Abstract
The surface of a material is rapidly covered with proteins once that material is placed in a biological environment. The structure and function of these bound proteins play a key role in the interactions and communications of the material with the biological environment. Thus, it is crucial to gain a molecular level understanding of surface bound protein structure. While X-ray diffraction and solution phase NMR methods are well established for determining the structure of proteins in the crystalline or solution phase, there is not a corresponding single technique that can provide the same level of structural detail about proteins at surfaces or interfaces. However, recent advances in sum frequency generation (SFG) vibrational spectroscopy have significantly increased our ability to obtain structural information about surface bound proteins and peptides. A multi-technique approach of combining SFG with (1) protein engineering methods to selectively introduce mutations and isotopic labels, (2) other experimental methods such as time-of-flight secondary ion mass spectrometry (ToF-SIMS) and near edge x-ray absorption fine structure (NEXAFS) to provide complementary information, and (3) molecular dynamic (MD) simulations to extend the molecular level experimental results is a particularly promising route for structural characterization of surface bound proteins and peptides. By using model peptides and small proteins with well-defined structures, methods have been developed to determine the orientation of both backbone and side chains to the surface.
1. Introduction
Information about protein structure and function at interfaces on the molecular level is crucial in drug design, biosensor applications and biomaterial engineering 1-5. Proteins on surfaces are an integral part of many biomedical applications (implanted biomedical devices, diagnostic arrays, tissue engineering scaffolds, cell cultures, etc.) and in biomimetic material design strategies (biomineralization 6-10 and surface functionalization 11, 12). This importance has stimulated research towards developing techniques to assess the structure, activity, and surface interactions of immobilized proteins. Appropriate tools to obtain this information can make the important difference between “trial and error” approaches and the next generation of structure-based design concepts.
To control the biological response to a material or sensor in biological environments, or the protein recognition mechanisms during biomineralization, it is important to understand the structure of the proteins immobilized onto those materials and the nature of protein – surface interactions 13. Large, globular proteins can undergo major structural changes at hydrophobic interfaces, unfolding to expose their hydrophobic cores. Even if a protein is not denatured by contact with a surface, it is very likely that the side chain structure will change depending on the chemical interaction with the surface. Membrane proteins, on the other hand, often fold into their active state only after binding to a lipid surface or incorporation into a bilayer. Protein - cell interactions are another important area where high-resolution information about protein structure on and within membranes and structural changes upon interaction with therapeutic agents will be crucial to enable the next generation of targeted drug delivery 14-16.
The development of techniques such as X-ray diffraction (XRD) and nuclear magnetic resonance (NMR) for determining the nanoscale or molecular structure of proteins in the crystalline or solution phase has been pursued for decades 17, 18. It is now possible to obtain angstrom level resolution of atomic positions in protein crystals. In contrast, we know much less about the structure of proteins on surfaces. The Protein Databank (PDB) provides a sobering picture of this situation: About 60,000 protein structures have been published, but not a single structure of a protein bound to an inorganic surface has been solved.
The small amount of protein found in monolayers on surfaces requires the use of techniques that, due to their sampling depths or selection rules, exclusively detect species present at the surface1, 2. This perspective article will first provide an overview of methodological advances in sum frequency generation (SFG) spectroscopy that can be used for investigation and understanding protein-interface interactions. We will then illustrate how these SFG methods can provide a detailed picture of proteins on surfaces using the example of synthetic peptides adsorbed onto model surfaces. In the third part of this article we will describe the promise of combining SFG measurements with near-edge X-ray absorption fine structure (NEXAFS) spectroscopy and time-of-flight secondary ion mass spectrometry (ToF-SIMS) and show the experimental data can be used to actually solve protein structures on surfaces.
2. Sum frequency generation spectroscopy for protein studies
Researchers are currently exploring possible routes to probe proteins on surfaces using a variety of surface analytical tools 19. Methods sensitive to the amount of protein adsorbed, such as scanned probe techniques, surface plasmon resonance (SPR), quartz crystal microbalance (QCM), radio labeling and X-ray photoelectron spectroscopy, have been successful in developing sophisticated models of protein film formation 20-26. The drawback of these techniques, however, is their lack of structure sensitivity. Molecular level details of proteins on surfaces have been studied using infrared and Raman spectroscopy, cryo-transmission electron microscopy, ToF-SIMS, NEXAFS and SFG spectroscopy 27-34. These techniques can provide information about the conformation, binding or orientation of surface bound proteins.
The unique capability of SFG in this context is that it is extremely surface sensitive and can probe proteins in situ in their native environments at interfaces (i.e., at the liquid-solid interface). SFG generates vibrational spectra of ordered species at interfaces and has been developed into an increasingly powerful technique for investigating protein films 35, 36. SFG is based on a coherent nonlinear optical process: Spectrally broad or tunable infrared and fixed visible laser pulses with high power are overlapped in time and space at an interface to generate photons at the sum of the incident beam frequencies. The signal is enhanced for IR frequencies in resonance with SFG-active vibrational modes at the interface yielding vibrational spectra. Since SFG is a second order process, no signal is generated in isotropic, randomly ordered or inversion symmetric media36, 37. Consequently, the bulk phase of many materials and liquids is SFG inactive and the signal is exclusively generated at the interface, where inversion symmetry is necessarily broken. According to the ‘selection rules’ of SFG, only molecular groups that have a net order will contribute to the measured signal36. Thus, even if at the surface, only molecules with a certain degree of order will generate a SFG signal. In addition, the orientation of molecular moieties can be probed by using different polarization combinations of the incident beams and the detected SFG signal.
Since surface interactions can introduce ordering in the binding regions of proteins and peptides, SFG is an excellent probe to identify the side chains involved in these binding events. This approach has been used to probe the adsorption of proteins such as BSA and fibrinogen onto hydrophobic polystyrene surfaces, as well as polar surfaces such as calcium fluoride and silicon dioxide.38-42 These early experiments laid the groundwork for later, more qualitative SFG methods and are discussed in detail in a number of research and review articles.43-45 The clear drawback of working with large proteins is their immense complexity and the redundancy of chemical groups making SFG spectra difficult to interpret.
Short, well-designed model peptides allow for more detailed analysis and an opportunity to understand protein-surface interactions on the molecular level. Model leucine-lysine (LK) peptides on a variety of surfaces ranging from different self-assembled monolayers on gold to spin-coated polymers to liquid surfaces have been successful investigated in this context. LK peptides are amphiphilic and comprised only of hydrophobic leucine (L) and hydrophilic lysine (K) side-chains. The beauty of this model system is that, depending on the hydrophobic periodicity of the amino acid sequence, α-helix, 310-helix and β-strand structures can be synthesized.46, 47
In SFG spectra of α-helical 14mer LK peptides (LKα14) the CH3 stretching modes of leucine became visible on hydrophobic polystyrene and fluorocarbon films, showing a net ordering of leucine upon peptide binding.48-50 On charged surfaces, conversely, strong N–H resonances became visible - a signature of lysines interacting with the surface.50, 51
In addition, we have employed phase sensitive SFG measurements to verify the pointing directions of the respective side chains. SFG is a coherent technique and the signal phase is related to molecular orientation with respect to the surface. The SFG phase can be measured directly using a reference beam52 or indirectly by observing the phase relative to a non-resonant background signal. Ward et al. have determined the relation of the relative SFG phase and the orientation of methyl groups on gold surfaces.53 Gold is an ideal substrate for phase sensitive measurements because it generates a very strong non-resonant background signal. For air-dried LKα14 films on hydrophobic and hydrophilic self-assembled monolayers on gold we have observed that the leucine chains were orienting towards hydrophobic surfaces, most likely binding the peptide via hydrophobic interactions. Contrarily, on negatively charged SAMs, the leucines were pointing away from the SAM-peptide interface.51, 54, 55
Air-drying can induce orientation and conformation changes. Therefore we here present data verifying the side chain orientations on SAMs in a biologically relevant aqueous environment. Figure 1 shows in situ C–H region SFG spectra recorded in PBS buffer for LKα14 adsorbed onto dodecanethiol (DoDT) SAM covered gold surfaces. The SAM was assembled on a gold layer deposited onto a calcium fluoride window. This allows SFG to probe the SAM-solution interface by going through the backside of the window (details can be found in ref 56). To avoid spectral confusion with the substrate, the peptide spectra were collected on a deuterated DoDT SAM (i.e., only the peptide will contribute to the C–H stretches. The data were fit using the following equation:
Figure 1.
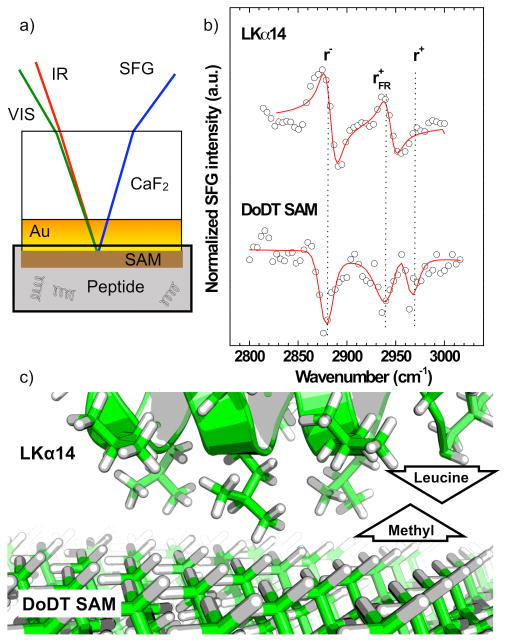
a) Experimental setup for in situ SFG measurements. A CaF2 window is coated with a 25 nm gold layer and then decorated with a DoDT SAM. b) SFG C–H spectra collected in situ through the backside of the window. For the LKα14 spectrum a deuterated DoDT SAM was used as substrate to avoid spectral confusion with the peptide CH modes. The peptide and SAM peaks have a phase difference of >π/2 showing the CH3 orientations of these two species have a predominately opposite orientation. c) Schematic showing the orientation of the leucine isopropanol groups binding to the DoDT methyl groups.
| (1) |
Here, χ(2)nr, Aq, ωq and Γq are the second order nonlinear optical susceptibility of the nonresonant background, the resonant oscillator strength, resonance frequency and damping constant of the qth vibration mode. Since χ(2)nr,Aq are complex numbers they have a modulus and a phase. The relative phase determines whether resonant modes q and the background signal will interfere constructively or destructively. In this analysis, the phase of the background was kept constant and the signal phase was determined relative to the background. The peak fits reveal that the relative phases of the peptide and SAM C–H modes are out of phase. This implies opposite orientations for the SAM methyl groups and the leucine isopropanol groups, i.e. the leucines are pointing towards the hydrophobic SAM surface. This experiment shows that, based on existing solution- or solid-phase protein structures from the Protein Database, in some cases, the orientation of entire peptides can already be estimated from the orientation of certain key amino acids.41, 48, 50, 51, 55, 57-60
The conformation of proteins can be monitored using amide I SFG spectra. The backbone-related modes are sensitive to conformation 28 and can be used to determine the secondary structure of adsorbed peptides.40, 61 For example, α-helix, β-sheet and turn structures have distinct resonance positions between 1620 cm-1 and 1750 cm-1. While amide I analysis by linear infrared spectroscopy has been a successful tool for many years, SFG provides a number of additional advantages. It is inherently surface specific, sensitive to molecular alignment and has the capability to probe interfaces in situ in aqueous environments without the need for special sample preparation, e.g. membrane stacking, which is sometimes used for infrared analysis.28, 62 Depending on the polarization of incident and detected light, chiral and achiral species can be interrogated independently.63, 64 SFG is also insensitive to bending modes of bulk water, which often interfere with IR amide I spectra.28 Since SFG is only sensitive to ordered species, proteins in solution do not generate a signal and films on surfaces can be probed even when covered by protein solutions.
Chen et al. have shown that SFG and infrared analysis are complementary tools for protein studies. Since infrared and SFG experiments probe different order parameters, the joint use of SFG and infrared spectra allows the determination of both the orientation and orientational distribution of interfacial protein layers and therefore improve the quantitative orientation analysis.65 A number of studies investigated the secondary structure of proteins and peptides on polymer surfaces 41, dielectrics66, 67 and model membrane surfaces44, 68-71. The influence of the chemical environment and the denaturing effect of detergents was monitored in situ in biotechnologically relevant environments.72 Real time SFG measurements can follow protein binding and the assembly process in situ. We have observed the in situ assembly of adsorbed LK peptide layers at hydrophobic fluorocarbon surfaces in a phosphate buffered saline (PBS) solution48 by recording resonance intensities related to ordering of the side chain and the backbone after peptide injection into the liquid cell. Interestingly, different time scales for side-chain binding and backbone assembly were observed indicating that the film orders in two steps. Since SFG intensities depend on a number of factors such as order, symmetry and surface density it is necessary to reference SFG kinetics to additional complementary data. Here, the direct comparison of SFG data with in situ mass sensitive methods such as SPR, QCM, surface pressure and radio labeling help to disentangle the factors related to order and symmetry from those related to the number density of molecules at an interface. We also compared the SFG measurements with ex situ XPS composition data taken after different adsorption times. XPS is mass sensitive and can help separate the coverage contribution to the SFG signal from order effects. It also can determine the elemental composition of the protein layer. The sensitivity of XPS for surface chemistry outbalances some of the disadvantages a vacuum-based technique has for protein studies. The results suggest an extended ordering process within the peptide film over a timescale of hours following the rapid initial adsorption process. Kinetic SFG measurements have been used to probe structural and orientational changes large proteins in membranes70 and when exposed to antibiotic compounds.73
Numerical procedures to quantify the orientations of helical and β-sheet structures using SFG amide I spectra have recently been published by the Chen and the Yan group.74, 75,72, 76 Polarization dependent SFG measurements, where the polarizations of the incident infrared and visible beams and the detected SFG photons are varied between s- and p-polarization, allow one to probe protein orientation in membranes and on surfaces 69, 74, 75, 77, 78.
Figure 2 shows, an example for a quantitative study of the orientation of LKα14 peptides adsorbed onto polystyrene surfaces. The analysis was performed in situ in a 0.5 mg/ml peptide solution in PBS with the photon beams entering and exiting through the backside of a prism. Details of the experimental geometry have been published elsewhere.60 SFG spectra recorded with ssp (s-polarized SFG, s-polarized visible and p-polarized IR, respectively), ppp and sps polarization combinations show a single peak near 1655 cm-1 related to an α-helical structure. The oscillator strengths were determined by fitting equation 1 to the spectra, and then used to calculate helix orientations according to the procedure described in ref 75. In short, the nonlinear optical susceptibilities (χ(2)RES) for ssp, ppp and sps resonances were determined from the oscillator strength 79 and then ratios of (χ(2)RES) for different polarizations were analyzed. These ratios are directly linked to the helix orientation. This procedure yielded a χ(2)ppp/χ(2)ssp ratio of 2.7 and a χ(2)ppp/χ(2)sps ratio of 5.7 related to helix orientations of 75° and 85°, respectively, thus suggesting an average of 80° orientation of the peptide background versus the surface normal. This analysis assumes a δ-distribution of peptide orientations and the helices are most likely not perfectly aligned on the surface, so the observed tilt angle represents a lower limit. This data confirms that the LK peptides are oriented almost parallel to the surface, as already suggested by the C–H region SFG experiments discussed above and ssNMR measurements of LK peptides on polystyrene micro beads published previously.54
Figure 2.
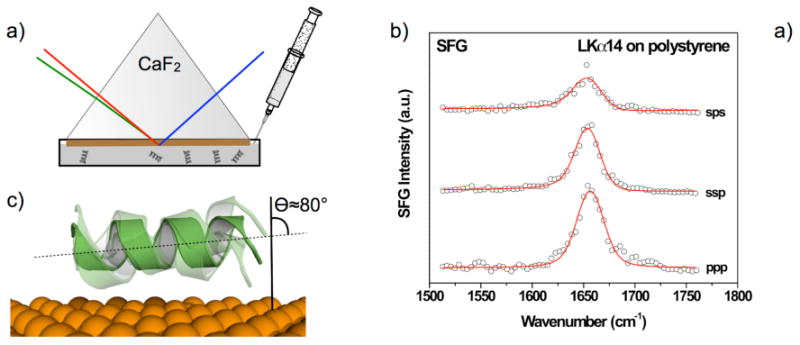
a) Experimental setup for in situ SFG measurements. One side of the prism is coated with a polystyrene film and then exposed to a protein solution. The interface is then interrogated going through the backside of the prism. b) SFG amide I spectra collected with different polarization combinations of the incident pump beams and the detected SFG photons. The single peak near 1655 cm-1 shows the peptides have an α-helical secondary structure when adsorbed onto a polystyrene surface. c) The polarization dependence of the amide I resonance can be used to determine the helix orientation vs. the surface normal.
The information about protein orientation and binding geometries discussed above can already can address important questions about the interaction of proteins with biomaterial surfaces such as: Are the proteins in their native, bioactive conformation after surface binding? What is the orientation of the proteins? Are the proteins well aligned or in a random state? However, here, the ‘resolution’ remains at the molecular level (i.e., overall shape and orientation of the entire protein). To go beyond this and probe details of side chain structure within proteins at high resolution (i.e. getting closer to actually solving a protein structure) isotope labels at specific protein sites are necessary since it is anticipated that more than one type of amino acid will show ordering when proteins adsorb onto surfaces. Also, for each ordered amino acid, it is likely that more than one residue contributes to observed spectra.
We have pioneered the combination of isotope labels for quantitative SFG orientation analysis. Based on earlier studies showing that SFG has the capability to detect individual deuterated residues within larger proteins, we have used isotope labels to probe the orientations of individual side chains in peptides and proteins 34, 51, 54, 55, 60. Deuterium substitution into the C–H groups leads to a red shift of ∼800 cm-1 for the C–H resonances. This allows SFG spectra of individual amino acids in the C-D stretching range (2000 -2350 cm-1) to be recorded without spectral confusion with chemically similar side chains. Established procedures for SFG orientation analysis of surface species can then be applied to probe the side chain orientation. Using a set of eight different peptide samples, each with deuterium labels at a different leucine residue, we have determined in situ the orientations of the entire set of leucine side chains in LK peptides bound to a polystyrene surface.60 The spectrum in Figure 3 shows a typical C–D region spectrum of a labeled LK peptide binding a polystyrene surface. The peaks near 2220 cm-1 and 2060 cm-1 are related to the asymmetric and symmetric CD3 stretching modes. Features near 2130 cm-1 and 2080 cm-1 are related to the CD3 Fermi resonance and the methine CD stretching mode of the carbon in the leucine isopropyl group, respectively. The tilt and torsion angles for the terminal isoleucine groups were obtained from comparing CD3 mode intensities at different polarization combinations of the incident probe beams and the detected SFG photons using a unified atom approach developed by Paul Cremer's group for isopropanol at the air-water interface.80
Figure 3.
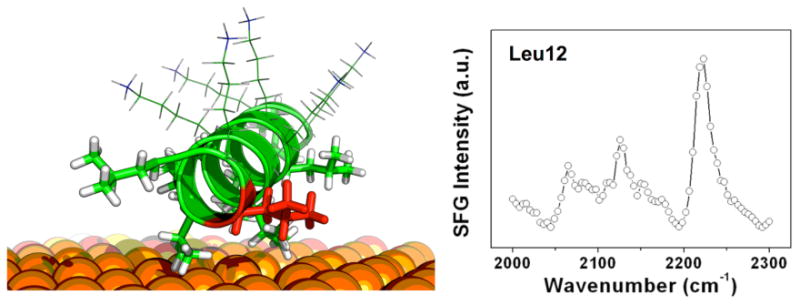
: Left: Schematic drawing of the model lysine-leucine peptide LKα14 adsorbed onto a polystyrene surface. The leucine side chains interact with the hydrophobic surface. Right: SFG spectrum of Leu 12 (labeled red) after that side chain has undergone site directed deuterium substitution for structural analysis studies. The orientations of all leucine side chains (sticks) were determined using a combination of SFG and solid-state NMR.60 The lysine residues (lines) are less ordered than the leucines and interact with the surrounding water molecules.
We have combined the SFG results with solid-state NMR (ssNMR) dynamics measurements.54, 60 While SFG can provide the overall orientation of the peptide backbone as well as the orientation of individual side chains, 2H ssNMR line shape simulations can determine side chain dynamics and, thus, the proximity of individual residues to the surface 54. Surface interactions dampen the side chain motions, which changes the respective NMR line shapes. By combining together the information obtained from the different experiments for LK peptides on polystyrene, i.e. secondary structure, backbone orientation, tilt and twist angles of the binding side chains as well as their surface proximities, we have developed a detailed structural model of a surface peptide (Figure 3). It is interesting to note, that the side chains near both ends of the sequence tend be oriented towards neighboring peptides and that leucines close to the surface show larger rotation angles, i.e. they bring one of the isopropyl methyl groups closer to the surface.
3. A strategy towards protein structure determination
The examples above demonstrate the potential of SFG to probe protein structure on surfaces. But its great advantage, its sensitivity to a range of different interfacial properties such as symmetry, order, orientation and surface density can also lead to ambiguous results and, in some cases, make data interpretation difficult if not impossible. For example, is the intensity of a SFG backbone resonance in a protein film deceasing because the protein is desorbing, disordering or even assembling into a centrosymmetric layer? Combining SFG with independent mass- or structure sensitive techniques can help answer this kind of question and can dramatically amplify the level of understanding gained from of SFG data. The usefulness of mass sensitive techniques is obvious and one example combining XPS with kinetic SFG data was discussed above. The combination of SFG with structure sensitive methods such as ssNMR, NEXAFS spectroscopy and ToF-SIMS can even be more powerful. However, the latter two methods use ultra-high vacuum conditions and can only probe dried protein layers. For larger proteins with a tendency to denature when dehydrated, Castner et al. have developed fixation methods for ToF-SIMS protein studies, which aim at maintaining the protein structure during the drying process.32
Examples for a successful combination of SFG backbone and side chain data with ssNMR dynamics and distance measurements have already been mentioned for the example of LK peptides on model surfaces. NEXAFS spectroscopy, on the other hand, provides a very different perspective on protein films. It can provide valuable information about interfacial species and help understand their orientation and order. NEXAFS spectra exhibit characteristic resonances related to electronic transitions from an atomic core levels to unoccupied molecular orbitals making this technique very sensitive to molecular bonds in general. For proteins studies, amide bonds can be probed using the nitrogen K-edge resonance near 400 eV. Evidence of protein order or insight into backbone structure is provided by the linear dichroism effect, i.e. the dependence of the intensity of amide resonances on the X-ray incidence angle. The cross section of the resonant photoexcitation process is determined by the orientation of the electric field vector of the incident synchrotron light relative to the transition dipole moment (TDM) of the respective molecular orbital.81 For ordered protein structures, the intensity of the amide resonance varies significantly with the X-ray incidence angle. The standard way of monitoring the linear dichroism is to analyze the difference between the spectra acquired at normal and glancing incidence. Figure 4 shows the nitrogen K-edge NEXAFS spectra of LKα14 and its β-strand counterpart, LKβ15, adsorbed onto negatively charged carboxylic acid terminated alkanethiol SAMs on gold to illustrate how this effect can be used to probe protein films (for experimental details see refs 55, 82). All the spectra exhibit an edge jump related to transitions into continuum states and an additional pronounced resonance near 401 eV related to transitions from N 1s core levels into unoccupied amide π* orbitals. Additional broader features at higher photon energies are assigned to N-H and N-C related transitions. While the LKα14 70° and 20° spectra show only very weak differences in amide π* intensity, the dichroic effect is much stronger for LKβ15. This can be explained by the inherently more aligned orientation of amide bonds in β-strand structures compared to helices.
Figure 4.
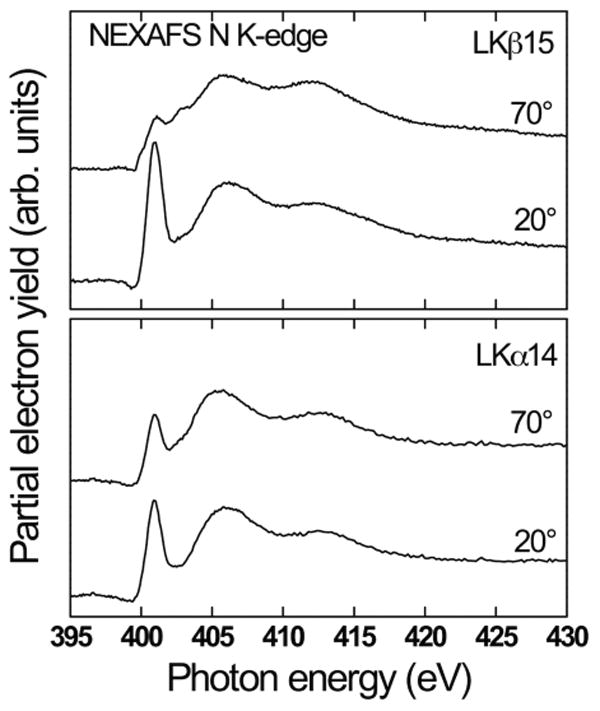
: NEXAFS nitrogen K-edge spectra for LKα14 and LKβ15 peptides adsorbed onto carboxylic acid termianted alkanethiol SAMs. The NEXAFS spectra were acquired at a near-normal angle of 70° and at a glancing angle of 20°. The significantly more pronounced intensity variation with the X-ray angle for β-strand peptides can be explained by a more aligned configuration of amide bonds in that film compared to the LKα14 film.
The complementary nature of SFG and NEXAFS spectroscopy becomes clear when comparing the α-helix and β-strand NEXAFS spectra with SFG data. SFG amide I spectra for the peptide samples are shown in Figure 5. For the LKα14 peptides, the amide I peak is readily detected by SFG while there is no signal visible for the LKβ15 sample. Most likely, the same symmetric arrangement of the amide bonds that leads to such a strong angle dependence in NEXAFS in the ordered LKβ15 films also results in a cancelation of signal from amide groups with opposite orientations and, thus, the suppression of a net SFG response.49, 55 Hence, the SFG data set would have been impossible to explain with certainty without comparison to the NEXAFS data. Recently, we have shown that NEXAFS can also provide information about individual side chains. By selectively labeling phenylalanine residues with fluorine, the respective ring orientations were determined in the binding domain of the human biomineralization protein statherin on hydroxyapatite, its native mineral surface.34 This method is complementary to SFG studies of individual side chains using isotope labels.
Figure 5.
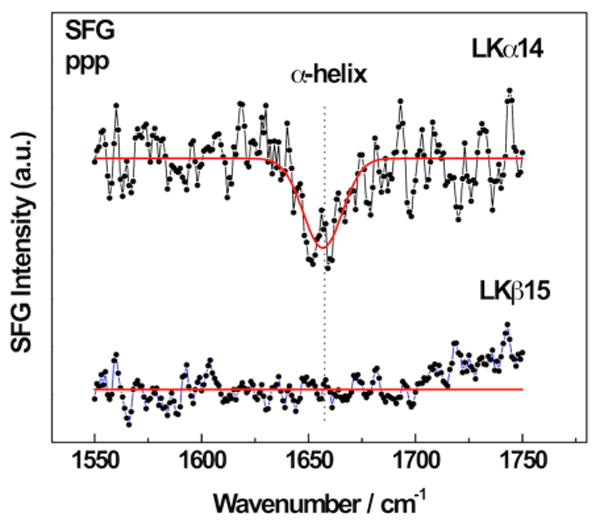
SFG spectra of the amide I region for α-helical and β-strand LK peptides adsorbed onto mercaptoundecanol SAMs on gold. The single peak near 1655 cm-1 for LKα14 shows the peptides are α-helical the surface. The lack of signal for the β-strand peptide can be explained by a symmetrical arrangement of the beta strands.
Another technique that can potentially provide a detailed picture of protein-surface interactions is ToF-SIMS. ToF-SIMS offers both high chemical specificity and surface sensitivity (typically ∼2 nm).83-85 For ToF-SIMS a surface is typically bombarded with a keV energy primary ion beam that sputters molecular fragments off the surface. A fraction of the sputtered fragments carries a net charge and can be extracted through a time-of-flight mass analyzer. The result is a mass spectrum of the secondary ions ejected from the interface.85, 86
The dynamics of the secondary ion ejection process are extremely complicated and dependent on experimental conditions such as the choice of primary ions.87 However, most amino acids have distinct spectral fingerprints83, 84 and reference tables with characteristic peak88 allow one to use ToF-SIMS to characterize protein films.83, 84 Taking advantage of the shallow sampling depth of ToF-SIMS makes it possible to probe the changes in conformation and orientation of surface immobilized proteins with ToF-SIMS.89-93 Ordering, unfolding and denaturation changes will change the relative concentrations of different side chains in the ToF-SIMS sampling region. Thus, changes in surface conformation can be tracked by examining intensity changes of secondary-ion signals from hydrophobic and hydrophilic amino acids.94 In previous studies, ToF-SIMS has also been shown to provide information about the orientation of surface immobilized proteins by recording the intensities of secondary ions originating from different amino acids asymmetrically located within the protein 3-D structure.89, 93, 95
We have also successfully begun to expand this multitechnique approach to complex proteins. The B1 domain of the protein G is a small protein (6 kDa) comprised only of two antiparallel β-sheets and one α-helix, which makes it an ideal model protein for these studies, exhibiting the next step in complexity compared to the LK peptides. In a combined SFG, NEXAFS and ToF-SIMS study of this protein immobilized on a variety of surfaces the protein secondary structure and backbone orientation were determined19, 27, 29, 58. ToF-SIMS was used to determine the overall protein orientation using selected peaks from asymmetrically distributed amino acids in the protein structure. SFG provided information about the secondary structure and could show, that the protein film exhibited a significant degree of backbone alignment that was induced from being bound to the surface. NEXAFS analysis provided an estimate of the β-sheet tilt angle with respect to the surface using a standard method to determine bond orientations from angle dependent NEXAFS spectra. Based on our previous LK peptide work this analysis assumed that helical structures do not contribute significantly to the angle dependence of NEXAFS data.
In summary, SFG is a very powerful tool to probe proteins on surfaces, but its full potential can only be unleashed if combined with complementary techniques. Since each of these methods has their strengths and weaknesses, determining protein structure on surfaces requires combining together information from a range of different approaches. The combination of surface analytical tools alone can still not provide atomic structures of entire proteins that would, for example, meet the accuracy and resolution criteria for a structure submission to the Protein Databank96. For solution- and solid-state protein structure determination the integration of computer modeling and experimental methods played an increasing role in recent years. Solution-state structures can be determined by combining sparse experimental NMR data with specialized computational methods such as RosettaNMR.97 For protein crystals, cryo-electron microscope density maps can be used to constrain in silico protein models. 98, 99 Such hybrid theoretical and experimental procedures also provide a promising approach for protein structure determination on surfaces. Figure 6 illustrates a possible route to such an atomic level analysis. Latour et al. have recently published benchmark test for the accuracy of different molecular dynamics force field for proteins on surfaces.100 Drobny, Gray et al. have taken an important step in this direction by showing that using ssNMR structural data to guide and constrain model calculations can lead to an improved understanding of the biomineralization protein statherin on hydroxyapatite surfaces.101 Pfaendtner and coworkers have compared interfacial peptide structures they had predicted using advanced MD sampling methods to experimentally determined structures102. Importantly, progress in simulating SFG spectra of proteins in the amide I region will lead to new approaches to combine simulations and experiments in a very direct way.103 The combination of specifically designed computer models guided and constrained by a broad variety of experimental data from a variety of complementary surface analytical techniques provides a viable path towards protein structure determination on surfaces.
Figure 6.
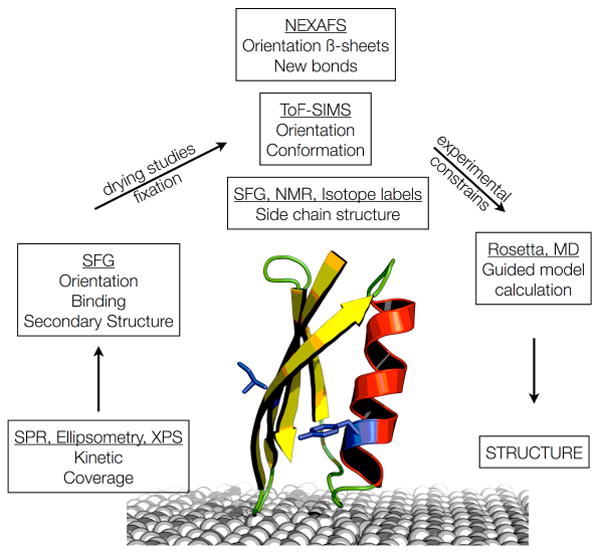
A flow diagram outlining a possible route towards determining the structure of proteins on surfaces.
Acknowledgments
The authors acknowledge support from NIH grants EB-002027, GM-074511 and DE-012554 during preparation of this manuscript as well as for some the results reported in it. TW thanks the EU Marie-Curie PEOPLE Programme for a Career Integration Grant (322124)
References
- 1.Castner DG, Ratner BD. Surf Sci. 2002;500:28–60. [Google Scholar]
- 2.Willem N, Thomas AH, John LB. Proteins at Interfaces III State of the Art, American Chemical Society. 2012;1120:1–34. [Google Scholar]
- 3.Nudelman F, Sommerdijk NAJM. Angew Chem Int Edit. 2012;51:6582–6596. doi: 10.1002/anie.201106715. [DOI] [PubMed] [Google Scholar]
- 4.Palmer LC, Newcomb CJ, Kaltz SR, Spoerke ED, Stupp SI. Chem Rev. 2008;108:4754–4783. doi: 10.1021/cr8004422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiner S, Addadi L. Annual Review of Materials Research. 2011;4141:21–40. [Google Scholar]
- 6.Hildebrand M. Chem Rev. 2008;108:4855–4874. doi: 10.1021/cr078253z. [DOI] [PubMed] [Google Scholar]
- 7.Kröger N, Deutzmann R, Sumper M. Science. 1999;286:1129–1132. doi: 10.1126/science.286.5442.1129. [DOI] [PubMed] [Google Scholar]
- 8.Wang B, Liu P, Tang R. BioEssays : news and reviews in molecular, cellular and developmental biology. 2010;32:698–708. doi: 10.1002/bies.200900120. [DOI] [PubMed] [Google Scholar]
- 9.Kim S, Park CB. Adv Funct Mater. 2013;23:10–25. [Google Scholar]
- 10.Mahamid J, Addadi L, Weiner S. Cells Tissues Organs. 2011;194:92–97. doi: 10.1159/000324229. [DOI] [PubMed] [Google Scholar]
- 11.Sarikaya M, Tamerler C, Jen AKY, Schulten K, Baneyx F. Nature Mater. 2003;2:577–585. doi: 10.1038/nmat964. [DOI] [PubMed] [Google Scholar]
- 12.Minelli C, Liew JX, Muthu M, Andresen H. Soft Matter. 2013;9:5119. [Google Scholar]
- 13.Ratner BD. Macromol Symp. 1997;130:327–335. [Google Scholar]
- 14.Chu DSH, Schellinger JG, Shi J, Convertine AJ, Stayton PS, Pun SH. Accounts Chem Res. 2012;45:1089–1099. doi: 10.1021/ar200242z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng Z, Al Zaki A, Hui JZ, Muzykantov VR, Tsourkas A. Science. 2012;338:903–910. doi: 10.1126/science.1226338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicolas J, Mura S, Brambilla D, Mackiewicz N, Couvreur P. Chem Soc Rev. 2013;42:1147–1235. doi: 10.1039/c2cs35265f. [DOI] [PubMed] [Google Scholar]
- 17.Rupp B. Biomolecular Crystallography: Principles, Practice and Application to Structural Biology. Garland Science; New York: 2009. [Google Scholar]
- 18.Cavanagh J, Fairbrother WJ, Palmer AG, III, Rance M, Skelton NJ. Protein NMR spectroscopy: principles and practice. 2. Academic Press; Boston: 2007. [Google Scholar]
- 19.Baio JE, Weidner T, Castner DG. Proteins at Interfaces III State of the Art, American Chemical Society. 2012;1120:761–779. [Google Scholar]
- 20.Baer DR, Engelhard MH. J Electron Spectrosc Relat Phenom. 2010;178:415–432. [Google Scholar]
- 21.Campbell CT, Kim G. Biomaterials. 2007;28:2380–2392. doi: 10.1016/j.biomaterials.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 22.Cheng F, Gamble LJ, Castner DG. Anal Chem. 2008;80:2564–2573. doi: 10.1021/ac702380w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dufrene YF, Marchal TG, Rouxhet PG. Appl Surf Sci. 1999;144-45:638–643. [Google Scholar]
- 24.Hook F, Kasemo B, Nylander T, Fant C, Sott K, Elwing H. Anal Chem. 2001;73:5796–5804. doi: 10.1021/ac0106501. [DOI] [PubMed] [Google Scholar]
- 25.Ray S, Shard AG. Anal Chem. 2011;83:8659–8666. doi: 10.1021/ac202110x. [DOI] [PubMed] [Google Scholar]
- 26.Servoli E, Maniglio D, Aguilar MR, Motta A, Roman JS, Belfiore LA, Migliaresi C. Macromol Biosci. 2008;8:1126–1134. doi: 10.1002/mabi.200800110. [DOI] [PubMed] [Google Scholar]
- 27.Baio JE, Weidner T, Baugh L, Gamble LJ, Stayton PS, Castner DG. Langmuir. 2012;28:2107–2112. doi: 10.1021/la203907t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh BR, editor. Infrared Analysis of peptides and Proteins. American Chemical Society; Washington DC: 2000. [Google Scholar]
- 29.Baugh L, Weidner T, Baio JE, Nguyen PCT, Gamble LJ, Slayton PS, Castner DG. Langmuir. 2010;26:16434–16441. doi: 10.1021/la1007389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim YP, Hong MY, Kim J, Oh E, Shon HK, Moon DW, Kim HS, Lee TG. Anal Chem. 2007;79:1377–1385. doi: 10.1021/ac0616005. [DOI] [PubMed] [Google Scholar]
- 31.Lawson CL, Baker ML, Best C, Bi CX, Dougherty M, Feng PW, Van Ginkel G, Devkota B, Lagerstedt I, Ludtke SJ, Newman RH, Oldfield TJ, Rees I, Sahni G, Sala R, Velankar S, Warren J, Westbrook JD, Henrick K, Kleywegt GJ, Berman HM, Chiu W. Nucleic Acids Res. 2011;39:D456–D464. doi: 10.1093/nar/gkq880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michel R, Castner DG. Surf Int Anal. 2006;38:1386–1392. [Google Scholar]
- 33.Muramoto S, Graham DJ, Wagner MS, Lee TG, Moon DW, Castner DG. J Phys Chem C. 2011;115:24247–24255. doi: 10.1021/jp208035x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weidner T, Dubey M, Breen NF, Ash J, Baio JE, Jaye C, Fischer DA, Drobny GP, Castner DG. J Amer Chem Soc. 2012;134:8750–8753. doi: 10.1021/ja301711w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen YR. Annu Rev Phys Chem. 1989;40:327–351. [Google Scholar]
- 36.Shen YR. The Principles of Nonlinear Optics. 1. John Wiley & Sons; New York: 1984. [Google Scholar]
- 37.Boyd RW. Nonlinear Optics. 1. Academic Press; London: 1992. [Google Scholar]
- 38.Wang J, Buck SM, Chen Z. Analyst. 2003;128:773–778. doi: 10.1039/b212551j. [DOI] [PubMed] [Google Scholar]
- 39.Chen Z, Ward R, Tian Y, Malizia F, Gracias DH, Shen YR, Somorjai GA. J Biomed Mater Res. 2002;62:254–264. doi: 10.1002/jbm.10075. [DOI] [PubMed] [Google Scholar]
- 40.Clarke ML, Wang J, Chen Z. J Phys Chem B. 2005;109:22027–22035. doi: 10.1021/jp054456k. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Chen XY, Clarke ML, Chen Z. J Phys Chem B. 2006;110:5017–5024. doi: 10.1021/jp0534683. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Clarke ML, Chen X, Even MA, Johnson WC, Chen Z. Surf Sci. 2005;587:1–11. [Google Scholar]
- 43.Chen Z, Shen YR, Somorjai GA. Annu Rev Phys Chem. 2002;53:437–465. doi: 10.1146/annurev.physchem.53.091801.115126. [DOI] [PubMed] [Google Scholar]
- 44.Chen XY, Chen Z. Biochim Biophys Acta - Biomembranes. 2006;1758:1257–1273. doi: 10.1016/j.bbamem.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 45.Chen XY, Clarke ML, Wang J, Chen Z. Int J Mod Phys B. 2005;19:691–713. [Google Scholar]
- 46.DeGrado WF, Lear JD. J Am Chem Soc. 1985;107:7684–7689. [Google Scholar]
- 47.Degrado WF, Wasserman ZR, Lear JD. Science. 1989;243:622–628. doi: 10.1126/science.2464850. [DOI] [PubMed] [Google Scholar]
- 48.Weidner T, Samuel NT, McCrea K, Gamble LJ, Ward RS, Castner DG. Biointerphases. 2010;5:9–16. doi: 10.1116/1.3317116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samuel NT. PhD Thesis. University of Washington; 2005. [Google Scholar]
- 50.Mermut O, Phillips DC, York RL, McCrea KR, Ward RS, Somorjai GA. J Amer Chem Soc. 2006;128:3598–3607. doi: 10.1021/ja056031h. [DOI] [PubMed] [Google Scholar]
- 51.Weidner T, Breen NF, Drobny GP, Castner DG. J Phys Chem B. 2009;113:15423–15426. doi: 10.1021/jp908773c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ostroverkhov V, Waychunas GA, Shen YR. Phys Rev Lett. 2005;94 doi: 10.1103/PhysRevLett.94.046102. [DOI] [PubMed] [Google Scholar]
- 53.Ward RN, Davies PB, Bain CD. J Phys Chem. 1993;97:7141–7143. [Google Scholar]
- 54.Breen NF, Weidner T, Li K, Castner DG, Drobny GP. J Amer Chem Soc. 2009;131:14148–14149. doi: 10.1021/ja905382m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weidner T, Apte JS, Gamble LJ, Castner DG. Langmuir. 2010;26:3433–3440. doi: 10.1021/la903267x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stein MJ, Weidner T, McCrea K, Castner DG, Ratner BD. J Phys Chem B. 2009;113:11550–11556. doi: 10.1021/jp9015867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jung SY, Lim SM, Albertorio F, Kim G, Gurau MC, Yang RD, Holden MA, Cremer PS. J Amer Chem Soc. 2003;125:12782–12786. doi: 10.1021/ja037263o. [DOI] [PubMed] [Google Scholar]
- 58.Baio JE, Weidner T, Samuel NT, McCrea KR, Baugh L, Stayton PS, Castner DG. J Vac Sci Technol B. 2010;28:C5D1–C5D8. doi: 10.1116/1.3456176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Samuel NT, McCrea KR, Gamble LJ, Ward RS, Castner DG. Abstr Pap Am Chem S. 2005;229:U671–U671. [Google Scholar]
- 60.Weidner T, Breen NF, Li K, Drobny GP, Castner DG. P Natl Acad Sci USA. 2010;107:13288–13293. doi: 10.1073/pnas.1003832107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen X, Wang J, Boughton AP, Kristalyn CB, Z C. J Amer Chem Soc. 2007;129:1420–1427. doi: 10.1021/ja067446l. [DOI] [PubMed] [Google Scholar]
- 62.Schuy S, Schäfer E, Yoder NC, Kumar K, Vogel R, Janshoff A. J Struct Biol. 2009;168:125–136. doi: 10.1016/j.jsb.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 63.Fu L, Liu J, Yan ECY. J Amer Chem Soc. 2011;133:8094–8097. doi: 10.1021/ja201575e. [DOI] [PubMed] [Google Scholar]
- 64.Wang J, Chen XY, Clarke ML, Chen Z. P Natl Acad Sci USA. 2005;102:4978–4983. doi: 10.1073/pnas.0501206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J, Lee SH, Chen Z. J Phys Chem B. 2008;112:2281–2290. doi: 10.1021/jp077556u. [DOI] [PubMed] [Google Scholar]
- 66.Wang J, Buck SM, Chen Z. J Phys Chem B. 2002;106:11666–11672. [Google Scholar]
- 67.Dreesen L, Sartenaer Y, Humbert C, Mani AA, Lemaire JJ, MÈthivier C, Pradier CM, Thiry PA, Peremans A. Thin Solid Films. 2004;464-465:373–378. [Google Scholar]
- 68.Chen XY, Boughton AP, Tesmer JJG, Chen Z. J Amer Chem Soc. 2007;129:12658–12659. doi: 10.1021/ja075542w. [DOI] [PubMed] [Google Scholar]
- 69.Yang P, Ramamoorthy A, Chen Z. Langmuir. 2011;27:7760–7767. doi: 10.1021/la201048t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ye S, Nguyen KT, Le Clair SV, Chen Z. J Struct Biol. 2009;168:61–77. doi: 10.1016/j.jsb.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ye SJ, Nguyen KT, Chen Z. J Phys Chem B. 2010;114:3334–3340. doi: 10.1021/jp911174d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xiao D, Fu L, Liu J, Batista VS, Yan ECY. J Mol Biol. 2012;421:537–547. doi: 10.1016/j.jmb.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen X, Tang H, Even MA, Wang J, Tew GN, Chen Z. J Amer Chem Soc. 2006;128:2711–2714. doi: 10.1021/ja057029t. [DOI] [PubMed] [Google Scholar]
- 74.Nguyen KT, King JT, Chen Z. J Phys Chem B. 2010;114:8291–8300. doi: 10.1021/jp102343h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nguyen KT, Le Clair SV, Ye SJ, Chen Z. J Phys Chem B. 2009;113:12169–12180. doi: 10.1021/jp904153z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boughton AP, Yang P, Tesmer VM, Ding B, Tesmer JJG, Chen Z. P Natl Acad Sci USA. 2011;108:E667–E673. doi: 10.1073/pnas.1108236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Le Clair SV, Nguyen K, Chen Z. J Adhesion. 2009;85:484–511. doi: 10.1080/00218460902996374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Han X, Soblosky L, Slutsky M, Mello CM, Chen Z. Langmuir. 2011;27:7042–7051. doi: 10.1021/la200388y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhuang X, Miranda PB, Kim D, Shen YR. Phys Rev B. 1999;59:12632–12640. [Google Scholar]
- 80.Kataoka S, Cremer PS. J Amer Chem Soc. 2006;128:5516–5522. doi: 10.1021/ja060156k. [DOI] [PubMed] [Google Scholar]
- 81.Stöhr J. NEXAFS Spectroscopy. Springer-Verlag; Berlin: 1992. [Google Scholar]
- 82.Apte JS, Collier G, Latour RA, Gamble LJ, Castner DG. Langmuir. 2009;26:3423–3432. doi: 10.1021/la902888y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wagner MS, Graham DJ, Ratner BD, Castner DG. Surf Sci. 2004;570:78–97. [Google Scholar]
- 84.Wagner MS, Castner DG. Appl Surf Sci. 2004;231-232:366–376. [Google Scholar]
- 85.Vickerman JC, Brown AA, Reed NM. Secondary ion mass spectrometry : principles and applications. Clarendon Press ; Oxford University Press, Oxford; Oxford; New York: 1989. [Google Scholar]
- 86.Belu AM, Graham DJ, Castner DG. Biomaterials. 2003;24:3635–3653. doi: 10.1016/s0142-9612(03)00159-5. [DOI] [PubMed] [Google Scholar]
- 87.Garrison BJ, Postawa Z. Mass Spectrom Rev. 2008;27:289–315. doi: 10.1002/mas.20165. [DOI] [PubMed] [Google Scholar]
- 88.Mantus DS, Ratner BD, Carlson BA, Moulder JF. Anal Chem. 1993;65:1431–1438. doi: 10.1021/ac00058a021. [DOI] [PubMed] [Google Scholar]
- 89.Lhoest JB, Detrait E, van den Bosch de Aguilar P, Bertrand P. J Biomed Mater Res A. 1998;41:95–103. doi: 10.1002/(sici)1097-4636(199807)41:1<95::aid-jbm12>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 90.Michel R, Pasche S, Textor M, Castner DG. Langmuir. 2005;21:12327–12332. doi: 10.1021/la051726h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xia N, Castner DG. J Biomed Mater Res A. 2003;67A:179–190. doi: 10.1002/jbm.a.10063. [DOI] [PubMed] [Google Scholar]
- 92.Xia N, May CJ, McArthur SL, Castner DG. Langmuir. 2002;18:4090–4097. [Google Scholar]
- 93.Wang H, Castner DG, Ratner BD, Jiang SY. Langmuir. 2004;20:1877–1887. doi: 10.1021/la035376f. [DOI] [PubMed] [Google Scholar]
- 94.Tidwell CD, Castner DG, Golledge SL, Ratner BD, Meyer K, Hagenhoff B, Benninghoven A. Surf Int Anal. 2001;31:724–733. [Google Scholar]
- 95.Lhoest JB, Wagner MS, Tidwell CD, Castner DG. J Biomed Mater Res. 2001;57:432–440. doi: 10.1002/1097-4636(20011205)57:3<432::aid-jbm1186>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 96.Schwede T, Sali A, Honig B, Levitt M, Berman HM, Jones D, Brenner SE, Burley SK, Das R, Dokholyan NV, Dunbrack RL, Fidelis K, Fiser A, Godzik A, Huang YJ, Humblet C, Jacobson MP, Joachimiak A, Krystek SR, Kortemme T, Kryshtafovych A, Montelione GT, Moult J, Murray D, Sanchez R, Sosnick TR, Standley DM, Stouch T, Vajda S, Vasquez M, Westbrook JD, Wilson IA. Structure. 2009;17:151–159. doi: 10.1016/j.str.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim DE, Chivian D, Baker D. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hryc CF, Chen DH, Chiu W. Curr Opin Virol. 2011;1:110–117. doi: 10.1016/j.coviro.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yahav T, Maimon T, Grossman E, Dahan I, Medalia O. Curr Opin Struct Biol. 2011;21:670–677. doi: 10.1016/j.sbi.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 100.Collier G, Vellore NA, Yancey JA, Stuart SJ, Latour RA. Biointerphases. 2012;7 doi: 10.1007/s13758-012-0024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Masica DL, Ash JT, Ndao M, Drobny GP, Gray JJ. Structure. 2010;18:1678–1687. doi: 10.1016/j.str.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Deighan M, Pfaendtner J. Langmuir. in press. [Google Scholar]
- 103.Roeters SJ, van Dijk CN, Torres Knoop A, Backus HG, Campen RK, Bonn M, Woutersen S. J Phys Chem A. doi: 10.1021/jp401159r. in press. [DOI] [PubMed] [Google Scholar]


