Abstract
Reprogramming of tumor cell metabolism contributes to disease progression and resistance to therapy, but how this process is regulated on the molecular level is unclear. Here we report that Heat Shock Protein 90 (Hsp90)-directed protein folding in mitochondria controls central metabolic networks in tumor cells, including the electron transport chain, citric acid cycle, fatty acid oxidation, amino acid synthesis, and cellular redox status. Specifically, mitochondrial Hsp90, but not cytosolic Hsp90, binds and stabilizes the electron transport chain Complex II subunit succinate dehydrogenase-B, maintaining cellular respiration under low-nutrient conditions, and contributing to hypoxia-inducible factor-1α-mediated tumorigenesis in patients carrying succinate dehydrogenase-B mutations. Thus, Hsp90-directed proteostasis in mitochondria regulates tumor cell metabolism, and may provide a tractable target for cancer therapy.
Reprogramming of tumor cell metabolism 1 is increasingly recognized as a multifaceted disease driver, enhancing biomass expansion 2, and promoting various mechanisms of oncogenic signaling 3. Although these processes have been mostly studied in the context of aerobic glycolysis, the so-called Warburg effect 3, there is evidence that mitochondria continue to play an important role in tumor metabolism 4,5, and organelle-driven oxidative phosphorylation has been associated with tumorigenic potential 6, drug resistance 7,8 and enhanced tumor cell survival 9. Harnessing these pathways may open new prospects for cancer therapy 10, but the regulators of mitochondrial homeostasis in tumors have remained largely elusive, and their potential suitability as drug candidates is unknown.
With a complex, multi-compartment topology, dependence on import of nuclear-encoded proteins, and production of protein-modifying reactive oxygen species (ROS), mitochondria must tightly control their protein folding environment 11. This is indispensable to maintain metabolic output 2, ensure organelle integrity 12, and prevent the consequences of an unfolded protein response, which may result in cell death 13. Buffering mitochondrial proteotoxic stress, especially in the protein-dense and energy-producing organelle matrix 14, relies on adaptive responses mediated by molecular chaperones and AAA proteases 15, and dysregulation of these mechanisms has been linked to human diseases, including neurodegeneration and cancer 14.
In this context, a pool of ATPase-directed molecular chaperones, including Heat Shock Protein-90 (Hsp90) 16 and its related homolog, TNF Receptor-Associate Protein-1 (TRAP-1) 17 localize to the mitochondria, almost exclusively in tumor cells 18. The molecular requirements for the selective accumulation of these chaperones in tumor mitochondria have not been completely elucidated. However, there is evidence that both Hsp90 and TRAP-1 form overlapping complexes with mitochondrial proteins, including cyclophilin D (CypD), a component of the permeability transition pore, and control their folding 19. Accordingly, inhibition of Hsp90 and TRAP-1 chaperone activity selectively in mitochondria triggered acute organelle dysfunction 20, defective hexokinase II (HK-II)-dependent 2 ATP production 21, and anticancer activity in preclinical tumor models, in vivo 19.
In this study, we examined the role of Hsp90-directed mitochondrial protein folding on cellular homeostasis. Using combined proteomics and metabolomics approaches, we found that mitochondrial Hsp90 and TRAP-1 are global regulators of tumor metabolic reprogramming, including oxidative phosphorylation, and are required for disease maintenance.
RESULTS
Identification of a mitochondrial Hsp90 proteome
We began this study by setting up a preliminary proteomics screen to identify regulators of mitochondrial protein homeostasis, or proteostasis, in tumors. For these experiments, we used non-cytotoxic concentrations of Gamitrinib (GA mitochondrial matrix inhibitor), a mitochondrial-targeted, small molecule ATPase antagonist that inhibits the chaperone activity of both Hsp90 and TRAP-1 in tumors 20.
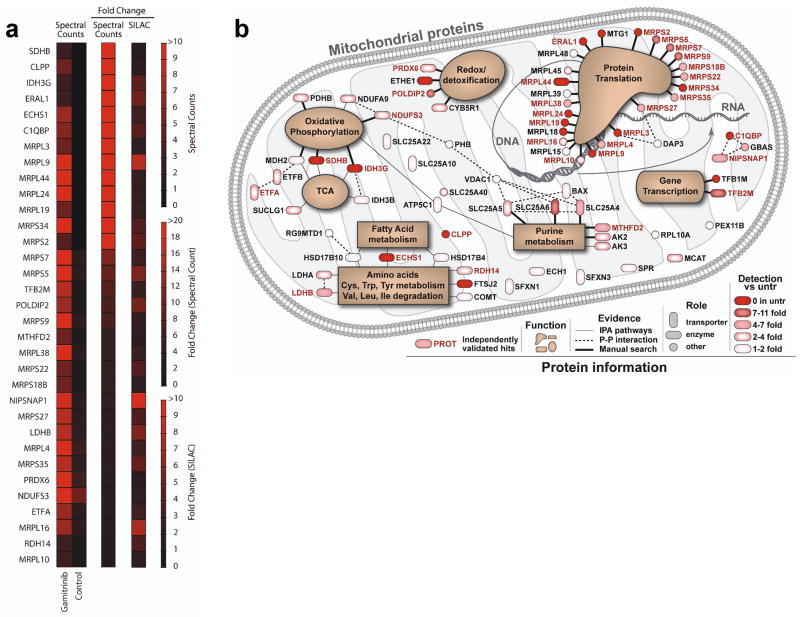
Treatment of glioblastoma LN229 cells with non-cytotoxic concentrations of Gamitrinib 21 caused the accumulation of aggregated and misfolded proteins, characterized by resistance to detergent solubilization (Supplementary Fig. S1). Preliminary mass spectrometry analysis of selected bands showing higher intensities with Gamitrinib treatment identified 96 mitochondrial proteins (Supplementary Data 1). Forty-four of these proteins based on spectral counts were elevated by more than 3-fold after Gamitrinib treatment, indicating a requirement of Hsp90 for their folding. Although gel-based comparison (Supplementary Figure S1) provides high detection sensitivity for specific bands, individual bands are not single proteins, and slight differences in band excision between control and Gamitrinib treatment can produce artificial differences. To minimize this concern, this experiment focused primarily on band differences at the 2% CHAPS condition, where protein complexity was the lowest, but not necessarily where the largest fold change occurred. To independently validate these initial results, we next performed unbiased proteomics studies using Stable Isotope Labeling by Amino Acids in Culture (SILAC) of control or Gamitrinib-treated cells. Of the original 44 proteins of the mitochondrial Hsp90 proteome identified by 1D mass spectrometry, 33 were independently confirmed for response to Gamitrinib in SILAC experiments (Fig. 1a). Of the remaining 11 proteins, 7 were below adequate detection levels for SILAC quantification, and 4 did not show significant changes.
Figure 1. Mitochondrial Hsp90 proteome.
(a) LN229 cells were treated with vehicle (Control) or non-cytotoxic concentrations of mitochondrial-targeted Hsp90 inhibitor, Gamitrinib, and detergent-insoluble mitochondrial proteins were identified by 1-D Mass Spectrometry (spectral counts), or, alternatively, by SILAC technology. The heat map quantifies changes in protein solubility (>3-fold cutoff) between the treatments assessed using the two independent proteomics approaches. (b) Schematic representation of the mitochondrial Hsp90 proteome. Proteins are annotated with functions based on literature search and information from Ingenuity software, which was also used to determine known direct protein-protein interactions. All proteins are color coded to reflect the magnitude of difference in detection between treated and untreated (untr) samples. Proteins marked in ‘red’ exhibited a >3-fold change difference after Gamitrinib treatment compared to control, and were independently confirmed by both 1-D Mass Spectrometry and SILAC technology.
These verified mitochondrial Hsp90-regulated proteins (Figure 1a and Supplementary Table S1) comprised transcription factors TFB1M and TFB2M involved in organelle gene expression 22, and glucose homeostasis 23, ribosomal proteins (MRPLs, MTG1, ERAL1) associated with RNA translation 24–26, regulators of purine biosynthesis and the methyl cycle (MTHFD2) 27, and effectors of oxidative phosphorylation 2, including SDHB, IDH3G, NDUFS3, PDHB, MDH2 28 (Fig. 1b). Mitochondrial proteins participating in redox status and detoxification pathways (PRDX6, POLDIP2, CYB5R1, ETHE1) 29–31 were also identified in the mitochondrial Hsp90 proteome (Fig. 1b).
Mitochondrial Hsp90 regulation of tumor metabolism
The impact of a mitochondrial Hsp90 proteome (Fig. 1b) on cellular homeostasis was next investigated. For these experiments, we quantified the level of 301 individual metabolites in prostate adenocarcinoma PC3 cells treated with non-cytotoxic concentrations of Gamitrinib 21 or, alternatively, silenced for expression of TRAP-1 by small interfering RNA (siRNA) 21. Both approaches produced global defects in tumor cell metabolism (Supplementary Data 2). Consistent with a requirement of Hsp90 for oxidative phosphorylation (Fig. 1b), Gamitrinib-treated cells exhibited aberrant accumulation of citric acid cycle metabolites, succinate, fumarate, and malate (Fig. 2a). This was associated with altered glutaminolysis (elevation in glutamine and α-ketoglutarate) (Supplementary Fig. S2) and deregulated fatty acid metabolism (Fig. 2b), leading to higher levels of palmitate and linoleate, increased long chain fatty acid transport into mitochondria (elevation of palmitoylcarnitine and stearoylcarnitine), and excess lipid oxidation (accumulation of the ketone body 3-hydroxybutyrate (Supplementary Fig. S3). Mitochondrial Hsp90-targeted cells also showed increased AMP/ATP ratio (Supplementary Fig. S3), indicative of cellular starvation, and consistent with the loss of ATP production, phosphorylation of the energy sensor, AMP-activated kinase (AMPK), and inhibition of mammalian target of Rapamycin complex (mTORC1) observed in response to Gamitrinib 21.
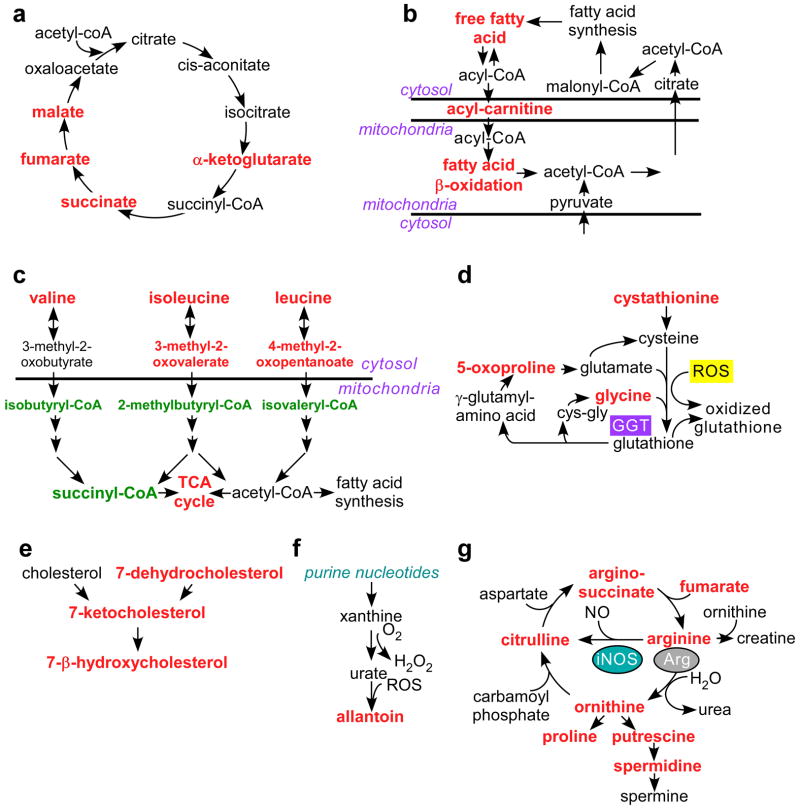
Figure 2. Mitochondrial Hsp90 control of tumor cell metabolism.
PC3 cells were transfected with control non-targeting siRNA or TRAP-1-directed siRNA, or, alternatively, treated with non-cytotoxic concentrations of 17-AAG (5 μM) or Gamitrinib (2.5–5 μM), and analyzed for changes in expression of 301 individual metabolites by Mass Spectrometry. The complete summary of metabolic changes induced by targeting mitochondrial Hsp90s is presented in Supplementary Data 2. The experiment was carried out once with 5 independent replicates per condition tested. The metabolic pathways affected under these conditions are depicted as follows: (a) Citric acid cycle; (b) Fatty acid oxidation; (c) Branched chain amino acid catabolism; (d) Redox status; (e) Cholesterol metabolism; (f) Purine nucleotide metabolism; and (g) Arginine metabolism. Significant (p<0.05) changes in metabolite levels within each group (Ctrl vs. TRAP-1 siRNA or vehicle vs. Gamitrinib) are indicated in red (increase) or green (decrease) using Welch’s two-sample t-test (n=5).
Targeting mitochondrial Hsp90s impaired the catabolism of branched chain amino acids (BCAA), with accumulation of valine, isoleucine and leucine (Fig. 2c), and decreased levels of BCAA catabolites, isobutyryl-carnitine, succinylcarnitine, 2-methylbutyryl-carnitine, and isovaleryl-carnitine (Fig. 2c, Supplementary Fig. S4). This was associated with defects in redox status (Fig. 2d), cholesterol homeostasis (Fig. 2e), and purine nucleotide metabolism (Fig. 2f), resulting in higher levels of cholesterol metabolites associated with lipid peroxidation, ROS-dependent allantoin generation (Fig. 2f, Supplementary Fig. S5), and increased oxidized glutathione, cysteine-glutathione disulfide, and the glutathione catabolic product, 5-oxoproline (Supplementary Fig. S6). Increased ROS production under these conditions may result from dysfunctional mitochondrial metabolism (see above), and/or increased nitric oxide generation from arginine, a possibility suggested by the accumulation of citrulline under these conditions (Fig. 2g, Supplementary Fig. S7).
Overall, Gamitrinib treatment produced more extensive changes in the tumor metabolome, compared to siRNA silencing of TRAP-1 (Fig. 2 and Supplementary Figs. S2–S7). This may reflect incomplete TRAP-1 knockdown by siRNA, or, alternatively, compensatory mechanisms provided by mitochondrial Hsp90, which is inhibited by Gamitrinib, but not by TRAP-1 knockdown. As a control, treatment of PC3 cells with 17-allylamino 17-demethoxygeldanamycin (17-AAG), which inhibits Hsp90 in the cytosol, but not mitochondria 20, or transfection of a control, non-targeting siRNA, had minimal effects on metabolic pathways (Supplementary Figs. S2–7). In previous experiments, addition of the triphenylphosphonium “mitochondriotropic” moiety, alone or in the presence of 17-AAG, had no effect on mitochondrial function 20.
Mechanism of mitochondrial Hsp90 control of tumor metabolism
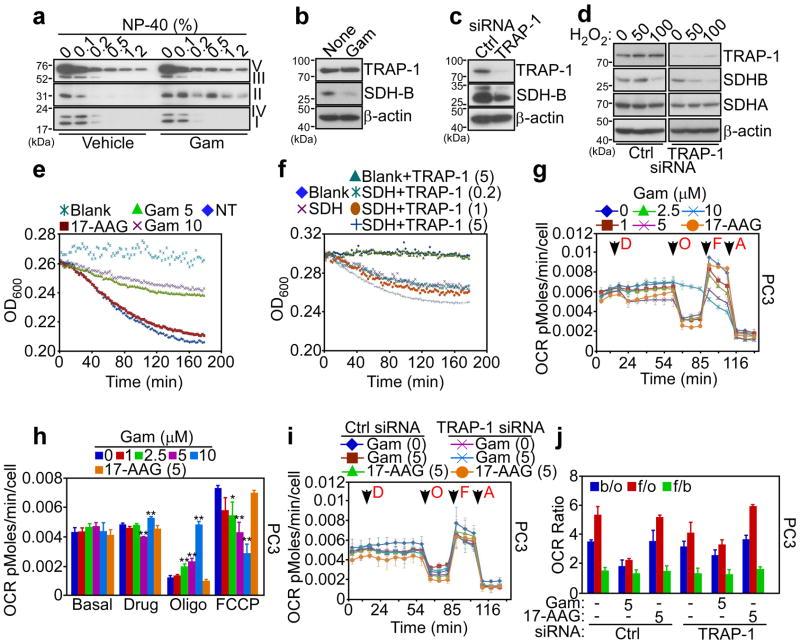
To elucidate how mitochondrial Hsp90s regulate tumor bioenergetics, we next focused on SDHB, the iron-sulfur subunit of ETC Complex II 32, which required Hsp90s for proper folding (Fig. 1a, b, Supplementary Tables S1, S2), and functional activity (Supplementary Fig. S2). Treatment of tumor cells with Gamitrinib caused insolubility of Complex II over a range of detergent concentrations (Fig. 3a and Supplementary Fig. S8a). In contrast, mitochondrial proteins comprising other ETC Complexes (I, IV, III and V) were minimally affected (Fig. 3a). Immune complexes precipitated from mitochondrial fractions of tumor cells with two independent antibodies to SDHB, but not control IgG, contained TRAP-1, in vivo (Supplementary Fig. S8b). In addition, immunoprecipitated SDHB associated with recombinant TRAP-1, in vitro (Supplementary Fig. S8c), demonstrating that these two proteins interact in tumor mitochondria. Suggestive of a chaperone-”client protein” recognition 33, this interaction was required to preserve SDHB stability, as Gamitrinib treatment (Fig. 3b), or siRNA silencing of TRAP-1 (Supplementary Fig. S8d) caused SDHB degradation in tumor cells (Fig. 3c).
Figure 3. Mitochondrial Hsp90 regulation of cellular respiration.
(a) PC3 cells were treated with vehicle or Gamitrinib (Gam), solubilized in the indicated increasing concentrations of detergent (NP-40), and insoluble proteins were analyzed by Western blotting with an antibody cocktail to the OXPHOS complex. (b, c) PC3 cells were treated with Gamitrinib (b) or transfected with control non-targeting siRNA (Ctrl) or TRAP-1-directed siRNA (c), and analyzed by Western blotting. None, untreated. (d) PC3 cells were transfected as in (c), treated with the indicated increasing concentrations of H2O2 (μM), and analyzed by Western blotting. (e) PC3 cells were treated with the indicated concentrations of Gamitrinib (Gam, μM) or 17-AAG (10 μM) and analyzed for SDHB activity at the indicated time intervals. NT, not treated. (f) Endogenous Complex II (SDH) was immunoprecipitated from PC3 cells, and analyzed for SDHB activity in the presence of increasing concentrations of recombinant TRAP-1 (μM). Data for panels (e, f) are from representative experiments out of at least two independent determinations. (g) PC3 cells were treated with 17-AAG (5 μM) or the indicated increasing concentrations of Gamitrinib (Gam, μM) and the oxygen consumption rate (OCR) was measured in real time under basal condition and in response to the indicated inhibitors. Arrows indicate the time of drug addition: D, Gamitrinib (Gam) or 17-AAG; O, oligomicyin (1.25 μM); F, FCCP (0.4 μM); A, antimycin (1.8 μM). (h) The OCR was normalized by the number of cells, and the extra-mitochondrial respiration after addition of antimycin was subtracted as background. * p<0.05; ** p<0.01 vs control sample at each state (two-sided unpaired t test). (i) PC3 cells were transfected with control (Ctrl) siRNA or TRAP-1-directed siRNA, treated with Gamitrinib (Gam, μM) or 17-AAG and analyzed for OCR as in (g). (j). Quantification of OCR ratio between: b/o, basal condition (before any addition) and after oligomycin addition; f/o, after FCCP and oligomycin addition; f/b, after FCCP addition and basal condition in PC3 cells transfected with control siRNA (Ctrl) or TRAP-1-directed siRNA. * p<0.05; ** p<0.01 (two-sided unpaired t test). For all OCR experiments, data are representative of two independent experiments carried out in triplicate, mean±sd
We next asked whether a TRAP-1-SDHB complex was important during cellular stress. In control experiments, exposure of tumor cells to concentrations >50 μM of the oxidative agent, hydrogen peroxide (H2O2), reduced SDHB levels (Fig. 3d). siRNA silencing of TRAP-1 exacerbated this response and induced nearly complete loss of SDHB expression at lower H2O2 concentrations (Fig. 3d). As a control, the expression of the flavoprotein subunit of Complex II, SDHA 28, was not affected (Fig. 3d). Functionally, treatment of tumor cells with Gamitrinib inhibited Complex II activity in a concentration-dependent manner, whereas 17-AAG had no effect (Fig. 3e). Reciprocally, addition of recombinant TRAP-1 to SDHB immuno-affinity isolated from mitochondrial extracts enhanced Complex II activity in a concentration-dependent manner, in vitro (Fig. 3f).
Mitochondrial Hsp90 regulation of bioenergetics stress
The results above have suggested that Hsp90-directed protein folding preserves the stability and function of SDHB in tumor cells. To determine whether this mechanism regulates oxidative phosphorylation, we next quantified the respiration rates of tumor cells in real time. At the same concentrations that induce SDHB misfolding (Fig. 3a), and impaired mitochondrial metabolism (Figs. 1, 2), Gamitrinib inhibited the oxygen consumption rate (OCR) in prostate PC3 cancer (Fig. 3g, h, Supplementary Fig. S9a), or glioblastoma LN229 (Supplementary Fig. S9b–d) cells, in a concentration-dependent manner. 17-AAG had no effect on OCR (Fig. 3g, h, Supplementary Fig. 9). siRNA knockdown of TRAP-1 in PC3 (Fig. 3i, j, Supplementary Fig. S10a), or LN229 (Supplementary Fig. S10b–d) cells, partially attenuated the inhibition of OCR mediated by Gamitrinib, compared to control transfectants. In contrast, transfection of tumor cells with non-targeting siRNA had no effect on OCR, with or without Gamitrinib (Fig. 3i, j, Supplementary Fig. S10a–d). The partial reduction in the respiratory capacity and SDHB inhibition produced by Gamitrinib when added after siRNA silencing of TRAP-1, as compared to the near complete inhibition observed when Gamitrinib is added without prior siRNA to TRAP-1, may reflect a compensatory protective response by the mitochondria as a result of the extended partial TRAP-1 inhibition produced by siRNA knockdown of TRAP-1, potentially involving organelle Hsp90.
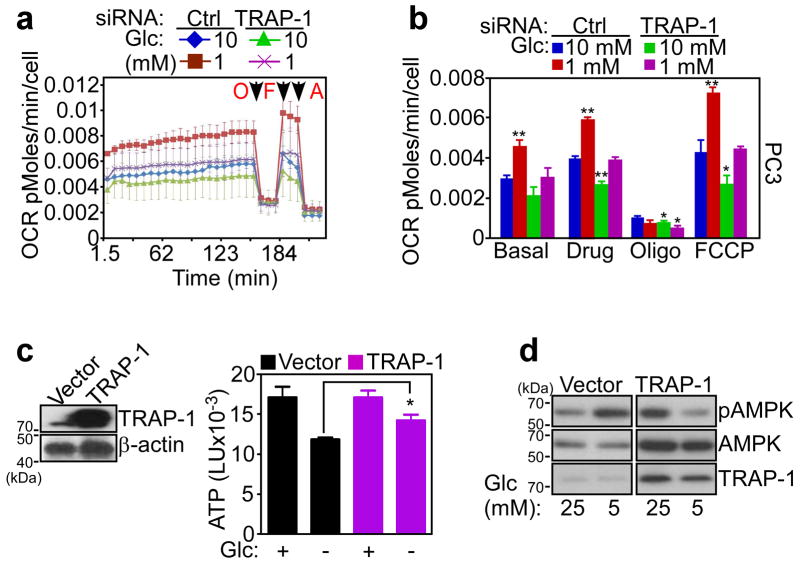
Most tumors undergo metabolic reprogramming, and utilize aerobic glycolysis as their main energy source 3. Therefore, we asked whether oxidative phosphorylation enabled by Hsp90-directed protein folding was important for tumor maintenance. Tumor cells transfected with control siRNA and maintained in abundant nutrients (10 mM glucose) exhibited normal cellular respiration (Fig. 4a, b, Supplementary Fig. S10e). This response was increased at lower glucose concentrations (1 mM), suggestive of a compensatory mechanism that elevates ATP output by oxidative phosphorylation during nutrient deprivation (Fig. 4a, b, Supplementary Fig. S10e). Under these experimental conditions, siRNA knockdown of TRAP-1 abolished the compensatory increase in OCR at limiting glucose concentrations (1 mM), whereas cellular respiration in 10 mM glucose was minimally affected (Fig. 4a, b, Supplementary Fig. S10e).
Figure 4. Mitochondrial Hsp90 regulation of stress bioenergetics.
(a) PC3 cells were transfected with control, non-targeting siRNA (Ctrl) or TRAP-1-directed siRNA and maintained in 1 or 10 mM glucose (Glc) for 3 h before analysis of OCR. Arrows indicate the time of drug addition: O, oligomicyin (1.25 μM); F, FCCP (0.4 μM); A, antimycin (1.8 μM). (b) OCR in a was quantified in siRNA-transfected cells in different glucose (Glc) concentrations. Data for panels (a, b) data are representative of two independent experiments carried out in triplicate, mean±sd. * p<0.05; ** p<0.01 vs control sample at each state (two-sided unpaired t test). (c) Normal NIH3T3 fibroblasts were transfected with control vector or TRAP-1 cDNA and analyzed by Western blotting (left) or ATP production in the presence (+) or absence (−) of glucose (Glc, 25 mM) (right). *, p=0.03 (two-sided unpaired t test). (d) NIH3T3 fibroblasts were transfected as in (c), incubated with the indicated concentrations of glucose (Glc, mM), and analyzed by Western blotting. p, phosphorylated.
In reciprocal experiments, we transfected a control plasmid or TRAP-1 cDNA in normal NIH3T3 fibroblasts (Fig. 4c), which have low endogenous levels of mitochondrial Hsp90 and TRAP-1 18. NIH3T3 fibroblasts transfected with control cDNA exhibited reduced ATP production (Fig. 4c), and phosphorylation of AMPK (Fig. 4d) at limiting glucose concentrations, consistent with cellular starvation. In contrast, transfection of TRAP-1 restored ATP production (Fig. 4c), and reduced AMPK phosphorylation (Fig. 4d) at low glucose concentrations.
Role of mitochondrial Hsp90s in SDH-mutant tumors
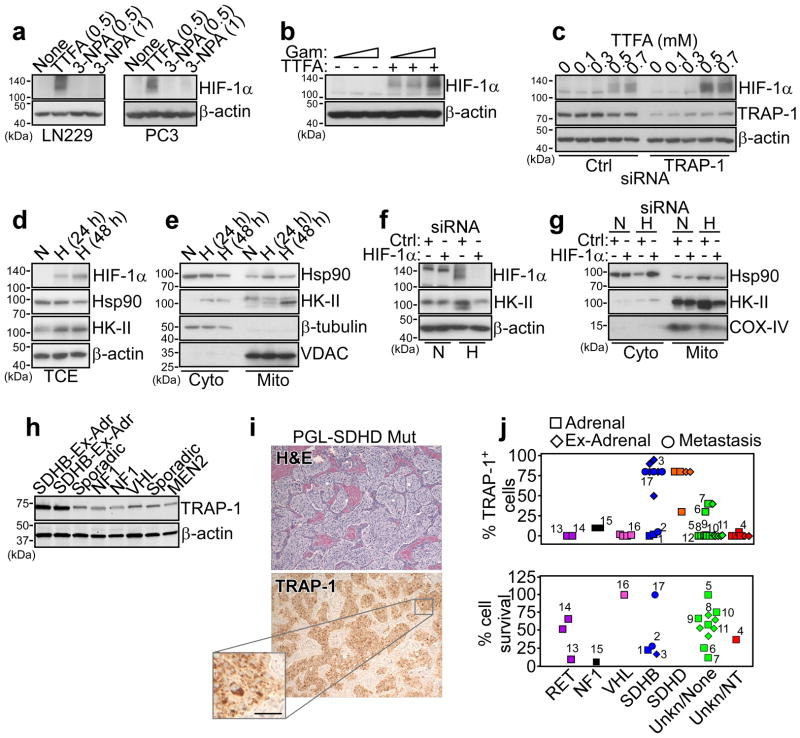
These experiments suggest that Hsp90 and TRAP-1 control multiple mitochondrial pathways of bioenergetics, and their role in oxidative phosphorylation may support energy production under conditions of nutrient deprivation. To test the implications of this model for tumor cell survival, we next targeted ETC Complex II function using pharmacologic inhibitors. Treatment of tumor cells with the Complex II inhibitor, TTFA, but not 3-NPA, increased the expression of hypoxia-inducible factor-1α (HIF-1α) (Fig. 5a), an oncogenic transcription factor implicated in adaptive responses to cellular stress 34. Inhibition of mitochondrial Hsp90s with Gamitrinib (Fig. 5b), or siRNA silencing of TRAP-1 (Fig. 5c), was insufficient, alone, to modulate HIF-1α levels, whereas both treatments strongly enhanced TTFA induction of HIF-1α in tumor cells (Fig. 5b, c). In parallel experiments, tumor cells exposed to hypoxia exhibited increased recruitment of Hsp90 to mitochondria, compared to cytosol (Fig. 5d, e), and this response was reversed by HIF-1α silencing by siRNA (Fig. 5f, g). The mitochondrial pool of HK-II was also increased under hypoxic conditions (Fig. 5d, e), and this response was also abolished by siRNA knockdown of HIF-1α (Fig. 5f, g). In contrast, normoxic conditions (Fig. 5d, e), or transfection of tumor cells with non-targeting siRNA (Fig. 5f, g) had no effect.
Figure 5. TRAP-1-SDHB complex regulates HIF-1α-directed tumorigenesis.
(a) The indicated tumor cell types (LN229 or PC3 cells) were treated with the various concentrations (mM) of the SDHB inhibitors, TTFA or 3-NPA and analyzed by Western blotting. (b) PC3 cells were treated with increasing concentrations of Gamitrinib (Gam, 0, 2.5, 5 μM) in the absence (−) or presence (+) of TTFA (0.3 mM) and analyzed by Western blotting. (c) LN229 cells were transfected with control siRNA (Ctrl) or TRAP-1-directed siRNA and analyzed by Western blotting in the presence of the indicated increasing concentrations of TTFA. (d, e) PC3 cells were maintained under conditions of hypoxia (H, 0.5% O2, 5% CO2, 94% N2 for 24 h) or normoxia (N), and analyzed by Western blotting in total cell extracts (TCE) (d), or fractionated cytosolic (Cyto) or mitochondrial (Mito) extracts (e). VDAC or β-tubulin was used as a mitochondrial or cytosolic marker, respectively. (f) PC3 cells were transfected with control siRNA (Ctrl) or HIF-1α-directed siRNA, maintained in normoxia (N) or hypoxia (H) conditions, and analyzed by Western blotting. (g) PC3 cells were transfected and treated as in (f), and isolated cytosolic (Cyto) or mitochondrial (Mito) fractions were analyzed by Western blotting. COX-IV was used as a mitochondrial marker. (h) Patient-derived tissue samples of PCC/PGL were analyzed by Western blotting. The mutational status of each tumor is indicated. Ex-Adr, extra-adrenal localization. (i) A tissue sample of extra-adrenal PGL with SDHD mutation, showing a typical nest-like (“Zellballen”) growth pattern was stained with hematoxylin/eosin (H&E, top) or TRAP-1 (bottom), by immunohistochemistry. Scale bar, 50 μm. (j) Quantification of immunohistochemical expression of TRAP-1 in PCC/PGL cases with the indicated mutational status (top). Cells from the various tumor samples were maintained in culture and analyzed for killing by Gamitrinib (10 μM for two weeks) (bottom) measured by counts of tyrosine hydroxylase-positive cells counted in an area defined by a randomly placed 22×22 mm square coverslips in 35 mm round culture dishes. Each point represents a single tumor. Paired samples of the same tumor were available in 12 instances and are indicated by matching numbers. Data are from a representative experiment.
Mutations in Complex II 35, including SDHB 36, have been linked to hereditary or sporadic pheochromocytoma (PCC) 37, and paraganglioma (PGL) 38, potentially through a mechanism of HIF-1α-dependent tumorigenesis 39. Consistent with HIF-1α-dependent accumulation of Hsp90 to mitochondria after Complex II inhibition (Fig. 5e, g), TRAP-1 was strongly expressed in PCC/PGL samples carrying SDHB and SDHD mutations, compared to tumors with mutations in RET, NF1, VHL, or of unknown genotype (Fig. 5h, i). Functionally, PCC/PGL tumors with Complex II mutations and high levels of TRAP-1 (Fig. 5j) were more sensitive to Gamitrinib-mediated killing, in vitro (Fig. 5j), suggesting a compensatory pro-survival role of mitochondrial Hsp90s in transformed cells with defective oxidative phosphorylation 39.
DISCUSSION
In this study, we have identified mitochondrial Hsp90s 18 as global regulators of tumor cell metabolism, including oxidative phosphorylation and redox networks. This pathway hinges on chaperone-directed protein folding in mitochondria 15, and affects a discrete Hsp90/TRAP-1 18 proteome intercalated in multiple, fundamental pathways of cellular homeostasis. This mechanism may be ideally suited to buffer the risk of proteotoxic stress in transformed cells with high biosynthetic needs 19, preserve organelle integrity against CypD-dependent apoptosis 20, and maintain multiple sources of energy production, including HK-II-dependent glycolysis 21, and oxidative phosphorylation (this study), especially under stress conditions of hypoxia and nutrient deprivation.
The considerable interest in aerobic glycolysis 3 as a central feature of tumor metabolic reprogramming 1, together with the signaling role of oncogenes in these responses 40, have brought into question the function of mitochondrial bioenergetics, and in particular oxidative phosphorylation, in tumor maintenance 28. However, recent studies have suggested that mitochondrial oxidative phosphorylation continues to remain critical for tumor cells 6, favoring resistance to therapy 7,8, and promoting cell survival 9. The data presented here provide a mechanistic framework in support of these observations, and identify Hsp90/TRAP-1-directed protein folding in mitochondria 18 as a key requirement of oxidative phosphorylation in tumors. This involved the formation of physical complex(es) between Hsp90/TRAP-1 and the iron-sulfur subunit of mitochondrial ETC Complex II, SDHB 32, preserving its folding, stability and enzymatic function under oxidative stress. Functionally, Hsp90/TRAP-1 regulation of SDHB maintained energy production under conditions of low nutrients and hypoxia, which are hallmarks of tumor growth, in vivo 41, and dampened biochemical signals of cellular starvation that are typically associated with tumor suppression 42.
SDHB 32 has attracted attention as a gene mutated in certain human neuroendocrine tumors 36. The molecular requirements of how these mutations contribute to malignancy are still being worked out 36, but one consequence of pharmacologic or genetic inactivation of SDHB observed here was an increased recruitment of Hsp90 to mitochondria 18. This pathway required HIF-1α, which is deregulated in SDHB-mutant tumors, and may potentially contribute to disease maintenance 39. The increased accumulation of mitochondrial Hsp90s under these conditions may help compensate for the impaired oxidative phosphorylation resulting from defective SDHB function 36, enhancing organelle integrity against CypD-mediated permeability transition 18 and energy production via HK-II-directed glycolysis 21. Consistent with this model, SDHB-mutant tumor cells were more sensitive to Gamitrinib-mediated killing than other neuroendocrine malignancies, suggesting that Hsp90-directed protein folding in mitochondria provides an adaptive and potentially “addictive” survival factor for these cells.
There is now intense interest in pursuing aberrant tumor cell metabolism for cancer therapeutics 10. However, inhibitors that can safely target these pathways in tumors, as opposed to normal tissues, especially with respect to oxidative phosphorylation 7,8, have not been clearly identified 43. As a mitochondrial-directed Hsp90 inhibitor 20, Gamitrinib may be ideally suited to function as a general antagonist of tumor cell metabolism. Supported by the differential targeting of tumor, as opposed to normal mitochondria 18, and a favorable safety profile in preclinical models 20, Gamitrinib inhibition of mitochondrial Hsp90s may simultaneously disable metabolic and survival adaptive networks in genetically heterogeneous tumors.
METHODS
Antibodies and reagents
The following antibodies to succinate dehydrogenase complex subunit B (SDHB, 1:500, Abcam), succinate dehydrogenase complex subunit A (SDHA, 1:3000, Abcam), hexokinase-II (HK-II, 1:1000, Cell Signaling), Cox-IV (1:1000, Cell Signaling), Hypoxia-Inducible Factor-1α (HIF1α, 1:500, Cell Signaling), Hsp90 (1:1000, BD Biosciences), Thr172-phosphorylated AMPKα (1:1000, Cell Signaling), AMPKα (1:1000, Cell Signaling), TRAP-1 (1:1000, BD Biosciences), and β-actin (1:5000, Sigma-Aldrich) were used. A total oxidative phosphorylation antibody cocktail (1:500, Mitoscience) directed against the 20-kD subunit of Complex I (20 kD), cytochrome C oxidase subunit II of Complex IV (22 kD), SDHB subunit of Complex II (30 kD), core 2 of complex III (~50 kD), and F1α (ATP synthase) of Complex V (~60 kD) was used. The complete chemical synthesis, HPLC profile, and mass spectrometry of mitochondrial-targeted small molecule Hsp90 antagonist, Gamitrinib (GA mitochondrial matrix inhibitors) has been reported 20. The Gamitrinib variant containing triphenylphosphonium as a mitochondrial-targeting moiety 20 was used in this study. Non-mitochondrially directed Hsp90 inhibitor, 17-allylamino-demethoxygeldanamycin (17-AAG) was obtained from LC-Laboratories. Oligomycin, carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP), antimycin A, 3-nitropropionic acid (3-NPA) and thenoyltrifluoroacetone (TTFA) were obtained from Sigma-Aldrich.
Transfections
For gene knockdown experiments, tumor cells were transfected using control, non-targeting small interfering RNA (siRNA) pool (Dharmacon, cat. no. D-001810) or specific ON-Target SMARTpool siRNAs to TRAP-1 (Dharmacon, cat. no. L-010104), or HIF1α (Dharmacon, cat. no. L-004018). The various siRNAs were transfected at 10–30 nM using Oligofectamine (Invitrogen). Transfection of plasmid DNA was carried out with Lipofectamine (Invitrogen), as described 20.
Subcellular fractionation
Mitochondrial fractions were isolated from Gamitrinib-treated LN229 cells (0–20 μM for 5 h) using an ApoAlert™ cell fractionation kit (CLONTECH), as described 20. Briefly, LN229 cells or PC3 cells were mechanically disrupted by 70 strokes with a Dounce homogenizer in isolation buffer containing 1 mM DTT plus protease inhibitor cocktail. Cell debris was removed by centrifugation at 700 g for 10 min. The supernatant was further centrifuged at 10,000 g for 25 min, and supernatants or mitochondrial pellets were processed for further analysis.
Mitochondrial protein folding
Mitochondrial fractions were isolated from vehicle- or Gamitrinib-treated LN229 cells (5 μM for 12 h), and suspended in equal volume of mitochondrial fractionation buffer containing increasing concentrations of CHAPS (0, 0.05, 0.1, 0.2, 0.5, 1 or 2%). Samples were incubated for 20 min on ice and detergent-insoluble protein aggregates were recovered by centrifugation (20,000 g) for 20 min. Pelleted proteins were separated by SDS-gel electrophoresis and visualized by silver staining (Sigma Aldrich).
Proteomics studies
To identify mitochondrial proteins that require organelle Hsp90s for proper folding and/or activity (mitochondrial Hsp90 proteome), individual silver-stained bands isolated from mitochondrial fractions of vehicle or Gamitrinib-treated LN229 cells were analyzed by 1D MS (see Supplementary Methods). As an independent experimental approach, global proteomics analysis of vehicle or Gamitrinib-treated LN229 cells was carried out by Stable Isotope Labeling by Amino acids in Culture (SILAC) technology (see Supplementary Methods). Changes in the expression of 301 metabolites were determined by Ultrahigh performance liquid chromatography/Mass Spectroscopy (UPLC/MS/MS) and Gas chromatography/Mass Spectroscopy (GC/MS) in PC3 cells treated with vehicle or Gamitrinib (2.5, 5 μM), non-mitochondrial targeted 17-AAG (5 μM), or alternatively, transfected with control non-targeting or TRAP-1-directed siRNA (see Supplementary Methods).
Purification of TRAP-1 Proteins
NIH3T3 cells were transfected with human TRAP1-Myc plasmid cDNA. After 48 h, cells were washed with PBS and lysed in PBS containing 1% TX-100 plus phosphatase inhibitor cocktail (Roche). Lysates were centrifuged at 14,000 × g for 10 min at 4°C, and c-Myc-tagged TRAP-1 proteins were isolated by immunoprecipitation with an antibody to c-Myc coupled to agarose beads (Sigma-Aldrich). Samples were then washed five times with lysis buffer, and TRAP-1-myc was eluted from the immune complex with 100 μg/ml c-Myc peptide (Sigma-Aldrich) in PBS. To eliminate free c-Myc peptide and further enrich eluted TRAP-1-containing material, samples were purified with centrifugal filter (30K, Millipore).
SDH activity assay
Tumor cells were analyzed for SDH complex activity as reduction of the dye 2,6-diclorophenolindophenol (DCPIP), which recycles the substrate ubiquinone using Complex II enzyme activity. Briefly, mitochondria isolated from PC3 or LN229 cells were lysed in enzyme assay buffer containing 1% n-dodecyl-β-D-maltopyranoside plus protease inhibitors (Roche) for 1 h at 4°C under constant agitation. After centrifugation at 15,000 g for 20 min at 4°C, supernatants were loaded on anti-Complex II antibody-coated 96-well plates, and incubated with increasing concentrations of recombinant TRAP-1 for 2 h. Enzyme activity was determined from SDH-dependent reduction of DCPIP, and quantified as changes in absorbance at 600 nm for 3 h at 2 min intervals using a plate reader (Beckman Coulter).
Cellular respiration
Oxygen consumption rates (OCR) were assayed using the Extracellular Flux System 24 Instrument (Seahorse Bioscience, Billerica, MD). PC3 or LN229 cells were grown in standard media and after trypsinization and re-suspension in growth media, 25,000 cells were plated in each well of a Seahorse XF24 cell culture plate (100 μl volume). After 4-h incubation to allow the cells to adhere to the plate, an additional 150 μl of media was added to each well, and the cells were grown for 24 h at 37°C with 5% CO2. The media was then exchanged with unbuffered DMEM XF assay media (Seahorse Bioscience) supplemented with 2 mM glutaMAX, 1 mM sodium pyruvate and 5 mM glucose (pH 7.4 at 37°C), and equilibrated for 30 min at 37°C and ~0.04 % CO2 before the experiment. Cellular oxygen consumption was monitored in basal condition (before any addition) and after addition of oligomycin (1.25 μM), carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone (FCCP) (0.4 μM), and antimycin (1.8 μM), all dissolved in DMSO. The three drugs were injected into the XF24 sequentially, and the oxygen consumption rates measured using the extra-cellular flux analyzer with three cycles of mixing (150 seconds), waiting (120 seconds), and measuring (210 seconds). This cycle was repeated following each injection 44. To test the effect of mitochondrial Hsp90s on cellular respiration, PC3 or LN229 cells were treated with non-cytotoxic concentrations of Gamitrinib (0–10 μM) or 17-AAG (2.5–5 μM) and continuously analyzed for OCR changes. Alternatively, cells were transfected with control or TRAP-1-directed siRNA and analyzed after 24–36 h.
Patient samples
All experiments involving patient-derived material were approved by the Tufts Medical Center Institutional Review Board following informed consent. A series of genetically characterized PCC/PGL with documented mutations of major susceptibility genes (n=10, SDHB; 6 SDHD; 4 VHL; 3 RET; 2 NF1), apparently sporadic PCC/PGL (n=22) and normal human adrenal medulla was examined in this study. All of the tumors with VHL, RET or NF1 mutations were intra-adrenal, while 10/13 with SDHB, 3/6 with SDHD, and 10/25 with no known mutations were extra-adrenal. Two of the extra-adrenal tumors with SDHD mutations were in the head or neck and the remainder retroperitoneal. For four of the tumors with SDHB mutations, tissue was available only from metastatic sites. One SDHB-mutated tumor was an adrenal bed recurrence of a primary malignancy that had given rise to metastases. All of the other specimens were primary tumors.
Statistical analysis
Data were analyzed using the two-sided unpaired t tests using a GraphPad software package (Prism 4.0) for Windows. Data are expressed as mean±SD or mean±SEM of multiple independent experiments. A p value of <0.05 was considered as statistically significant. For pair-wise comparisons in metabolite screening studies, the Welch’s t-tests, Wilcoxon’s rank sum tests or ANOVA were performed. For classification studies, random forest analyses were performed. Statistical analyses are performed with the program “R” http://cran.r-project.org/.
Supplementary Material
Acknowledgments
We gratefully acknowledge the assistance of The Wistar Institute Proteomics Core for performing LC-MS/MS analyses, Tony Chang-Wong for assistance in computational processing of proteomics data, and Sira Sriswasdi for preparing heat maps. This work was supported by the PheoPara Alliance, National Institutes of Health (NIH) grants CA140043, CA78810, HL54131 and CA118005 to DCA, NS021328 to DCW and Department of Defense grant PR100171 to AST. Support for Core Facilities utilized in this study was provided by Cancer Center Support Grant (CCSG) CA010815 to The Wistar Institute.
Footnotes
AUTHOR CONTRIBUTIONS
Y.C.C., A.A., A.S.T., R.D.M., D.C.W., L.R.L., D.W.S., and D.C.A. designed research; Y.C.C., A.A., K.D.S., H.W., J.F.P., and E.D.K. performed research; S.L., K.P., S.F. and R.D.M. contributed new reagents/analytical tools; Y.C.C., A.A., A.S.T., R.D.M., D.C.W., L.R.L., D.W.S., and D.C.A. analyzed data, and Y.C.C., A.A., A.S.T., D.C.W., D.W.S., and D.C.A. wrote the paper.
COMPETING FINANCIAL INTEREST
The authors declare no competing financial interest.
References
- 1.Kroemer G, Pouyssegur J. Tumor Cell Metabolism: Cancer’s Achilles’ Heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 4.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matoba S, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 6.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haq R, et al. Oncogenic BRAF Regulates Oxidative Metabolism via PGC1alpha and MITF. Cancer Cell. 2013;23:302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caro P, et al. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell. 2012;22:547–560. doi: 10.1016/j.ccr.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vazquez F, et al. PGC1alpha Expression Defines a Subset of Human Melanoma Tumors with Increased Mitochondrial Capacity and Resistance to Oxidative Stress. Cancer Cell. 2013;23:287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang JB, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haynes CM, Ron D. The mitochondrial UPR - protecting organelle protein homeostasis. J Cell Sci. 2010;123:3849–3855. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- 12.Green DR, Kroemer G. The pathophysiology of mitochondrial cell death. Science. 2004;305:626–629. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- 13.Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011;13:184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging and cancer: A dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker BM, Haynes CM. Mitochondrial protein quality control during biogenesis and aging. Trends Biochem Sci. 2011;36:254–261. doi: 10.1016/j.tibs.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 17.Leskovar A, Wegele H, Werbeck ND, Buchner J, Reinstein J. The ATPase cycle of the mitochondrial Hsp90 analog Trap1. J Biol Chem. 2008;283:11677–11688. doi: 10.1074/jbc.M709516200. [DOI] [PubMed] [Google Scholar]
- 18.Kang BH, et al. Regulation of tumor cell mitochondrial homeostasis by an organelle-specific Hsp90 chaperone network. Cell. 2007;131:257–270. doi: 10.1016/j.cell.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 19.Siegelin MD, et al. Exploiting the mitochondrial unfolded protein response for cancer therapy in mice and human cells. J Clin Invest. 2011;121:1349–1360. doi: 10.1172/JCI44855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang BH, et al. Combinatorial drug design targeting multiple cancer signaling networks controlled by mitochondrial Hsp90. J Clin Invest. 2009;119:454–464. doi: 10.1172/JCI37613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chae YC, et al. Control of tumor bioenergetics and survival stress signaling by mitochondrial HSP90s. Cancer Cell. 2012;22:331–344. doi: 10.1016/j.ccr.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Litonin D, et al. Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J Biol Chem. 2010;285:18129–18133. doi: 10.1074/jbc.C110.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koeck T, et al. A common variant in TFB1M is associated with reduced insulin secretion and increased future risk of type 2 diabetes. Cell Metab. 2011;13:80–91. doi: 10.1016/j.cmet.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Barrientos A, et al. MTG1 codes for a conserved protein required for mitochondrial translation. Mol Biol Cell. 2003;14:2292–2302. doi: 10.1091/mbc.E02-10-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christian BE, Spremulli LL. Mechanism of protein biosynthesis in mammalian mitochondria. Biochim Biophys Acta. 2012;1819:1035–1054. doi: 10.1016/j.bbagrm.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uchiumi T, et al. ERAL1 is associated with mitochondrial ribosome and elimination of ERAL1 leads to mitochondrial dysfunction and growth retardation. Nucleic Acids Res. 2010;38:5554–5568. doi: 10.1093/nar/gkq305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pike ST, Rajendra R, Artzt K, Appling DR. Mitochondrial C1-tetrahydrofolate synthase (MTHFD1L) supports the flow of mitochondrial one-carbon units into the methyl cycle in embryos. J Biol Chem. 2010;285:4612–4620. doi: 10.1074/jbc.M109.079855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eismann T, et al. Peroxiredoxin-6 protects against mitochondrial dysfunction and liver injury during ischemia-reperfusion in mice. Am J Physiol Gastrointest Liver Physiol. 2009;296:G266–274. doi: 10.1152/ajpgi.90583.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyle AN, et al. Poldip2, a novel regulator of Nox4 and cytoskeletal integrity in vascular smooth muscle cells. Circ Res. 2009;105:249–259. doi: 10.1161/CIRCRESAHA.109.193722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tiranti V, et al. Loss of ETHE1, a mitochondrial dioxygenase, causes fatal sulfide toxicity in ethylmalonic encephalopathy. Nat Med. 2009;15:200–205. doi: 10.1038/nm.1907. [DOI] [PubMed] [Google Scholar]
- 32.Yankovskaya V, et al. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- 33.Taipale M, et al. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 2012;150:987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shay JE, Celeste Simon M. Hypoxia-inducible factors: crosstalk between inflammation and metabolism. Semin Cell Dev Biol. 2012;23:389–394. doi: 10.1016/j.semcdb.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 35.Neumann HP, et al. Germ-line mutations in nonsyndromic pheochromocytoma. N Engl J Med. 2002;346:1459–1466. doi: 10.1056/NEJMoa020152. [DOI] [PubMed] [Google Scholar]
- 36.King A, Selak MA, Gottlieb E. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene. 2006;25:4675–4682. doi: 10.1038/sj.onc.1209594. [DOI] [PubMed] [Google Scholar]
- 37.Astuti D, et al. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am J Hum Genet. 2001;69:49–54. doi: 10.1086/321282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gimenez-Roqueplo AP, Tischler AS. Pheochromocytoma and Paraganglioma: progress on all fronts. Endocr Pathol. 2012;23:1–3. doi: 10.1007/s12022-011-9190-7. [DOI] [PubMed] [Google Scholar]
- 39.Selak MA, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 40.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laderoute KR, et al. 5′-AMP-activated protein kinase (AMPK) is induced by low-oxygen and glucose deprivation conditions found in solid-tumor microenvironments. Mol Cell Biol. 2006;26:5336–5347. doi: 10.1128/MCB.00166-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheong H, Lu C, Lindsten T, Thompson CB. Therapeutic targets in cancer cell metabolism and autophagy. Nat Biotechnol. 2012;30:671–678. doi: 10.1038/nbt.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu M, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol - Cell Physiol. 2007;292:C125–C136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.