Abstract
The pandemic influenza AH1N1 (2009) caused an outbreak of human infection that spread to the world. Neuraminidase (NA) is an antigenic surface glycoprotein, which is essential to the influenza infection process, and is the target of anti-flu drugs oseltamivir and zanamivir. Currently, NA inhibitors are the pillar pharmacological strategy against seasonal and global influenza. Although mutations observed after NA-inhibitor treatment are characterized by changes in conserved amino acids of the enzyme catalytic site, it is possible that specific amino acid substitutions (AASs) distant from the active site such as H274Y, could confer oseltamivir or zanamivir resistance. To better understand the molecular distribution pattern of NA AASs, we analyzed NA AASs from all available reported pandemic AH1N1 NA sequences, including those reported from America, Africa, Asia, Europe, Oceania, and specifically from Mexico. The molecular distributions of the AASs were obtained at the secondary structure domain level for both the active and catalytic sites, and compared between geographic regions. Our results showed that NA AASs from America, Asia, Europe, Oceania and Mexico followed similar molecular distribution patterns. The compiled data of this study showed that highly conserved amino acids from the NA active site and catalytic site are indeed being affected by mutations. The reported NA AASs follow a similar molecular distribution pattern worldwide. Although most AASs are distributed distantly from the active site, this study shows the emergence of mutations affecting the previously conserved active and catalytic site. A significant number of unique AASs were reported simultaneously on different continents.
Keywords: Influenza, Pandemic H1N1 (2009), Neuraminidase, Molecular distribution pattern, Geographic regions, amino acid substitution
Background
In the last century, many different deadly influenza epidemics have affected humanity. The 1918 Spanish flu epidemic killed approximately 20 to 50 million people worldwide, and the Asian flu epidemic (1957), the Hong Kong flu epidemic (1968), and the Russian flu epidemic (1977) had severe fatal consequences for human populations [1]. According to WHO, influenza pandemic occurs when a newly mutated influenza virus appears in a human population with no immunity, resulting in worldwide spread with high morbidity and mortality. In 2009, the global pandemic of AH1N1 influenza, or “swine flu,” emerged in Mexico and the United States, arising from a genome reassortment of multiple known influenza viruses (swine, avian and humans) [2, 3]. By February 5, 2010, WHO statistics reported that the AH1N1 (2009) virus had been confirmed across 209 countries and killed at least 15,174 people. Given Mexico's status as one of the countries where the virus first emerged [2], the high morbidity and mortality there [4], and the increased vigilance of the Latin American epidemiological surveillance network, Mexico is clearly an important epidemiological site to compare with the rest of the world. The lack of adequate influenza vaccines resulting from the continuous seasonal viral reassortment process [5, 6] makes anti-influenza drugs, including NA-inhibitors, a crucial weapon against influenza infection. Hemagglutinin and NA are important antigenic surface proteins from influenza A virus. Influenza A NA activity facilitates the release of progeny virions from infected cells [7]. NA-inhibitors block NA activity by targeting the highly conserved enzyme active site (AS) and catalytic site (CS). Based on NA crystal structure for 2009 pandemic AH1N1 human influenza [8], six amino acids (R156, W178, I222, E227, E277 and N294) compose and serve as framework residues (FR) in the stabilization of the AS structure, and another nine amino acids (R118, E119, D151, R152, R224, E276, R292, R371 and Y406) act as catalytic residues in direct contact with the substrate [9, 10]. NA-inhibitors currently constitute the most important anti-viral agents for influenza A and B viruses. Recently, strains of seasonal and pandemic AH1N1 virus have been associated with resistance to first-line NA-inhibitor oseltamivir [4, 11]. Specific AASs in AS, and particularly in CS residues have been shown to confer viral resistance to NA-inhibitors [12, 13]. The association between AH1N1 NA AASs and resistance to oseltamivir has revealed the importance of specific AASs.
A well-characterized case is the amino acid substitution H274Y (crystallographic nomenclature: PDB code 3NSS), implicated in a distant effect of a structural change at the AS level [14]. Since the first report of oseltamivir-resistant AH1N1 in June 2009 (Oseltamivir resistance in immunocompromised hospital patients: pandemic H1N1 2009 briefing note by WHO), clinical studies have been published about the use of oseltamivir and its impact on the emergence of NA-inhibitor resistant influenza [15–19]. Several NA sequences and crystallographic structures for multiple strains are currently available in public databases, and the launch of the Influenza Genome Sequencing Project [20] has led to a rapid increase in the availability of sequence information, as well as epidemiological and clinical data. The contribution of different influenza virus databases to the scientific community has been notably important [21, 22]. Nevertheless, recent studies of NA sequencing and protein structure analysis have mostly been applied to drug development [23]. To date, the rapid large-scale sequencing, data sharing in influenza and the use of analytical and visualization tools, such as GENGIS [24] and SUPRAMAP [25], have let the integration of phylogenetic data and geographic information. Global geographic maps of NA AASs, phylogenetic relationships and geographical location are frequently reported. However, the molecular distribution patterns of NA AASs at the sequence and structural level and its relationship to geographic regions or NA-inhibitor resistance have never been reported. The present study aims to fill this gap, showing the molecular distribution patterns of NA AASs in different sequences of 2009 pandemic AH1N1 human influenza virus at protein sequence levels, in Mexico and global geographic regions.
Methodology
NA sequences:
NA sequences from 2009 pandemic AH1N1 influenza virus strains were obtained from the Influenza Virus Sequence Database [22] (April 26, 2011) using the following filters: selected sequence type (protein), type (A), host (human), country/region (America, Asia, Oceania, Europa, Africa and Mexico), protein (NA), subtype (H1N1), full-length only, required segments (NA), get sequences from (only pandemic H1N1 2009 and include The Flu project). To prevent bias when estimating the distribution of AASs, only complete sequences (469 amino acids length) were included.
Detection of NA amino acid substitutions:
A multiple sequence alignment of the NA protein sequences was performed using ClustalW-MPI [26]. Amino acid substitutions were automatically determined using a python script developed for us (http://code.google.com/p/neuraminidase-scripts/) for this specific purpose. All substitutions were compared to the native reference pandemic NA amino acid sequence (id: ACQ73395) reported by the Mexican health authorities. A random sample of 100 sequences was manually analyzed to determine NA AASs. Manual inspection correlated 100% with the automatic process. NA AASs associated with oseltamivir and zanamivir resistance were obtained from the Drug resistance prediction tool, [27, 28], implemented in Influenza Virus Resource. NA AASs associated with oseltamivir and zanamivir resistance reported in different subtypes of seasonal influenza and pandemic AH1N1 (2009) influenza were also included [19, 29–31]. In addition, based on the scientific literature AASs related with resistance were classified in confirmed [15–18, 25] and potential (confirmed in different subtypes). The positions of these AASs were named as RRAP (reported resistance-associated position) and PRAP (potentially resistance-associated position), respectively. The amino acids found in FR of the AS, CS, RRAP and PRAP were extracted, joined in pseudo-sequences and aligned using CLC Main Workbench 6.8.2. Sequence logos were made with these using no duplicated pseudo-sequences and the software WebLogo 3.3 [32].
Molecular distribution patterns of NA amino acid substitutions.:
The molecular distribution patterns of NA AASs were determined by mapping the AASs in the protein sequence. To prevent selection bias, only unique AASs were considered (i.e. only one AAS of a particular type was used regardless the number of times the AAS was reported). Mexican and global AASs were compiled for different continents (Africa, America, Asia, Europe and Oceania). The mapping of AASs was determined according to the localization of the substitutions within each of the 59 domains of the secondary structure. Secondary structure was determined according to the reference NA crystal structure for 2009 pandemic AH1N1 human influenza (PDB code 3nss) [8]. The secondary structure of NA is comprised of two alpha helices (a1, a2), 27 beta sheets (β1, β2, β3, …, β27)) and 30 loops (L1, L2, L3, …, L30). The transmembrane and linker regions were included as independent domains. Given the importance of the AS and CS in enzymatic function, AASs occurring particularly in the AS (R156, W178, I222, E227, and N294), and in the CS (R118, E119, D151, R152, R224, E276, R292, and R371), were recognized separately. Equivalence position of amino acids between crystallographic and sequence lineal nomenclature is shown in Table S1, (available at http://code.google.com/p/neuraminidasescripts/).
Statistical analysis of the molecular distribution patterns:
The molecular distributions of AASs at the level of secondary structure domain were compared with AASs reported from America, Europe, Asia and Oceania, and Africa (i.e. America with Europe, America with Asia and Oceania, America with Africa). In addition, we compared AASs reported in Mexico with those reported in the rest of the world. The frequency of AASs in each domain was calculated as the accumulated number of AASs per normalized domain by the total number of AASs. Using statistical software STATA® 10, the equality of the distribution functions between continents, and between Mexico and the rest of the world, was verified with the Kolmogorov-Smirnov test. The independence of the frequencies of AASs between continents, and between Mexico and rest of the world, were tested using Spearman's rank correlation test.
Results and Discussion
This study shows for the first time the compilation and comparison of all globally reported NA AASs from the 2009 pandemic AH1N1 influenza virus in different geographic areas. We compared the molecular distribution pattern of AASs in the NA secondary structure and active and catalytic site levels between continents and particularly with Mexico.
Pandemic human influenza (AH1N1) NA sequences:
A total of 3740 NA protein sequences were downloaded from the Influenza Virus Sequence Database corresponding to the 2009 pandemic AH1N1 human influenza virus (Dataset S1, available at http://code.google.com/p/neuraminidasescripts/). Of these, 112 were from Mexico and 3628 were from the rest of the world. In addition, sequences were classified according to the continent of origin: 59 from Africa, 2298 from America, 521 from Asia, 772 from Europe and 89 from Oceania. Excluded from the analysis were 1254 incomplete sequences, the majority of which lacked the amino and carboxy-terminal extremes.
Amino acid substitutions in 2009 human AH1N1 NA:
A total of 530 unique AASs were detected. From these, 312 were reported in America, 204 in Asia and Oceania, 219 in Europe, 26 in Africa, and 38 in Mexico. According to the Influenza Virus Sequence Database, NA AASs H274Y and N294S were associated with resistance to oseltamivir (seasonal H3N2). Based on clinical data, substitutions H274Y and I222R were potentially associated with oseltamivir and zanamivir resistance (pandemic AH1N1 2009). AASs reported in NAs from related viruses that confer resistance to oseltamivir were D198N (seasonal B), S248N (seasonal H1N1) and K261R (seasonal H1N1), and those that confer resistance to oseltamivir and zanamivir were Y155H and S246N (seasonal H1N1). AASs specifically located in residues of NA AS or CS were D151N and I222T, and are potentially associated with resistance to zanamivir and both oseltamivir and zanamivir, respectively. The occurrence of these variations is showed in Figure S1 (see supplementary material). Reported variants were found (D151N, Y155H, D198N, I222R, I222T, S246N, K261R, H274Y). But, new variants were found: E119K and Y406H (Asia, in CS), D198G and D198Y (Europe), G248R and G248E (Asia and Mexico). Each pseudo-sequence represents a group of NA sequences by its most important reported amino acids. According these, we found only one type of strain in Africa, Central America and Oceania. This sequence corresponds to the predominant amino acids of the pseudo-sequences in the other regions studied (Asia, Europe, Mexico and South America). This would be due the low amount of sequences analyzed for these regions (59 in Africa, 89 in Oceania and 153 in Central America) compared with the others (112 in Mexico and >520 in each of the others). Most amino acid variations were found in Europe and Asia.
Molecular distribution patterns of NA amino acid substitutions.:
The AASs in NA were non-uniformly distributed across the protein sequence. The molecular distributions for continents appeared qualitatively very similar, showing an identical clustering pattern shown in Figure S2 & Figure S3 (see supplementary material). The molecular distribution of AASs reported in America and Asia-Oceania were found to be significantly different (P=0.005, Kolmogorov-Smirnov nonparametric test). The same result was found between America and Africa, and between Mexico and rest of the world (P<0.001 and p=0.001 respectively, Kolmogorov-Smirnov nonparametric test). There was no evidence to reject the null hypothesis that the molecular distribution pattern of AASs reported from America and Europe were different (P=0.432, Kolmogorov-Smirnov nonparametric test). On the other hand, Spearman correlation tests in all cases rejected the null hypotheses that the molecular distribution frequencies were independent (p<0.001), thus the evidence favors a similar pattern of AASs distribution.
Thus, the molecular distribution of AASs were, qualitatively and quantitatively, very similar between all continents with the exception of Africa, likely due to a lack of AH1N1 NA sequences reported from this region. In the particular case of Mexico, only 38 unique AASs were available, therefore despite the qualitative similarity in the molecular distribution of AASs when compared to the rest of the world, there was a lack of statistical significance. Since the type of AASs take relevance in the study, it is important to mention that the study did not include the frequency of each type of AAS at continental level because it is directly related to the number of NA sequences available, which could be associated to a reporting bias. In addition, the remarkable similarity between the worldwide molecular distribution of NA AASs stratified by continent, and the presence of identical hot-spot regions, suggest the existence of a global pattern of NA AASs associated to the 2009 pandemic originating in Mexico and United States.
The molecular distribution of AASs between Mexico and the rest of the world is shown in Figure S2 (see supplementary material). In all regions, the highest incidence of unique AAS was localized on the trans-membrane and linker domains (TM, L1, a1, Ll5, L17 and L30), which are not part of the CS or AS. AASs reported in Mexico affected the domains TM, L1, a1, b2, b4, L13, L15, b15, L17, b17, L21, b20, L23, L24, b23, b26 and L30. Domains b2 and L13 included CS and AS residues, respectively. AASs reported in the rest of the world affected the domains TM, L1, b1, L2, a1, L3, b2, L4, b3, L5, b4, b5, L7, b6, b7, b8, L10, b9, L11, b10, b11, L13, b12, b13, L15, b14, L16, b15, L17, b16, L18, b17, L19, b18, L20 b19, L21, b20, L22, b21, L23, b22, L24, b23, L25, b24, b25, a2, L28, b26, L29, b27 and L30. From these regions β2, L4 and L7 included AASs affecting the CS, while the b6, L13 and b25 domains included AASs affecting the AS. Only domain L19 had reported AASs in both the CS and the AS. The most conserved domains in Mexico and the rest of the world included L6, L8, L9, L12, L14, L26 and L27.
"The molecular distribution of AASs stratified by continents is shown in Figure S3 (see supplementary material). All distributions appeared qualitatively similar showing an identical clustering pattern. The stratified distributions for continents let us identify nine domains with the highest incidence of AAS. These were TM, L1, L7, L15, b14, L21, b22, L24 and L30. America did not report AASs in domains L2, b6, b18 regions, unlike Europe, Asia and Oceania. Europe did not present AASs in domains b1, L3, L4, b3, b5, b8, b20 in contrast to Asia, Oceania, and America. For Asia and Oceania, domains β 12, L18, β 17 and L19 did not present AASs unlike America and Europe. Africa presented a small number of AASs probably due to underreporting.
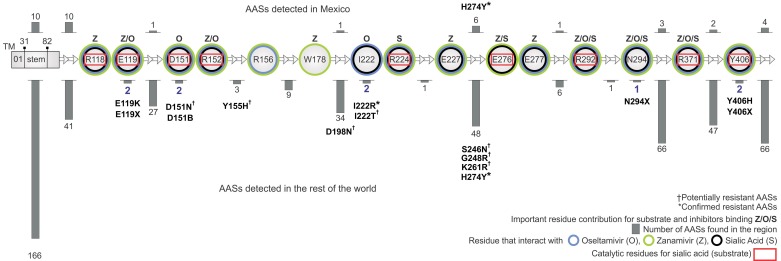
AASs that affected the AS and CS were non-uniformly distributed across the protein sequence in samples from both Mexico and the rest of the world (Figure 1). AASs occurring particularly in the AS and CS were recognized separately. Three AASs were reported to affect the AS, and none of these were reported in Mexico. Six AASs were reported to affect the catalytic residues, and none of these were reported in Mexico. Amino acids of the AS affected by substitutions were I222 and N294. Amino acids of the CS affected by substitutions were E119, D151, and Y406. AASs reported were: E119K, E119X, D151N, D151B, I222R, I222T, N294X, Y406H, and Y406X. AASs H274Y and I222R were clinically reported to be resistant to oseltamivir and oseltamivir/zanamivir respectively.
Figure 1.
Distribution of amino acid substitutions (AASs) from 2009 pandemic AH1N1 human influenza virus NA by active site and catalytic site. (┼) Potentially resistant AASs. (*) Confirmed resistant AASs. The number of AASs reported in each section is indicated with a gray bar. Circle color indicates residues that interact with oseltamivir (O, light blue), zanamivir (Z, light green) and sialic acid (S, black). Catalytic residues shown in red rectangles indicate direct contact with sialic acid (substrate). Based on Liu et al [34], the principal binding energy contribution for each site associated with an influenza drug or substrate is indicated with letters Z, O, and S.
It is important to highlight that all AH1N1 influenza NA AASs reported at this time are primarily distributed in enzymatic hot-spot regions that do not affect the AS or CS. This finding partially substantiates the previous results reported by Maurer- Stroh [33] that until 2009 there was a strict conservation of the NA CS region and the drug-binding pocket, leaving these regions free of AASs. Our study shows AASs affecting the AS and CS directly, indicating that the hydrophobic core is no longer intact. However, the region comprised of domains L6, L8, L9, L12, L14, L26 and L27 has remained free of AASs, and thus remains a potential zone for the design of an epitope vaccine due to the low variability of its amino acids. Besides, most AASs analyzed in this study are from clinical strains, thus the few AASs associated with oseltamivir/zanamivir resistance may have clinical significance with regard to future resistance patterns and mechanisms. The distribution of AASs for total data was reported by amino acid and continent Figure S2 (available at http://code.google.com/p/neuraminidasescripts/). Each continent reported exclusive AASs (America 165, Africa 5, Asia 91, Oceania 10, and Europa 94). AASs appearing simultaneously in different regions are also reported.
Amino acid substitutions and its possible relationship with a pharmacological selection pressure:
Understanding the molecular distribution pattern of AASs associated with drug resistance will help guide strategies to prevent the emergence of resistance. Although more in-depth research is needed, a first discussion of a possible relationship between AASs and pharmacological selection pressure could be made. In the 2009 AH1N1 influenza pandemic Mexico reported few cases of oseltamivir/zanamivir resistance, an AAS pattern that contrast with other pathogens under pharmacological pressure such as Mycobacterium tuberculosis [35], HIV [36] and Plasmodium falciparum [37]. The lack of a large number of AASs in the AS and CS relative to other areas of the enzyme could be a good evidence for a lack of a past pharmacological selection pressure, which is consistent with the historically narrow use of NA-inhibitors as anti-influenza treatment. However, the new incidence of NA AASs associated with resistance to zanamivir and/or oseltamivir could suggest that a neuraminidase-inhibitor pharmacological selection pressure is beginning to emerge.
Zanamivir and oseltamivir were introduced in the market around 1999-2002 [29], and were used as effective alternative anti-influenza drugs for the AH5N1 in 2003 and 2004, which was resistant to amantandine and rimantadine (M2 proteininhibitors). This success led to further reinforcement of their use during the 2009 pandemic. With the continuous and widespread use of these anti-influenza drugs, it is very likely that a future AH1N1 pandemic will yield more predominant zanamivir- and/or oseltamivir-resistant strains, in which NA AASs tend to accumulate in hot-spots associated with the active and catalytic sites. Given this possibility, it may be of benefit to identify potential NA AASs in the AS and CS that may cause drug-resistance in order to design effective alternate anti-influenza drugs. Although AASs may evolve spontaneously in reservoir populations, our compiled global data show a significant number of unique NA AASs that were reported simultaneously on different continents. For example, among the 38 unique AASs reported in Mexico, 29 are also reported elsewhere. Although not conclusive, this evidence favors the hypothesis of global transmission of AH1N1 strains carrying specific NA AASs. It is interesting to note the emergence of new AAS in worldwide circulating strains that are selected and share the same NAAAS.
Conclusion
NA AASs associated with secondary structure domains in pandemic AH1N1 influenza suggest a conserved molecular distribution pattern present worldwide. The majority of unique AH1N1 NA AASs have incidence on multiple continents, suggesting human transmission as an important factor in the spread of new AASs. The majority of present NA AASs continues to occur in sites distant from the AS and CS suggesting a historic lack of pharmacological selection pressures, however the recent identification of AASs affecting the AS and CS may be evidence of emerging pharmacological selection pressures associated with increased NA-inhibitor use.
Supplementary material
Acknowledgments
The authors thank PhD(c) Myra E. Flores for her constructive criticisms at the moment to write the manuscript. The work was partially supported by grants from ICyTDF (PIRIVE09-9), CONACYT and PIFI-SIP-COFAA-IPN.
Footnotes
Citation:Quiliano et al, Bioinformation 9(13): 673-679 (2013)
References
- 1.Tumpey TM, et al. Science. 2005;310:77. [Google Scholar]
- 2.Smith GJ, et al. Nature. 2009;459:1122. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 3.Christman MC, et al. Infect Genet Evol. 2011;11:803. doi: 10.1016/j.meegid.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramirez-Gonzalez JE, et al. l Emerg Infect Dis. 2011;17:283. doi: 10.3201/eid1702.100897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hurt C, et al. Expert Rev Anti Infect Ther. 2006;4:795. doi: 10.1586/14787210.4.5.795. [DOI] [PubMed] [Google Scholar]
- 6.Yin JK, et al. Influenza Other Respi Viruses. 2011;5:299. doi: 10.1111/j.1750-2659.2011.00229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu C, et al. J Virol. 1995;69:1099. doi: 10.1128/jvi.69.2.1099-1106.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q, et al. Nat Struct Mol Biol. 2010;17:1266. doi: 10.1038/nsmb.1909. [DOI] [PubMed] [Google Scholar]
- 9.Kim U, et al. Antivir Chem Chemother. 1999;10:141. doi: 10.1177/095632029901000401. [DOI] [PubMed] [Google Scholar]
- 10.von Itzstein M, et al. Nat Rev Drug Discov. 2007;7:967. doi: 10.1038/nrd2400. [DOI] [PubMed] [Google Scholar]
- 11.Renaud , et al. J Clin Virol. 2011;52:70. doi: 10.1016/j.jcv.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 12.McKimm-Breschkin JL, et al. Antiviral Res. 2000;47:1. doi: 10.1016/s0166-3542(00)00103-0. [DOI] [PubMed] [Google Scholar]
- 13.Brookes W, et al. J Antimicrob Chemother. 2011;66:466. doi: 10.1093/jac/dkq486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins PJ, et al. Nature. 2008;453:1258. doi: 10.1038/nature06956. [DOI] [PubMed] [Google Scholar]
- 15.Chan PN, et al. Clin Microbiol Infect. 2010;16:1576. doi: 10.1111/j.1469-0691.2010.03212.x. [DOI] [PubMed] [Google Scholar]
- 16.Couturier BA, et al. Influenza Other Respi Viruses. 2010;4:199. doi: 10.1111/j.1750-2659.2010.00144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esposito S, et al. J Clin Virol. 2010;48:62. doi: 10.1016/j.jcv.2010.02.019. [DOI] [PubMed] [Google Scholar]
- 18.Janies DA, et al. Int J Health Geogr. 2010;9:13. doi: 10.1186/1476-072X-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen HT, et al. Clin Infect Dis. 2010;51:983. doi: 10.1086/656439. [DOI] [PubMed] [Google Scholar]
- 20.Fauci S, et al. Nature. 2005;435:423. doi: 10.1038/435423a. [DOI] [PubMed] [Google Scholar]
- 21.Squires B, et al. Nucleic Acids Res. 2008;36:D497. doi: 10.1093/nar/gkm905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao Y, et al. J Virol. 2008;82:596. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Itzstein M, Thomson R. Handb Exp Pharmacol. 2009;111:54. doi: 10.1007/978-3-540-79086-0_5. [DOI] [PubMed] [Google Scholar]
- 24.Donovan P, et al. PLoS Curr. 2009;1:RRN1014. doi: 10.1371/currents.RRN1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janies DA, et al. Anim Health Res Rev. 2010;11:73. doi: 10.1017/S1466252310000083. [DOI] [PubMed] [Google Scholar]
- 26.Li KB. Bioinformatics. 2003;19:1585. doi: 10.1093/bioinformatics/btg192. [DOI] [PubMed] [Google Scholar]
- 27.Tang JW, et al. J Med Virol. 2008;80:895. doi: 10.1002/jmv.21155. [DOI] [PubMed] [Google Scholar]
- 28.Yen HL, et al. J Virol. 2006;80:8787. doi: 10.1128/JVI.00477-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monto S, et al. Antimicrob Agents Chemother. 2006;50:2395. doi: 10.1128/AAC.01339-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferraris O, Lina B. J Clin Virol. 2008;41:13. doi: 10.1016/j.jcv.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 31.Sheu TG, et al. Antimicrob Agents Chemother. 2008;52:3284. doi: 10.1128/AAC.00555-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crooks E, et al. Genome Res. 2004;14:1188. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maurer-Stroh S, et al. Biol Direct. 2009;4:18. doi: 10.1186/1745-6150-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H, et al. Mol Pharm. 2010;7:894. doi: 10.1021/mp100041b. [DOI] [PubMed] [Google Scholar]
- 35.Zimic M, et al. Infect Genet Evol. 2010;10:346. doi: 10.1016/j.meegid.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price H, et al. Antivir Ther. 2010;15:203. doi: 10.3851/IMP1511. [DOI] [PubMed] [Google Scholar]
- 37.McCollum M, et al. Malar J. 2012;11:77. doi: 10.1186/1475-2875-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.