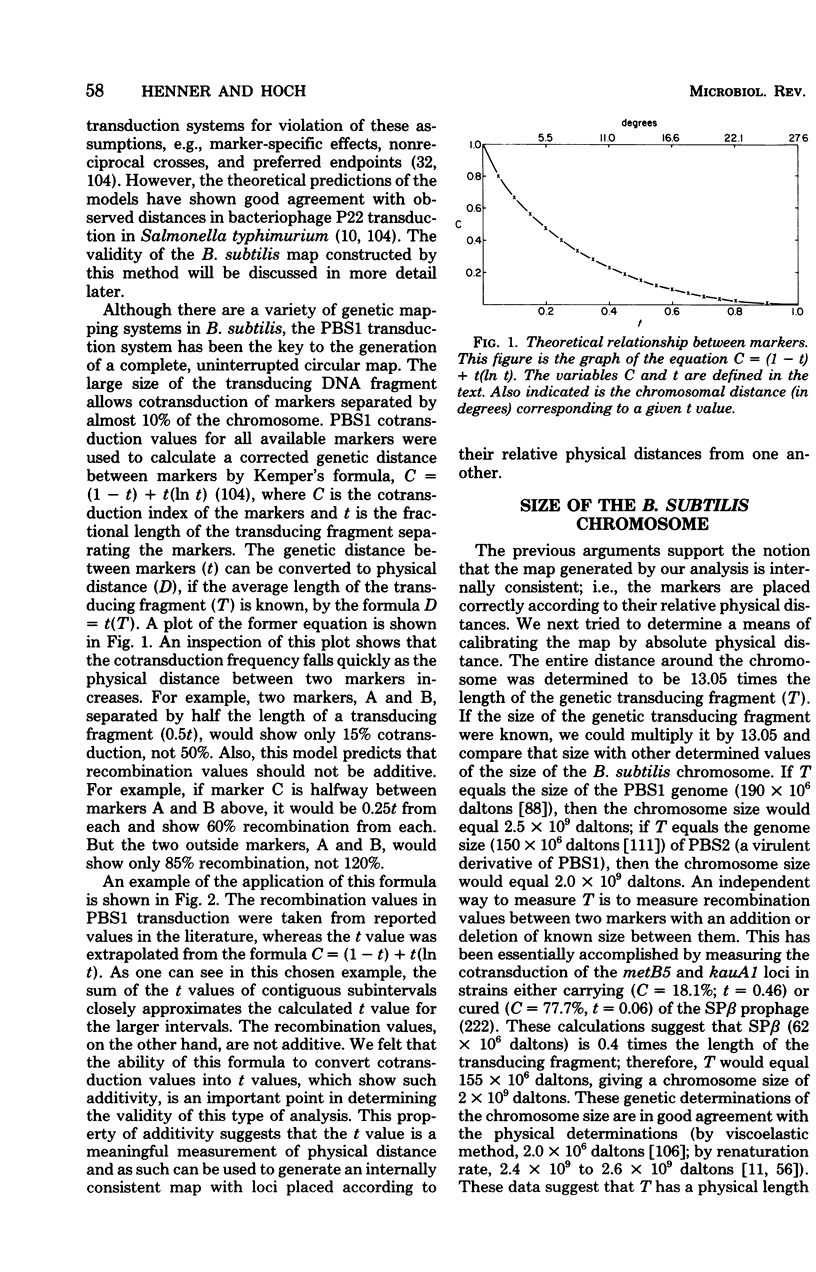
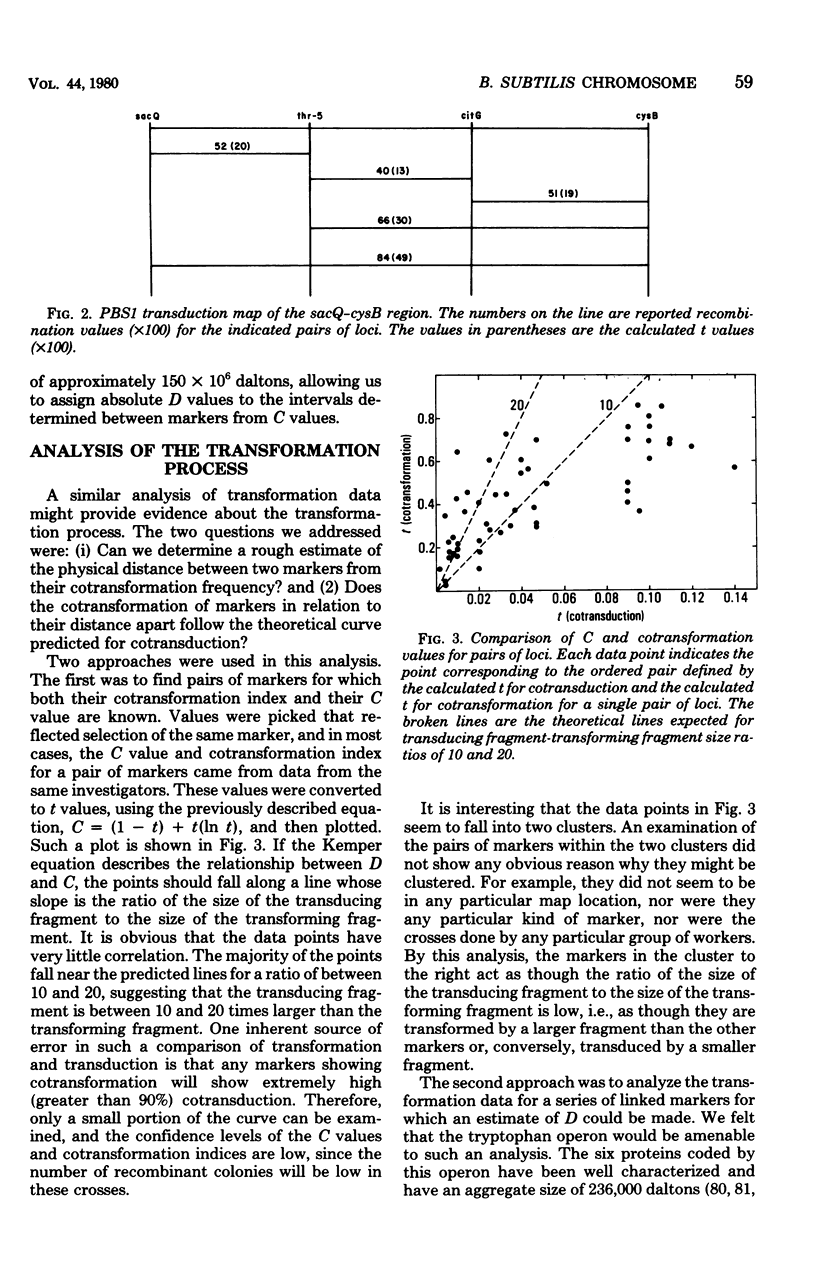
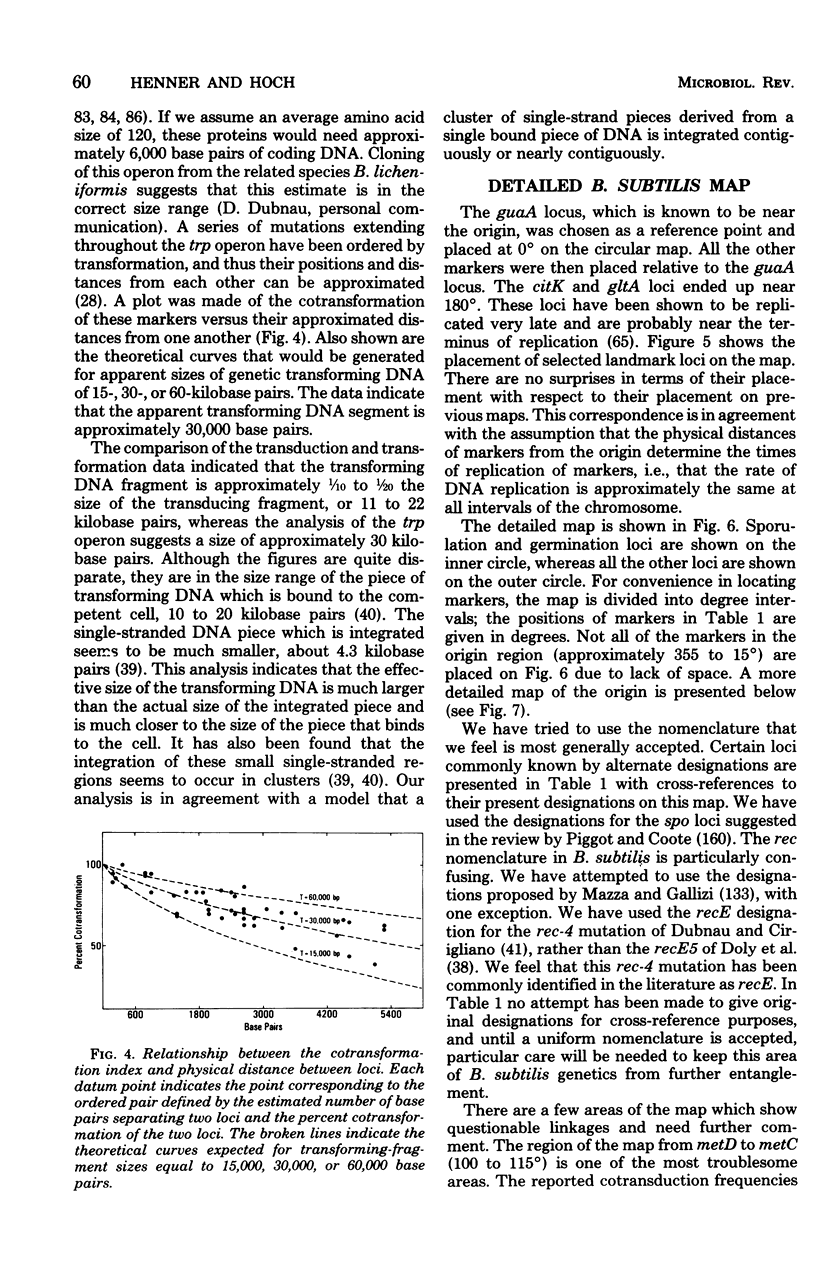
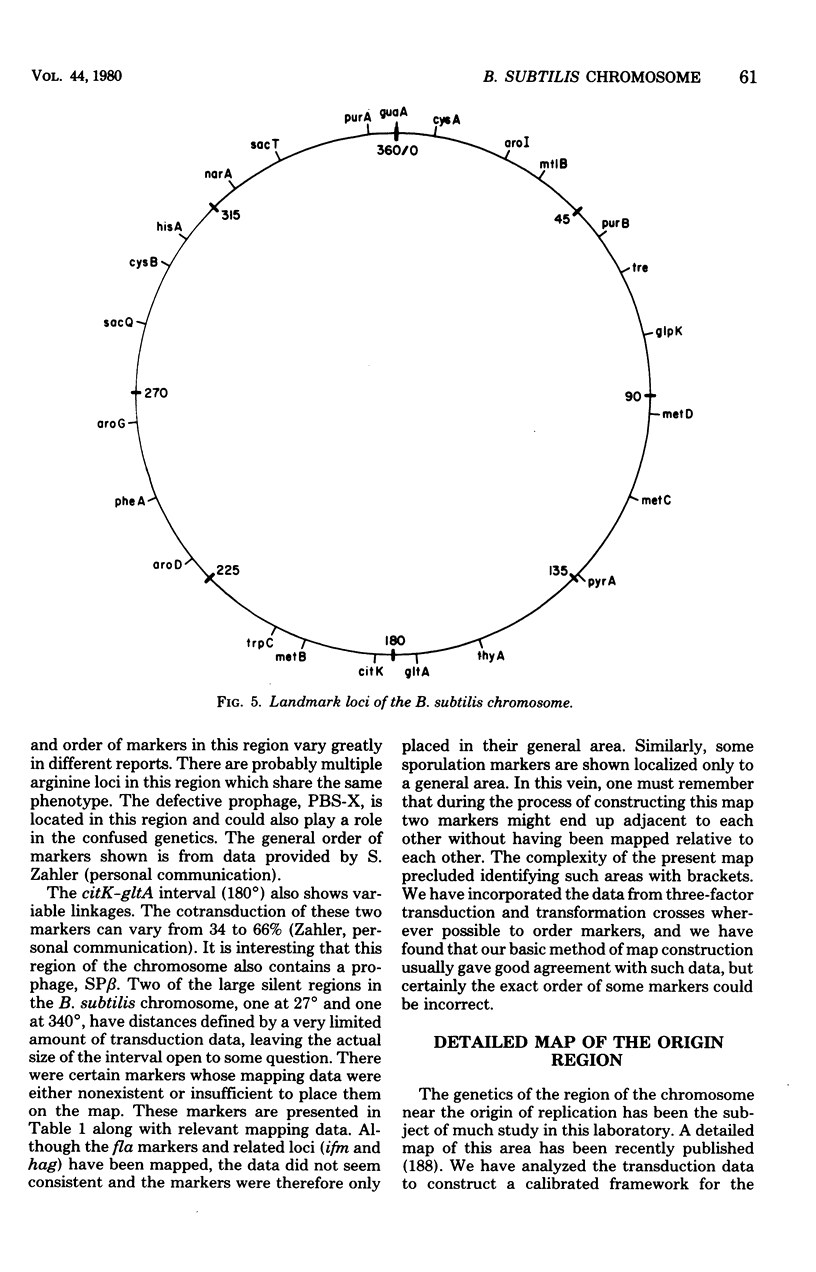
Full text
PDF
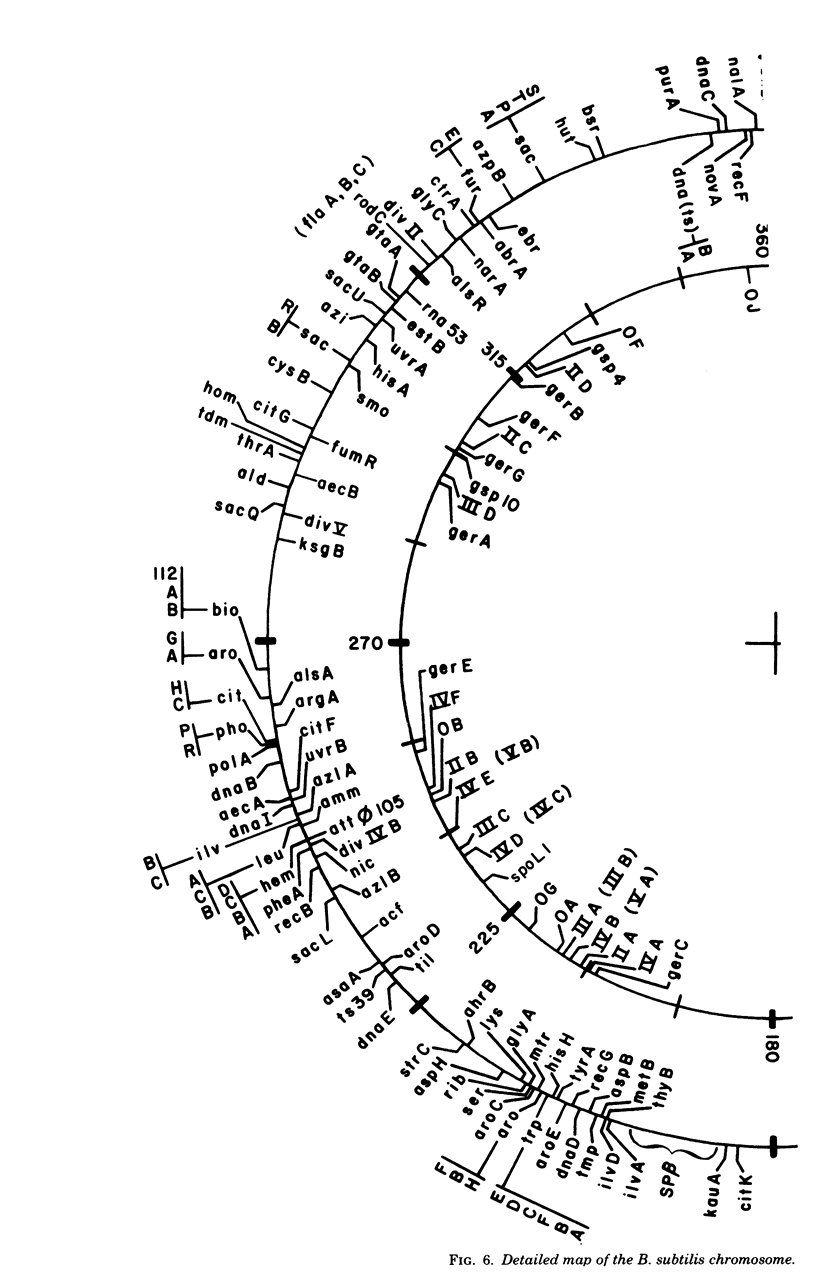
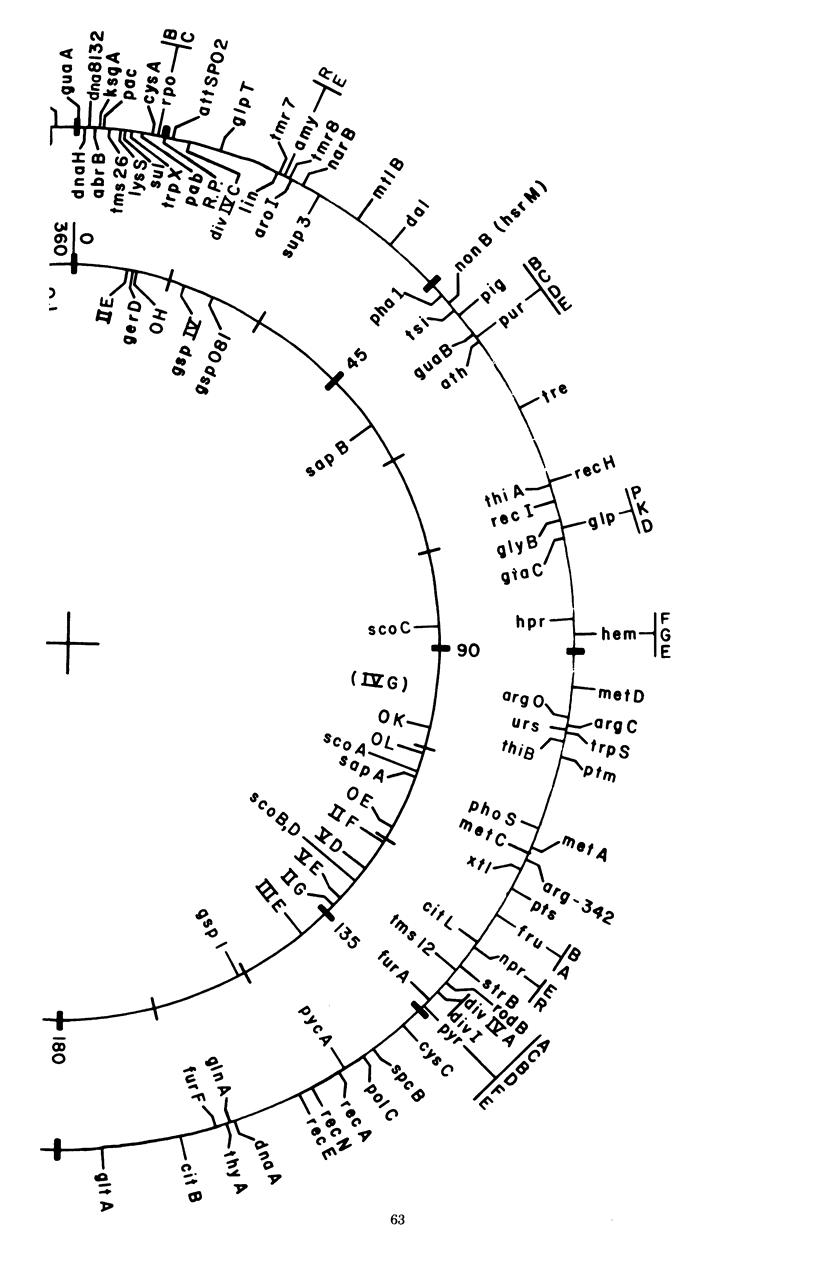


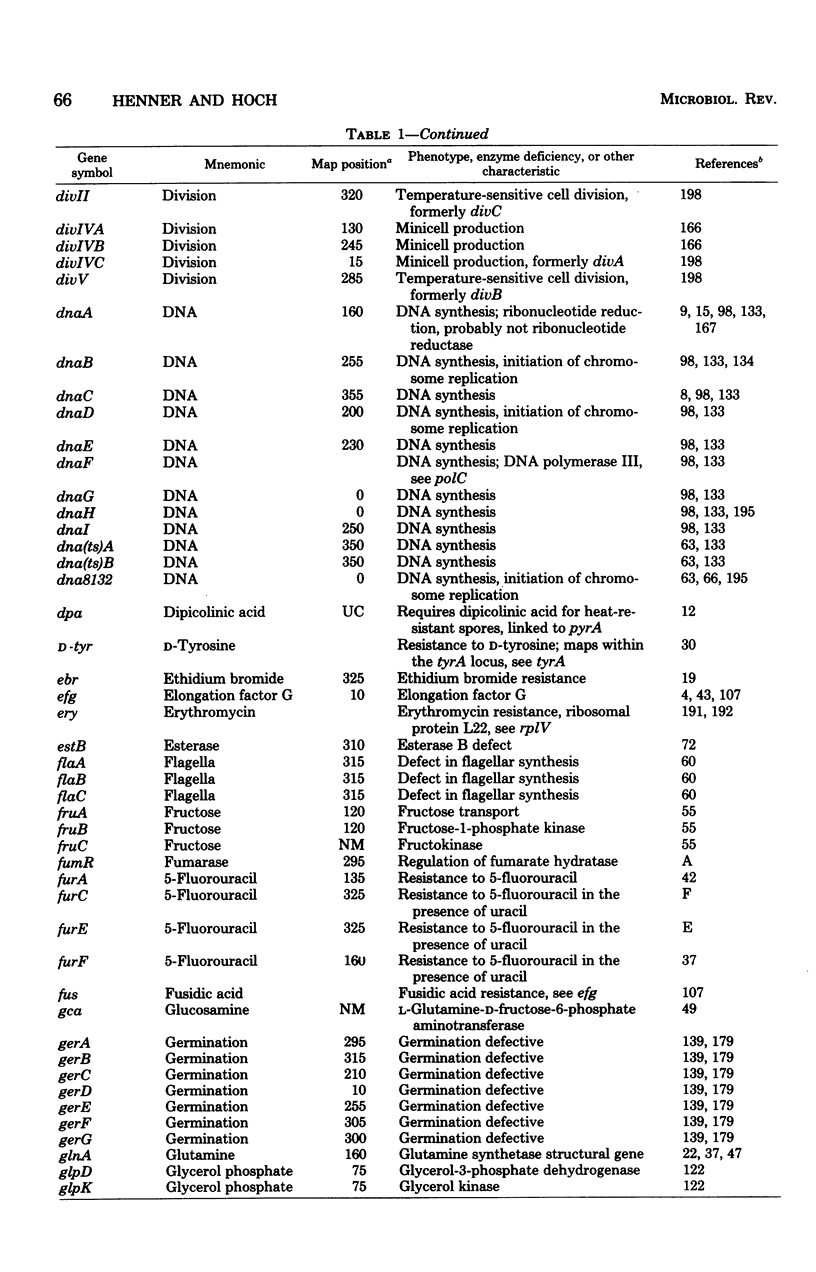
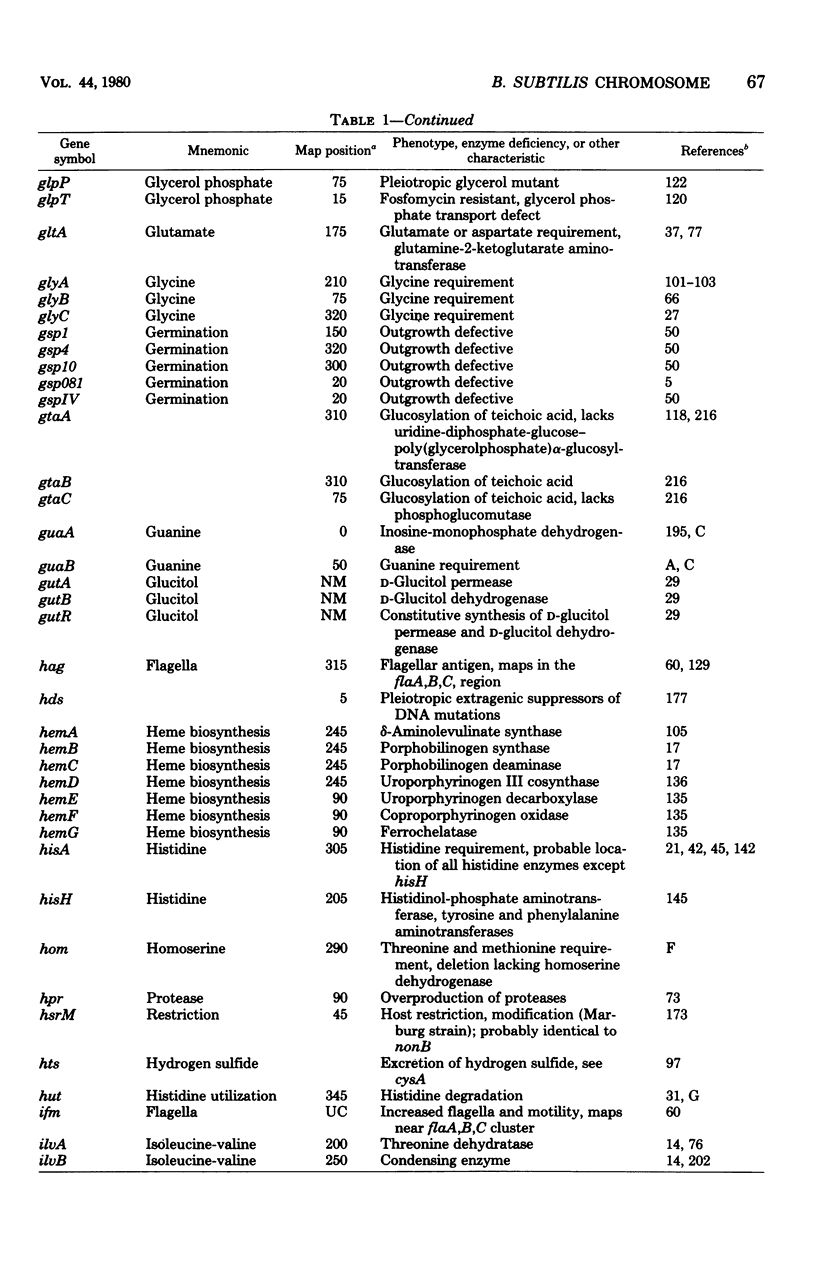











Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ANAGNOSTOPOULOS C., CRAWFORD I. P. Transformation studies on the linkage of markers in the tryptophan pathway in Bacillus subtilis. Proc Natl Acad Sci U S A. 1961 Mar 15;47:378–390. doi: 10.1073/pnas.47.3.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A., Oishi M. Genetic properties of arsenate sensitive mutants of Bacillus subtilis 168. Mol Gen Genet. 1972;118(4):295–310. doi: 10.1007/BF00333565. [DOI] [PubMed] [Google Scholar]
- Adams A. Transformation and transduction of a large deletion mutation in Bacillus subtilis. Mol Gen Genet. 1972;118(4):311–320. doi: 10.1007/BF00333566. [DOI] [PubMed] [Google Scholar]
- Aharonowitz Y., Ron E. Z. A temperature sensitive mutant in Bacillus subtilis with an altered elongation factor G. Mol Gen Genet. 1972;119(2):131–138. doi: 10.1007/BF00269132. [DOI] [PubMed] [Google Scholar]
- Albertini A. M., Galizzi A. Mutant of Bacillus subtilis with a temperature-sensitive lesion in ribonucleic acid synthesis during germination. J Bacteriol. 1975 Oct;124(1):14–25. doi: 10.1128/jb.124.1.14-25.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anagnostopoulos C., Schneider-Champagne A. M. Déterminisme génétique de l'exigence en thymine chez certains mutants de Bacillus subtilis. C R Acad Sci Hebd Seances Acad Sci D. 1966 Mar 14;262(11):1311–1314. [PubMed] [Google Scholar]
- Andersen J. J., Ganesan A. T. Temperature-sensitive deoxyribonucleic acid replication in a dnaC mutant of Bacillus subtilis. J Bacteriol. 1975 Jan;121(1):173–183. doi: 10.1128/jb.121.1.173-183.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attolini C., Mazza G., Fortunato A., Ciarrocchi G., Mastromei G., Riva S., Falaschi A. On the identity of dnaP and dnaF genes of Bacillus subtilis. Mol Gen Genet. 1976 Oct 18;148(1):9–17. doi: 10.1007/BF00268540. [DOI] [PubMed] [Google Scholar]
- BARAT M., ANAGNOSTOPOULOS C., SCHNEIDER A. M. LINKAGE RELATIONSHIPS OF GENES CONTROLLING ISOLEUCINE, VALINE, AND LEUCINE BIOSYNTHESIS IN BACILLUS SUBTILIS. J Bacteriol. 1965 Aug;90:357–369. doi: 10.1128/jb.90.2.357-369.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak A. L., Christiansen C., Stenderup A. Bacterial genome sizes determined by DNA renaturation studies. J Gen Microbiol. 1970 Dec;64(3):377–380. doi: 10.1099/00221287-64-3-377. [DOI] [PubMed] [Google Scholar]
- Balassa G., Milhaud P., Raulet E., Silva M. T., Sousa J. C. A Bacillus subtilis mutant requiring dipicolinic acid for the development of heat-resistant spores. J Gen Microbiol. 1979 Feb;110(2):365–379. doi: 10.1099/00221287-110-2-365. [DOI] [PubMed] [Google Scholar]
- Balassa G., Milhaud P., Sousa J. C., Silva M. T. Decadent sporulation mutants of Bacillus subtilis. J Gen Microbiol. 1979 Feb;110(2):381–392. doi: 10.1099/00221287-110-2-381. [DOI] [PubMed] [Google Scholar]
- Bazill G. W., Karamata D. Temperature-sensitive mutants of B. subtilis defective in deoxyribonucleotide synthesis. Mol Gen Genet. 1972;117(1):19–29. [PubMed] [Google Scholar]
- Bazzicalupo M., Parisi B., Pirali G., Polsinelli M., Sala F. Genetic and biochemical characterization of a ribosomal mutant of Bacillus subtilis resistant to sporangiomycin. Antimicrob Agents Chemother. 1975 Dec;8(6):651–656. doi: 10.1128/aac.8.6.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berek I., Miczák A., Ivánovics G. Mapping the delta-aminolaevulinic acid dehydrase and porphobilinogen deaminase loci in Bacillus subtilis. Mol Gen Genet. 1974;132(3):233–239. doi: 10.1007/BF00269396. [DOI] [PubMed] [Google Scholar]
- Bettinger G. E., Young F. E. Transformation of Bacillus subtilis using gently lysed L-forms; a new mapping technique. Biochem Biophys Res Commun. 1973 Dec 19;55(4):1105–1111. doi: 10.1016/s0006-291x(73)80009-9. [DOI] [PubMed] [Google Scholar]
- Bishop P. E., Brown L. R. Ethidium bromide-resistant mutant of Bacillus subtilis. J Bacteriol. 1973 Sep;115(3):1077–1083. doi: 10.1128/jb.115.3.1077-1083.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop R. J., Sueoka N. 5-Bromouracil-tolerant mutants of Bacillus subtilis. J Bacteriol. 1972 Nov;112(2):870–876. doi: 10.1128/jb.112.2.870-876.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borenstein S., Ephrati-Elizur E. Spontaneous release of DNA in sequential genetic order by Bacillus subtilis. J Mol Biol. 1969 Oct 14;45(1):137–152. doi: 10.1016/0022-2836(69)90216-2. [DOI] [PubMed] [Google Scholar]
- Bott K. F., Reysset G., Gregoire J., Islert D., Aubert J. P. Characterization of glutamine requiring mutants of Bacillus subtilis. Biochem Biophys Res Commun. 1977 Dec 7;79(3):996–1003. doi: 10.1016/0006-291x(77)91208-6. [DOI] [PubMed] [Google Scholar]
- Boylan R. J., Mendelson N. H., Brooks D., Young F. E. Regulation of the bacterial cell wall: analysis of a mutant of Bacillus subtilis defective in biosynthesis of teichoic acid. J Bacteriol. 1972 Apr;110(1):281–290. doi: 10.1128/jb.110.1.281-290.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramucci M. G., Keggins K. M., Lovett P. S. Bacteriophage PMB12 conversion of the sporulation defect in RNA polymerase mutants of Bacillus subtilis. J Virol. 1977 Oct;24(1):194–200. doi: 10.1128/jvi.24.1.194-200.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton R. S. A heat-sensitive lysis mutant of Bacillus subtilis 168 with a low activity of pyruvate carboxylase. J Gen Microbiol. 1978 Apr;105(2):175–185. doi: 10.1099/00221287-105-2-175. [DOI] [PubMed] [Google Scholar]
- Cannon J. G., Bott K. F. Spectinomycin-resistant mutants of Bacillus subtilis with altered sporulation properties. Mol Gen Genet. 1979 Jul 13;174(2):149–162. doi: 10.1007/BF00268352. [DOI] [PubMed] [Google Scholar]
- Canosi U., Siccardi A. G., Falaschi A., Mazza G. Effect of deoxyribonucleic acid replication inhibitors on bacterial recombination. J Bacteriol. 1976 Apr;126(1):108–121. doi: 10.1128/jb.126.1.108-121.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton B. C. Fine-structure mapping by transformation in the tryptophan region of Bacillus subtilis. J Bacteriol. 1966 May;91(5):1795–1803. doi: 10.1128/jb.91.5.1795-1803.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalumeau H., Delobbe A., Gay P. Biochemical and genetic study of D-glucitol transport and catabolism in Bacillus subtilis. J Bacteriol. 1978 Jun;134(3):920–928. doi: 10.1128/jb.134.3.920-928.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champney W. S., Jensen R. A. D-Tyrosine as a metabolic inhibitor of Bacillus subtilis. J Bacteriol. 1969 Apr;98(1):205–214. doi: 10.1128/jb.98.1.205-214.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasin L. A., Magasanik B. Induction and repression of the histidine-degrading enzymes of Bacillus subtilis. J Biol Chem. 1968 Oct 10;243(19):5165–5178. [PubMed] [Google Scholar]
- Chelala C. A., Margolin P. Effects of deletions on cotransduction linkage in Salmonella typhimurium: evidence that bacterial chromosome deletions affect the formation of transducing DNA fragments. Mol Gen Genet. 1974;131(2):97–112. doi: 10.1007/BF00266146. [DOI] [PubMed] [Google Scholar]
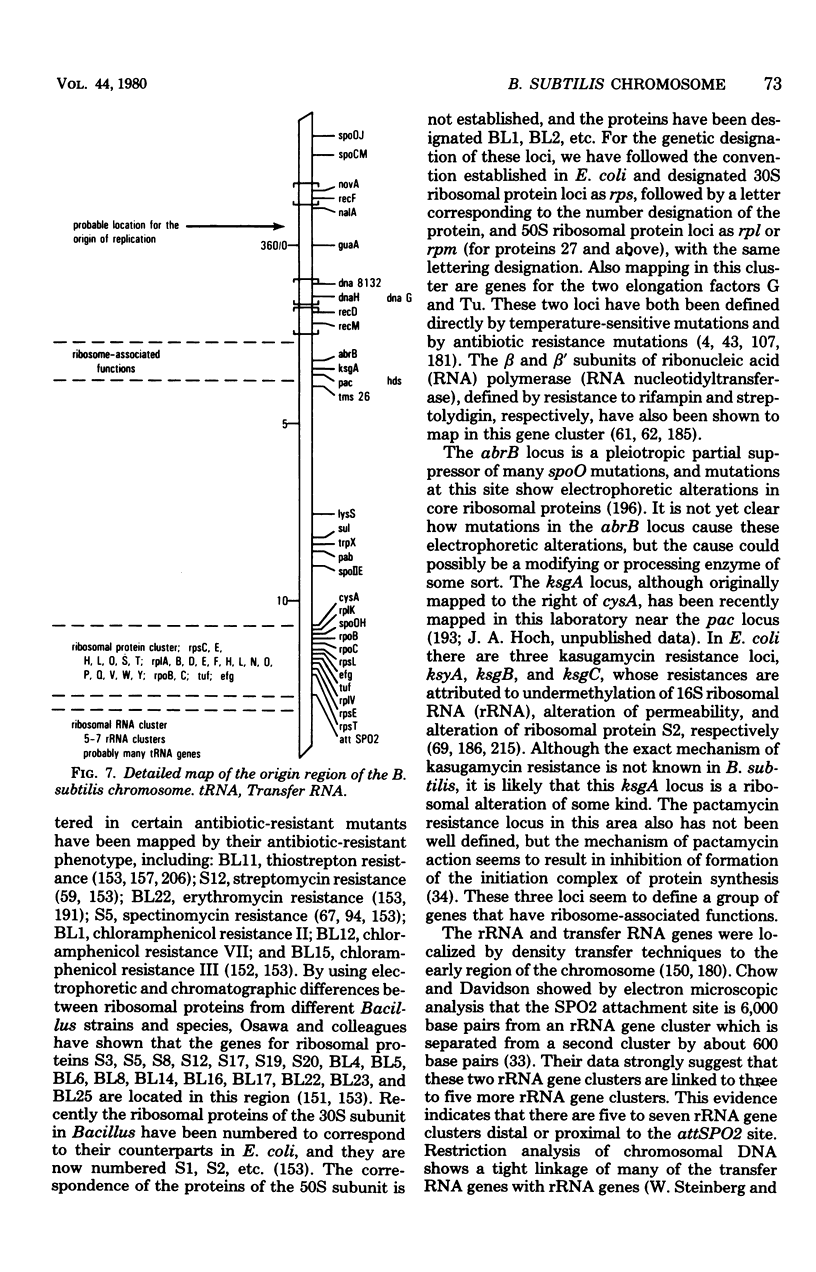
- Chow L. T., Davidson N. Electron microscope mapping of the distribution of ribosomal genes of the Bacillus subtilis chromosome. J Mol Biol. 1973 Apr 5;75(2):265–279. doi: 10.1016/0022-2836(73)90020-x. [DOI] [PubMed] [Google Scholar]
- Cohen L. B., Herner A. E., Goldberg I. H. Inhibition by pactamycin of the initiation of protein synthesis. Binding of N-acetylphenylalanyl transfer ribonucleic acid and polyuridylic acid to ribosomes. Biochemistry. 1969 Apr;8(4):1312–1326. doi: 10.1021/bi00832a004. [DOI] [PubMed] [Google Scholar]
- Copeland J. C., Marmur J. Identification of conserved genetic functions in Bacillus by use of temperature-sensitive mutants. Bacteriol Rev. 1968 Dec;32(4 Pt 1):302–312. [PMC free article] [PubMed] [Google Scholar]
- Cozzarelli N. R., Low R. L. Mutational alteration of Bacillus subtilis DNA polymerase 3 to hydroxyphenylazopyrimidine resistance: polymerase 3 is necessary for DNA replication. Biochem Biophys Res Commun. 1973 Mar 5;51(1):151–157. doi: 10.1016/0006-291x(73)90521-4. [DOI] [PubMed] [Google Scholar]
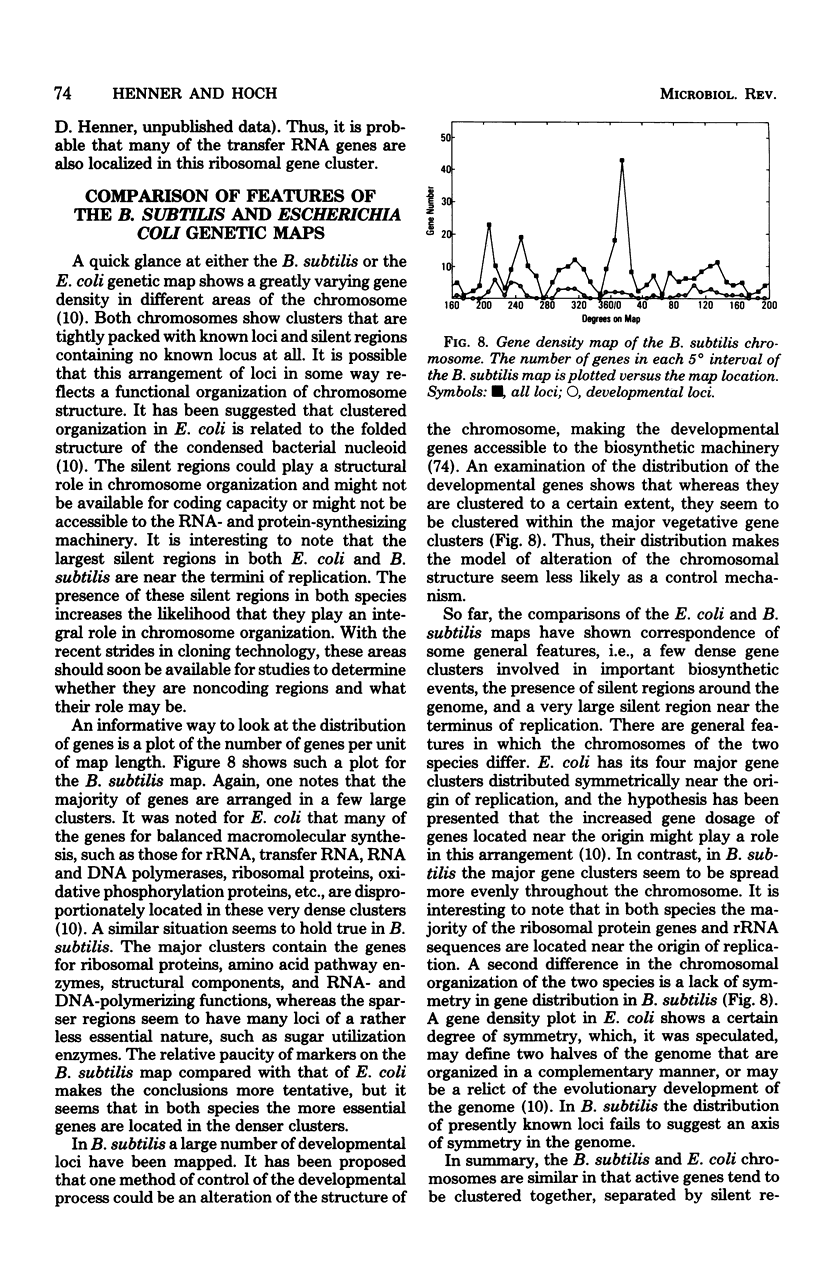
- Dean D. R., Hoch J. A., Aronson A. I. Alteration of the Bacillus subtilis glutamine synthetase results in overproduction of the enzyme. J Bacteriol. 1977 Sep;131(3):981–987. doi: 10.1128/jb.131.3.981-987.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doly J., Sasarman E., Anagnostopoulos C. ATP-dependent deoxyribonuclease in Bacillus subtilis and a mutant deficient in this activity. Mutat Res. 1974 Jan;22(1):15–23. doi: 10.1016/0027-5107(74)90003-7. [DOI] [PubMed] [Google Scholar]
- Dubnau D., Cirigliano C. Fate of transforming DNA following uptake by competent Bacillus subtilis. Formation and properties of products isolated from transformed cells which are derived entirely from donor DNA. J Mol Biol. 1972 Feb 28;64(1):9–29. doi: 10.1016/0022-2836(72)90318-x. [DOI] [PubMed] [Google Scholar]
- Dubnau D., Cirigliano C. Fate of transforming deoxyribonucleic acid after uptake by competent Bacillus subtilis: size and distribution of the integrated donor segments. J Bacteriol. 1972 Aug;111(2):488–494. doi: 10.1128/jb.111.2.488-494.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D., Cirigliano C. Genetic characterization of recombination-deficient mutants of Bacillus subtilis. J Bacteriol. 1974 Feb;117(2):488–493. doi: 10.1128/jb.117.2.488-493.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D., Goldthwaite C., Smith I., Marmur J. Genetic mapping in Bacillus subtilis. J Mol Biol. 1967 Jul 14;27(1):163–185. doi: 10.1016/0022-2836(67)90358-0. [DOI] [PubMed] [Google Scholar]
- Dubnau E., Pifko S., Sloma A., Cabane K., Smith I. Conditional mutations in the translational apparatus of Bacillus subtils. Mol Gen Genet. 1976 Aug 10;147(1):1–12. doi: 10.1007/BF00337929. [DOI] [PubMed] [Google Scholar]
- Dul M. J., Young F. E. Genetic mapping of a mutant defective in D,L-alanine racemase in Bacillus subtilis 168. J Bacteriol. 1973 Sep;115(3):1212–1214. doi: 10.1128/jb.115.3.1212-1214.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPHRATI-ELIZUR E., SRINIVASAN P. R., ZAMENHOF S. Genetic analysis, by means of transformation, of histidine linkage groups in Bacillus subtilis. Proc Natl Acad Sci U S A. 1961 Jan 15;47:56–63. doi: 10.1073/pnas.47.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FREESE E., PARK S. W., CASHEL M. THE DEVELOPMENTAL SIGNIFICANCE OF ALANINE DEHYDROGENASE IN BACILLUS SUBTILIS. Proc Natl Acad Sci U S A. 1964 Jun;51:1164–1172. doi: 10.1073/pnas.51.6.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fein J. E., Rogers H. J. Autolytic enzyme-deficient mutants of Bacillus subtilis 168. J Bacteriol. 1976 Sep;127(3):1427–1442. doi: 10.1128/jb.127.3.1427-1442.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S. H., Sonenshein A. L. Glutamine-requiring mutants of Bacillus subtilis. Biochem Biophys Res Commun. 1977 Dec 7;79(3):987–995. doi: 10.1016/0006-291x(77)91207-4. [DOI] [PubMed] [Google Scholar]
- Freese E. B., Cole R. M., Klofat W., Freese E. Growth, sporulation, and enzyme defects of glucosamine mutants of Bacillus subtilis. J Bacteriol. 1970 Mar;101(3):1046–1062. doi: 10.1128/jb.101.3.1046-1062.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galizzi A., Siccardi A. G., Albertini A. M., Amileni A. R., Meneguzzi G., Polsinelli M. Properties of Bacillus subtilis mutants temperature sensitive in germination. J Bacteriol. 1975 Feb;121(2):450–454. doi: 10.1128/jb.121.2.450-454.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garro A. J., Leffert H., Marmur J. Genetic mapping of a defective bacteriophage on the chromosome of Bacillus subtilis 168. J Virol. 1970 Sep;6(3):340–343. doi: 10.1128/jvi.6.3.340-343.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garro A. J., Sprouse C., Wetmur J. G. Association of the recombination-deficient phenotype of Bacillus subtilis recC strains with the presence of an SPO2 prophage. J Bacteriol. 1976 Apr;126(1):556–558. doi: 10.1128/jb.126.1.556-558.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass K. B., Cozzarelli N. R. Further genetic and enzymological characterization of the three Bacillus subtilis deoxyribonucleic acid polymerases. J Biol Chem. 1973 Nov 25;248(22):7688–7700. [PubMed] [Google Scholar]
- Gay P., Cordier P., Marquet M., Delobbe A. Carbohydrate metabolism and transport in Bacillus subtilis. A study of ctr mutations. Mol Gen Genet. 1973 Mar 19;121(4):355–368. doi: 10.1007/BF00433234. [DOI] [PubMed] [Google Scholar]
- Gay P., Delobbe A. Fructose transport in Bacillus subtilis. Eur J Biochem. 1977 Oct 3;79(2):363–373. doi: 10.1111/j.1432-1033.1977.tb11817.x. [DOI] [PubMed] [Google Scholar]
- Gillis M., De Ley J., De Cleene M. The determination of molecular weight of bacterial genome DNA from renaturation rates. Eur J Biochem. 1970 Jan;12(1):143–153. doi: 10.1111/j.1432-1033.1970.tb00831.x. [DOI] [PubMed] [Google Scholar]
- Goldstein B. J., Zahler S. A. Uptake of branched-chain alpha-keto acids in Bacillus subtilis. J Bacteriol. 1976 Jul;127(1):667–670. doi: 10.1128/jb.127.1.667-670.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldthwaite C., Dubnau D., Smith I. Genetic mapping of antibiotic resistance in markers Bacillus subtilis. Proc Natl Acad Sci U S A. 1970 Jan;65(1):96–103. doi: 10.1073/pnas.65.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldthwaite C., Smith I. Genetic mapping of aminoglycoside and fusidic acid resistant mutations in Bacillus subtilis. Mol Gen Genet. 1972;114(3):181–189. doi: 10.1007/BF01788887. [DOI] [PubMed] [Google Scholar]
- Grant G. F., Simon M. I. Synthesis of bacterial flagella. II. PBS1 transduction of flagella-specific markers in Bacillus subtilis. J Bacteriol. 1969 Jul;99(1):116–124. doi: 10.1128/jb.99.1.116-124.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halling S. M., Burtis K. C., Doi R. H. Reconstitution studies show that rifampicin resistance is determined by the largest polypeptide of Bacillus subtilis RNA polymerase. J Biol Chem. 1977 Dec 25;252(24):9024–9031. [PubMed] [Google Scholar]
- Halling S. M., Burtis K. C., Doi R. H. beta' subunit of bacterial RNA polymerase is responsible for streptolydigin resistance in Bacillus subtilis. Nature. 1978 Apr 27;272(5656):837–839. doi: 10.1038/272837a0. [DOI] [PubMed] [Google Scholar]
- Hara H., Yoshikawa H. Asymmetric bidirectional replication of Bacillus subtilis chromosome. Nat New Biol. 1973 Aug 15;244(137):200–203. doi: 10.1038/newbio244200a0. [DOI] [PubMed] [Google Scholar]
- Harford N. Bidirectional chromosome replication in Bacillus subtilis 168. J Bacteriol. 1975 Mar;121(3):835–847. doi: 10.1128/jb.121.3.835-847.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harford N. Genetic analysis of rec mutants of Bacillus subtilis. Evidence for at least six linkage groups. Mol Gen Genet. 1974 Mar 27;129(3):269–274. doi: 10.1007/BF00267919. [DOI] [PubMed] [Google Scholar]
- Harford N., Sueoka N. Chromosomal location of antibiotic resistance markers in Bacillus subtilis. J Mol Biol. 1970 Jul 28;51(2):267–286. doi: 10.1016/0022-2836(70)90142-7. [DOI] [PubMed] [Google Scholar]
- Harwood C. R., Baumberg S. Arginine hydroxamate-resistant mutants of Bacillus subtilis with altered control of arginine metabolism. J Gen Microbiol. 1977 May;100(1):177–188. doi: 10.1099/00221287-100-1-177. [DOI] [PubMed] [Google Scholar]
- Helser T. L., Davies J. E., Dahlberg J. E. Mechanism of kasugamycin resistance in Escherichia coli. Nat New Biol. 1972 Jan 5;235(53):6–9. doi: 10.1038/newbio235006a0. [DOI] [PubMed] [Google Scholar]
- Henkin T. M., Campbell K. M., Chambliss G. H. Spectinomycin dependence in Bacillus subtilis. J Bacteriol. 1979 Mar;137(3):1452–1455. doi: 10.1128/jb.137.3.1452-1455.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henner D. J., Steinberg W. Genetic location of the Bacillus subtilis sup-3 suppressor mutation. J Bacteriol. 1979 Aug;139(2):668–670. doi: 10.1128/jb.139.2.668-670.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higerd T. B., Hoch J. A., Spizizen J. Hyperprotease-producing mutants of Bacillus subtilis. J Bacteriol. 1972 Nov;112(2):1026–1028. doi: 10.1128/jb.112.2.1026-1028.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higerd T. B. Isolation of acetyl esterase mutants of Bacillus subtilis 168. J Bacteriol. 1977 Feb;129(2):973–977. doi: 10.1128/jb.129.2.973-977.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J. A., Anagnostopoulos C. Chromosomal location and properties of radiation sensitivity mutations in Bacillus subtilis. J Bacteriol. 1970 Aug;103(2):295–301. doi: 10.1128/jb.103.2.295-301.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J. A., Coukoulis H. J. Genetics of the alpha-ketoglutarate dehydrogenase complex of Bacillus subtilis. J Bacteriol. 1978 Jan;133(1):265–269. doi: 10.1128/jb.133.1.265-269.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J. A., Nester E. W. Gene-enzyme relationships of aromatic acid biosynthesis in Bacillus subtilis. J Bacteriol. 1973 Oct;116(1):59–66. doi: 10.1128/jb.116.1.59-66.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch S. O., Anagnostopoulos C., Crawford I. P. Enzymes of the tryptophan operon of Bacillus subtilis. Biochem Biophys Res Commun. 1969 Jun 27;35(6):838–844. doi: 10.1016/0006-291x(69)90700-1. [DOI] [PubMed] [Google Scholar]
- Hoch S. O. Isolation and characterization of two tryptophan biosynthetic enzymes, indoleglycerol phosphate synthase and phosphoribosyl anthranilate isomerase, from Bacillus subtilis. J Bacteriol. 1979 Aug;139(2):362–368. doi: 10.1128/jb.139.2.362-368.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch S. O. Mapping of the 5-methyltryptophan resistance locus in Bacillus subtilis. J Bacteriol. 1974 Jan;117(1):315–317. doi: 10.1128/jb.117.1.315-317.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch S. O., Roth C. W., Crawford I. P., Nester E. W. Control of tryptophan biosynthesis by the methyltryptophan resistance gene in Bacillus subtilis. J Bacteriol. 1971 Jan;105(1):38–45. doi: 10.1128/jb.105.1.38-45.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch S. O. Tryptophan synthetase from Bacillus subtilis. Purification and characterization of the component. J Biol Chem. 1973 May 10;248(9):2999–3003. [PubMed] [Google Scholar]
- Holmes W. M., Kane J. F. Anthranilate synthase from Bacillus subtilis. The role of a reduced subunit X in aggregate formation and amidotransferase activity. J Biol Chem. 1975 Jun 25;250(12):4462–4469. [PubMed] [Google Scholar]
- Hori H. Molecular evolution of 5S RNA. Mol Gen Genet. 1976 May 7;145(2):119–123. doi: 10.1007/BF00269583. [DOI] [PubMed] [Google Scholar]
- Hunter B. I., Yamagishi H., Takahashi I. Molecular weight of bacteriophage PBS 1 deoxyribonucleic acid. J Virol. 1967 Aug;1(4):841–842. doi: 10.1128/jvi.1.4.841-842.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima T., Diesterhaft M. D., Freese E. Sodium effect of growth on aspartate and genetic analysis of a Bacillus subtilis mutant with high aspartase activity. J Bacteriol. 1977 Mar;129(3):1440–1447. doi: 10.1128/jb.129.3.1440-1447.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionesco H., Michel J., Cami B., Schaeffer P. Symposium on bacterial spores: II. Genetics of sporulation in Bacillus subtilis Marburg. J Appl Bacteriol. 1970 Mar;33(1):13–24. doi: 10.1111/j.1365-2672.1970.tb05230.x. [DOI] [PubMed] [Google Scholar]
- Isolation and characterization of a Bacillus subtilis mutant with a defective N-glycosidase activity for uracil-containing deoxyribonucleic acid. J Bacteriol. 1977 Aug;131(2):438–445. doi: 10.1128/jb.131.2.438-445.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J., Mildner G., Spizizen J. Early blocked asporogenous mutants of Bacillus subtilis 168. I. Isolation and characterization of mutants resistant to antibiotic(s) produced by sporulating Bacillus subtilis 168. Mol Gen Genet. 1971;112(2):104–109. doi: 10.1007/BF00267488. [DOI] [PubMed] [Google Scholar]
- Ito J. Pleiotropic nature of bacteriophage tolerant mutants obtained in early-blocked asporogenous mutants of Bacillus subtilis 168. Mol Gen Genet. 1973 Aug 10;124(2):97–106. doi: 10.1007/BF00265143. [DOI] [PubMed] [Google Scholar]
- Itoh T. Amino acid replacement in the protein S5 from a spectinomycin resistant mutant of Bacillus subtilis. Mol Gen Genet. 1976 Feb 27;144(1):39–42. doi: 10.1007/BF00277301. [DOI] [PubMed] [Google Scholar]
- Iyer V. N. Mutations determining mitomycin resistance in Bacillus subtilis. J Bacteriol. 1966 Dec;92(6):1663–1669. doi: 10.1128/jb.92.6.1663-1669.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane J. F., Goode R. L., Wainscott J. Multiple mutations in cysA 14 MUTANTS OF Bacillus subtilis. J Bacteriol. 1975 Jan;121(1):204–211. doi: 10.1128/jb.121.1.204-211.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane J. F. Regulation of a common amidotransferase subunit. J Bacteriol. 1977 Nov;132(2):419–425. doi: 10.1128/jb.132.2.419-425.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamata D., Gross J. D. Isolation and genetic analysis of temperature-sensitive mutants of B. subtilis defective in DNA synthesis. Mol Gen Genet. 1970;108(3):277–287. doi: 10.1007/BF00283358. [DOI] [PubMed] [Google Scholar]
- Karamata D., McConnell M., Rogers H. J. Mapping of rod mutants of Bacillus subtilis. J Bacteriol. 1972 Jul;111(1):73–79. doi: 10.1128/jb.111.1.73-79.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly M. S. Physical and mapping properties of distant linkages between genetic markers in transformation of Bacillus subtilis. Mol Gen Genet. 1967;99(4):333–349. doi: 10.1007/BF00330909. [DOI] [PubMed] [Google Scholar]
- Kelly M. S. The causes of instability of linkage in transformation of Bacillus subtilis. Mol Gen Genet. 1967;99(4):350–361. doi: 10.1007/BF00330910. [DOI] [PubMed] [Google Scholar]
- Kemper J. Gene order and co-transduction in the leu-ara-fol-pyrA region of the Salmonella typhimurium linkage map. J Bacteriol. 1974 Jan;117(1):94–99. doi: 10.1128/jb.117.1.94-99.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss I., Berek I., Ivánovics G. Mapping the -aminolaevulinic acid synthetase locus in Bacillus subtilis. J Gen Microbiol. 1971 May;66(2):153–159. doi: 10.1099/00221287-66-2-153. [DOI] [PubMed] [Google Scholar]
- Klotz L. C., Zimm B. H. Size of DNA determined by viscoelastic measurements: results on bacteriophages, Bacillus subtilis and Escherichia coli. J Mol Biol. 1972 Dec 30;72(3):779–800. doi: 10.1016/0022-2836(72)90191-x. [DOI] [PubMed] [Google Scholar]
- Kobayashi H., Kobayashi K., Kobayashi Y. Isolation and characterization of fusidic acid-resistant, sporulation-defective mutants of Bacillus subtilis. J Bacteriol. 1977 Oct;132(1):262–269. doi: 10.1128/jb.132.1.262-269.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst F., Pascal M., Lepesant-Kejzlarova J., Lepesant J. A., Billault A., Dedonder R. Pleiotropic mutations affecting sporulation conditions and the syntheses of extracellular enzymes in Bacillus subtilis 168. Biochimie. 1974;56(11-12):1481–1489. doi: 10.1016/s0300-9084(75)80270-7. [DOI] [PubMed] [Google Scholar]
- Kunst F., Steinmetz M., Lepesant J. A., Dedonder R. Presence of a third sucrose hydrolyzing enzyme in Bacillus subtilis: constitutive levanase synthesis by mutants of Bacillus subtilis Marburg 168. Biochimie. 1977;59(3):289–292. [PubMed] [Google Scholar]
- Laipis P. J., Ganesan A. T. A deoxyribonucleic acid polymerase I-deficient mutant of Bacillus subtilis. J Biol Chem. 1972 Sep 25;247(18):5867–5871. [PubMed] [Google Scholar]
- Lauer G. D., Klotz L. C. Molecular weight of bacteriophage PBS2 DNA. J Virol. 1976 Jun;18(3):1163–1164. doi: 10.1128/jvi.18.3.1163-1164.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hegarat J. C., Anagnostopoulos C. Localisation chromosomique d'un gène gouvernant la synthèse d'une phosphatase alcaline chez Bacillus subtilis. C R Acad Sci Hebd Seances Acad Sci D. 1969 Nov 17;269(20):2048–2050. [PubMed] [Google Scholar]
- Le Hégarat J. C., Anagnostopoulos C. Purification, subunit structure and properties of two repressible phosphohydrolases of Bacillus subtilis. Eur J Biochem. 1973 Nov 15;39(2):525–539. doi: 10.1111/j.1432-1033.1973.tb03151.x. [DOI] [PubMed] [Google Scholar]
- Leibovici J., Anagnostopoulos C. Propriétés de la thréonine désaminase de la souche sauvage et d'un mutant sensible a la valine de Bacillus subtilis. Bull Soc Chim Biol (Paris) 1969 Sep 18;51(4):691–707. [PubMed] [Google Scholar]
- Leighton T. J., Dor R. H., Warren R. A., Kelln R. A. The relationship of serine protease activity to RNA polymerase modification and sporulation in Bacillus subtilis. J Mol Biol. 1973 May 5;76(1):103–122. doi: 10.1016/0022-2836(73)90083-1. [DOI] [PubMed] [Google Scholar]
- Lepesant-Kejzlarová J., Lepesant J. A., Walle J., Billault A., Dedonder R. Revision of the linkage map of Bacillus subtilis 168: indications for circularity of the chromosome. J Bacteriol. 1975 Mar;121(3):823–834. doi: 10.1128/jb.121.3.823-834.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepesant-Kejzlarová J., Walle J., Billault A., Kunst F., Lepesant J. A., Dedonder R. Etablissement de la carte génétique de Bacillus subtilis: réexamen de la localisation du segment chromosomique compris entre les marqueurs sacQ36 et gtaA12. C R Acad Sci Hebd Seances Acad Sci D. 1974 Apr 1;278(14):1911–1914. [PubMed] [Google Scholar]
- Lepesant J. A., Billault A., Kejzlarová-Lepesant J., Pascal M., Kunst F., Dedonder R. Identification of the structural gene for sucrase in Bacillus subtilis Marburg. Biochimie. 1974;56(11-12):1465–1470. doi: 10.1016/s0300-9084(75)80268-9. [DOI] [PubMed] [Google Scholar]
- Lepesant J. A., Kunst F., Lepesant-Kejzlarová J., Dedonder R. Chromosomal location of mutations affecting sucrose metabolism in Bacillus subtilis Marburg. Mol Gen Genet. 1972;118(2):135–160. doi: 10.1007/BF00267084. [DOI] [PubMed] [Google Scholar]
- Lindgren V., Holmgren E., Rutberg L. Bacillus subtilis mutant with temperature-sensitive net synthesis of phosphatidylethanolamine. J Bacteriol. 1977 Nov;132(2):473–484. doi: 10.1128/jb.132.2.473-484.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren V. Mapping of a genetic locus that affects glycerol 3-phosphate transport in Bacillus subtilis. J Bacteriol. 1978 Feb;133(2):667–670. doi: 10.1128/jb.133.2.667-670.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren V., Rutberg L. Genetic control of the glp system in Bacillus subtilis. J Bacteriol. 1976 Sep;127(3):1047–1057. doi: 10.1128/jb.127.3.1047-1057.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren V., Rutberg L. Glycerol metabolism in Bacillus subtilis: gene-enzyme relationships. J Bacteriol. 1974 Aug;119(2):431–442. doi: 10.1128/jb.119.2.431-442.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez M. E., Ferrari F. A., Siccardi A. G., Mazza G., Polsinelli M. New purine markers in Bacillus subtilis. J Bacteriol. 1976 Apr;126(1):533–535. doi: 10.1128/jb.126.1.533-535.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorence J. H., Nester E. W. Multiple molecular forms of chorismate mutase in Bacillus subtillis. Biochemistry. 1967 May;6(5):1541–1553. doi: 10.1021/bi00857a041. [DOI] [PubMed] [Google Scholar]
- Love E., D'Ambrosio D., Brown N. C. Mapping of the gene specifying DNA polymerase III of Bacillus subtilis. Mol Gen Genet. 1976 Mar 30;144(3):313–321. doi: 10.1007/BF00341730. [DOI] [PubMed] [Google Scholar]
- MAHLER I., NEUMANN I. M., MARMUR J. Studies of genetic-units controlling arginine biosynthesis in Bacillus subtilis. Biochim Biophys Acta. 1963 May 28;72:69–79. [PubMed] [Google Scholar]
- Martinez R. J., Ichiki A. T., Lundh N. P., Tronick S. R. A single amino acid substitution responsible for altered flagellar morphology. J Mol Biol. 1968 Jun 28;34(3):559–564. doi: 10.1016/0022-2836(68)90180-0. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Shibata T., Saito H. Genetic mapping in Bacillus subtilis by 5-bromouracil sensitization to ultraviolet inactivation of transforming activities. J Bacteriol. 1974 Sep;119(3):666–671. doi: 10.1128/jb.119.3.666-671.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza G., Fortunato A., Ferrari E., Canosi U., Falaschi A., Polsinelli M. Genetic and enzymic studies on the recombination process in Bacillus subtilis. Mol Gen Genet. 1975;136(1):9–30. doi: 10.1007/BF00275445. [DOI] [PubMed] [Google Scholar]
- Mendelson N. H., Gross J. D. Characterization of a temperature-sensitive mutant of Bacillus subtilis defective in deoxyribonucleic acid replication. J Bacteriol. 1967 Nov;94(5):1603–1608. doi: 10.1128/jb.94.5.1603-1608.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miczák A., Berek I., Ivanovics G. Mapping the uroporphyrinogen decarboxylase, coproporphyrinogen oxidase and ferrochelatase loci in Bacillus subtilis. Mol Gen Genet. 1976 Jul 5;146(1):85–87. doi: 10.1007/BF00267986. [DOI] [PubMed] [Google Scholar]
- Miczák A., Prágai B., Berek I. Mapping the uroporphyrinogen III cosynthase locus in Bacillus subtilis. Mol Gen Genet. 1979 Jul 24;174(3):293–295. doi: 10.1007/BF00267802. [DOI] [PubMed] [Google Scholar]
- Miki T., Minami Z., Ikeda Y. The genetics of alkaline phosphatase formation in Bacillus subtilis. Genetics. 1965 Nov;52(5):1093–1100. doi: 10.1093/genetics/52.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir A., Lafferty E., Smith D. A. Genetics analysis of spore germination mutants of Bacillus subtilis 168: the correlation of phenotype with map location. J Gen Microbiol. 1979 Mar;111(1):165–180. doi: 10.1099/00221287-111-1-165. [DOI] [PubMed] [Google Scholar]
- Munakata N., Ikeda Y. A mutant of Bacillus subtilis producing ultraviolet-sensitive spores. Biochem Biophys Res Commun. 1968 Nov 8;33(3):469–475. doi: 10.1016/0006-291x(68)90597-4. [DOI] [PubMed] [Google Scholar]
- Nasser D., Henderson G., Nester E. W. Regulated enzymes of aromatic amino acid synthesis: control, isozymic nature, and aggregation in Bacillus subtilis and Bacillus licheniformis. J Bacteriol. 1969 Apr;98(1):44–50. doi: 10.1128/jb.98.1.44-50.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasser D., Nester E. W. Aromatic amino acid biosynthesis: gene-enzyme relationships in Bacillus subtilis. J Bacteriol. 1967 Nov;94(5):1706–1714. doi: 10.1128/jb.94.5.1706-1714.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumov L. S., Savchenko G. V., Prozorov A. A. Kartirovanie oblasti khromosomy Bacillus subtilis, nesushchei mutatsiiu rec 342 (mutatsiia, snizhaiushchaia aktivnost' ATF-zavisimoi dezoksiribonukleazy) Genetika. 1974 Feb;10(2):126–131. [PubMed] [Google Scholar]
- Nester E W, Schafer M, Lederberg J. Gene Linkage in DNA Transfer: A Cluster of Genes Concerned with Aromatic Biosynthesis in Bacillus Subtilis. Genetics. 1963 Apr;48(4):529–551. doi: 10.1093/genetics/48.4.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nester E. W., Montoya A. L. An enzyme common to histidine and aromatic amino acid biosynthesis in Bacillus subtilis. J Bacteriol. 1976 May;126(2):699–705. doi: 10.1128/jb.126.2.699-705.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhard J., Price A. R., Schack L., Thomassen E. Two thymidylate synthetases in Bacillus subtilis. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1194–1198. doi: 10.1073/pnas.75.3.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niaudet B., Gay P., Dedonder R. Identification of the structural gene of the PEP-phosphotransferase enzyme I in Bacillus subtilis Marburg. Mol Gen Genet. 1975;136(4):337–349. doi: 10.1007/BF00341718. [DOI] [PubMed] [Google Scholar]
- Nomura S., Yamane K., Sasaki T., Yamasaki M., Tamura G., Maruo B. Tunicamycin-resistant mutants and chromosomal locations of mutational sites in Bacillus subtilis. J Bacteriol. 1978 Nov;136(2):818–821. doi: 10.1128/jb.136.2.818-821.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan A., Howard K., Sueoka N. Location of a unique replication terminus and genetic evidence for partial bidirectional replication in the Bacillus subtilis chromosome. J Mol Biol. 1975 Jan 5;91(1):15–38. doi: 10.1016/0022-2836(75)90369-1. [DOI] [PubMed] [Google Scholar]
- O'Sullivan A., Sueoka N. Sequential replication of the Bacillus subtilis chromosome. IV. Genetic mapping by density transfer experiment. J Mol Biol. 1967 Jul 28;27(2):349–368. doi: 10.1016/0022-2836(67)90025-3. [DOI] [PubMed] [Google Scholar]
- Oishi M., Oishi A., Sueoka N. Location of genetic loci of soluble RNA on Bacillus subtilis chromosome. Proc Natl Acad Sci U S A. 1966 May;55(5):1095–1103. doi: 10.1073/pnas.55.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa S., Takata R., Tanaka K., Tamaki M. Chloramphenicol resistant mutants of Bacillus subtilis. Mol Gen Genet. 1973 Dec 20;127(2):163–173. doi: 10.1007/BF00333664. [DOI] [PubMed] [Google Scholar]
- Osawa S., Tokui A., Saito H. Mapping by interspecies transformation experiments of several ribosomal protein genes near the replication origin of Bacillus subtilis chromosome. Mol Gen Genet. 1978 Aug 17;164(2):113–129. doi: 10.1007/BF00267376. [DOI] [PubMed] [Google Scholar]
- Pai C. H. Genetics of biotin biosynthesis in Bacillus subtilis. J Bacteriol. 1975 Jan;121(1):1–8. doi: 10.1128/jb.121.1.1-8.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S., Weiss D., Vince R., Wienen B., Stöffler G., Smith I. Thiostrepton-resistant mutants of Bacillus subtilis: localization of resistance to the 50S subunit. Mol Gen Genet. 1976 Mar 30;144(3):235–241. doi: 10.1007/BF00341721. [DOI] [PubMed] [Google Scholar]
- Peterson A. M., Rutberg L. Linked transformation of bacterial and prophage markers in Bacillus subtilis 168 lysogenic for bacteriophage phi 105. J Bacteriol. 1969 Jun;98(3):874–877. doi: 10.1128/jb.98.3.874-877.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot P. J., Coote J. G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976 Dec;40(4):908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot P. J., De Lencastre H. A rapid method for constructing multiply marked strains of Bacillus subtilis. J Gen Microbiol. 1978 May;106(1):191–194. doi: 10.1099/00221287-106-1-191. [DOI] [PubMed] [Google Scholar]
- Piggot P. J. Short communications. Characterization of a cym mutant of Bacillus subtilis. J Gen Microbiol. 1975 Aug;89(2):371–374. doi: 10.1099/00221287-89-2-371. [DOI] [PubMed] [Google Scholar]
- Piggot P. J., Taylor S. Y. New types of mutation affecting formation of alkaline phosphatase by Bacillus subtilis in sporulation conditions. J Gen Microbiol. 1977 Sep;102(1):69–80. doi: 10.1099/00221287-102-1-69. [DOI] [PubMed] [Google Scholar]
- Potvin B. W., Kelleher R. J., Jr, Gooder H. Pyrimidine biosynthetic pathway of Baccillus subtilis. J Bacteriol. 1975 Aug;123(2):604–615. doi: 10.1128/jb.123.2.604-615.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine F. M., Steinberg W. Genetic location of two mutations affecting the lysyl-transfer ribonucleic acid synthetase of Bacillus subtilis. J Bacteriol. 1974 Oct;120(1):384–389. doi: 10.1128/jb.120.1.384-389.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve J. N., Mendelson N. H., Coyne S. I., Hallock L. L., Cole R. M. Minicells of Bacillus subtilis. J Bacteriol. 1973 May;114(2):860–873. doi: 10.1128/jb.114.2.860-873.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rima B. K., Takahashi I. Deoxyribonucleoside-requiring mutants of Bacillus subtilis. J Gen Microbiol. 1978 Jul;107(1):139–145. doi: 10.1099/00221287-107-1-139. [DOI] [PubMed] [Google Scholar]
- Rima B. K., Takahashi I. Synthesis of thymidine nucleotides in Bacillus subtilis. Can J Biochem. 1978 Mar;56(3):158–160. doi: 10.1139/o78-027. [DOI] [PubMed] [Google Scholar]
- Riva S., Villani G., Mastromei G., Mazza G. Bacillus subtilis mutant temperature sensitive in the synthesis of ribonucleic acid. J Bacteriol. 1976 Aug;127(2):679–690. doi: 10.1128/jb.127.2.679-690.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva S., van Sluis C., Mastromei G., Attolini C., Mazza G., Polsinelli M., Falaschi A. A new mutant of Bacillus subtilis altered in the initiation of chromosome replication. Mol Gen Genet. 1975;137(3):185–202. doi: 10.1007/BF00333015. [DOI] [PubMed] [Google Scholar]
- Rogolsky M. Genetic mapping of a locus which regulates the production of pigment associated with spores of Bacillus subtilis. J Bacteriol. 1968 Jun;95(6):2426–2427. doi: 10.1128/jb.95.6.2426-2427.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutberg B., Hoch J. A. Citric acid cycle: gene-enzyme relationships in Bacillus subtilis. J Bacteriol. 1970 Nov;104(2):826–833. doi: 10.1128/jb.104.2.826-833.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutberg L. Mapping of a temperate bacteriophage active on Bacillus subtilis. J Virol. 1969 Jan;3(1):38–44. doi: 10.1128/jvi.3.1.38-44.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H., Shibata T., Ando T. Mapping of genes determining nonpermissiveness and host-specific restriction to bacteriophages in Bacillus subtilis Marburg. Mol Gen Genet. 1979 Feb 26;170(2):117–122. doi: 10.1007/BF00337785. [DOI] [PubMed] [Google Scholar]
- Sekiguchi J., Takada N., Okada H. Genes affecting the productivity of alpha-amylase in Bacillus subtilis Marburg. J Bacteriol. 1975 Feb;121(2):688–694. doi: 10.1128/jb.121.2.688-694.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siccardi A. G., Ferrari F. A., Mazza G., Galizzi A. Identification of coreplicating chromosomal sectors in Bacillus subtilis by nitrosoguanidine-induced comutation. J Bacteriol. 1976 Mar;125(3):755–761. doi: 10.1128/jb.125.3.755-761.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siccardi A. G., Ottolenghi S., Fortunato A., Mazza G. Pleiotropic, extragenic suppression of dna mutants in Bacillus subtilis. J Bacteriol. 1976 Oct;128(1):174–181. doi: 10.1128/jb.128.1.174-181.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel E. C., Marmur J. Temperature-sensitive induction of bacteriophage in Bacillus subtilis 168. J Virol. 1969 Nov;4(5):610–618. doi: 10.1128/jvi.4.5.610-618.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I., Dubnau D., Morrell P., Marmur J. Chromosomal location of DNA base sequences complementary to transfer RNA and to 5 s, 16 s and 23 s ribosomal RNA in Bacillus subtilis. J Mol Biol. 1968 Apr 14;33(1):123–140. doi: 10.1016/0022-2836(68)90285-4. [DOI] [PubMed] [Google Scholar]
- Smith I., Paress P. Genetic and biochemical characterization of kirromycin resistance mutations in Bacillus subtilis. J Bacteriol. 1978 Sep;135(3):1107–1117. doi: 10.1128/jb.135.3.1107-1117.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I., Smith H. Location of the SPO2 attachment site and the bryamycin resistance marker on the Bacillus subtilis chromosome. J Bacteriol. 1973 Jun;114(3):1138–1142. doi: 10.1128/jb.114.3.1138-1142.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I., Weiss D., Pestka S. A micrococcin-resistant mutant of Bacillus subtilis: localization of resistance to the 50s subunit. Mol Gen Genet. 1976 Mar 30;144(3):231–233. doi: 10.1007/BF00341720. [DOI] [PubMed] [Google Scholar]
- Sonenshein A. L., Alexander H. B., Rothstein D. M., Fisher S. H. Lipiarmycin-resistant ribonucleic acid polymerase mutants of Bacillus subtilis. J Bacteriol. 1977 Oct;132(1):73–79. doi: 10.1128/jb.132.1.73-79.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshein A. L., Cami B., Brevet J., Cote R. Isolation and characterization of rifampin-resistant and streptolydigin-resistant mutants of Bacillus subtilis with altered sporulation properties. J Bacteriol. 1974 Oct;120(1):253–265. doi: 10.1128/jb.120.1.253-265.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparling P. F., Ikeya Y., Elliot D. Two genetic loci for resistance to kasugamycin in Escherichia coli. J Bacteriol. 1973 Feb;113(2):704–710. doi: 10.1128/jb.113.2.704-710.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal S. P., Hoch J. A. Conditional dihydrostreptomycin resistance in Bacillus subtilis. J Bacteriol. 1972 Apr;110(1):202–207. doi: 10.1128/jb.110.1.202-207.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg W., Anagnostopoulos C. Biochemical and genetic characterization of a temperature-sensitive, tryptophanyl-transfer ribonucleic acid synthetase mutant of Bacillus subtilis. J Bacteriol. 1971 Jan;105(1):6–19. doi: 10.1128/jb.105.1.6-19.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M., Kunst F., Dedonder R. Mapping of mutations affecting synthesis of exocellular enzymes in Bacillus subtilis. Identity of the sacUh, amyB and pap mutations. Mol Gen Genet. 1976 Nov 17;148(3):281–285. doi: 10.1007/BF00332902. [DOI] [PubMed] [Google Scholar]
- Swanton M., Edlin G. Isolation and characterization of an RNA relaxed mutant of B. subtilis. Biochem Biophys Res Commun. 1972 Jan 31;46(2):583–588. doi: 10.1016/s0006-291x(72)80179-7. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Tamaki M., Osawa S., Kimura A., Takata R. Erythromycin resistant mutants of Bacillus subtilis. Mol Gen Genet. 1973 Dec 20;127(2):157–161. doi: 10.1007/BF00333663. [DOI] [PubMed] [Google Scholar]
- Tipper D. J., Johnson C. W., Ginther C. L., Leighton T., Wittmann H. G. Erythromycin resistant mutations in Bacillus subtilis cause temperature sensitive sporulation. Mol Gen Genet. 1977 Jan 18;150(2):147–159. doi: 10.1007/BF00695395. [DOI] [PubMed] [Google Scholar]
- Tominaga A., Kobayashi Y. Kasugamycin-resistant mutants of Bacillus subtilis. J Bacteriol. 1978 Sep;135(3):1149–1150. doi: 10.1128/jb.135.3.1149-1150.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautner T. A., Pawlek B., Bron S., Anagnostopoulos C. Restriction and modification in B. subtilis. Biological aspects. Mol Gen Genet. 1974;131(3):181–191. doi: 10.1007/BF00267958. [DOI] [PubMed] [Google Scholar]
- Trowsdale J., Chen S. M., Hoch J. A. Genetic analysis of a class of polymyxin resistant partial revertants of stage O sporulation mutants of Bacillus subtilis: map of the chromosome region near the origin of replication. Mol Gen Genet. 1979 May 23;173(1):61–70. doi: 10.1007/BF00267691. [DOI] [PubMed] [Google Scholar]
- Trowsdale J., Sheflett M., Hoch J. A. New cluster of ribosomal genes in Bacillus subtilis with regulatory role in sporulation. Nature. 1978 Mar 9;272(5649):179–181. doi: 10.1038/272179a0. [DOI] [PubMed] [Google Scholar]
- Uehara H., Yamane K., Maruo B. Thermosensitive, extracellular neutral proteases in Bacillus subtilis: isolation, characterization, and genetics. J Bacteriol. 1979 Aug;139(2):583–590. doi: 10.1128/jb.139.2.583-590.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Alstyne D., Simon M. I. Division mutants of Bacillus subtilis: isolation and PBS1 transduction of division-specific markers. J Bacteriol. 1971 Dec;108(3):1366–1379. doi: 10.1128/jb.108.3.1366-1379.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnek D., Greer S. Minor threonine dehydratase encoded within the threonine synthetic region of Bacillus subtilis. J Bacteriol. 1971 Jun;106(3):983–993. doi: 10.1128/jb.106.3.983-993.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnek D., Greer S. Suppression by derepression in threonine dehydratase-deficient mutants of Bacillus subtilis. J Bacteriol. 1971 May;106(2):615–625. doi: 10.1128/jb.106.2.615-625.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. B., Jr, Zahler S. A. Genetic studies of leucine biosynthesis in Bacillus subtilis. J Bacteriol. 1973 Nov;116(2):719–726. doi: 10.1128/jb.116.2.719-726.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. B., Jr, Zahler S. A. Regulation of leucine biosynthesis in Bacillus subtilis. J Bacteriol. 1973 Nov;116(2):727–735. doi: 10.1128/jb.116.2.727-735.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K., Sueoka N. Temperature-sensitive DNA synthesis mutants of Bacillus subtilis--appendix: theory of density transfer for symmetric chromosome replication. Genetics. 1973 Feb;73(2):185–214. doi: 10.1093/genetics/73.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitt D. D., Carlton B. C. Characterization of mutants with single and multiple defects in the tryptophan biosynthetic pathway in Bacillus subtilis. J Bacteriol. 1968 Oct;96(4):1273–1280. doi: 10.1128/jb.96.4.1273-1280.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienen B., Ehrlich R., Stöffler-Meilicke M., Stöffler G., Smith I., Weiss D., Vince R., Pestka S. Ribosomal protein alterations in thiostrepton- and Micrococcin-resistant mutants of Bacillus subtilis. J Biol Chem. 1979 Aug 25;254(16):8031–8041. [PubMed] [Google Scholar]
- Williams M. T., Young F. E. Temperate Bacillus subtilis bacteriophage phi 3T: chromosomal attachment site and comparison with temperate bacteriophages phi 105 and SPO2. J Virol. 1977 Feb;21(2):522–529. doi: 10.1128/jvi.21.2.522-529.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. C., Farmer J. L., Rothman F. Thymidylate synthesis and aminopterin resistance in Bacillus subtilis. J Bacteriol. 1966 Jul;92(1):186–196. doi: 10.1128/jb.92.1.186-196.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOSHIKAWA H., SUEOKA N. Sequential replication of the Bacillus subtilis chromosome. II. Isotopic transfer experiments. Proc Natl Acad Sci U S A. 1963 Jun;49:806–813. doi: 10.1073/pnas.49.6.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K., Maruo B. Alkaline phosphatase possessing alkaline phosphodiesterase activity and other phosphodiesterases in Bacillus subtilis. J Bacteriol. 1978 Apr;134(1):108–114. doi: 10.1128/jb.134.1.108-114.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E. C., Steinberg W. The effect of gene position, gene dosage and a regulatory mutation on the temporal sequence of enzyme synthesis accompanying outgrowth of Bacillus subtilis spores. Mol Gen Genet. 1978 Jan 17;158(3):287–296. doi: 10.1007/BF00267200. [DOI] [PubMed] [Google Scholar]
- Yoneda Y., Maruo B. Mutation of Bacillus subtilis causing hyperproduction of alpha-amylase and protease, and its synergistic effect. J Bacteriol. 1975 Oct;124(1):48–54. doi: 10.1128/jb.124.1.48-54.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Okuyama A., Tanaka N. A third kasugamycin resistance locus, ksgC, affecting ribosomal protein S2 in Escherichia coli K-12. J Bacteriol. 1975 May;122(2):796–797. doi: 10.1128/jb.122.2.796-797.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young F. E., Smith C., Reilly B. E. Chromosomal location of genes regulating resistance to bacteriophage in Bacillus subtilis. J Bacteriol. 1969 Jun;98(3):1087–1097. doi: 10.1128/jb.98.3.1087-1097.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. Genetic mapping of sporulation operons in Bacillus subtilis using a thermosensitive sporulation mutant. J Bacteriol. 1975 Jun;122(3):1109–1116. doi: 10.1128/jb.122.3.1109-1116.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuki S. On the gene controlling the rate of amylase production in Bacillus subtilis. Biochem Biophys Res Commun. 1968 Apr 19;31(2):182–187. doi: 10.1016/0006-291x(68)90727-4. [DOI] [PubMed] [Google Scholar]
- Zahler S. A., Korman R. Z., Rosenthal R., Hemphill H. E. Bacillus subtilis bacteriophage SPbeta: localization of the prophage attachment site, and specialized transduction. J Bacteriol. 1977 Jan;129(1):556–558. doi: 10.1128/jb.129.1.556-558.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]


