presented in this article. We also thank Matthew Slattery for his contribution to the design of Figure 2C.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/fly/article/24162
References
- 1.Lott SE, Ludwig MZ, Kreitman M. Evolution and inheritance of early embryonic patterning in Drosophila simulans and D. sechellia. Evolution. 2011;65:1388–99. doi: 10.1111/j.1558-5646.2010.01206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gregor T, Wieschaus EF, McGregor AP, Bialek W, Tank DW. Stability and nuclear dynamics of the bicoid morphogen gradient. Cell. 2007;130:141–52. doi: 10.1016/j.cell.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He F, Wen Y, Deng J, Lin X, Lu LJ, Jiao R, et al. Probing intrinsic properties of a robust morphogen gradient in Drosophila. Dev Cell. 2008;15:558–67. doi: 10.1016/j.devcel.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lucchetta EM, Lee JH, Fu LA, Patel NH, Ismagilov RF. Dynamics of Drosophila embryonic patterning network perturbed in space and time using microfluidics. Nature. 2005;434:1134–8. doi: 10.1038/nature03509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Namba R, Pazdera TM, Cerrone RL, Minden JS. Drosophila embryonic pattern repair: how embryos respond to bicoid dosage alteration. Development. 1997;124:1393–403. doi: 10.1242/dev.124.7.1393. [DOI] [PubMed] [Google Scholar]
- 6.Lott SE, Kreitman M, Palsson A, Alekseeva E, Ludwig MZ. Canalization of segmentation and its evolution in Drosophila. Proc Natl Acad Sci U S A. 2007;104:10926–31. doi: 10.1073/pnas.0701359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–6. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 8.Bar-Even A, Paulsson J, Maheshri N, Carmi M, O’Shea E, Pilpel Y, et al. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–43. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- 9.Zhou S, Campbell TG, Stone EA, Mackay TF, Anholt RR. Phenotypic plasticity of the Drosophila transcriptome. PLoS Genet. 2012;8:e1002593. doi: 10.1371/journal.pgen.1002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466:490–3. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol. 2010;20:1562–7. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perry MW, Boettiger AN, Levine M. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc Natl Acad Sci U S A. 2011;108:13570–5. doi: 10.1073/pnas.1109873108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunipace L, Ozdemir A, Stathopoulos A. Complex interactions between cis-regulatory modules in native conformation are critical for Drosophila snail expression. Development. 2011;138:4075–84. doi: 10.1242/dev.069146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halfon MS, Carmena A, Gisselbrecht S, Sackerson CM, Jiménez F, Baylies MK, et al. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell. 2000;103:63–74. doi: 10.1016/S0092-8674(00)00105-7. [DOI] [PubMed] [Google Scholar]
- 15.Rebay I, Rubin GM. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell. 1995;81:857–66. doi: 10.1016/0092-8674(95)90006-3. [DOI] [PubMed] [Google Scholar]
- 16.Rogge R, Green PJ, Urano J, Horn-Saban S, Mlodzik M, Shilo BZ, et al. The role of yan in mediating the choice between cell division and differentiation. Development. 1995;121:3947–58. doi: 10.1242/dev.121.12.3947. [DOI] [PubMed] [Google Scholar]
- 17.Schober M, Rebay I, Perrimon N. Function of the ETS transcription factor Yan in border cell migration. Development. 2005;132:3493–504. doi: 10.1242/dev.01911. [DOI] [PubMed] [Google Scholar]
- 18.Lai ZC, Fetchko M, Li Y. Repression of Drosophila photoreceptor cell fate through cooperative action of two transcriptional repressors Yan and Tramtrack. Genetics. 1997;147:1131–7. doi: 10.1093/genetics/147.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai ZC, Rubin GM. Negative control of photoreceptor development in Drosophila by the product of the yan gene, an ETS domain protein. Cell. 1992;70:609–20. doi: 10.1016/0092-8674(92)90430-K. [DOI] [PubMed] [Google Scholar]
- 20.Scholz H, Sadlowski E, Klaes A, Klämbt C. Control of midline glia development in the embryonic Drosophila CNS. Mech Dev. 1997;62:79–91. doi: 10.1016/S0925-4773(96)00652-1. [DOI] [PubMed] [Google Scholar]
- 21.O’Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–47. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- 22.Weber U, Pataki C, Mihaly J, Mlodzik M. Combinatorial signaling by the Frizzled/PCP and Egfr pathways during planar cell polarity establishment in the Drosophila eye. Dev Biol. 2008;316:110–23. doi: 10.1016/j.ydbio.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gabay L, Scholz H, Golembo M, Klaes A, Shilo BZ, Klämbt C. EGF receptor signaling induces pointed P1 transcription and inactivates Yan protein in the Drosophila embryonic ventral ectoderm. Development. 1996;122:3355–62. doi: 10.1242/dev.122.11.3355. [DOI] [PubMed] [Google Scholar]
- 24.Graham TG, Tabei SM, Dinner AR, Rebay I. Modeling bistable cell-fate choices in the Drosophila eye: qualitative and quantitative perspectives. Development. 2010;137:2265–78. doi: 10.1242/dev.044826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolff T, Ready D. Pattern formation in the Drosophila retina. In: Bate M, Martinez-Arias A, eds. The Development of Drosophila melanogaster. Plainview, NY: Cold Spring Harbor Press, 1993:1277-325. [Google Scholar]
- 26.Qiao F, Song H, Kim CA, Sawaya MR, Hunter JB, Gingery M, et al. Derepression by depolymerization; structural insights into the regulation of Yan by Mae. Cell. 2004;118:163–73. doi: 10.1016/j.cell.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 27.Tran HH, Kim CA, Faham S, Siddall MC, Bowie JU. Native interface of the SAM domain polymer of TEL. BMC Struct Biol. 2002;2:5. doi: 10.1186/1472-6807-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim CA, Phillips ML, Kim W, Gingery M, Tran HH, Robinson MA, et al. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J. 2001;20:4173–82. doi: 10.1093/emboj/20.15.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karim FD, Urness LD, Thummel CS, Klemsz MJ, McKercher SR, Celada A, et al. The ETS-domain: a new DNA-binding motif that recognizes a purine-rich core DNA sequence. Genes Dev. 1990;4:1451–3. doi: 10.1101/gad.4.9.1451. [DOI] [PubMed] [Google Scholar]
- 30.Nye JA, Petersen JM, Gunther CV, Jonsen MD, Graves BJ. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992;6:975–90. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- 31.Webber JL, Zhang J, Cote L, Vivekanand P, Ni X, Zhou J, et al. The Relationship Between Long-Range Chromatin Occupancy and Polymerization of the Drosophila ETS Family Transcriptional Repressor Yan. Genetics. 2013;193:633–49. doi: 10.1534/genetics.112.146647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhoumik A, Takahashi S, Breitweiser W, Shiloh Y, Jones N, Ronai Z. ATM-dependent phosphorylation of ATF2 is required for the DNA damage response. Mol Cell. 2005;18:577–87. doi: 10.1016/j.molcel.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romeijn RJ, Gorski MM, van Schie MA, Noordermeer JN, Mullenders LH, Ferro W, et al. Lig4 and rad54 are required for repair of DNA double-strand breaks induced by P-element excision in Drosophila. Genetics. 2005;169:795–806. doi: 10.1534/genetics.104.033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kooistra R, Pastink A, Zonneveld JB, Lohman PH, Eeken JC. The Drosophila melanogaster DmRAD54 gene plays a crucial role in double-strand break repair after P-element excision and acts synergistically with Ku70 in the repair of X-ray damage. Mol Cell Biol. 1999;19:6269–75. doi: 10.1128/mcb.19.9.6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu D, Woodfield SE, Lee TV, Fan Y, Antonio C, Bergmann A. Genetic control of programmed cell death (apoptosis) in Drosophila. Fly (Austin) 2009;3:78–90. doi: 10.4161/fly.3.1.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lam D, Shah S, de Castro IP, Loh SH, Martins LM. Drosophila happyhour modulates JNK-dependent apoptosis. Cell Death Dis. 2010;1:e66. doi: 10.1038/cddis.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee G, Chung J. Discrete functions of rictor and raptor in cell growth regulation in Drosophila. Biochem Biophys Res Commun. 2007;357:1154–9. doi: 10.1016/j.bbrc.2007.04.086. [DOI] [PubMed] [Google Scholar]
- 38.Fullard JF, Kale A, Baker NE. Clearance of apoptotic corpses. Apoptosis. 2009;14:1029–37. doi: 10.1007/s10495-009-0335-9. [DOI] [PubMed] [Google Scholar]
- 39.Tsuda M, Ootaka R, Ohkura C, Kishita Y, Seong KH, Matsuo T, et al. Loss of Trx-2 enhances oxidative stress-dependent phenotypes in Drosophila. FEBS Lett. 2010;584:3398–401. doi: 10.1016/j.febslet.2010.06.034. [DOI] [PubMed] [Google Scholar]
- 40.Melkani GC, Lee CF, Cammarato A, Bernstein SI. Drosophila UNC-45 prevents heat-induced aggregation of skeletal muscle myosin and facilitates refolding of citrate synthase. Biochem Biophys Res Commun. 2010;396:317–22. doi: 10.1016/j.bbrc.2010.04.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang YY, Neufeld TP. Autophagy takes flight in Drosophila. FEBS Lett. 2010;584:1342–9. doi: 10.1016/j.febslet.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perrimon N, Pitsouli C, Shilo BZ. Signaling mechanisms controlling cell fate and embryonic patterning. Cold Spring Harb Perspect Biol. 2012;4:a005975. doi: 10.1101/cshperspect.a005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J, Graham TG, Vivekanand P, Cote L, Cetera M, Rebay I. Sterile alpha motif domain-mediated self-association plays an essential role in modulating the activity of the Drosophila ETS family transcriptional repressor Yan. Mol Cell Biol. 2010;30:1158–70. doi: 10.1128/MCB.01225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 45.Nègre N, Hennetin J, Sun LV, Lavrov S, Bellis M, White KP, et al. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 2006;4:e170. doi: 10.1371/journal.pbio.0040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–5. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 47.Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, et al. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet. 2006;38:694–9. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- 48.Qiao F, Bowie JU. The many faces of SAM. Sci STKE. 2005;2005:re7. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- 49.Golembo M, Schweitzer R, Freeman M, Shilo BZ. Argos transcription is induced by the Drosophila EGF receptor pathway to form an inhibitory feedback loop. Development. 1996;122:223–30. doi: 10.1242/dev.122.1.223. [DOI] [PubMed] [Google Scholar]
- 50.Flores GV, Duan H, Yan H, Nagaraj R, Fu W, Zou Y, et al. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/S0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- 51.Vivekanand P, Tootle TL, Rebay I. MAE, a dual regulator of the EGFR signaling pathway, is a target of the Ets transcription factors PNT and YAN. Mech Dev. 2004;121:1469–79. doi: 10.1016/j.mod.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 52.Agelopoulos M, McKay DJ, Mann RS. Developmental regulation of chromatin conformation by Hox proteins in Drosophila. Cell Rep. 2012;1:350–9. doi: 10.1016/j.celrep.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44:148–56. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 54.Splinter E, de Laat W. The complex transcription regulatory landscape of our genome: control in three dimensions. EMBO J. 2011;30:4345–55. doi: 10.1038/emboj.2011.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–72. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 56.Silver SJ, Rebay I. Signaling circuitries in development: insights from the retinal determination gene network. Development. 2005;132:3–13. doi: 10.1242/dev.01539. [DOI] [PubMed] [Google Scholar]
- 57.Kumar JP. The molecular circuitry governing retinal determination. Biochim Biophys Acta. 2009;1789:306–14. doi: 10.1016/j.bbagrm.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salzer CL, Elias Y, Kumar JP. The retinal determination gene eyes absent is regulated by the EGF receptor pathway throughout development in Drosophila. Genetics. 2010;184:185–97. doi: 10.1534/genetics.109.110122. [DOI] [PMC free article] [PubMed] [Google Scholar]