Abstract
Background: Increasing evidence suggests that the DNA repair gene XRCC6 (Ku70) may be critically involved in the aetiology of the human carcinogenesis. Many studies have investigated the association between the rs2267437 polymorphism and cancer susceptibility. However, the results of these studies have been controversial. This meta-analysis was conducted to quantitatively summarize the evidence for a relationship between the rs2267437 polymorphism and cancer risk. Methods: Electronic databases, including PUBMED and EMBASE, were searched for publications that met the inclusion criteria. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to evaluate the strength of the association between the XRCC6 promoter rs2267437 polymorphism and cancer risk in a fixed-effects model (the Mantel-Haenszel method) or a random-effects model (the DerSimonian and Laird method), as appropriate. Results: A total of 13 case–control studies, involving 3675 cases and 4247 controls, investigating the XRCC6 rs2267437 polymorphism and cancer susceptibility were identified for the meta-analysis. The pooled analysis showed that there is a significant relationship between the XRCC6 rs2267437 polymorphism and cancer susceptibility (GG vs. CC: OR=1.28, 95% CI=1.03–1.60). Subgroup analyses based on the cancer type, ethnicity, and source of the controls were also performed, and these results indicated that the XRCC6 promoter rs2267437 polymorphism was associated with cancer risk in breast cancer studies (GG vs. CC: OR=1.79, 95% CI=1.25–2.56; GG vs. CG+CC: OR=1.40, 95% CI=1.01–1.95), in Asian populations (GG vs. CC: OR=1.33, 95% CI=1.01–1.74) and in population-based studies (GG vs. CC: OR=1.57, 95% CI=1.12–2.22; CG vs. CC: OR=1.35, 95% CI=1.11–1.64; GG+CG vs. CC: OR=1.37, 95% CI=1.14–1.65). Conclusion: This meta-analysis suggests that the XRCC6 rs2267437 polymorphism may affect breast cancer susceptibility and increase the risk of cancer in Asian populations and in the general population. It is critical that further large-scale and well-designed studies be conducted to confirm the association between the rs2267437 genotype and cancer risk.
Introduction
Cancer is a multifactorial disease that is the result of complex interactions between environmental and genetic factors (Pharoah et al., 2004). DNA double-strand breaks (DSBs) are considered the most lethal DNA lesions for eukaryotic cells; DSBs can be caused by a variety of factors and constitute a serious threat to cell viability and genome stability. Genetic polymorphisms in genes involved in DSB repair may alter the function of the DNA DSB repair machinery and affect an individual's cancer susceptibility.
In mammalian species, DSBs can be repaired by two mechanisms: homologous recombination and nonhomologous end joining (NHEJ) (Yano et al., 2009). In the NHEJ repair process, the Ku70/80 heterodimer (encoded by XRCC6 and XRCC5) binds to the ends of DSBs and activates the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs). In the final step, the LIG4 and XRCC4 proteins are recruited to perform the end-joining reaction (Mahaney et al., 2009). NHEJ deficiencies have been shown to increase genome instability (Gilley et al., 2001; Goytisolo et al., 2001) and promote tumorigenesis (Difilippantonio et al., 2000; Gao et al., 2000; Ferguson and Alt, 2001; Zhu et al., 2002). XRCC6-deficient mice are growth retarded, radiosensitive, and display inefficient V(D)J recombination (Gu et al., 1997). The inactivation of Ku70 in mice and derived cell lines promotes malignant transformation both in vitro and in vivo (Li et al., 1998).
The rs2267437 polymorphism is located in the promoter of the XRCC6 gene. The promoter region has been implicated in the regulation of transcription and mRNA stability (Wilkie et al., 2003). To date, many studies have evaluated the role of rs2267437 in cancer development, including breast cancer (Fu et al., 2003; Willems et al., 2008, 2009; He et al., 2012), glioma (Liu et al., 2007), bladder cancer (Wang et al., 2008), oral cancer (Bau et al., 2008), squamous cell carcinomas of the head and neck (Werbrouck et al., 2008), acute myeloid leukemia (Wang et al., 2009), lung cancer (Tseng et al., 2009), gastric cancer (Yang et al., 2011), hepatocellular carcinoma (Li et al., 2011), and esophageal cancer (Li et al., 2012). However, the results of these studies remain inconclusive. Considering the important role of XRCC6 in carcinogenesis, we performed a meta-analysis of all eligible case–control studies to estimate the overall cancer risk associated with XRCC6 promoter rs2267437 genotypes.
Materials and Methods
Identification and selection of relevant studies
We searched the literatures in PubMed and Embase (last searched on February 5, 2012) for all articles on the association between the XRCC6 polymorphism and cancer risk, using the keywords (“Ku70” or “XRCC6”) and (“polymorphism” or “variant” or “variation”). The search was limited to English language articles. Additional studies were identified by a manual search of the bibliographies of all included studies. In our meta-analysis, the studies had to conform to the following inclusion criteria: (1) the article focused on the XRCC6 rs2267437 polymorphism and cancer susceptibility, (2) the study had a case–control design, and (3) the article provided sufficient data for estimating an odds ratio (OR) with 95% confidence interval (CI). Exclusion criteria for this meta-analysis were (1) not designed as case–control studies, (2) duplicate of previous publications, (3) based on incomplete data, and (4) systemic reviews, case series report, or review or editorial.
Data extraction
Two investigators independently extracted the data from published studies using a standardized form and reached a consensus on all items. The following information was extracted from each study: the first author's name, year of publication, country, patient ethnicity, cancer type, source of control groups, numbers of cases and controls, genotype distributions in cases and controls, and p value for the Hardy–Weinberg equilibrium (HWE) test in controls.
Data synthesis
Statistical analyses were performed with STATA version 10 (StataCorp LP, College Station, TX), using two-sided p-values. In the control groups of each study, HWE was tested by the chi-square test for goodness of fit, and p<0.05 was considered to represent significant disequilibrium. The strength of the association between the rs2267437 polymorphism and cancer susceptibility was evaluated by the ORs with 95% CIs. Pooled OR and 95% CI were assessed in a codominant model (GG vs. CC; CG vs. CC), dominant model (GG+CG vs. CC), and recessive model (GG vs. CG+CC). The significance of the pooled OR was determined using the Z test, with p<0.05 considered statistically significant. Subgroup analyses were performed by the cancer type, ethnicity, and source of controls. The chi-squared-based Q-statistic test was used to assess heterogeneity. When the result of the heterogeneity test was p<0.05, the random-effects model was used (the DerSimonian and Laird method) (DerSimonian and Laird, 1986). Otherwise, the fixed-effects model was selected (the Mantel-Haenszel method) (Mantel and Haenszel, 1959). The I2 value, ranging from 0–100%, was used to quantitatively estimate heterogeneity, with 0% and 100% representing low and high degrees of inconsistency, respectively (Higgins and Thompson, 2002; Higgins et al., 2003). Sensitivity analyses were performed to assess the stability of the results of the meta-analysis. Begg's funnel plots and the Egger's regression test were used to investigate the potential publication bias; p<0.05 was considered statistically significant (Egger et al., 1997).
Results
Selection and characteristics of eligible studies
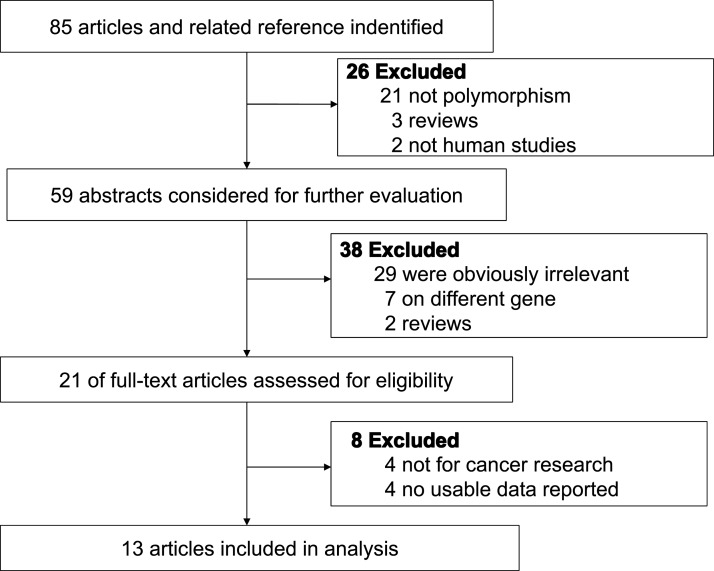
We identified 85 articles using the above search terms. Sixty four articles were excluded because of obvious irrelevance by screening the title and abstract (50 did not study the relevant single-nucleotide polymorphisms (SNP), 7 investigated different genes, 5 were not case–control studies, and 2 were not conducted in humans). Eight studies were excluded after reading through the full texts of the remaining articles, four articles were not cancer research, and four did not report usable data.
Ultimately, a total of 13 case–control studies, involving 3547 cases and 4133 controls, concerning the XRCC6 rs2267437 polymorphism and cancer susceptibility were included in this meta-analysis (Fig. 1).
FIG. 1.
Studies identified with criteria for inclusion and exclusion.
Among the 13 studies included, 4 investigated breast cancer, and 9 investigated other cancers. Ten of these studies were conducted in Asian populations, and 3 were conducted in European populations. Of the 13 studies, 10 used hospital-based controls and 3 used controls derived from healthy populations. The characteristics of the selected studies are presented in Table 1.
Table 1.
Study Characteristics from 13 Studies Included in the Meta-Analysis
| |
|
|
|
|
|
|
Case |
Control |
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| First author | Year | Ethnicity | Cancer type | SNP | Source of controls | Total | CC/AA | CG/AG | GG/GG | CC/AA | CG/AG | GG/GG | HWE |
| Fu | 2003 | Asian | Breast cancer | rs2267437 | HB | 633 | 192 | 55 | 7 | 261 | 106 | 12 | 0.76 |
| Willems | 2008 | European | Breast cancer | rs2267437 | PB | 288 | 44 | 94 | 31 | 45 | 54 | 20 | 0.48 |
| Willems | 2009 | European | Breast cancer | rs2267437 | PB | 377 | 59 | 107 | 40 | 71 | 73 | 27 | 0.26 |
| He | 2012 | Asian | Breast cancer | rs2267437 | HB | 594 | 141 | 127 | 25 | 179 | 113 | 9 | 0.08 |
| Liu | 2007 | Asian | Glioma | rs2267437 | HB | 1478 | 475 | 245 | 25 | 483 | 227 | 23 | 0.56 |
| 2007 | Asian | Glioma | rs132770 | HB | 1488 | 612 | 132 | 13 | 613 | 109 | 9 | 0.10 | |
| Wang | 2008 | Asian | Bladder cancer | rs2267437 | HB | 448 | 129 | 71 | 13 | 149 | 74 | 12 | 0.48 |
| Bau | 2008 | Asian | Oral cancer | rs2267437 | HB | 636 | 227 | 83 | 8 | 214 | 98 | 6 | 0.17 |
| 2008 | Asian | Oral cancer | rs132770 | HB | 636 | 18 | 35 | 265 | 13 | 37 | 268 | 0.00 | |
| Werbrouck | 2008 | European | Squamous cell carcinoma | rs2267437 | HB | 308 | 67 | 71 | 13 | 59 | 74 | 24 | 0.92 |
| Wang | 2009 | Asian | Acute myeloid leukemia | rs2267437 | HB | 330 | 83 | 35 | 2 | 116 | 86 | 8 | 0.10 |
| Tseng | 2009 | Asian | Lung cancer | rs2267437 | HB | 301 | 140 | 9 | 1 | 138 | 11 | 2 | 0.01 |
| Yang | 2011 | Asian | Gastric cancer | rs2267437 | HB | 696 | 95 | 37 | 4 | 383 | 167 | 10 | 0.09 |
| 2011 | Asian | Gastric cancer | rs132770 | HB | 696 | 8 | 19 | 109 | 28 | 73 | 459 | 0.00 | |
| Li | 2011 | Asian | Hepatocellular carcinoma | rs2267437 | PB | 1342 | 433 | 207 | 35 | 457 | 184 | 26 | 0.17 |
| 2011 | Asian | Hepatocellular carcinoma | rs132770 | PB | 1348 | 4 | 103 | 565 | 4 | 88 | 584 | 0.73 | |
| Li | 2012 | Asian | Esophageal cancer | rs2267437 | HB | 249 | 76 | 40 | 1 | 89 | 42 | 1 | 0.10 |
SNP, single-nucleotide polymorphism; PB, population based; HB, hospital based; HWE, Hardy–Weinberg equilibrium.
Quantitative synthesis
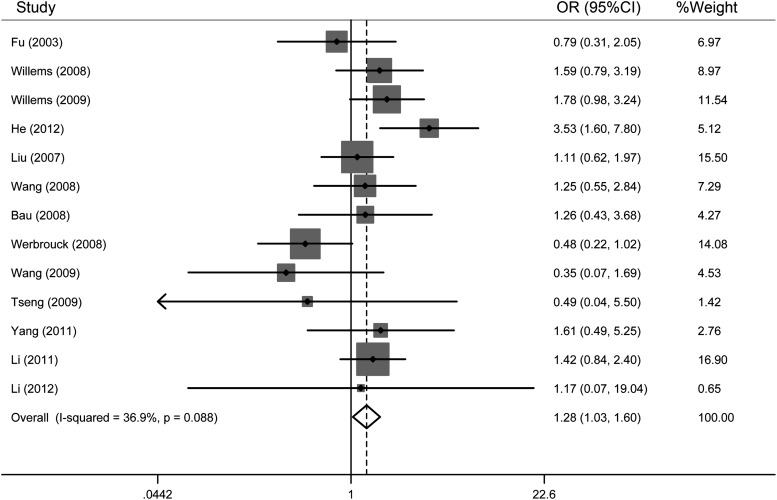
As shown in Table 2, when all the eligible studies were pooled, the GG genotype was associated with a significantly increased risk of all types of cancers compared with the CC genotype (OR=1.28, 95% CI=1.03–1.60, I2=36.9%). The forest plots of the meta-analysis are shown in Figure 2.
Table 2.
Results of the Pooled Data Analyses for XRCC6 rs2267437 and rs132770 Polymorphisms on Cancer Risk in the Meta-Analysis
| |
GG vs. CC |
CG vs. CC |
Dominant model |
Recessive model |
||||
|---|---|---|---|---|---|---|---|---|
| rs2267437 | OR (95% CI) | pa | OR (95% CI) | pa | OR (95% CI) | pa | OR (95% CI) | pa |
| Total | 1.28 (1.03, 1.60) | 0.088 | 1.04 (0.88, 1.23)b | 0.006 | 1.05 (0.88, 1.25)b | 0.001 | 1.17 (0.95, 1.45) | 0.302 |
| Cancer type | ||||||||
| Breast cancer | 1.79 (1.25, 2.56) | 0.125 | 1.31 (0.84, 2.03)b | 0.004 | 1.35 (0.86, 2.11)b | 0.002 | 1.40 (1.01, 1.95) | 0.146 |
| Other cancer | 1.04 (0.78, 1.37) | 0.385 | 1.00 (0.89, 1.12) | 0.207 | 1.01 (0.90, 1.13) | 0.096 | 1.03 (0.78, 1.36) | 0.525 |
| Source of control | ||||||||
| Population based | 1.57 (1.12, 2.22) | 0.855 | 1.35 (1.11, 1.64) | 0.176 | 1.37 (1.14, 1.65) | 0.199 | 1.26 (0.91, 1.73) | 0.893 |
| Hospital based | 1.11 (0.83, 1.48) | 0.059 | 0.97 (0.86, 1.09) | 0.062 | 0.94 (0.77, 1.13)b | 0.010 | 1.10 (0.83, 1.46) | 0.147 |
| Ethnicity | ||||||||
| Asian | 1.33 (1.01, 1.74) | 0.279 | 0.98 (0.83, 1.15)b | 0.036 | 0.99 (0.83, 1.19)b | 0.007 | 1.30 (0.99, 1.70) | 0.492 |
| European | 1.13 (0.52, 2.49)b | 0.019 | 1.38 (0.85, 2.25)b | 0.049 | 1.32 (0.76, 2.30)b | 0.013 | 0.98 (0.69, 1.39) | 0.128 |
| |
GG vs. AA |
AG vs. AA |
Dominant model |
Recessive model |
||||
|---|---|---|---|---|---|---|---|---|
| rs132770 | OR (95% CI) | pa | OR (95% CI) | pa | OR (95% CI) | pa | OR (95% CI) | pa |
| Total | 0.93 (0.60, 1.43) | 0.661 | 1.13 (0.88, 1.45) | 0.614 | 1.12 (0.88, 1.42) | 0.490 | 0.90 (0.73, 1.11) | 0.725 |
| Source of control | ||||||||
| Hospital based | 0.92 (0.58, 1.46) | 0.452 | 1.13 (0.88, 1.45) | 0.406 | 1.12 (0.88, 1.43) | 0.303 | 0.96 (0.72, 1.29) | 0.647 |
p-Value of Q-test for heterogeneity test.
Random-effects model was used when p-value for heterogeneity test <0.05; otherwise, fixed-effects model was used. The results which are in bold type show statistical significance.
FIG. 2.
Forest plots of 13 individual studies of the XRCC6 rs2267437 polymorphism and cancer risk in a codominant model (GG vs. CC). The squares and horizontal lines correspond to the study-specific OR and 95% confidence interval (CI). The area of the squares indicates the study-specific weight. The diamond represents the pooled OR and 95% CI.
In an analysis stratified by ethnicity, significantly increased risk was found in the Asian population (GG vs. CC: OR=1.33, 95% CI=1.01–1.74).
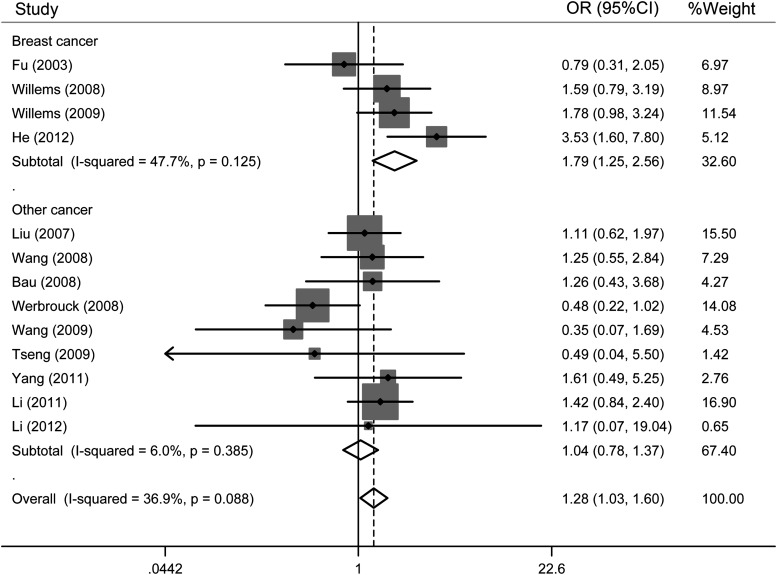
In an analysis stratified by the cancer type, statistically significant effects were observed for breast cancer (GG vs. CC: OR=1.79, 95% CI=1.25–2.56; GG vs. CG+CC: OR=1.40, 95% CI=1.01–1.95) (Fig. 3), but not for other cancers.
FIG. 3.
Forest plots of studies stratified by study type (GG vs. CC). The squares and horizontal lines correspond to the study-specific OR and 95% confidence interval (CI). The diamond reflects the pooled OR and 95% CI.
After analysis stratified by the source of controls, significantly increased risks were found in population-based studies. The pooled ORs for GG versus CC, CG versus CC, and the dominant genetic model were 1.57 (95% CI=1.12–2.22), 1.35 (95% CI=1.11–1.64), and 1.37 (95% CI=1.14–1.65), respectively.
Test of heterogeneity
There was significant heterogeneity to allow heterozygote comparison across the studies (CG vs. CC: pheterogeneity=0.006) and dominant model comparisons (GG+CG vs. CC: pheterogeneity=0.001), but not the other two comparisons. In the subgroup analysis of the cancer type, heterogeneity disappeared in the subgroup analysis of other cancers (CG vs. CC: pheterogeneity=0.207; GG+CG vs. CC: pheterogeneity=0.096). In addition, when patients were stratified based on the source of control, heterogeneity disappeared in population-based studies (CG vs. CC: pheterogeneity=0.176; GG+CG vs. CC: pheterogeneity=0.199) and hospital-based studies (CG vs. CC: pheterogeneity=0.062).
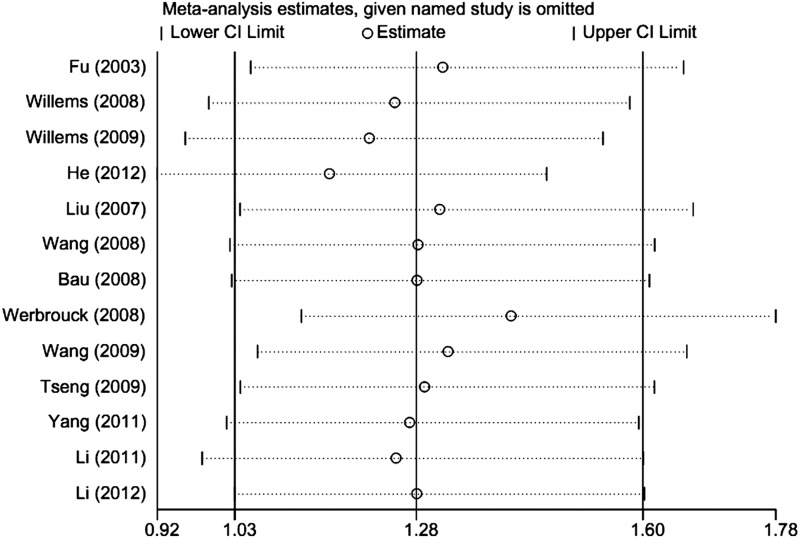
Sensitivity analysis
We performed sensitivity analyses after the sequential removal of each eligible study to assess the influence of each individual study on the pooled OR. Sensitivity analyses revealed that two independent studies were the main source of heterogeneity (Werbrouck et al., 2008; He et al., 2012) (Fig. 4). The heterogeneity was effectively decreased by the exclusion of these two studies (GG vs. CC: OR=1.28, 95% CI=1.00–1.64, I2=0.0%, pheterogeneity=0.791). In addition, no single study changed the pooled ORs qualitatively, suggesting that the results of this meta-analysis were statistically reliable.
FIG. 4.
Influence analysis of GG versus CC in this meta-analysis.
Publication bias
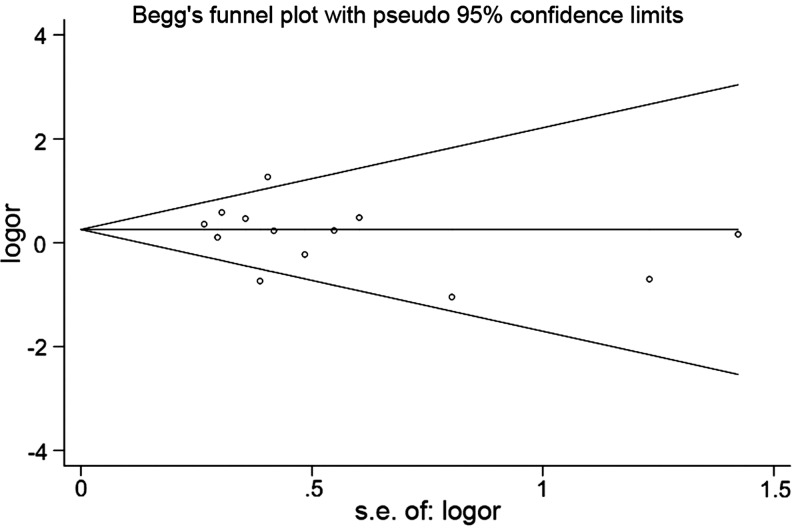
The publication bias in the literature was assessed by the Begg's funnel plot and Egger's test (Fig. 5). The shape of the funnel plots did not reveal any evidence of obvious asymmetry. Then, the Egger's test was used to provide statistical evidence of funnel plot symmetry. The results still showed no evidence of publication bias (t=− 1.01, p=0.335 for GG vs. CC), and the 95% CI=− 2.93, 1.09 included zero, demonstrating a lack of publication bias.
FIG. 5.
The Begg's funnel plot to detect publication bias.
Discussion
It is well known that there is individual variation in cancer susceptibility. Genetic susceptibility to cancer has attracted great interest in the scientific community, and there have been many studies of the genetic polymorphisms involved in carcinogenesis. Given the pivotal roles of XRCC6 in carcinogenesis, it is possible that XRCC6 gene variants may modulate cancer risk. Numerous epidemiological studies have been performed in recent years to evaluate the association between the XRCC6 rs2267437 polymorphism and cancer risk. However, the results of these studies are not fully conclusive. Hence, a meta-analysis was performed to help us derive a more precise estimation of the relationship between XRCC6 rs2267437 polymorphisms and cancer risk. This meta-analysis included 3547 cases and 4133 controls, giving it a greater statistical power than all previous studies. We found that the GG genotype was associated with increased cancer risks, especially for breast cancer, in Asian populations and in population-based studies.
DSBs are considered to be the most harmful form of DNA damage. NHEJ is the predominant DSB response pathway in mammalian cells (Kanaar et al., 2008). If left unrepaired or misrepaired, DSBs can cause cell death, chromosomal translocations, and genomic instability, which can contribute to cancer progression in higher eukaryotic organisms (Khanna and Jackson, 2001). The NHEJ pathway involves XRCC4, XRCC5 (Ku80), XRCC6 (Ku70), DNA-PKcs, DNA ligase IV, Artemis, and XLF (Bassing et al., 2002; Shrivastav et al., 2008). Ku is a multifunctional protein that plays a key role in multiple DNA damage responses, such as DSB repair, apoptosis, telomere maintenance, and V(D)J recombination. SNPs are the most common sources of human genetic variation and may contribute to an individual's susceptibility to cancer (Wu et al., 2009). The XRCC6 rs2267437 polymorphism is adjacent to the first putative CACCC box of the Ku70 promoter in a sequence that acts as a binding site for Sp1 and other Kruppel-like transcription factors (Hosoi et al., 2004). Single-nucleotide substitutions within the Sp1/Kruppel-like binding sites or the adjacent CACCC box sequences have a profound effect on the binding activity of these transcription factors (Hasan and MacDonald, 2002). Given the important roles of XRCC6 in cancer aetiology, it is biologically plausible that the XRCC6 rs2267437 polymorphism modulates cancer risk by modifying its transcription and ultimately DSB repair activity.
Because tumors of different origins could have different susceptibility conferred by XRCC6 polymorphisms, we performed subgroup analyses by the cancer type. Interestingly, an association between XRCC6 rs2267437 and breast cancer risk was found. In the analysis stratified by ethnicity, statistically significantly elevated cancer risks were observed in Asians, but not in Europeans. There are several factors that could contribute to this discrepancy. First, different cancers may have different mechanisms of pathogenesis, and the XRCC6 rs2267437 polymorphism might play a different role in various types of cancers. Second, different underlying genetic backgrounds and environments might contribute to the discrepancy. Third, selection bias and different matching criteria may play an important role. Larger numbers of studies are warranted to confirm our findings in the future.
There was no substantial between-study heterogeneity of the polymorphism among the 13 studies, except for He et al. (2012) and Werbrouck et al. (2008). The subgroup analysis showed that the major source of heterogeneity was from ethnicity, suggesting that ethnicity plays an important role in the frequency of this polymorphism.
In the current study, publication bias was analyzed using the Begg's funnel plot and Egger's test. Both the shape of funnel plots and statistical results demonstrated a lack of publication bias, indicating the strength of the results of our meta-analysis.
In interpreting the current results, some limitations of our meta-analysis should be discussed. First, we included only articles written in English and excluded studies published in other languages, which thus may bias the results of our meta-analysis. Second, we were unable to examine the gene–environment interactions that may be an important component of association between the XRCC6 rs2267437 polymorphism and cancer risk. Third, our results were based on unadjusted effect estimates because insufficient data for these analyses were available from most of the literature. Fourth, the results of our subgroup analyses should be interpreted with caution because of the number of subjects in the specific subgroups were relatively small. In spite of these caveats, our meta-analysis has several advantages. First, there was no evidence for heterogeneity among the studies in the codominant model (GG vs. CC) or recessive model (GG vs. CG+CC). Second, the distribution of genotypes was consistent with the Hardy–Weinberg equilibrium (p>0.01) in all studies. Third, no publication bias was observed in our meta-analysis, indicating that the pooled results should be unbiased.
Conclusions
In summary, this meta-analysis convincingly demonstrated that the XRCC6 rs2267437 polymorphism is associated with increased cancer risk, especially in breast cancer, in Asian populations and in population-based studies. Larger well-designed studies should be performed to further confirm our results. Moreover, gene–environment and gene–gene interaction analyses will be needed to clarify the role of the XRCC6 rs2267437 polymorphism in cancer risk.
Acknowledgments
We thank Dr. Meilin Wang for the scientific design. “This work is supported by the National Natural Science Foundation of China (grant 30901534 and 81172694); the Jiangsu Province's Natural Science Foundation (Proj. no. BK2009444); the Grant for the 135 Key Medical Project of Jiangsu Province (No. XK201117); the National high technology research and development program 863 (No. 2012AA02A508), and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions”.
Author Disclosure Statement
The authors declare that there are no competing interests.
References
- Bassing CH. Chua KF. Sekiguchi J, et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci U S A. 2002;99:8173–8178. doi: 10.1073/pnas.122228699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bau DT. Tseng HC. Wang CH, et al. Oral cancer and genetic polymorphism of DNA double strand break gene Ku70 in Taiwan. Oral Oncol. 2008;44:1047–1051. doi: 10.1016/j.oraloncology.2008.02.008. [DOI] [PubMed] [Google Scholar]
- DerSimonian R. Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Difilippantonio MJ. Zhu J. Chen HT, et al. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger M. Davey Smith G. Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson DO. Alt FW. DNA double strand break repair and chromosomal translocation: lessons from animal models. Oncogene. 2001;20:5572–5579. doi: 10.1038/sj.onc.1204767. [DOI] [PubMed] [Google Scholar]
- Fu YP. Yu JC. Cheng TC, et al. Breast cancer risk associated with genotypic polymorphism of the nonhomologous end-joining genes: a multigenic study on cancer susceptibility. Cancer Res. 2003;63:2440–2446. [PubMed] [Google Scholar]
- Gao Y. Ferguson DO. Xie W, et al. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- Gilley D. Tanaka H. Hande MP, et al. DNA-PKcs is critical for telomere capping. Proc Natl Acad Sci U S A. 2001;98:15084–15088. doi: 10.1073/pnas.261574698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goytisolo FA. Samper E. Edmonson S, et al. The absence of the dna-dependent protein kinase catalytic subunit in mice results in anaphase bridges and in increased telomeric fusions with normal telomere length and G-strand overhang. Mol Cell Biol. 2001;21:3642–3651. doi: 10.1128/MCB.21.11.3642-3651.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y. Seidl KJ. Rathbun GA, et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 1997;7:653–665. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- Hasan NM. MacDonald MJ. Sp/Kruppel-like transcription factors are essential for the expression of mitochondrial glycerol phosphate dehydrogenase promoter B. Gene. 2002;296:221–234. doi: 10.1016/s0378-1119(02)00865-x. [DOI] [PubMed] [Google Scholar]
- He W. Luo S. Huang T, et al. The Ku70-1310C/G promoter polymorphism is associated with breast cancer susceptibility in Chinese Han population. Mol Biol Rep. 2012;39:577–583. doi: 10.1007/s11033-011-0773-7. [DOI] [PubMed] [Google Scholar]
- Higgins JP. Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- Higgins JP. Thompson SG. Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoi Y. Watanabe T. Nakagawa K, et al. Up-regulation of DNA-dependent protein kinase activity and Sp1 in colorectal cancer. Int J Oncol. 2004;25:461–468. [PubMed] [Google Scholar]
- Kanaar R. Wyman C. Rothstein R. Quality control of DNA break metabolism: in the ‘end’, it's a good thing. EMBO J. 2008;27:581–588. doi: 10.1038/emboj.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna KK. Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- Li GC. Ouyang H. Li X, et al. Ku70: a candidate tumor suppressor gene for murine T cell lymphoma. Mol Cell. 1998;2:1–8. doi: 10.1016/s1097-2765(00)80108-2. [DOI] [PubMed] [Google Scholar]
- Li R. Yang Y. An Y, et al. Genetic polymorphisms in DNA double-strand break repair genes XRCC5, XRCC6 and susceptibility to hepatocellular carcinoma. Carcinogenesis. 2011;32:530–536. doi: 10.1093/carcin/bgr018. [DOI] [PubMed] [Google Scholar]
- Li T. Suo Q. He D, et al. Esophageal cancer risk is associated with polymorphisms of DNA repair genes MSH2 and WRN in Chinese population. J Thorac Oncol. 2012;7:448–452. doi: 10.1097/JTO.0b013e31823c487a. [DOI] [PubMed] [Google Scholar]
- Liu Y. Zhang H. Zhou K, et al. Tagging SNPs in non-homologous end-joining pathway genes and risk of glioma. Carcinogenesis. 2007;28:1906–1913. doi: 10.1093/carcin/bgm073. [DOI] [PubMed] [Google Scholar]
- Mahaney BL. Meek K. Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel N. Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- Pharoah PD. Dunning AM. Ponder BA, et al. Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer. 2004;4:850–860. doi: 10.1038/nrc1476. [DOI] [PubMed] [Google Scholar]
- Shrivastav M. De Haro LP. Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- Tseng RC. Hsieh FJ. Shih CM, et al. Lung cancer susceptibility and prognosis associated with polymorphisms in the nonhomologous end-joining pathway genes: a multiple genotype-phenotype study. Cancer. 2009;115:2939–2948. doi: 10.1002/cncr.24327. [DOI] [PubMed] [Google Scholar]
- Wang G. Wang S. Shen Q, et al. Polymorphisms in XRCC5, XRCC6, XRCC7 genes are involved in DNA double-strand breaks (DSBs) repair associated with the risk of acute myeloid leukemia (AML) in Chinese population. J Nanjing Med Univ. 2009;23:93–99. [Google Scholar]
- Wang SY. Peng L. Li CP, et al. Genetic variants of the XRCC7 gene involved in DNA repair and risk of human bladder cancer. Int J Urol. 2008;15:534–539. doi: 10.1111/j.1442-2042.2008.02049.x. [DOI] [PubMed] [Google Scholar]
- Werbrouck J. De Ruyck K. Duprez F, et al. Single-nucleotide polymorphisms in DNA double-strand break repair genes: association with head and neck cancer and interaction with tobacco use and alcohol consumption. Mutat Res. 2008;656:74–81. doi: 10.1016/j.mrgentox.2008.07.013. [DOI] [PubMed] [Google Scholar]
- Wilkie GS. Dickson KS. Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Willems P. Claes K. Baeyens A, et al. Polymorphisms in nonhomologous end-joining genes associated with breast cancer risk and chromosomal radiosensitivity. Genes Chromosomes Cancer. 2008;47:137–148. doi: 10.1002/gcc.20515. [DOI] [PubMed] [Google Scholar]
- Willems P. De Ruyck K. Van den Broecke R, et al. A polymorphism in the promoter region of Ku70/XRCC6, associated with breast cancer risk and oestrogen exposure. J Cancer Res Clin Oncol. 2009;135:1159–1168. doi: 10.1007/s00432-009-0556-x. [DOI] [PubMed] [Google Scholar]
- Wu GY. Hasenberg T. Magdeburg R, et al. Association between EGF, TGF-beta1, VEGF gene polymorphism and colorectal cancer. World J Surg. 2009;33:124–129. doi: 10.1007/s00268-008-9784-5. [DOI] [PubMed] [Google Scholar]
- Yang MD. Wang HC. Chang WS, et al. Genetic polymorphisms of DNA double strand break gene Ku70 and gastric cancer in Taiwan. BMC Cancer. 2011;11:174. doi: 10.1186/1471-2407-11-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K. Morotomi-Yano K. Adachi N, et al. Molecular mechanism of protein assembly on DNA double-strand breaks in the non-homologous end-joining pathway. J Radiat Res (Tokyo) 2009;50:97–108. doi: 10.1269/jrr.08119. [DOI] [PubMed] [Google Scholar]
- Zhu C. Mills KD. Ferguson DO, et al. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]