Abstract
It has been observed that mycobacterial species has high content of cardiolipin (CL) in their cell membranes more so pathogenic mycobacteria and in bacteria CL activates polymerases, gyrases by removing the bound ADP. Therefore, in the present study cardiolipin synthase (cls) which catalyses the formation of CL was isolated purified and characterized from the cell membrane of Mycobacterium phlei. The purified cls obtained from C-18 RP-HPLC column had a molecular weight of 58 kDa with an isoelectric point of 4.5. The enzyme activity (11.5+0.15 µM of CL phosphorous. ml-1 minute-1 for PG as substrate and 14+0.35µM of CL phosphorous. ml-1 minute-1 for CDP-DG as substrate) was optimal at pH 4.8 and showed KM values of 55+0.05µM and 2.56+0.04µM for phosphatidyl glycerol and CDP-diacylglycerol, respectively, with an absolute requirement of Mg2+ and Mn2+ ions for its activity however, Ca2+ ions inhibited the activity of the cls. The partial amino acid sequence of cls showed significant homology with pgsA3 gene of M. tuberculosis and in this organism the CL biosynthesis is very high having three genes coding for PLs biosynthesis therefore, enzymes involved in CL biosynthesis may be an attractive drug target in the development of new antimycobacterial drugs.
Keywords: Cardiolipin, Cardiolipin synthase, CDP-diacylglycerol, phosphatidyl glycerol
Background
Mycobacteria has high amount of phospholipids in their cell membranes. The mycobacterial phospholipids include cardiolipin (CL), phosphatidyl ethanolamine (PE), phosphatidyl inositol (PI) and phosphatidylinositolmannosides (PIMS). Among the phospholipids of mycobacteria, cardiolipin is often the most abundant (30-50%) of total phospholipids present in both pathogenic as well as in saprophytic species of mycobacteria. Virulent strains of mycobacteria contain a higher percentage of phospholipids as compared to the avirulent strains [1, 2]. High content of CL in pathogenic mycobacteria is further, confirmed by the presence of three functional pgsA genes in the M. tuberculosis [3]. The past studies have revealed that crude phosphatide fractions of mycobacteria when injected intraperitoneally into rabbits elicited typical tuberculosis lesions. Antibodies to phospholipids have been demonstrated in patients suffering from tuberculosis. Recent studies have demonstrated that, during infection of macrophages by mycobacteria, phospholipids (PLs) are released from the mycobacterial cell wall within infected macrophages and transported out of this compartment into intracellular vesicles. The release of these PLs may have functions that influence the outcome of mycobacterial infections Owing to their serological, antigenic and physiological properties, phospholipids of mycobacteria are well studied [4, 5].
The cell membranes of E. coli, S. aureus, M. lysodeikticus, L. plantarum, Bacillus stearothermophillus, Bacillus firmus, and Mycobacterium smegmatis contains CL which is synthesized by Cardiolipin synthase (cls) and the enzyme is located on the cell membrane of these organisms [4, 6–8] Kornberg and coworkers reported that CL plays an important role in DNAreplication in E. coli by displacing the bound ADP from dnaA protein thereby, reactivating the enzyme required for replication, and also activates gyrases in E. coli. Thus, mutation in cls gene makes E. coli susceptible to novobiocin. A 10 fold increased activity of cls was observed in E coli in stationary growth phase while the activity of other enzymes in phospholipid biosynthesis decreased, accordingly, CL should play a role in survival of the E. coli [8, 11]. Presence of large content of CL in the mycobacteria in particular in pathogenic mycobacteria and its physiological importance of CL in the prokaryotes, it is imperative to study the Cardiolipin synthase involved in the biosynthesis of CL in the mycobacteria. In this report, we describe the isolation, purification and physical and chemical properties of cls enzyme isolated from Mycobacterium phlei. The molecular weight, isoelectric point, KM value of the enzyme, the optimal pH for activity, requirement of various divalent cations for its optimal activity, and the partial amino acid sequence of the enzyme are also established.
Methodology
Bacterial Strains and culturing Methods:
Mycobacterium phlei TMC 1548 was cultured in TYE medium containing 0.2% Tween-20 at pH-7.4. The pelleted cells were washed with 50 ml of 50mM Tris-HCl, pH-7.5, and these cells were used for the isolation of cardiolipin synthase [12].
Isolation and Purification of the cardiolipin synthase (cls) Preparation of Membrane Fraction:
The cells of M. phlei were sonicated then centrifuged in a ultracentrifuge Beckman L7-35. The pellet was washed with 50mM Tris- HCl, pH 7.5 and was used as the starting material for purification and isolation of Cardiolipin synthase enzyme.
Assay of Cardiolipin synthase activity:
The enzyme assay was carried out in 50µM of acetate buffer pH 4.8. Each test tube contained 30µM of phosphatidyl glycerol (PG), 10µM of ATP, 100µM of MgCl2, 80µM of MnCl2, and 0.1 ml of enzyme fraction (crude or pure). For CDP-diacylglycerol pathway 0.4µM of CDP-diacylglycerol was used instead of PG. The reaction mixture was incubated at 37°C for 1 hour, and stopped by addition of 2.5 ml methanol. The solution in the tubes was suspended in chloroform-methanol (2:1 v/v) mixture and stirred for 4 hours at room temperature. The homogenate was filtered and the residue was again stirred for 2 hours in chloroform-methanol (2:1 v/v) and vaccum dried at 45°C in rotatory vaccum evaporator. The dry mass was dissolved in a 1 ml of chloroform, and washed with one fifth volume of KCl and left overnight at 4°C. The upper phase was discarded and lower layer was evaporated to dry in a rotatory vaccum evaporator at 45°C. The residue was dissolved in 0.5 ml of chloroform. This sample was now subjected to thin layer chromatography (TLC). To the activated chromatoplates samples were spotted along with blank and developed for 90 minutes in the Chloroform : methanol : ammonia (115:45:7.5, v/v/v). Phospholipid standards were also run alongwith mycobacterial lipid samples for identification. The plates were air dried and sprayed with 50% H2SO4 and heated for 15 minutes at 110°C till charred spots appeared. The spot corresponding to the cardiolipin was identified in all the samples and scrapped into test tubes and used for the estimation of phospholipid phosphorous. Presence of phosphorous in phospholipids was estimated by Marinetti (1962) method [13]. The KM values of the purified Cls for both CDP-Diacylglycerol (CDP-DG) and Phosphatidyl Glycerol (PG) were determined. The effect of substrate concentration was first determined by varying concentration of PG between 20 to 100 µmoles. The concentration of CDP-DG was kept constant at 0.4µmoles. The concentration of CDP-DG was varied from 0.2 to 4µmoles. The concentration of PG was kept constant at 0.5µmoles. The enzyme activity of cls was expressed as µM of CL phosphorous. ml-1 minute-1. Km and Vmax were calculated by plotting Hanes–Woolf plot ([S/V] vs [S]) [4, 10, 14, 15]. By taking varying concentrations of MgCl2, CaCl2 as 50 to 150µmoles and MnCl2 as 40 to 150µmoles, the effect of Mg2+, Ca2+ and Mn2+ ions on enzyme assay was determined as described in earlier section [4, 5, 14, 15]. The M. phlei membrane fractions were solubilized in 2M Urea. After solubilization the suspension was dialysed to remove urea [4, 12].
Protein sequencing of cls:
The peak fraction from reverse phase C-18 (4.6 × 150× 5 microns) column was subjected to amino acid analysis. For Nterminal sequence analysis, cls of M. phlei 10µg in 25 µl of 10 mM Tris-HCl, pH-7.5 was applied to prospin device (Applied Biosystems) according to manufacturer's protocol. The resulting PVDF blot was washed twice with 20% methanol prior to sequencing in a Blot cartridge (Applied Biosystems) on a model 477 A pulsed liquid protein sequencer equipped with a model 120APTH analyzer (Applied Biosystems) using methods and cycles supplied by the manufacturer. Data was collected and analyzed on a model 610A data analysis system (Applied Biosystems).
Enzymatic digestion and internal Amino acid se:
20µg of cls in 10mM Tris-HCl, pH-7.5 was dried in a speed vac. (Savant, Formingdale, NJ). The resulting residue was taken up in 25µl of 8M urea and 0.4M NH4 HCO3 and subjected to reduction, alkylation and proteolytic digestion with 0.3µg modified sequencing grade trypsin [17]. The resulting digest was separated by reversed phase HPLC on a narrow bore (21.1 × 250mm) Vydac 218TP52 column and gaurd column (separations group, Hesponia, CA) and eluted at 0.25 ml /min at 35oC utilizing a gradient on a system Gold HPLC equipped with a model 168 diode array detector (Beckman, Fullerton, CA). Solvent A was 0.1 %TFA in water and solvent B was 0.1 % TFA in acetonitrile. The column effluent was monitored at 210 and 280 nm. Fractions were collected at 30 seconds interval and stored at -20°C. Fractions (200µl) containing tryptic peptides were applied to Biobrene (Applied Biosystems) treated glass fiber filter and dried prior to amino acid sequencing on the previously described sequencer [17].
Results & Discussion
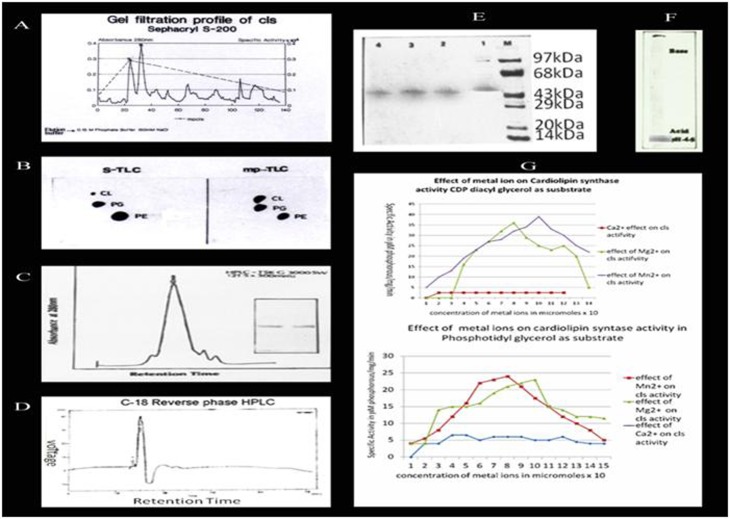
The 2M urea was observed to be a useful solubilizer of M. phlei membranes while maintaining the enzymatic activity of cls (Figure 1A). The solubilized membranes of M. phlei were used initially to study the synthesis of CL using both CDPdiacylglycerol (CDP-DG) and Phosphatidyl Glycerol (PG) as substrates and the enzyme activity was observed to be 4.5+0.25 µM of CL phosphorous.ml-1 minute-1 and 6.5+0.25 µM of CL phosphorous.ml-1 minute-1 and the optimal activity of the enzyme was achieved at pH 4.8. This urea solubilized fraction was further used in the purification of cls the increased enzyme activity was observed with increased purification (Figure 1B) Table 1(see supplementary material). The cls enzyme eluted on GPC-HPLC (TSK G-3000 SW) at a retention time of 42 minutes (Figure 1C) was concentrated and fractionated on C-18 RPHPLC which was eluted as single peak at a retention time of 15 minutes (Figure 1D) gave single band on SDS-PAGE with a molecular weight of 58kDa (Figure 1E) and IEF analysis showed an isoelectric pH 4.5 (Figure 1F) which confirms that cls of Mycobacterium phlei is an acidic protein.
Figure 1.
A) Gel filtration profile of cardiolipin synthetase (cls) isolated from Mycobacterium phlei; B) Thin layer chromatogram of cardiolipin synthesized by the cls isolated from Mycobacterium phlei. S-TLC: Phospholipids, Phosphatidyl ethanolamine (PE), Phosphatidyl glycerol (PG) and cardiolipin (CL) obtained from sigma, mp-TLC: Phospholipids, PG and CL obtained from the synthesis of CL by cls of M. phlei.; C) HPLC profile of cls obtained from Sephacryl S-200 column. 2.5 mg of cls purified on Sephacryl S-200 column was applied on HPLC-Gel permeation column [TSK G-3000SW (21.5 × 300 mm)]; D) RP-HPLC profile of cls obtained from HPLC-GPC column. 1 mg of cls purified from HPLC-GPC column was applied on C-18 RP-HPLC (Waters); E) 10% SDS-PAGE analysis of cls purified on HPLC. Lane 1. 5 µg of cls purified on Sephacryl S-200, lanes 2 and 3 5 µg of cls fractionated on HPLC-GPC lane 4 cls purified on C 18 RP HPLC, lane M. molecular size markers obtained from Bangalore genei pvt ltd.; F) Isoelectric focussing of gel pattern of purified cls; G) Effects of Mg2+, Mn2+ and Ca2+ on cls activity by using CDP-diacyl glycerol and phosphotidyl glycerol as substrates.
The pure cls showed an enzyme activity of 11.5+0.15 µM of CL phosphorous.ml-1 minute-1 for PG as substrate and 14+0.35 µM of CL phosphorous.ml-1 minute-1 for CDP-DG as substrate with absolute requirement for divalent cations with Mg2+ and Mn2+ in particular however, Ca2+ ions completely inactivated the cls enzyme (Figure 1G). At an optimal concentration of phospholipid the KM vlaues were 55+0.05µM and 2.56+0.04µM for PG and CDP-DG, respectively. The cls appeared to be highly thermostable as it retained its 60% activity even at 550C when kept for one hour. Attempts to detect the amino terminal residue were unsuccessful, indicating that the amino terminal was blocked therefore, the protein was digested with trypsin and the fragments were resolved by C-18 RP-HPLC. The peptide obtained at 72 min had ITDFMDGYIARKYGLKTIAGTILDPLADKL sequence and BLAST-P search analysis indicated that the sequence had resemblance with the cls reported so far (SWISS -PROT data bank with Accession Number-P82260) also the sequence showed significant homology with the pgsA3 (Rv2746c) gene of M. tuberculosis H37Rv and pgsA genes of E. coli and Bacillus subtilis.
The complete Mycobacterium tuberculosis genome was recently sequenced and annotated. However, of the 3,924 putative gene products, only a minority are assigned experimentally established physiological functions [3]. Thus, continued analyses of the M. tuberculosis proteome are required to fully elucidate the biology of this intracellular pathogen. In this context researcher's all over the world are concentrating and studying various genes in the closely related organisms like Mycobacterium smegmatis and Mycobacterium phlei in understanding the physiological functions of these genes and implicating their role in M.tuberculosis. This would help in understanding quickly the physiological importance of these enzymes/ proteins in the pathogenicity of the organism, further that are required for the formation of successful pathogen [18].
The partial sequence of cardiolipin synthase (cls) of Mycobacterium phlei (Acc.No P-82260) showed significant homology with the pgsA3 (Rv2746c) gene of the M. tuberculosis H37Rv shows the physiological importance of this enzyme in the mycobacteria which has high amount of cardiolipin in their cell membranes. The high content of cardiolipin (CL) appears to have an important role on the stability of the mycobacteria particularly in the pathogenic mycobacteria, cardiolipin is often the most abundant (30-50%) of total phospholipids. This is further corroborated by the fact that there are three pgsA genes in M tuberculosis, where as only one cls gene is found in E coli [1–4]. Expression of cls is highest in the stationary phase of E coli, since mycobacterial species grows even in low oxygen conditions thus; the requirement of CL is very high in this genus. Thus, probably the CL-biosynthesis is pivotal requirement in the mycobacteria especially in pathogenic mycobacteria [8, 9].
The purified cls showed an absolute requirement for Mg2+ and Mn2+ions however, Ca2+ ions inactivated cls enzyme. The SDSPAGE analysis indicated that cls had a molecular weight of 58 kDa while the same cls isolated from E. coli, Bacillus firmus had a mol .wt of 45 and 57 kDa respectively [7, 9]. The isoelectric point of cls was achieved at pH 4.5 indicating acidic in nature (Figure 1F) which is in agreement that optimum activity of cls was observed at 4.8 pH. Similarly cls of S. aureus and L. plantarum exhibited optimum activity at pH-4.4 [6, 10]. Temperature dependent analysis showed the cls of M. phlei retained its 60 % activity even at 55°C which is consistent with nature of M. phlei which can even grow at 55°C. The low KM values of 2.56µM and 55µM for CDP-DG and PG respectively, suggests that the cls of M. phlei has very high affinity for its substrates. Similar properties were observed for cls present in E. coli and in human cardiac cells and in rat liver mitochondrial membranes indicating [8, 9, 14, 15].
It has been observed by Nigou and Besra [5] that phospholipids are released from the mycobacteria within the infected macrophages plays an important role in the outcome of mycobacterial infections. The important factor in understanding the virulence of M. tuberculosis is its survival inside the host cell phagosome. The pathogen is able to convert the hostile environment of the phagosome by inhibiting the acidification and phagosome-lysosome fusion [19, 20]. It is very much clear that in CL in bacteria reactivates polymerases and gyrases [11]; it is therefore, possible to predict that the requirement of CL is very pivotal in pathogenic mycobacteria. These findings open up plausible explanations for the presence of high amount of CL in the cell membranes of M. tuberculosis for its survival in the macrophages. Therefore; enzymes involved in the biosynthesis of phospholipids may be attractive drug targets in the development of new anti-mycobacterial drugs.
Conclusion
Cardiolipin synthase which catalyses the formation of cardiolipin, was purified from the membranes of Mycobacterium phlei. The purified enzyme showed a molecular weight of 58KD and pI of 4.5. The enzyme showed absolute requirement of Mg2+ and Mn2+ ions for its activity however, Ca2+ ions inhibited the cls activity. The partial amino acid sequence of cls showed significant homology with pgsA3 gene of M. tuberculosis. Cardiolipin in bacteria activates polymerases, gyrases and presence of high amount of cardiolipin in the memebranes of M. tuberculosis possibly explains its survival in the macrophages therefore; enzymes involved in the biosynthesis of CL may be an attractive drug targets in the development of antimycobacterial drugs.
Supplementary material
Acknowledgments
This paper dedicated to late Dr. P.S. MURTHY, who inspired me a lot and ignited many innovative thoughts among scientific entities. I wish his soul to be rest in peace.
Footnotes
Citation:Sarma et al, Bioinformation 9(13): 690-695 (2013)
References
- 1.Akamatsu Y, Nojima S. J Biochem. 1965;57:430. doi: 10.1093/oxfordjournals.jbchem.a128097. [DOI] [PubMed] [Google Scholar]
- 2.Akamatsu Y, et al. J Biochem. 1966;59:176. doi: 10.1093/oxfordjournals.jbchem.a128279. [DOI] [PubMed] [Google Scholar]
- 3.Cole ST, et al. Nature. 1998;393:537. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 4.Mathur AK, et al. Can J Microbiol. 1976;22:354. doi: 10.1139/m76-054. [DOI] [PubMed] [Google Scholar]
- 5.Nigou J, Besra GS. Biochem J. 2002;367:157. doi: 10.1042/BJ20020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burritt MF, Henderson TO. J Bacteriol. 1975;123:972. doi: 10.1128/jb.123.3.972-977.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo D, Tropp BE. Biochim Biophys Acta. 1998;1389:34. doi: 10.1016/s0005-2760(97)00086-6. [DOI] [PubMed] [Google Scholar]
- 8.Hiraoka S, et al. FEBS Lett. 1993;336:221. doi: 10.1016/0014-5793(93)80807-7. [DOI] [PubMed] [Google Scholar]
- 9.Nishijima S, et al. J Bacteriol. 1988;170:775. doi: 10.1128/jb.170.2.775-780.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Short SA, White DC. J Bacteriol. 1972;109:820. doi: 10.1128/jb.109.2.820-826.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekimizu K, Kornberg A. J Biol Chem. 188;263:7131. [PubMed] [Google Scholar]
- 12.Sarma PV, et al. FEMS Microbiol Lett. 1998;159:27. doi: 10.1111/j.1574-6968.1998.tb12837.x. [DOI] [PubMed] [Google Scholar]
- 13.Marinetti GV. J Lipid Res. 1962;3:120. [Google Scholar]
- 14.Schlame M, Hostetler KY. J Biol Chem. 1991;266:22398. [PubMed] [Google Scholar]
- 15.Schlame M, et al. Lipids. 1995;30:633. doi: 10.1007/BF02537000. [DOI] [PubMed] [Google Scholar]
- 16.Morrissey JH, et al. Anal Biochem. 1981;117:307. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez J, et al. Anal Biochem. 1992;201:255. doi: 10.1016/0003-2697(92)90336-6. [DOI] [PubMed] [Google Scholar]
- 18.Alekshun MN. Nat Biotechnol. 2001;19:1124. doi: 10.1038/nbt1201-1124. [DOI] [PubMed] [Google Scholar]
- 19.Deretic V, Fratti RA. Mol Microbiol. 1999;31:1603. doi: 10.1046/j.1365-2958.1999.01279.x. [DOI] [PubMed] [Google Scholar]
- 20.Sturgill-Koszycki S, et al. Science. 1994;263:678. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.