Abstract
Hepatitis C virus (HCV) is a major causative agent of chronic hepatitis and hepatocellular carcinoma worldwide and thus poses a significant public health threat. A hallmark of HCV infection is the extraordinary ability of the virus to persist in a majority of infected people. Innate immune responses represent the front line of defense of the human body against HCV immediately after infection. They also play a crucial role in orchestrating subsequent HCV-specific adaptive immunity that is pivotal for viral clearance. Accumulating evidence suggests that the host has evolved multifaceted innate immune mechanisms to sense HCV infection and elicit defense responses, while HCV has developed elaborate strategies to circumvent many of these. Defining the interplay of HCV with host innate immunity reveals mechanistic insights into hepatitis C pathogenesis and informs approaches to therapy. In this review, we summarize recent advances in understanding innate immune responses to HCV infection, focusing on induction and effector mechanisms of the interferon antiviral response as well as the evasion strategies of HCV.
1. Introduction
Infection with hepatitis C virus (HCV) is a significant cause of chronic hepatitis globally. It is estimated that 130 million people are infected with HCV, and that the number of newly infected individuals is on the rise. HCV has a restricted host range, infecting only humans and chimpanzees. The virus replicates predominantly in hepatocytes, which constitute 80% of the liver mass. HCV infection is usually subclinical and typically asymptomatic. However, the virus replicates in the liver continuously and progressively, establishing life-long, intrahepatic persistent infection in up to 70% of infected individuals, putting patients at risk for cirrhosis and hepatocellular carcinoma. As such, HCV-associated end-stage liver disease represents the leading indication for liver transplantation (95). There is no vaccine available to prevent HCV infection. Until recently, pegylated interferon (IFN)-α2a or -α2b plus ribavirin (PegIFN/RBV) was the standard-of-care treatment for chronic hepatitis C, but eliminated the virus in no more than 50% of those infected with genotype 1 virus who received the therapy. Various direct-acting antiviral drugs (DAAs) are in the development pipeline and have demonstrated promising efficacy in clinical trials. Of note, two viral protease inhibitors have recently been approved by FDA for clinical use and have been shown to improve the efficacy of PegIFN/RBV therapy in these difficult-to-cure patients. However, because of rapid selection of resistant virus, these protease inhibitors must be administered in combination with PegIFN/RBV (94). This 3-drug combination has recently become the standard-of-care in the U.S., but will likely be replaced in the future by IFN-sparing drug combinations incorporating more potent DAAs.
HCV is a small, enveloped RNA virus classified within the family Flaviviridae. The virus possesses a ∼10-kb long, positive-sense RNA genome packed with the viral capsid protein (core) inside a 50∼60 nm enveloped virion. Its genome encodes a large polyprotein of about 3000 amino acids that is processed by a combination of host and viral proteases into 10 individual proteins. The structural proteins, core, envelope 1 (E1) and E2 are located in the amino-terminal one-third, while the remaining carboxyl terminal two-thirds comprises the nonstructural (NS) proteins, p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B. Among these, the NS3 to NS5B proteins are absolutely required for viral RNA replication (120). A frameshift (F, a.k.a., ARF) protein may be expressed from the core-coding region as a result of translation from an alternative reading frame, but its role in the HCV life cycle, if any, is uncertain (19). The known functions of HCV proteins are shown in Fig. 1. Flanking the HCV polyprotein-coding sequence are the 5′- and 3′-nontranslating regions (NTRs), which contain complex secondary structures that serve as cis-replication signals interacting with viral and host factors. The 5′-NTR also contains an internal ribosome entry site (IRES), which directs the cap-independent translation of the HCV polyprotein (95, 120). HCV is highly heterogeneous in its genomic sequence – it comprises 6 major genotypes and dozens of subtypes that exhibit different geographical distribution and differential responses to PegIFN/RBV therapy (40).
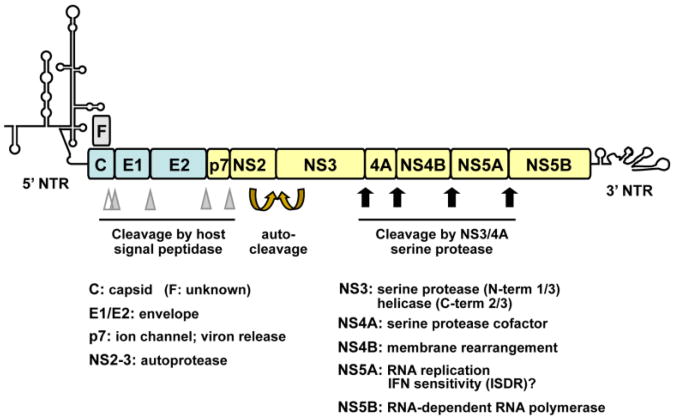
Figure 1. Organization of the HCV genome and the proteins HCV encodes.
The ∼10-kb positive-sense, single-stranded RNA genome encodes a large polyprotein that is processed into 10 individual proteins by a combination of viral and cellular proteases. Flanking the HCV polyprotein are the highly structured, 5′- and 3′-nontranslated regions (NTRs) important for viral RNA replication and/or protein translation. A frameshift (F, a.k.a., ARF) protein may be expressed from the core-coding region as a result of translation from an alternative reading frame. The junctions between the structural proteins, C, E1 and E2 (shaded in light blue) and those between E2 and p7, p7 and NS2 are cleaved by cellular signal peptidase (filled triangles). An additional processing proximal to the C-terminus of core is mediated by cellular signal peptide peptidase (empty triangle). NS2, along with the N-terminal portion of NS3, constitute the NS2-3 autoprotease that cleaves the NS2-3 junction. The downstream NS proteins are processed by the NS3 serine protease with the aid of the cofactor NS4A. Among the NS proteins (shaded in light yellow), only NS3 to NS5B are required for HCV RNA replication. In addition to its role in viral replication, NS5A contains an IFN sensitivity-determining region (ISDR, spanning codons 2209 to 2248), which has been associated with efficacy of IFN therapy in genotype 1b HCV-infected Japanese patients.
Since the identification of HCV in 1989, research on the virus has been impeded by a lack of efficient cell culture systems and small convenient animal models. In 1997, replication of the first infectious molecular clone of the H77 strain (genotype 1a) of HCV in the chimpanzee model was described (88). The first HCV RNA replicon derived from the Con1 strain (genotype 1b) was established in hepatoma Huh-7 cells in 1999 (105). Complete replication of HCV in cell culture became possible in 2005 (102, 175, 187), owing to isolation of the unique, highly replication-competent JFH-1 strain (genotype 2a) from a Japanese patient with fulminant hepatitis C (80). A workable infectious culture system based on H77 has also been described (183), and minimal amounts of infectious virus have been produced from Con1 RNA in cell culture (128), but both are less robust than JFH-1. With these systems, a better picture of the HCV life cycle is emerging, details of which were reviewed recently (103, 120). Briefly, following attachment, HCV enters host cells by receptor-mediated endocytosis. Upon uncoating, the incoming messenger-sense viral RNA serves as the template for translation of HCV proteins via the IRES in 5′NTR. The NS proteins, along with the genomic RNA template, assemble viral replication complexes in cytoplasmic vesicles that result from extensive membrane re-arrangements induced by NS4B. Acting as the catalytic core of a large macromolecular replicase complex, the NS5B polymerase then directs the transcription of the anti-sense HCV RNA strands, followed by synthesis of nascent sense strands in an asymmetric fashion. As with many positive-stranded RNA viruses (180), HCV RNA replication process generates double-stranded (ds) RNA intermediates (165), which present a major viral pathogen associated molecular pattern (PAMP) that is sensed by various innate immune sensors, known as pattern recognition receptors (PRRs, see details in Section 3). The nascent messenger-sense strand HCV RNAs can serve as templates for translation of more viral protein or synthesis of negative strand RNAs, or can be packaged within new virions that exit the cells through a secretory pathway that appears to usurp aspects of the lipoprotein secretion apparatus of the hepatocyte.
Antigen-specific adaptive T cell immunity plays an important role in determining the outcome of acute HCV infection. The development of early, vigorous and multi-epitope CD4+ and CD8+ T cell responses is crucial for HCV clearance (18, 118, 140). Because adaptive immunity requires weeks and even months to develop, early host control of HCV infection relies on the innate immune response, an evolutionarily ancient, intrinsic set of defenses that host cell use to combat invading pathogens immediately after infection. These target invaders in an antigen-independent, but often pathogen class-specific fashion. Not only does innate immunity buy time for the host to develop adaptive immunity, it is also instrumental in shaping the latter. Apart from the participation of innate immune cells such as natural killer (NK), NKT and dendritic cells (DCs) (40, 137), a major component of the innate immune response to HCV infection is the induction of IFNs and inflammatory cytokines and chemokines. The IFNs act directly against HCV replication in infected cells and protect uninfected neighboring cells from infection by inducing the expression of antiviral IFN-stimulated genes (ISGs). No less important, IFNs activate a variety of immune effector cells important for HCV surveillance and clearance. Chemokines and inflammatory cytokines are central to T cell homing to the liver in HCV infection (176, 185). Taken together, IFNs and chemokines are critical mediators that bridge innate and adaptive immunity, and their induction likely plays an important role in HCV pathogenesis and infection outcome. Recent evidence suggests that HCV infection both induces and disrupts signaling pathways leading to IFN and cytokine expression. In this review, we summarize recent advances in host innate immunity to HCV infection, focusing on the induction and effector mechanisms of the IFN-driven antiviral response as well as the evasion strategies evolved by HCV.
2. Overview of the innate immune responses to HCV
A hallmark of the innate immune responses of mammalian hosts to viral infection is the rapid induction of IFNs and cytokines. IFNs inhibit viral replication in infected cells and establish an antiviral state in uninfected neighboring cells by inducing the expression of ISGs with broad antiviral activities. IFNs also play an important role in activation of various immune effector cells, thereby linking innate and adaptive immunity (16, 148). Pro-inflammatory cytokines and chemokines regulate the maturation of innate and adaptive immune cells and are crucial for their recruitment to the site of infection (16). There are 3 types of IFNs that are defined by the receptors they utilize. The type I IFNs comprise IFN-β and a number of IFN-α subtypes. These can be produced by most cell types in the body and act through an equally broadly-expressed receptor. Type II IFN includes only a single molecule, IFN-γ, and its production is confined to NK cells and activated T cells. Type III IFNs, the most recently identified group of IFNs, include 3 members, IL29 (IFN-λ.1), IL28A (IFN-λ2) and IL28B (IFN-λ3) (16). The IFN-λs can be produced by many but not all cell types, and target a receptor that has primarily an epithelial distribution (that includes hepatocytes).
The IFNs activate downstream signaling pathways, known as Jak-Stat signaling, by binding to and activating their class-specific cell surface receptors (16, 148). The type I IFN receptor is composed of two subunits, IFNAR1 and IFNAR2, which are ubiquitously expressed in all cell types. Engagement of type I IFNs triggers the hetero-dimerization of IFNAR1 and IFNAR2, leading to the activation of intracellular Janus kinases, JAK1 and TYK2. Subsequently, the JAKs phosphorylate the latent transcription factors, Stat1 and Stat2, which then recruit IFN-regulatory factor (IRF) 9 to form a hetero- trimeric complex, IFN stimulated gene factor-3 (ISGF3). ISGF3 moves to the nucleus where it binds to the IFN-stimulated response element (ISRE) present in promoters of ISGs, thereby activating their transcription. Several hundreds of genes are known to be transcriptionally-regulated by type I IFNs, but the biological functions of most of these ISGs remain undefined. The type III IFNs activate the same Jak-Stat pathway as type I IFNs, but do so through an entirely different hetero-dimeric receptor comprised of IL28R1 and IL10R2. IL28R1 has a very limited distribution, and is expressed primarily by epithelial cells as discussed above. For the most part, type I and type III IFNs induce similar sets of ISGs, although the kinetics of these responses may differ.
Signaling induced by type II IFN, IFN-γ, is different from that of type I and III IFNs in many ways. It utilizes a receptor comprised of IFNGR1 and IFNGR2 to activate JAK1 and JAK2 kinases. This triggers the formation of a phosphorylated Stat1 homo-dimer, which binds to a different element in target promoters known as the IFN--γ activated site (GAS). Thus, the ISG profile induced by IFN-γ is different from those induced by type I and III IFNs. It should be noted that human hepatocytes express functional receptors for all 3 IFN types. Consistent with this, all three types of IFNs exhibit antiviral activity against HCV replication in vitro (47, 58, 111, 139, 189).
Microarray analysis has demonstrated that HCV infection is generally associated with induction of a strong ISG response in the liver in vivo (11, 158). In addition, the expression of Intrahepatic chemokines such as the CCR5 ligands, RANTES, MIP-1α and MIP-1β, and the CXCR3 ligand, IP-10, I-Tac and MIG is often elevated in hepatitis C, and the levels of some of these correlate with the outcome of HCV infection or severity of liver inflammation (64, 176, 185). However, the specific cellular sources of IFN(s) and cytokines/chemokines in the HCV-infected liver are not known, and the signaling pathways responsible for their induction are not defined. Apart from replicating in hepatocytes, circumstantial evidence suggests that HCV may have a limited capacity to infect specific immune cell sets. However, cell-culture derived JFH-1 virus does not infect any type of circulating immune cells, including B and T lymphocytes, monocytes, macrophages and DCs (114). At the same time, in vitro data suggest that human hepatocytes have the ability to induce type I and type III IFNs and cytokines/chemokines (73, 96, 98, 178). Relatively little is known, however, how they respond to HCV infection. A recent study showed that infection of primary human fetal liver cells (HFLC) with HCV led to the induction of type III IFNs and ISGs with little evidence of type I IFN expression, raising the possibility that type III IFNs may play an important role in the innate antiviral responses to HCV (113). While this finding would be consistent with the recent discovery in genome-wide association studies that IL28B gene polymorphisms are associated with the outcome of HCV infection and Peg-IFN/RBV therapy, the IL28B SNP genotype did not correlate with levels of IL29, IP-10 or I-Tac expressed in these HCV-infected HFLC cultures (113), nor did it correlate with IL28A/B expression levels in liver biopsies of hepatitis C patients (170). Exactly how the induction of different IFNs, and SNPs near the IL28B gene contribute to innate immune responses to HCV infection in the liver remains elusive. As discussed further below, high levels of ISG expression within the liver correlate generally with poor response to Peg/IFN therapy.
With the exception of IFN-γ for which transcription is regulated differently, viral induction of the type I and III IFNs as well as other cytokines shares overlapping regulatory mechanisms, and is dependent on coordinated activation of the latent cytosolic transcription factors, IRF3 and/or IRF7, and NF- κB(16), following signal transduction from upstream PRRs (see Section 3). Among these, IRF3 and NF-κB are constitutively expressed, while IRF7 is expressed at low levels except in plasmacytoid DCs (pDCs) and requires IFN autocrine/paracrine signaling to increase its expression. Activation of IRF3 and IRF7 depends on specific phosphorylation within their C-terminal domains by the IRF3/7 kinases, Tank binding kinase 1 (TBK1) or IKKε. This results in homo- or hetero-dimerization of IRF3 and/or IRF7 and subsequent nuclear translocation to bind to target promoters. In contrast, activation of NF-κB relies on phosphorylation and subsequent proteasomal degradation of IκB, the repressor unit that normally sequesters NF-κB in the cytoplasm. This is mediated by the classical IKK complex composed of IKKα, IKKβ and IKKγ. In general, IFN-β and IFN-λ1 are transcriptionally controlled by IRF3, while transcription of the IFN-αs and IFN-λ2/3 is more dependent upon IRF7 (122, 124, 149). Induction of the cytokines/chemokines, on the other hand, is due more to NF-κB activation. Very little is known about the activation status of these different transcription factors in different cell types within the HCV-infected liver.
3. PRRs and innate immune pathways sensing HCV infection
Upon infection of host cells, viruses present various types of PAMPs that are recognized as “non-self” by the host. These PAMPs are engaged by different PRRs that trigger distinct signaling pathways leading to production of IFNs and inflammatory cytokines and chemokines. Depending on their subcellular localization, PRRs are classified into two groups, i.e., the cytoplasmic retinoic inducible gene-I (RIG-I)-like receptors (RLRs) and the membrane-bound, usually endosomal, Toll-like receptors (TLRs) (84). Although viral proteins can be sensed by certain host PRRs, viral nucleic acids have distinct features and represent the major PAMP that activates innate antiviral immunity. We discuss here the PRRs and pathways that have been implicated in innate sensing of HCV infection by the host (Fig. 2).
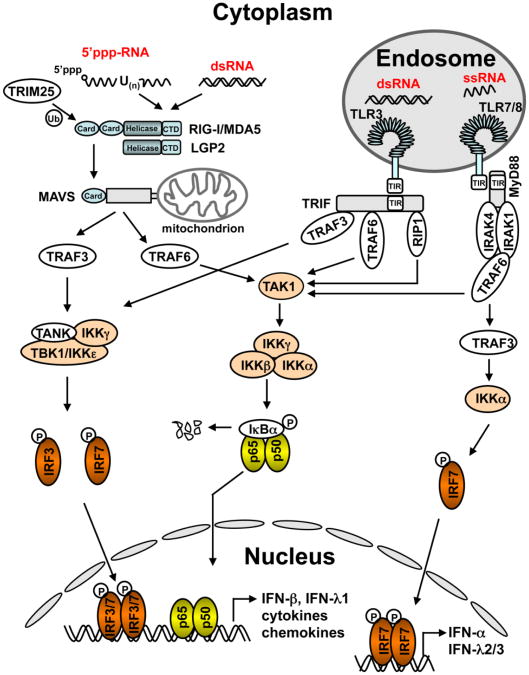
Figure 2. Innate intracellular signaling pathways that sense RNA virus infections and lead to expression of type I and III IFNs, cytokines and chemokines.
Viral dsRNAs and 5′-triphosphate-bearing, poly-U rich viral RNAs are recognized by the RLRs (RIG-I, MDA5 and LGP2) that are constitutively expressed in the cytoplasm and that initiate MAVS-dependent signaling leading to activation of IRF3/7 and NF-κB, and ultimately, synthesis of type I and III IFNs and inflammatory cytokines/chemokines. Viral dsRNAs can also be sensed by TLR3 in late endosomes and trigger a TRIF-dependent pathway activating innate immune responses. Both the RIG-I and TLR3 pathways are capable of sensing HCV infection in hepatocytes. Viral ssRNAs can be sensed by TLR7 or TLR8 in endosomes and activate MyD88-dependent innate immune responses. TLR7 operates exclusively in pDCs and mainly results in IRF7 activation and subsequent induction of IFN-α and IFN-λs, while TLR8 operates in mDCs and mainly leads to expression of NF-κB-dependent cytokines. pDCs use the TLR7 pathway to sense co-cultured HCV-infected hepatocytes, resulting in IFN-α production.
3.1. RLRs
The RLRs are a group of three cytosolic RNA helicases: RIG-I (a.k.a., DDX58), melanoma differentiation associated gene 5 (MDA5, a.k.a., IFIH1 or helicard), and laboratory of genetics and physiology 2 (LGP2, a.k.a., DHX58). They share a similar structure composition, containing tandem caspase activation and recruitment (CARD) domains at the N-terminus (except LGP2) that activate downstream signaling, a central helicase domain, and a C-terminal domain (CTD) that plays a regulatory role (77). The RLRs are present in an auto-repressed state with little activity in resting cells. During viral infection, the CTD of RIG-I/MDA5 binds to its preferred ligand, resulting in a conformational change that exposes the N-terminal CARDs, which can then interact with the CARD domain of mitochondrial antiviral signaling protein (MAVS, a.k.a, Cardif, IPS-1 and VISA), the adaptor for RLRs. Interestingly, MAVS was found to localize on both peroxisomes and mitochondrial outer membrane (34, 150), which are platforms for downstream signaling for early and late ISG expression, respectively. The peroxisome MAVS induces rapid, albeit temporary induction of a small subset of ISGs through IRF1 and IRF3, independent of IFN induction, while mitochondrial MAVS associates with TRAF3, leading to the activation of TBK1 or IKKε, kinases that phosphorylate and activate IRF3/IRF7, resulting in a somewhat later, full-blown IFN response. In addition, MAVS activates the classical IKK complex leading to NF-κB activation, possibly with the participation of FADD and caspase 8/10 (84). Other evidence suggests that the endoribonuclease, RNase L (see also in section 4.1), cleaves cellular and viral RNAs and amplifies RLR signaling (110).
The RLR pathway functions in both somatic cells as well cells of the immune system, except for pDCs in which the TLR7 pathway is dominant for sensing viral RNAs (see Section 3.3). The RLRs are basally expressed at low levels; however they are strongly induced by IFN stimulation or virus infection, providing a positive feedback mechanism of innate immune signaling.
The PAMP for RIG-I has been defined as short, blunt-ended dsRNAs (<300 bp) or single-stranded (ss) RNAs bearing 5′ triphosphates and rich in polyuridine runs (77, 107). Gene knockout studies in mice have shown that RIG-I is essential for innate antiviral responses to negative-strand RNA viruses, such as influenza and vesicular stomatitis virus, Sendai virus, as well as flaviviruses, such as Japanese encephalititis virus, dengue virus and West Nile virus (WNV)(46, 79, 106). The latter two viruses are also known to be sensed by MDA5 (see below). HCV RNA is also sensed by RIG-I in Huh-7 cells and in the liver of mice transfected with synthetic RNA by hydrodynamic injection (141). One HCV PAMP for RIG-I was mapped to the poly-U/UC track RNA in 3′NTR, along with its 5′-end triphosphate moiety (141, 172). Deletion of this sequence motif from HCV RNA diminished its ability to activate RIG-I dependent IFN response when transfected into mice (141). Huh-7.5 cells, which are derived from Huh-7 cells and highly permissive for HCV replication, are defective for RIG-I signaling due to a lethal mutation in the RIG-I CARD. Although other cellular factors are likely to contribute to the high permissiveness of Huh-7.5 cells, reconstitution of RIG-I rendered Huh-7.5 cells relatively non-permissive for HCV replication (159). Thus, there are several lines of evidence supporting a role for RIG-I in sensing and controlling HCV infection.
MDA5 preferentially recognizes long dsRNAs generated during infection with picornaviruses such as EMCV, mengovirus and Theiler's virus, and high molecular mass synthetic poly-I:C, a dsRNA surrogate (78, 79). Whether MDA5 serves as a PRR for HCV infection is unclear, although two flaviviruses closely related to HCV, dengue virus and West Nile virus, activate MDA5 in addition to RIG-I (46, 79, 106). Of note, both RIG-I and MDA5 were identified in a recent ISG screen for antiviral effectors restricting HCV replication in Huh-7.5 cells (145). Because transient expression of MDA5 by itself induces greater IFN promoter activation than RIG-I (184), it is possible that the anti-HCV phenotype may have been due to the IFN-inducing effect of MDA5 over-expression per se, independent of MDA5 recognition of HCV dsRNA. Although there are differences in the recognition of viral RNAs by RIG-I and MDA5, it is clear that there is considerable overlap.
Relatively little is known about the function of the third RLR, LGP2, in signaling. Studies in knockout mice suggest that LGP2 plays a regulatory role for RIG-I/MDA5 recognition of certain RNA viruses such as EMCV (144, 173). Because LGP2 is dispensable for cytokine induction following stimulation of synthetic RNA ligands, it is postulated that LGP2 may help strip off proteins from viral ribonucleoprotein complexes, allowing access of viral RNAs to RIG-I/MDA5 (84). No information is available as to whether LGP2 contributes to RLR sensing of HCV infection.
3.2. TLR3
Toll-like receptor (TLR)-3 was the first identified TLR that recognizes viral nucleic acids (3). TLR3 is expressed in myeloid DCs (mDCs), macrophages, fibroblasts, epithelial cells, hepatocytes and cells in the central nervous system and (96, 115, 178). A type I transmembrane protein, TLR3 is comprised of an amino-terminal horse shoe-shaped ectodomain that contains leucine rich repeats recognizing dsRNA, a transmembrane region, and a cytosolic carboxy-terminal Toll-IL1 receptor homology (TIR) domain that activates downstream signaling (84). TLR3 senses genomes of dsRNA viruses, dsRNAs derived from replication intermediates of positive-strand RNA viruses, and overlapping bidirectional transcripts of DNA viruses, as well as poly-I:C. A minimal length of 45 bp is required for dsRNA to bind to a dimeric unit of two TLR3 ectodomains to trigger downstream signaling (17). In contrast to the cytoplasmic RLRs, TLR3 engages dsRNA in late endosomes and/or lysosomes, where it localizes in most cell types (except fibroblasts in which TLR3 can sense dsRNA on the cell surface). Consistent with this, TLR3 signaling is abrogated by inhibitors of vacuolar acidification (48), such as bafilomycin A1 and chloroquine.
Among the 10 (12 in mice) known human TLRs, TLR3 is only TLR that does not use the myeloid differentiation primary response gene 88 (MyD88) adaptor. Instead, it recruits the TIR-domain containing adaptor inducing IFN-β (TRIF, a.k.a., TICAM1). Upon ligation of dsRNAs, TLR3 associates with TRIF through homotypic TIR-TIR interactions. TRIF interacts with TRAF3 and TBK1 through its N-terminal motifs, leading to IRF3 activation. TRIF can also recruit TRAF6 and RIP1 through N- and C-terminal motifs, respectively, leading to activation of the IKK complex and NF-κB (82). TLR3 has been shown to mediate innate immunity to a number of positive-strand RNA viruses, such as WNV (30), EMCV (60), poliovirus (123), and HCV (178), in addition to some DNA viruses (84).
In the liver, TLR3 is expressed in stellate cells, Kupffer cells (liver-resident macrophages), mDCs and billiary endothelial cells (147). TLR3 is also expressed at low levels in normal human hepatocytes (178). When stimulated with extracellular poly-I:C, primary human hepatocytes (PHH) strongly induce expression of ISGs, including TLR3 itself and numerous pro-inflammatory cytokines (73, 98, 178). As is the case in mDCs, TLR3 signaling in hepatocytes also depends on acidification of the late endosome/lysosome compartment (98). However, TLR3 is not expressed by Huh-7 hepatoma cells, which are typically used to propagate HCV in vitro (96). The mechanism responsible for this defect is unclear, but it may be related to mutation of p53, which regulates TLR3 expression (167). p53 is mutated in Huh-7 cells and many other hepatoma cell lines.
A role for TLR3 in mediating innate immunity to HCV infection has been demonstrated in TLR3-reconstituted Huh-7 and Huh-7.5 cells (98, 178). When TLR3 signaling is restored, the ability of these hepatoma cells to support HCV replication was substantially reduced. This was associated with activation of IRF3 and induction of ISGs in infected cells. Remarkably, point mutations in TLR3 that disrupt the dsRNA binding ability of TLR3 ablate the antiviral effect, confirming that TLR3 sensing of HCV dsRNA, and not TLR3 over-expression, is responsible for the inhibition of HCV replication. Interestingly, when infected at high multiplicity, HCV was able to overwhelm the TLR3-mediated antiviral effect, consistent with the ability of the major HCV protease, NS3/4A, to inhibit signaling through this pathway (see Section 5.2.1). In agreement with the fact that TLR3 signaling activates both IRF3 and NF-κB, reconstitution of TLR3 signaling in Huh-7 cells also conferred the ability to activate NF-κB and induce pro-inflammatory cytokines, such as RANTES, MIP-1α, MIP-1β and IP-10 etc. These cytokines are often strongly induced in HCV-infected liver, possibly due to TLR3 signaling.
The activation of TLR3 by HCV infection requires viral replication that produces dsRNA duplexes of ≥ ∼80-100bp. However, in contrast to RIG-I, activation of TLR3 is not dependent on the nucleotide composition or genome position from which HCV dsRNAs derive (98). The highly structured 3′NTR RNA, which contains a poly-U/UC track that is highly potent for activating RIG-I (141, 172), was unable to trigger TLR3 signaling unless annealed with its negative-sense, complementary strand to form dsRNA duplexes. This suggests that structured regions (such as the stem-loops) in either strand of HCV RNA do not present the extent of dsRNA structure required for TLR3 signaling or do not form stable complexes with TLR3 (98). Precisely how HCV dsRNAs generated during viral replication engage and activate TLR3 in the late endosome/lysosomes is unknown and warrants further investigation.
3.3. TLR7/8
Both TLR7 and TLR8 recognize single-stranded (ss) RNAs derived from ssRNA viruses and synthetic imidazoquinoline antiviral compounds such as R848. While not an exclusive requirement, TLR7 and TLR8 prefers GU-rich and AU-rich RNA sequences, respectively (84). Similar to TLR3, TLR7 and TLR8 reside in late endosomal compartments and require endosomal acidification for signaling.
TLR8 is expressed in mDCs and monocytes, and signals mainly to induce NF-κB-dependent inflammatory cytokines such as TNF-α, IL-6 and IL-10. The TLR8 signaling pathway depends on recruitment of the MyD88 adaptor, TRAF6, and activation of the classical IKK complex (83, 104). Little is known about how the TLR8 pathway functions in HCV infection.
TLR7 is expressed mainly, if not exclusively, in pDCs. These cells sense viral RNAs upon viral entry/uptake or delivery of the RNA to lysosomes from the cytoplasm through autophagy (as shown with viruses replicating in pDCs such as VSV). Activated pDCs initiate signaling cascades leading to rapid production of type I IFNs (83), at levels 100-1000 times higher than that produced by any other blood cell type (104). This process involves the recruitment of MyD88, TRAF6, TRAF3, and the kinases IRAK1 and IKKα, which phosphorylate and activate IRF7. Unlike other immune and non-immune cell types, pDCs constitutively express IRF-7, explaining why they have the unique capacity to rapidly produce extraordinarily high levels of type I IFNs upon sensing viral RNAs through the TLR7 pathway, without the need for IFN-mediated autocrine/paracrine signaling (83, 104).
TLR7 agonists are effective in reducing viremia in hepatitis C patients, and this correlates with the induction of endogenous IFN production (68). This supports the relevance of TLR7 signaling to host defenses against HCV. Because pDCs are “professional” IFN producing cells that utilize the TLR7 pathway (104) and infiltrate HCV-infected liver (91), the role of TLR7 in sensing HCV infection has been studied mainly in pDCs. When transfected with HCV RNA, pDCs produce significant amounts of IFN-α (162, 186). More physiologically relevant, freshly isolated pDCs placed in co-culture with Huh-7.5 cells infected with JFH-1 virus or containing replicating RNA replicons (i.e., the stimulator cells) demonstrate robust production of type I IFNs. This response is dependent on HCV RNA replication in the stimulator cells, close proximity of the stimulator cells to pDCs, and signaling through TLR7. The pDCs are not actively infected by HCV, and the mechanism underlying the transfer of HCV RNA from stimulator cells to pDCs remains to be elucidated (162). A puzzling fact is that HepG2, HEK293, HeLa or mouse hepatoma cells harboring HCV replicons failed to activate co-cultured pDCs to produce IFN when similarly stimulated. Whether the TLR7 pathway functions in human hepatocytes to induce antiviral responses to HCV infection is unclear. TLR7 mRNA is expressed at low level in hepatocytes (92). The stimulation of Huh-7 replicon-bearing cells by a potent TLR7 ligand, SM360320, moderately reduced HCV replication without inducing type I IFNs (92). However, when PHH cultures were treated with R848, a well-characterized TLR7/8 ligand, there was no IFN or ISG induction (178).
3.4. PKR
The dsRNA-activated Protein kinase R (PKR, a.k.a., EIF2AK2) is a serine/threonine kinase that regulates protein synthesis, cell proliferation, apoptosis, and signal transduction. PKR is comprised of tandem N-terminal dsRNA binding motifs (DRBM) and a C-terminal catalytic domain. dsRNAs longer than 30-bp or short imperfect stem-loop RNAs bearing 5′-triphosphates can bind to the DRBM of PKR, triggering PKR dimerization and auto-phosphorylation. This converts PKR into an active kinase capable of phosphorylating protein substrates such as the translation initiation factor eIF2α, which results in shut-off of protein synthesis (61, 129). PKR also interacts with various TRAFs and activates NF-κB, although the mechanism of this remains poorly characterized (129). PKR was the first PRR shown to recognize dsRNA. However, the original proposal that PKR serves as a PRR mediating IFN induction to viruses has been largely dismissed since the discovery of the RLRs. Nonetheless, it is puzzling that PKR is essential for IFN production to certain but not all viral infections (7, 21, 55). This may be partially explained by a recent study demonstrating that PKR contributes to responses against RNA viruses that are primarily sensed by MDA5 by regulating the stability of mRNAs for IFN-β and IFN-α. In PKR deficient cells infected with EMCV, TMEV and SFV, the induction of type I IFN mRNAs was normal, but no IFN-β/α proteins were made because the IFN mRNAs lacked poly(A) tails (146). PKR is phosphorylated in Huh-7 cells upon transfection of JFH-1 genomic RNA, but not a replication-defective counterpart (76), and upon infection with JFH-1 virus (5, 53). This suggests that dsRNAs generated during HCV replication activate PKR. In HCV-infected Huh-7 cells, PKR was found to bind to HCV RNA (although it was unclear whether PKR acted as a PRR or an adaptor) early after infection, and to recruit MAVS and TRAF3 prior to RIG-I, resulting in activation of IRF3 and the induction of a small subset of ISGs (5). Paradoxically, this early activation of PKR-dependent host responses seemed to benefit the virus rather than the host, as it delayed activation of RIG-I signaling (see details in section 5.2.1). Overall, the role played by PKR in innate antiviral response to HCV is uncertain. Depending on experimental conditions, PKR has been found to act with either antiviral or proviral effect (see Section 4.4).
3.5. TLR2
TLR2 is expressed on plasma membrane of monocytes and some other cell types. It acts in concert with TLR1 or TLR6 in recognizing bacterial and fungal cell wall components as well as certain viral proteins. Upon engagement, TLR2 recruits MyD88 through a bridging adaptor, MyD88 adaptor-like (MAL, a.k.a., TIRAP), and relays signals to IRAK kinases and TRAF6, leading to activation of the IKK complex and NF-κB and production of inflammatory cytokines (83). TLR2 was shown to mediate a response to recombinant HCV core and NS3 proteins by human monocytes, resulting in activation of NF-κB, MAP kinase and production of IL-10 and TNFα. This innate recognition mechanism, however, contributes to inflammatory responses and impairs DC differentiation rather than facilitates anti-HCV immunity (35, 36). Of note, the TLR2 pathway has been shown to be functional in inducing TNFα expression following stimulation by synthetic TLR2 ligands in various hepatocyte cell lines (132). Hepatic expression of TLR2 and TNF-α correlates with hepatic inflammatory activity in hepatitis C patients, as does TLR2 expression in peripheral blood monocytes (9, 138).
4. ISGs that demonstrate antiviral activity against HCV
More than 300 ISGs are transcriptionally regulated by IFNs through the Jak-Stat signaling pathway, but only a small fraction of them have been experimentally examined for antiviral actions (16). Among these, some have demonstrated antiviral activity against HCV in vitro and are discussed below. In addition to these ISG proteins, a small number of microRNAs (miRNAs) can be also induced by IFN and may be capable of targeting the HCV genome and inhibiting HCV replication (127). Overall, how the effector functions of the innate immune system control HCV infection represents a robust area of current investigation. While there are a number of examples described below in which results from different studies have been contradictory or inconsistent, this is likely to be due to differences in the specific cells studied or perhaps levels of expression of HCV proteins that counteract these mechanisms (see Section 5). There is also a high degree of redundancy in innate immune control mechanisms targeting HCV.
4.1. The OAS-RNase L system
Transcriptionally upregulated by IFNs, the 2′,5′-oligoadenylate synthetases (OASs) catalyze the production of 3-6-base-long, 2′-to-5′ linked oligoadenylates (2-5A) from ATP. During viral infections, viral dsRNA activates one of the 3 functional human OASs (OAS1-3) to synthesize 2-5A, which in turn, binds to and activates the latent RNase L enzyme. Upon dimerization, RNase L becomes a potent endoribonuclease that cleaves single-stranded regions of RNA substrates at UA and UU dinucleotides. In addition to targeting viral RNAs, RNase L also processes and degrades cellular mRNAs and rRNAs, halting protein synthesis and inducing apoptosis of infected cells (153). Evidence suggesting the OAS-RNAse L system acts as an IFN-induced antiviral effector against HCV came from a study conducted by Han and Barton (59), who demonstrated that HCV RNA was detected and destroyed by RNase L in cytoplasmic extracts of HeLa cells. Interestingly, HCV RNAs from the more IFN-resistant genotypes, 1a and 1b, were found to have fewer UA and UU dinucleotides than those from relatively IFN-sensitive genotypes (genotype 2 and 3), offering a potential explanation for the different sensitivity of HCV genotypes to IFN therapy seen in clinics.
4.2. ISG20
The IFN-stimulated gene 20 kDa protein (ISG20) has recently emerged as a second IFN-regulated RNase that suppresses RNA virus replication (33). Classified within the DEDDh exonuclease family, ISG20 acts as a 3′-5′ exonuclease that has a preference for ssRNA over ssDNA (121). Ectopically expressed ISG20 strongly inhibited replication of subgenomic, genotype 1b HCV RNA replicons in HEK293 cells (71) and propagation of cell culture-derived genotype 2a JFH-1 virus in hepatoma cells (188). Although the underlying mechanism remains poorly understood, ISG20 acts via its exonuclease activity to exert an anti-HCV effect (71, 188). A catalytically inactive mutant of ISG20 exerted a dominant negative (DN) effect on the antiviral activity of IFN-α against subgenomic JFH-1 replicons, suggesting ISG20 is a significant contributor to the overall antiviral effects of IFN (188).
4.3. RSAD2
Accumulating evidence suggests the radical S-adenosyl methionine (SAM) domain containing 2 (RSAD2, a.k.a. viperin or cig5) is a potent antiviral factor against a broad range of RNA viruses from distinct families. Initially identified as a cytomegalovirus-induced protein, RSAD2 is now known to be strongly induced by dsRNA and lipopolysaccharides, as well as type I IFN and, to a lesser extent, by IFN-γ (116). Highly conserved from vertebrates to mammals, RSAD2 has an N-terminal amphipathic helix domain that directs RSAD2 to the ER and also lipid droplets (65), a central region containing 4 conserved SAM motifs whose function is ill-defined, and a C-terminal portion important for RSAD2 dimerization (116). When overexpressed, RSAD2 potently inhibited HCV replication in HCV-N replicon-bearing Huh-7 (63) and HEK293 cells (71), and in JFH-1-infected Huh-7 cells (62, 179). The mechanism by which RSAD2 inhibits HCV replication is poorly understood. RSAD2 associates with the vesicle-associated membrane protein-associated protein A (VAP-A) (62, 179), and disrupts the interaction of the latter with NS5A (179), which is required for assembly of the HCV replication complex on lipid membranes (52). However, Jiang et al. (71) showed that the SAM domain was important for the anti-HCV activity of RSAD2 in HEK293 cells, while Wang et al. (179) and Helbig et al. (62) found the C-terminal region and not the SAM domain to be important in HCV-infected Huh-7 cells. Helbig et al. (62) also found the N-terminal amphipathic helix to be indispensable. In addition, Helbig et al. (62) found that RSAD2 interacted with both NS5A and VAP-A by FRET analysis, while Wang et al. (179) reported that RASD2 associated with VAP-A but not NS5A by co-immunoprecipitation. These inconsistencies could be attributed to differences in experimental approaches and/or cell types.
4.4. PKR
In addition to its possible dsRNA sensor functions discussed above, PKR may act as an effector of the innate immune response. However, studies examining the impact of PKR on the HCV life cycle have provided mixed results. Direct evidence for an antiviral role in HCV replication came from a study by Chang et al. (23) who demonstrated that subgenomic JFH-1 strain RNAs replicated more efficiently in PKR−/− knockout MEFs than in wild-type (WT) MEFs. Subsequently, overexpression of PKR was shown to inhibit HCV 1b replicons in HEK293 (71) and Huh-7 cells (70) and to suppress HCV antigen expression in JFH-1-infected Huh-7 cells (76). In Huh-7 cells with stable knockdown of PKR, transfection of genomic JFH-1 RNA yielded more infectious progeny viruses than in control Huh-7 cells (22, 76). PKR depletion also up-regulated HCV RNA abundance in JFH-1 replicon-bearing WT MEFs (23). Precisely how PKR exerts its antiviral function is unclear, but the kinase activity of PKR appears to be important (71, 76). It is likely to involve phosphorylation of eIF2α, which results in a global suppression of protein synthesis. HCV replication was inhibited by over-expression of a phospho-mimetic eIF2α mutant (76), while expression of PKR was associated with modest retardation in cell growth (71). However, translation initiation mediated by the HCV IRES is relatively insensitive to eIF2α phosphorylation (6, 152). Thus, it is possible that PKR inhibits HCV replication in part by limiting the expression of proviral cellular factors, the translation of which are eIF2α-dependent. In contrast, others have reported that transient knockdown of PKR has no effect on HCV RNA replication or progeny virus production in Huh-7 cells (53), or is actually detrimental to replication and virus production in Huh-7.5 cells (135). Thus, while PKR may help to control of HCV infection, its overall role in HCV pathogenesis is uncertain. As discussed below in Section 5.2.3, PKR activation may actually be counterproductive from the host perspective, as it may limit the synthesis of ISG effector proteins (53).
4.5. ADAR1
Adenosine deaminase acting on RNA (ADAR) 1 catalyzes the adenosine (A) to inosine (I) editing of viral and cellular RNAs possessing double-stranded structures. Because I is recognized as G by ribosomes and RNA polymerases, RNA editing by ADAR1 results in nucleotide substitutions that could alter mRNA translation, RNA structure and/or stability, and RNA-protein interactions (54). All of these can ultimately impact viral replication. IFN-α promotes ADAR1 editing of HCV RNAs in genotype 1b replicon-bearing Huh-7 cells, while HCV RNA replication is enhanced following siRNA knockdown of ADAR1 or inhibition of ADAR1 function by adenovirus associated VA RNAI (168).
4.6. IRF1 and IRF7
Kanazawa et al. (75) found that IRF1 basal expression was lower in Huh-7 cells selected to support the replication of an HCV-N replicon than in parental Huh-7 cells. Upon reconstitution of IRF1 expression, the highly permissive phenotype of IFN-cured replicon cells was reversed. Consistent with this, over-expression of IRF1 strongly inhibited HCV replication in Huh-7 replicon cells as well as in JFH-1-infected Huh-7.5.1 cells (69, 70). A role for IRF7 in limiting HCV replication has also been suggested. Aly et al. (4) reported that inhibition of IRF7 function by over-expression of DN-IRF7 or by siRNA-mediated depletion of IRF7 enhanced the permissiveness of HPV18/E6E7-immortalized human hepatocytes for infection by JFH-1 virus or by serum-derived HCV isolates (genotype 2b and 4a). The antiviral effect of IRF1 and IRF7 against HCV infection was confirmed in a recent ISG screening study (145). Given the known role of IRF1 and IRF7 as transcription factors controlling IFN and ISG expression (163), it is not a complete surprise that both would have antiviral activity against HCV. Although direct effects on HCV cannot be ruled out, these IRFs likely execute their antiviral function through inducing IFNs and/or other antiviral ISGs. This notion is supported by the fact that IRF1 and IRF7 were found to be more potent in inhibiting HCV replication than other modestly inhibitory ISGs (69, 145).
4.7. IFIT1
The IFN-induced protein with tetratricopeptide repeats 1 (IFIT1, a.k.a. p56, ISG56) binds to the p48 subunit of the eIF3 complex and inhibits its function in initiation of protein synthesis (57). Wang et al. (177) showed that IFIT1 associated with eIF3 in the active HCV RNA translation initiation complex, thereby suppressing HCV IRES-mediated viral RNA translation. Raychoudhuri et al. (136) found overexpression of IFIT1 inhibited the replication of con1 and JFH-1 subgenomic HCV RNA replicons in Huh-7 cells, and reduced HCV RNA levels in Huh-7 and immortalized human hepatocytes (IHHs) infected with JFH-1 virus. However, others reported that ectopic expression of IFIT1 did not influence colony formation efficiency in HEK293 cells transfected with a selectable, subgenomic HCV-N replicon that has been adapted in HeLa cells (71). Since the HCV IRES sequence is highly conserved among different genotypes, it is possible that this discrepancy reflects a cell-type specific effect of IFIT1 on IRES function.
4.8. IFITM1
Raychoudhuri et al. (136) reported that transient expression of the IFN induced transmembrane protein 1 (IFITM1, a.k.a., 9-27) suppressed HCV RNA accumulation in Huh-7 cells bearing subgenomic con1 and JFH-1 replicons. IFITM1 over-expression also reduced HCV RNA levels in Huh-7 and IHH cells infected with JFH-1 virus, while knockdown of IFITM1 had the opposite effect . In contrast, Jiang et al. (71) did not observe an effect of IFITM1 on replication of HCV-N replicon in HEK293 cells.
4.9. GBP1
Classified within the dynamin superfamily of large GTPases, the guanylate binding protein 1 (GBP1) is a type I and II IFN-inducible GTPase that exhibits antiviral and antibacterial activities (174). When ectopically expressed, GBP1 moderately inhibited HCV replication in Huh-7 cells bearing HCV-N or JFH-1 replicons, and restricted JFH-1 virus replication in Huh-7.5.1 cells. On the other hand, knockdown of GBP1 enhanced HCV replication. The antiviral mechanism of GBP1 is unknown. While the GTPase activity was shown to be essential, GBP1 did not inhibit HCV IRES activity, nor did it activate ISG expression or affect signaling pathways such as NF-κB and AP-1 (69, 70). In contrast to these findings, however, forced expression of GBP1 had no apparent impact on replication of an HCV-N replicon in HEK293 cells (71).
4.10. IFITM3
The antiviral effect of the IFN induced transmembrane protein 3 (IFITM3, a.k.a., 1-8U) against HCV was first reported by Zhu et al. (190) while studying the antiviral effector mechanism of a proinflammatory cytokine, IL1, against the replication of subgenomic HCV-N RNA replicon in Huh-7 cells. These authors showed that IFITM3 was induced by IL1 as well as by IFN-α and that over-expression of IFITM3 reduced HCV RNA abundance in replicon-bearing cells. How IFITM3 exerts its anti-HCV effect remains elusive, but it was recently reported that IFITM3 suppresses both cellular and HCV IRES-dependent translation (182). It should be noted, however, IFITM3 was reported not to inhibit HCV-N replicon in HEK293 cells (71).
4.11. Other ISGs
Other less well-characterized ISGs implicated in restricting HCV replication include the IFN alpha-inducible protein 6 (IFI6, a.k.a, 6-16) and IFN alpha-inducible protein 27 (IFI27, a.k.a, ISG12). Itsui et al. (69, 70) have reported that both proteins, when over-expressed, moderately reduce HCV replication in HCV-N replicon-bearing Huh-7 cells and JFH-1-infected Huh-7.5.1 cells. However, Jiang et al. (71) documented a lack of antiviral effect of IFI27 on HCV-N replicon in HEK293 cells.
5. Evasion and counteraction of innate immunity by HCV
A growing body of evidence suggests that HCV has developed multilayered strategies to not only evade but also actively counteract host innate defenses. These are likely to contribute to the extraordinary ability of HCV to establish persistent infection.
5.1. Stealth
In the blood of hepatitis C patients, HCV virions are associated with host lipids and proteins such as the lipoproteins and circulate as heterogeneous particles, which physiochemically resemble very low density lipoprotein particles (160). This may disguise HCV from recognition by the host immune system including the innate immune cells.
In infected hepatocytes, HCV replicates its RNAs within rearranged cytoplasmic membrane vesicles, usually derived from the ER (41, 56). This highly compartmentalized microenvironment is typical of positive-strand RNA viruses, and may shield HCV replication complexes from attacks by cellular nucleases and proteinases as well as from sensing by the cytoplasmic RLRs. In addition, various groups have reported that HCV induces autophagy, an evolutionally conserved, ancient process by which cells recycle cytoplasmic constituents through the lysosomal pathway under stress conditions, to facilitate its own life cycle (2, 37, 154, 164). Importantly, the induction of autophagy may help HCV evade RIG-I-mediated innate antiviral immunity, as knockdown of autophagy components enhanced induction of IFN-β and ISGs in Huh-7 cells following stimulation by HCV 3′UTR RNA, a PAMP sensed by RIG-I (85). Certain autophagy-related proteins, such as Atg5-Atg12 conjugates, can associate with the CARDs of RIG-I and MAVS and interfere with RIG-I signaling (74). Autophagosome formation may also sequester viral PAMPs from recognition by cytoplasmic RLRs. Supporting the latter notion, HCV RNAs have been found to be concealed in autophagosome-like structures - double membrane vesicles decorated with LC3-II, a specific marker of autophagic membranes, in Huh-7.5 cells replicating subgenomic JFH-1 replicons (43).
HCV is known to hijack miR-122, a liver specific miRNA, to physically stabilize and facilitate replication of its genome (72, 151). This involves binding of miR-122 to the HCV 5′NTR at two sites near the 5′ end of the genome in association with Ago2. This appears to protect HCV RNAs from cellular 5′ exonucleases (151), but it could also limit recognition of the 5′ triphosphate of the genome by RIG-I. However, this mechanism of evasion, if proven, would only impair cellular sensing of positive-strand HCV RNAs, as any 5′ triphosphate present in the negative-strand RNA would also be recognized by RIG-I.
Because the NS5B RNA-dependent RNA polymerase lacks proofreading activity, HCV replication is error-prone. Thus, under the continuous selection pressure imposed by the host immune system, HCV exists as a heterogeneous population comprising genetically related viral variants known as “quasispecies” (112). Not only does this provide a mechanism by which HCV accumulates mutations allowing escape from cytotoxic T-cells and neutralizing antibodies, but it also may aid HCV RNA to evade OAS-RNase L-mediated antiviral responses. Notably, in hepatitis C patients treated with IFN, HCV RNAs were found to accumulate silent mutations over time preferentially at UA and UU dinucleotides, the cleavage sites for RNase L (59).
5.2. Counteraction
5.2.1. Inhibition of the induction of IFNs
HCV has been reported to activate a PKR-MAVS-TRAF3-IRF3 pathway early after infection in Huh-7.25.CD81 cells and prior to its recognition by RIG-I, leading to upregulation of a small subset of ISGs (5). One of the most highly induced genes was ISG15, which negatively affected RIG-I ubiquitylation by TRIM25, a prerequisite for interaction of RIG-I and its adaptor MAVS (49). Silencing of ISG15 expression restored RIG-I ubiquitylation and early IFN-β induction, and correlated with a reduction in HCV replication (5). This study thus suggests that HCV may subvert a PKR-mediated immediate host response to inhibit innate immune signaling at the PRR level.
HCV is known to interfere with activation of IFN response via both MAVS- and TRIF-dependent pathways (20, 73, 97, 125). The NS3/4A protease of HCV has been shown to target both MAVS and TRIF for proteolysis. The evidence for HCV hydrolysis of MAVS is especially strong. NS3/4A cleaves MAVS at Cys508 immediately before the C-terminal transmembrane domain, dislodging MAVS from the mitochondrial outer membrane and/or mitochondrial associated membrane, the platform for RLR signaling (67, 99, 108, 119). Remarkably, MAVS cleavage was also observed in the liver of hepatitis C patients, although the reported frequency varied, and how MAVS cleavage relates to the reduced activation of endogenous IFN remains debated (8, 108, 157). The cleavage of MAVS likely contributes to HCV pathogenesis, but is unlikely to be the proximate cause of viral persistence (93). Two other hepatotropic positive-strand RNA viruses, GB virus B and hepatitis A virus (a picornavirus), also encode a protease or protease precursor that cleaves MAVS (26, 181). Although disruption of MAVS signaling is as efficient as with HCV, both of these viruses typically cause self-limiting infections in vivo.
TRIF is cleaved by purified NS3/4A protease in cell-free reactions at Cys372, a site bearing remarkable sequence homology to the NS4B/5A cleavage site in HCV polyprotein, separating the N-terminal TBK1 and TRAF binding domains from the C-terminal TIR domain (44, 97). TRIF protein abundance was substantially reduced in HeLa cells bearing HCV replicons (97) and in Huh-7.5 cells infected with JFH-1 virus (178), and this could be partially reversed by short term treatment of a specific NS3/4A inhibitor. Consistent with this, HCV infection substantially blunted the induction of ISGs in response to extracellular poly-I:C (178). However, the reduction of TRIF abundance by NS3/4A has not been seen in some other experimental settings (31, 73). It is possible that this reflects differences in TRIF or NS3/4A localization within the cell, as well as technical difficulties in detecting this relatively low abundance protein.
HCV NS5A has been reported to interfere with MyD88-dependent TLR signaling pathways when stably expressed in the Raw264.7 mouse macrophage cell line (1). NS5A from different HCV genotypes bound to MyD88 in co-transfected HEK-293T cells, and this interaction disrupted the recruitment of IRAK1 to MyD88, ultimately inhibiting cytokine production following stimulation by various TLR ligands. Interestingly, this interaction was mapped to a previously recognized “interferon sensitivity-determining region” (ISDR) region in NS5A (42), which associated with the death domain of MyD88. It should be noted, however, this mechanism of immune evasion is operative only if active infection and replication of HCV takes place in DCs and monocytes/macrophages, which remains a subject of debate.
At the kinase level, it has been reported that HCV NS3 interacts directly with TBK1 to inhibit the activation of IFN-β-promoter by TRIF and TBK1 (125). The C-terminal helicase domain of NS3 competed with IRF3 for interacting with TBK1, thereby interfering with IRF3 activation by TBK1. Such an effect would be likely to impact both RIG-I and TLR3 signaling. In addition, Tasaka et al. (166) reported that HCV NS4B inhibited the activation of IFN response by overexpression of RIG-I or MAVS through an undefined mechanism. However, others have found that NS4B does not impair RIG-I signaling (45, 81).
Despite all of the evidence that HCV proteins are able to disrupt the IFN induction pathways in vitro, there is strong evidence that ISG expression is induced in HCV-infected liver in vivo (11, 142, 158). However, this may be explained by the finding that HCV only infects a small proportion of hepatocytes in the liver (100, 157). IFNs may still be induced in newly infected hepatocytes at an early stage of infection prior to abundant HCV proteins being made, or in uninfected pDCs infiltrating the liver that sense HCV-infected hepatocytes. Nonparenchymal liver cells may also sample viral nucleic acids released from infected cells undergoing necrotic death to produce IFNs. Collectively, the IFNs produced could act in a paracrine/autocrine fashion to upregulate ISG expression in the liver. Significantly, high-level endogenous expression of ISG transcripts within the liver correlates with both poor response to treatment with Peg-IFN/RBV and the unfavorable IL28B haplotype (66, 142). However, the mechanism underlying this association is unknown.
5.2.2. Inhibition of IFN signaling downstream of the IFN receptors
A large body of data suggests that HCV also impairs IFN signaling downstream of the IFN receptors via numerous mechanisms (Fig. 3). In transfected cell lines, both core and NS5A appear to interact with Stat1 and impair IFN-induced Stat1 phosphorylation, thereby inhibiting downstream ISG transcription (89, 101). Core was also found to interfere with Stat1 nuclear translocation (14, 117) or binding of ISGF3 to the ISRE in ISG promoters (32). Over-expression of core was associated with the induction of suppressor of cytokine signaling-3 (SOCS3) (14), which targets Janus kinases (Jaks) to inhibit Stat1 phosphorylation.
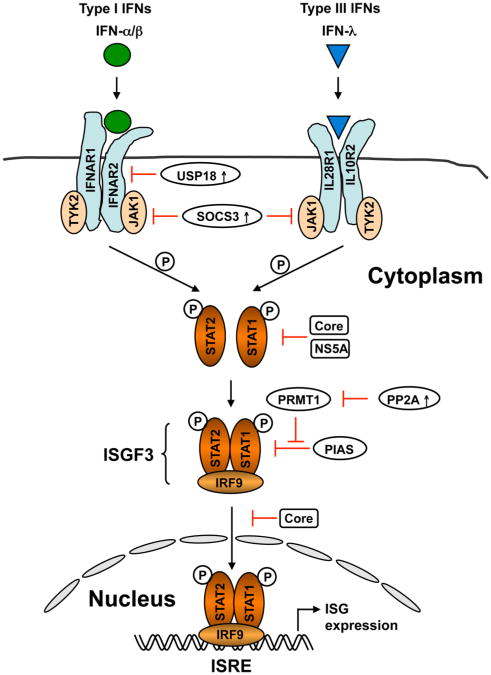
Figure 3. Induction of ISGs by type I and III IFNs through the JAK-STAT signaling pathway and its inhibition by HCV.
Type I and III IFNs bind to distinct, heterodimeric cell surface receptors, but then activate a similar intracellular signaling pathway involving recruitment and activation of the Janus kinases, JAK1 and TYK2, phosphorylation of STAT1 and STAT2, and the formation of the heterotrimeric transcription factor complex, IFN-stimulated gene factor-3 (ISGF3) comprising phosphorylated STAT1 and STAT2 and IRF9. ISGF3 migrates into the nucleus and induces the transcription of hundreds of ISGs by binding to the IFN-stimulated response element (ISRE) in ISG promoters. HCV core and NS5A associate with STAT1, suppressing its phosphorylation and/or nuclear translocation. Core can also induce SOCS3 expression, which negatively regulates JAK. HCV-induced endoplasmic reticulum stress up-regulates expression of protein phosphatase 2A (PP2A), which inhibits the transcriptional activity of STAT1 by suppressing protein arginine methyltransferase 1 (PRMT1), thereby promoting STAT1 hypomethylation and STAT1 association with the protein inhibitor of activated STAT1 (PIAS1). Up-regulation of ubiquitin-like specific protease 18 (USP18) in HCV infection also attenuates JAK-STAT signaling at the receptor level through direct binding to IFNAR2.
IFN-α-induced Jak-Stat signaling was impaired in transgenic mice expressing the HCV polyprotein (12) and in living cells from liver biopsies from chronic hepatitis C patients (39). IFN-induced Stat1 phosphorylation was intact, but the binding of Stat1 to ISG promoters was significantly compromised. Protein phosphatase 2A (PP2A) was up-regulated, presumably as a result of ER stress, and this was associated with hypomethylation of Stat1 on Arg 31, probably due to the inhibition of PP2A on protein arginine methyltransferase 1 (PRMT1). The hypomethylation of Stat1 promotes its association with protein inhibitor of activated STAT1 (PIAS1), a negative regulator of the transcriptional activity of Stat1 (28, 38).
Expression of NS5A has been shown to induce IL8 mRNA and protein, which, in turn, attenuates IFN's antiviral action in HeLa cells (130). Interestingly, not only were serum IL8 levels higher in hepatitis C patients than in healthy control subjects, but IL8 levels were significantly elevated in patients who did not respond to IFN therapy compared with those who did respond to therapy (131).
Despite the reported inhibitory effects of HCV proteins on IFN signaling, genome-length HCV RNA replicons are sensitive to IFN treatment. Replicon-bearing cells can be cured by either type I or II IFNs. However, HCV-infected cells have been shown to be significantly more resistant to IFN than replicon cells (53). Nonetheless, there are data from HCV-infected hepatoma cells that do not support the notion that HCV interferes with Jak-Stat signaling. In Huh-7 cells infected with JFH-1 virus, neither transcription from the ISRE promoter nor induction of ISG mRNAs was impaired following stimulation with IFN (27, 53). Thus, more studies in the context of HCV infection in vivo will be required to reconcile this inconsistency.
There are yet other views on why some hepatitis C patients fail to respond to PegIFN/RBV therapy. In chimpanzees chronically infected HCV, the intrahepatic ISG expression was already maximally induced. As a result, there was no further up-regulation of ISGs when exogenous IFN was administered (90). Consistent with this, it was found that in chronic hepatitis C patients undergoing PegIFN/RBV therapy, a poor response was associated with higher expression levels of ISGs prior to therapy (66, 142). Others reported that pre-activation of the ISGylation/de-ISGylation pathway involving the ubiquitin-like modifier, ISG15 and its deconjugating enzyme, ubiquitin-like specific protease 18 (USP18, a.k.a., Ubp43) in hepatocytes, not only promoted HCV replication but correlated with poor response to IFN therapy in hepatitis C patients (25). An ISG that is significantly elevated in non-responders compared with responders, USP18 is known to dampen IFN signaling to the Jak-Stat pathway via de-ISGylation as well as by disrupting the association between IFNAR2 and JAK1 in a protease-independent fashion (109). In cell culture, siRNA-mediated depletion of USP18 enhanced ISG induction as well as the antiviral effect of IFN-α against HCV replication (133).
5.2.3. Inhibition/subversion of the function of antiviral effector ISGs
Both NS5A and E2 of HCV have been reported to bind to PKR and inhibit its effector function. Gale et al. (50, 51) reported that the NS5A of HCV genotype 1b directly bound to the PKR catalytic domain and inhibited PKR dimerization and phosphorylation of its substrate, eIF2α. The ISDR along with a stretch of 26 amino acids carboxyl to it were responsible for both PKR association and repression. In the case of E2, Taylor and colleagues (169) found that the E2 proteins from genotypes 1a and 1b contain a 12-amino acid sequence homologous to the PKR autophosphorylation site and the phosphorylation site of eIF2α. They demonstrated that E2 bound to PKR mainly through this site and that this inhibited PKR's kinase activity and effect on protein synthesis. A cytosolic, unglycosylated form of E2 was found to mediate the PKR interaction (126). However, the reported effects of NS5A and E2 on PKR have varied depending on HCV isolates and experimental systems used, and have not been consistently demonstrated by different research groups. Attempts at clinical correlations have not reached a consensus (13), and these findings have yet to be validated using the infectious HCV culture systems.
Two recent studies revealed that HCV hijacks PKR function to inhibit host protein synthesis, thereby antagonizing RIG-I-mediated IFN production (6) or IFN-induced antiviral action (53). Both studies demonstrated HCV infection of Huh-7 cells activated PKR auto-phosphorylation and eIF2α phosphorylation, resulting in inhibition of host protein synthesis. These effects were more pronounced in cells also treated with IFN (53). However, HCV replication was not affected because HCV IRES-mediated translation was relatively resistant to eIF2α phosphorylation. Surprisingly, PKR activation suppressed HCV-induced IFN production (6) and blocked ISG protein expression from mRNA (53). Inhibition of PKR function in HCV-infected cells restored IFN induction (6) and IFN-induced ISG protein expression, and enhanced the antiviral effects of IFN (53). Of note, these results are in contrast to earlier studies showing PKR to be an anti-HCV factor in various cellular systems with replicons or transfection of JFH-1 genomic RNA (22, 23, 70, 71, 76).
There is also evidence that genotype 1b HCV NS5A physically interacts with OAS1 via its N-terminal one-third when both proteins are ectopically co-expressed in HeLa cells (161). The NS5A fragment capable of binding to OAS1 was as effective as the full-length NS5A in antagonizing IFN action against EMCV. Whether NS5A expressed from replicating HCV RNA could also associate with endogenous OAS1 was not studied, but a genotype 1b replicon with a point mutation in NS5A that disrupted the NS5A-OAS1 interaction had reduced fitness when transfected into Huh-7 cells.
Finally, HCV NS5B has been reported to bind to the N-terminal GTPase domain of GBP1 via its fingers domain when ectopically expressed in HEK-293T cells. This interaction suppressed the GTPase activity of GBP1 and was able to relieve the moderate inhibitory effect of GBP1 on HCV replication in genotype 1b replicon-bearing Huh-7 cells (69).
6.0 Perspective and Concluding Remarks
Although many lines of evidence suggest that HCV infection disrupts IFN responses at multiple levels as described above, it is clear that ISGs are often prominently expressed in the liver in both acute and chronic infections (10, 142, 158). High-level expression of ISGs within the liver prior to treatment is associated with high serum levels of IP10 and is strongly predictive of a poor therapeutic outcome (142). In contrast, patients who experience rapid virologic responses to PegIFN/RBV therapy have low pre-therapy expression of ISGs within the liver, but demonstrate impressive up-regulation of these genes when treated. This is a mystery that is yet to be resolved. The pattern of ISG expression within PBMCs differs substantially from that in the liver, and does not correlate with treatment outcome. The underlying reason for the liver-specific nature of pre-treatment ISG expression is unknown, but is consistent with it being driven primarily by IFN-λ,or alternatively by IFN-α/β produced locally within the liver. Exactly why this pre-treatment response occurs in some patients (IFN non-responders) and not in others (rapid responders) is not known, nor is it understood why patients that have high endogenous antiviral responses do not spontaneously clear their infection. There are clues that can be deduced from the work described in the preceding sections, most of which was done using in vitro systems, but just as many gaps in our knowledge. Underlying all of this is the fact that we simply do not understand the mechanism responsible for the profound association of IL28B genotype with both infection outcome and response to Peg-IFN/RBV therapy.
Thus, although much has been learned about the functions of various HCV proteins and the mechanism of HCV replication since identification of the virus more than two decades ago, we have only a limited understanding of how the host immune system, and in particular the innate immune system, interacts with HCV. To no small part, this can be attributed to the lack of a convenient, immunocompetent small animal model and efficient cell culture systems possessing uncompromised innate immune systems. Hopefully, this area will advance with the development of mouse models that fully recapitulate the HCV life cycle and the improvement of HCV culture systems employing primary cultures of normal human hepatocytes. New knowledge of the interplay between HCV and innate immune responses would undoubtedly help our understanding of HCV pathogenesis and the development of effective immunotherapy for chronic hepatitis C.
Acknowledgments
The authors gratefully acknowledge Zongdi Feng for expert review of this manuscript. Work in the authors' laboratories was supported by grants from the National Institutes of Health: AI-069285, AI-095690, AI-040035 and DA-018054. The authors apologize to colleagues whose contributions could not be cited because of space constraints.
Footnotes
This article is published as part of the Special Issue on Immunopathology of viral hepatitis [34:4]
References Cited
- 1.Abe T, Kaname Y, Hamamoto I, Tsuda Y, Wen X, Taguwa S, Moriishi K, Takeuchi O, Kawai T, Kanto T, Hayashi N, Akira S, Matsuura Y. Hepatitis C virus nonstructural protein 5A modulates the toll-like receptor-MyD88-dependent signaling pathway in macrophage cell lines. J Virol. 2007;81:8953–8966. doi: 10.1128/JVI.00649-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ait-Goughoulte M, Kanda T, Meyer K, Ryerse JS, Ray RB, Ray R. Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol. 2008;82:2241–2249. doi: 10.1128/JVI.02093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 4.Aly HH, Watashi K, Hijikata M, Kaneko H, Takada Y, Egawa H, Uemoto S, Shimotohno K. Serum-derived hepatitis C virus infectivity in interferon regulatory factor-7-suppressed human primary hepatocytes. J Hepatol. 2007;46:26–36. doi: 10.1016/j.jhep.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 5.Arnaud N, Dabo S, Akazawa D, Fukasawa M, Shinkai-Ouchi F, Hugon J, Wakita T, Meurs EF. Hepatitis C virus reveals a novel early control in acute immune response. PLoS Pathog. 2011;7:e1002289. doi: 10.1371/journal.ppat.1002289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnaud N, Dabo S, Maillard P, Budkowska A, Kalliampakou KI, Mavromara P, Garcin D, Hugon J, Gatignol A, Akazawa D, Wakita T, Meurs EF. Hepatitis C virus controls interferon production through PKR activation. PLoS One. 2010;5:e10575. doi: 10.1371/journal.pone.0010575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barry G, Breakwell L, Fragkoudis R, Attarzadeh-Yazdi G, Rodriguez-Andres J, Kohl A, Fazakerley JK. PKR acts early in infection to suppress Semliki Forest virus production and strongly enhances the type I interferon response. J Gen Virol. 2009;90:1382–1391. doi: 10.1099/vir.0.007336-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellecave P, Sarasin-Filipowicz M, Donze O, Kennel A, Gouttenoire J, Meylan E, Terracciano L, Tschopp J, Sarrazin C, Berg T, Moradpour D, Heim MH. Cleavage of mitochondrial antiviral signaling protein in the liver of patients with chronic hepatitis C correlates with a reduced activation of the endogenous interferon system. Hepatology. 2010;51:1127–1136. doi: 10.1002/hep.23426. [DOI] [PubMed] [Google Scholar]
- 9.Berzsenyi MD, Roberts SK, Preiss S, Woollard DJ, Beard MR, Skinner NA, Bowden DS, Visvanathan K. Hepatic TLR2 & TLR4 expression correlates with hepatic inflammation and TNF-alpha in HCV & HCV/HIV infection. J Viral Hepat. 2011;18:852–860. doi: 10.1111/j.1365-2893.2010.01390.x. [DOI] [PubMed] [Google Scholar]
- 10.Bigger CB, Brasky KM, Lanford RE. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J Virol. 2001;75:7059–7066. doi: 10.1128/JVI.75.15.7059-7066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bigger CB, Brasky KM, Lanford RE. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J Virol. 2001;75:7059–7066. doi: 10.1128/JVI.75.15.7059-7066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blindenbacher A, Duong FH, Hunziker L, Stutvoet ST, Wang X, Terracciano L, Moradpour D, Blum HE, Alonzi T, Tripodi M, La Monica N, Heim MH. Expression of hepatitis c virus proteins inhibits interferon alpha signaling in the liver of transgenic mice. Gastroenterology. 2003;124:1465–1475. doi: 10.1016/s0016-5085(03)00290-7. [DOI] [PubMed] [Google Scholar]
- 13.Bode JG, Brenndorfer ED, Haussinger D. Subversion of innate host antiviral strategies by the hepatitis C virus. Arch Biochem Biophys. 2007;462:254–265. doi: 10.1016/j.abb.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 14.Bode JG, Ludwig S, Ehrhardt C, Albrecht U, Erhardt A, Schaper F, Heinrich PC, Haussinger D. IFN-alpha antagonistic activity of HCV core protein involves induction of suppressor of cytokine signaling-3. Faseb J. 2003;17:488–490. doi: 10.1096/fj.02-0664fje. [DOI] [PubMed] [Google Scholar]
- 15.Bode JG, Ludwig S, Ehrhardt C, Albrecht U, Erhardt A, Schaper F, Heinrich PC, Haussinger D. IFN-alpha antagonistic activity of HCV core protein involves induction of suppressor of cytokine signaling-3. FASEB J. 2003;17:488–490. doi: 10.1096/fj.02-0664fje. [DOI] [PubMed] [Google Scholar]
- 16.Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Botos I, Liu L, Wang Y, Segal DM, Davies DR. The toll-like receptor 3:dsRNA signaling complex. Biochim Biophys Acta. 2009;1789:667–674. doi: 10.1016/j.bbagrm.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowen DG, Walker CM. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature. 2005;436:946–952. doi: 10.1038/nature04079. [DOI] [PubMed] [Google Scholar]
- 19.Branch AD, Stump DD, Gutierrez JA, Eng F, Walewski JL. The hepatitis C virus alternate reading frame (ARF) and its family of novel products: the alternate reading frame protein/F-protein, the double-frameshift protein, and others. Semin Liver Dis. 2005;25:105–117. doi: 10.1055/s-2005-864786. [DOI] [PubMed] [Google Scholar]
- 20.Breiman A, Grandvaux N, Lin R, Ottone C, Akira S, Yoneyama M, Fujita T, Hiscott J, Meurs EF. Inhibition of RIG-I-dependent signaling to the interferon pathway during hepatitis C virus expression and restoration of signaling by IKKepsilon. J Virol. 2005;79:3969–3978. doi: 10.1128/JVI.79.7.3969-3978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carpentier PA, Williams BR, Miller SD. Distinct roles of protein kinase R and toll-like receptor 3 in the activation of astrocytes by viral stimuli. Glia. 2007;55:239–252. doi: 10.1002/glia.20450. [DOI] [PubMed] [Google Scholar]
- 22.Chang JH, Kato N, Muroyama R, Taniguchi H, Guleng B, Dharel N, Shao RX, Tateishi K, Jazag A, Kawabe T, Omata M. Double-stranded RNA-activated protein kinase inhibits hepatitis C virus replication but may be not essential in interferon treatment. Liver Int. 2010;30:311–318. doi: 10.1111/j.1478-3231.2009.02144.x. [DOI] [PubMed] [Google Scholar]
- 23.Chang KS, Cai Z, Zhang C, Sen GC, Williams BR, Luo G. Replication of hepatitis C virus (HCV) RNA in mouse embryonic fibroblasts: protein kinase R (PKR)-dependent and PKR-independent mechanisms for controlling HCV RNA replication and mediating interferon activities. J Virol. 2006;80:7364–7374. doi: 10.1128/JVI.00586-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L, Borozan I, Feld J, Sun J, Tannis LL, Coltescu C, Heathcote J, Edwards AM, McGilvray ID. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology. 2005;128:1437–1444. doi: 10.1053/j.gastro.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 25.Chen L, Li S, McGilvray I. The ISG15/USP18 ubiquitin-like pathway (ISGylation system) in hepatitis C virus infection and resistance to interferon therapy. Int J Biochem Cell Biol. 2011;43:1427–1431. doi: 10.1016/j.biocel.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z, Benureau Y, Rijnbrand R, Yi J, Wang T, Warter L, Lanford RE, Weinman SA, Lemon SM, Martin A, Li K. GB Virus B Disrupts RIG-I Signaling by NS3/4A-Mediated Cleavage of the Adaptor Protein MAVS. J Virol. 2007;81:964–976. doi: 10.1128/JVI.02076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng G, Zhong J, Chisari FV. Inhibition of dsRNA-induced signaling in hepatitis C virus-infected cells by NS3 protease-dependent and -independent mechanisms. Proc Natl Acad Sci U S A. 2006;103:8499–8504. doi: 10.1073/pnas.0602957103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christen V, Treves S, Duong FH, Heim MH. Activation of endoplasmic reticulum stress response by hepatitis viruses up-regulates protein phosphatase 2A. Hepatology. 2007;46:558–565. doi: 10.1002/hep.21611. [DOI] [PubMed] [Google Scholar]
- 29.Coccia EM, Severa M, Giacomini E, Monneron D, Remoli ME, Julkunen I, Cella M, Lande R, Uze G. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur J Immunol. 2004;34:796–805. doi: 10.1002/eji.200324610. [DOI] [PubMed] [Google Scholar]
- 30.Daffis S, Samuel MA, Suthar MS, Gale M, Jr, Diamond MS. Toll-like receptor 3 has a protective role against West Nile virus infection. J Virol. 2008;82:10349–10358. doi: 10.1128/JVI.00935-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dansako H, Ikeda M, Kato N. Limited suppression of the interferon-beta production by hepatitis C virus serine protease in cultured human hepatocytes. Febs J. 2007;274:4161–4176. doi: 10.1111/j.1742-4658.2007.05942.x. [DOI] [PubMed] [Google Scholar]
- 32.de Lucas S, Bartolome J, Carreno V. Hepatitis C virus core protein down-regulates transcription of interferon-induced antiviral genes. J Infect Dis. 2005;191:93–99. doi: 10.1086/426509. [DOI] [PubMed] [Google Scholar]
- 33.Degols G, Eldin P, Mechti N. ISG20, an actor of the innate immune response. Biochimie. 2007;89:831–835. doi: 10.1016/j.biochi.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 34.Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, Nibert ML, Superti-Furga G, Kagan JC. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dolganiuc A, Kodys K, Kopasz A, Marshall C, Do T, Romics L, Jr, Mandrekar P, Zapp M, Szabo G. Hepatitis C virus core and nonstructural protein 3 proteins induce pro- and anti-inflammatory cytokines and inhibit dendritic cell differentiation. J Immunol. 2003;170:5615–5624. doi: 10.4049/jimmunol.170.11.5615. [DOI] [PubMed] [Google Scholar]
- 36.Dolganiuc A, Oak S, Kodys K, Golenbock DT, Finberg RW, Kurt-Jones E, Szabo G. Hepatitis C core and nonstructural 3 proteins trigger toll-like receptor 2-mediated pathways and inflammatory activation. Gastroenterology. 2004;127:1513–1524. doi: 10.1053/j.gastro.2004.08.067. [DOI] [PubMed] [Google Scholar]
- 37.Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duong F, Christen HV, Filipowicz M, Heim MH. S-Adenosylmethionine and betaine correct hepatitis C virus induced inhibition of interferon signaling in vitro. Hepatology. 2006;43:796–806. doi: 10.1002/hep.21116. [DOI] [PubMed] [Google Scholar]
- 39.Duong FH, Filipowicz M, Tripodi M, La Monica N, Heim MH. Hepatitis C virus inhibits interferon signaling through up-regulation of protein phosphatase 2A. Gastroenterology. 2004;126:263–277. doi: 10.1053/j.gastro.2003.10.076. [DOI] [PubMed] [Google Scholar]
- 40.Dustin LB, Rice CM. Flying under the radar: the immunobiology of hepatitis C. Annu Rev Immunol. 2007;25:71–99. doi: 10.1146/annurev.immunol.25.022106.141602. [DOI] [PubMed] [Google Scholar]
- 41.Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N. Interferon sensitivity determining sequence of the hepatitis C virus genome. Hepatology. 1996;24:460–460. doi: 10.1053/jhep.1996.v24.ajhep0240460. [DOI] [PubMed] [Google Scholar]
- 43.Ferraris P, Blanchard E, Roingeard P. Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J Gen Virol. 2010;91:2230–2237. doi: 10.1099/vir.0.022186-0. [DOI] [PubMed] [Google Scholar]
- 44.Ferreon JC, Ferreon AC, Li K, Lemon SM. Molecular determinants of TRIF proteolysis mediated by the hepatitis C virus NS3/4A protease. J Biol Chem. 2005;280:20483–20492. doi: 10.1074/jbc.M500422200. [DOI] [PubMed] [Google Scholar]
- 45.Foy E, Li K, Wang C, Sumpter R, Jr, Ikeda M, Lemon SM, Gale M., Jr Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300:1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- 46.Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale M., Jr Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1. J Virol. 2008;82:609–616. doi: 10.1128/JVI.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frese M, Schwarzle V, Barth K, Barth K, Krieger N, Lohmann V, Mihm S, Haller O, Bartenschlager R. Interferon-gamma inhibits replication of subgenomic and genomic hepatitis C virus RNAs. Hepatology. 2002;35:694–703. doi: 10.1053/jhep.2002.31770. [DOI] [PubMed] [Google Scholar]
- 48.Funami K, Matsumoto M, Oshiumi H, Akazawa T, Yamamoto A, Seya T. The cytoplasmic ‘linker region’ in Toll-like receptor 3 controls receptor localization and signaling. Int Immunol. 2004;16:1143–1154. doi: 10.1093/intimm/dxh115. [DOI] [PubMed] [Google Scholar]
- 49.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 50.Gale M, Jr, Blakely CM, Kwieciszewski B, Tan SL, Dossett M, Tang NM, Korth MJ, Polyak SJ, Gretch DR, Katze MG. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18:5208–5218. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gale MJ, Jr, Korth MJ, Tang NM, Tan SL, Hopkins DA, Dever TE, Polyak SJ, Gretch DR, Katze MG. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;30:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 52.Gao L, Aizaki H, He JW, Lai MM. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J Virol. 2004;78:3480–3488. doi: 10.1128/JVI.78.7.3480-3488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garaigorta U, Chisari FV. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe. 2009;6:513–522. doi: 10.1016/j.chom.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.George CX, Gan Z, Liu Y, Samuel CE. Adenosine deaminases acting on RNA, RNA editing, and interferon action. J Interferon Cytokine Res. 2011;31:99–117. doi: 10.1089/jir.2010.0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gilfoy FD, Mason PW. West Nile virus-induced interferon production is mediated by the double-stranded RNA-dependent protein kinase PKR. J Virol. 2007;81:11148–11158. doi: 10.1128/JVI.00446-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol. 2003;77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo J, Hui DJ, Merrick WC, Sen GC. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 2000;19:6891–6899. doi: 10.1093/emboj/19.24.6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo JT, Bichko VV, Seeger C. Effect of alpha interferon on the hepatitis C virus replicon. J Virol. 2001;75:8516–8523. doi: 10.1128/JVI.75.18.8516-8523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han JQ, Barton DJ. Activation and evasion of the antiviral 2′-5′ oligoadenylate synthetase/ribonuclease L pathway by hepatitis C virus mRNA. RNA. 2002;8:512–525. doi: 10.1017/s1355838202020617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hardarson HS, Baker JS, Yang Z, Purevjav E, Huang CH, Alexopoulou L, Li N, Flavell RA, Bowles NE, Vallejo JG. Toll-like receptor 3 is an essential component of the innate stress response in virus-induced cardiac injury. Am J Physiol Heart Circ Physiol. 2007;292:H251–258. doi: 10.1152/ajpheart.00398.2006. [DOI] [PubMed] [Google Scholar]
- 61.He B. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 2006;13:393–403. doi: 10.1038/sj.cdd.4401833. [DOI] [PubMed] [Google Scholar]
- 62.Helbig KJ, Eyre NS, Yip E, Narayana S, Li K, Fiches G, McCartney EM, Jangra RK, Lemon SM, Beard MR. The antiviral protein viperin inhibits hepatitis C virus replication via interaction with nonstructural protein 5A. Hepatology. 2011;54:1506–1517. doi: 10.1002/hep.24542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Helbig KJ, Lau DT, Semendric L, Harley HA, Beard MR. Analysis of ISG expression in chronic hepatitis C identifies viperin as a potential antiviral effector. Hepatology. 2005;42:702–710. doi: 10.1002/hep.20844. [DOI] [PubMed] [Google Scholar]
- 64.Helbig KJ, Ruszkiewicz A, Lanford RE, Berzsenyi MD, Harley HA, McColl SR, Beard MR. Differential expression of the CXCR3 ligands in chronic hepatitis C virus (HCV) infection and their modulation by HCV in vitro. J Virol. 2009;83:836–846. doi: 10.1128/JVI.01388-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hinson ER, Cresswell P. The antiviral protein, viperin, localizes to lipid droplets via its N-terminal amphipathic alpha-helix. Proc Natl Acad Sci U S A. 2009;106:20452–20457. doi: 10.1073/pnas.0911679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Honda M, Sakai A, Yamashita T, Nakamoto Y, Mizukoshi E, Sakai Y, Nakamura M, Shirasaki T, Horimoto K, Tanaka Y, Tokunaga K, Mizokami M, Kaneko S. Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology. 2010;139:499–509. doi: 10.1053/j.gastro.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 67.Horner SM, Liu HM, Park HS, Briley J, Gale M., Jr Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci U S A. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Horsmans Y, Berg T, Desager JP, Mueller T, Schott E, Fletcher SP, Steffy KR, Bauman LA, Kerr BM, Averett DR. Isatoribine, an agonist of TLR7, reduces plasma virus concentration in chronic hepatitis C infection. Hepatology. 2005;42:724–731. doi: 10.1002/hep.20839. [DOI] [PubMed] [Google Scholar]
- 69.Itsui Y, Sakamoto N, Kakinuma S, Nakagawa M, Sekine-Osajima Y, Tasaka-Fujita M, Nishimura-Sakurai Y, Suda G, Karakama Y, Mishima K, Yamamoto M, Watanabe T, Ueyama M, Funaoka Y, Azuma S, Watanabe M. Antiviral effects of the interferon-induced protein guanylate binding protein 1 and its interaction with the hepatitis C virus NS5B protein. Hepatology. 2009;50:1727–1737. doi: 10.1002/hep.23195. [DOI] [PubMed] [Google Scholar]
- 70.Itsui Y, Sakamoto N, Kurosaki M, Kanazawa N, Tanabe Y, Koyama T, Takeda Y, Nakagawa M, Kakinuma S, Sekine Y, Maekawa S, Enomoto N, Watanabe M. Expressional screening of interferon-stimulated genes for antiviral activity against hepatitis C virus replication. J Viral Hepat. 2006;13:690–700. doi: 10.1111/j.1365-2893.2006.00732.x. [DOI] [PubMed] [Google Scholar]
- 71.Jiang D, Guo H, Xu C, Chang J, Gu B, Wang L, Block TM, Guo JT. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J Virol. 2008;82:1665–1678. doi: 10.1128/JVI.02113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 73.Jouan L, Melancon P, Rodrigue-Gervais IG, Raymond VA, Selliah S, Boucher G, Bilodeau M, Grandvaux N, Lamarre D. Distinct antiviral signaling pathways in primary human hepatocytes and their differential disruption by HCV NS3 protease. J Hepatol. 2010;52:167–175. doi: 10.1016/j.jhep.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 74.Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, Okuda K. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl Acad Sci U S A. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kanazawa N, Kurosaki M, Sakamoto N, Enomoto N, Itsui Y, Yamashiro T, Tanabe Y, Maekawa S, Nakagawa M, Chen CH, Kakinuma S, Oshima S, Nakamura T, Kato T, Wakita T, Watanabe M. Regulation of hepatitis C virus replication by interferon regulatory factor 1. J Virol. 2004;78:9713–9720. doi: 10.1128/JVI.78.18.9713-9720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kang JI, Kwon SN, Park SH, Kim YK, Choi SY, Kim JP, Ahn BY. PKR protein kinase is activated by hepatitis C virus and inhibits viral replication through translational control. Virus Res. 2009;142:51–56. doi: 10.1016/j.virusres.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 77.Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev. 2011;243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 78.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 80.Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, Wakita T. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology. 2003;125:1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 81.Kaukinen P, Sillanpaa M, Kotenko S, Lin R, Hiscott J, Melen K, Julkunen I. Hepatitis C virus NS2 and NS3/4A proteins are potent inhibitors of host cell cytokine/chemokine gene expression. Virol J. 2006;3:66. doi: 10.1186/1743-422X-3-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci. 2008;1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- 83.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 84.Kawasaki T, Kawai T, Akira S. Recognition of nucleic acids by pattern-recognition receptors and its relevance in autoimmunity. Immunol Rev. 2011;243:61–73. doi: 10.1111/j.1600-065X.2011.01048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ke PY, Chen SS. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011;121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kelly C, Klenerman P, Barnes E. Interferon lambdas: the next cytokine storm. Gut. 2011;60:1284–1293. doi: 10.1136/gut.2010.222976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim KA, Lin W, Tai AW, Shao RX, Weinberg E, De Sa Borges CB, Bhan AK, Zheng H, Kamegaya Y, Chung RT. Hepatic SOCS3 expression is strongly associated with non-response to therapy and race in HCV and HCV/HIV infection. J Hepatol. 2009;50:705–711. doi: 10.1016/j.jhep.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kolykhalov AA, Agapov EV, Blight KJ, Mihalik K, Feinstone SM, Rice CM. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 89.Lan KH, Lan KL, Lee WP, Sheu ML, Chen MY, Lee YL, Yen SH, Chang FY, Lee SD. HCV NS5A inhibits interferon-alpha signaling through suppression of STAT1 phosphorylation in hepatocyte-derived cell lines. J Hepatol. 2007;46:759–767. doi: 10.1016/j.jhep.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 90.Lanford RE, Guerra B, Bigger CB, Lee H, Chavez D, Brasky KM. Lack of response to exogenous interferon-alpha in the liver of chimpanzees chronically infected with hepatitis C virus. Hepatology. 2007;46:999–1008. doi: 10.1002/hep.21776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lau DT, Fish PM, Sinha M, Owen DM, Lemon SM, Gale M., Jr Interferon regulatory factor-3 activation, hepatic interferon-stimulated gene expression, and immune cell infiltration in hepatitis C virus patients. Hepatology. 2008;47:799–809. doi: 10.1002/hep.22076. [DOI] [PubMed] [Google Scholar]
- 92.Lee J, Wu CC, Lee KJ, Chuang TH, Katakura K, Liu YT, Chan M, Tawatao R, Chung M, Shen C, Cottam HB, Lai MM, Raz E, Carson DA. Activation of anti-hepatitis C virus responses via Toll-like receptor 7. Proc Natl Acad Sci U S A. 2006;103:1828–1833. doi: 10.1073/pnas.0510801103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lemon SM. Induction and evasion of innate antiviral responses by hepatitis C virus. J Biol Chem. 2010;285:22741–22747. doi: 10.1074/jbc.R109.099556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lemon SM, McKeating JA, Pietschmann T, Frick DN, Glenn JS, Tellinghuisen TL, Symons J, Furman PA. Development of novel therapies for hepatitis C. Antiviral Res. 2010;86:79–92. doi: 10.1016/j.antiviral.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 95.Lemon SM, Walker C, Alter MJ, Yi M. Hepatitis C viruses. In: Knipe D, Howley P, Griffin DE, Martin MA, Lamb RA, editors. Fields' virology. 5th. 2007. pp. 1253–1304. [Google Scholar]
- 96.Li K, Chen Z, Kato N, Gale M, Jr, Lemon SM. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J Biol Chem. 2005;280:16739–16747. doi: 10.1074/jbc.M414139200. [DOI] [PubMed] [Google Scholar]
- 97.Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M, Jr, Lemon SM. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci U S A. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li K, Li NL, Wei D, Pfeffer SR, Fan M, Pfeffer LM. Activation of chemokine and inflammatory cytokine response in HCV-infected hepatocytes depends on TLR3 sensing of HCV dsRNA intermediates. Hepatology. 2012 doi: 10.1002/hep.24763. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liang Y, Shilagard T, Xiao SY, Snyder N, Lau D, Cicalese L, Weiss H, Vargas G, Lemon SM. Visualizing hepatitis C virus infections in human liver by two-photon microscopy. Gastroenterology. 2009;137:1448–1458. doi: 10.1053/j.gastro.2009.07.050. [DOI] [PubMed] [Google Scholar]
- 101.Lin W, Kim SS, Yeung E, Kamegaya Y, Blackard JT, Kim KA, Holtzman MJ, Chung RT. Hepatitis C virus core protein blocks interferon signaling by interaction with the STAT1 SH2 domain. J Virol. 2006;80:9226–9235. doi: 10.1128/JVI.00459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 103.Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- 104.Liu YJ. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu Rev Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 105.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 106.Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, Gale M., Jr Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida M, Yoneyama T, Fujita T, Saito T, Lee WM, Hagedorn CH, Lau DT, Weinman SA, Lemon SM, Gale M., Jr Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci U S A. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Malakhova OA, Kim KI, Luo JK, Zou W, Kumar KG, Fuchs SY, Shuai K, Zhang DE. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006;25:2358–2367. doi: 10.1038/sj.emboj.7601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, Rice CM. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 112.Martell M, Esteban JI, Quer J, Genesca J, Weiner A, Esteban R, Guardia J, Gomez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Marukian S, Andrus L, Sheahan TP, Jones CT, Charles ED, Ploss A, Rice CM, Dustin LB. Hepatitis C virus induces interferon-lambda and interferon-stimulated genes in primary liver cultures. Hepatology. 2011;54:1913–1923. doi: 10.1002/hep.24580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Marukian S, Jones CT, Andrus L, Evans MJ, Ritola KD, Charles ED, Rice CM, Dustin LB. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology. 2008;48:1843–1850. doi: 10.1002/hep.22550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Matsumoto M, Oshiumi H, Seya T. Antiviral responses induced by the TLR3 pathway. Rev Med Virol. 2011;21:67–77. doi: 10.1002/rmv.680. [DOI] [PubMed] [Google Scholar]
- 116.Mattijssen S, Pruijn GJ. Viperin, a key player in the antiviral response. Microbes Infect. 2012 doi: 10.1016/j.micinf.2011.11.015. in press. [DOI] [PubMed] [Google Scholar]
- 117.Melen K, Fagerlund R, Nyqvist M, Keskinen P, Julkunen I. Expression of hepatitis C virus core protein inhibits interferon-induced nuclear import of STATs. J Med Virol. 2004;73:536–547. doi: 10.1002/jmv.20123. [DOI] [PubMed] [Google Scholar]
- 118.Mengshol JA, Golden-Mason L, Rosen HR. Mechanisms of Disease: HCV-induced liver injury. Nat Clin Pract Gastroenterol Hepatol. 2007;4:622–634. doi: 10.1038/ncpgasthep0961. [DOI] [PubMed] [Google Scholar]
- 119.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 120.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 121.Nguyen LH, Espert L, Mechti N, Wilson DM., 3rd The human interferon-and estrogen-regulated ISG20/HEM45 gene product degrades single-stranded RNA and DNA in vitro. Biochemistry. 2001;40:7174–7179. doi: 10.1021/bi010141t. [DOI] [PubMed] [Google Scholar]
- 122.Onoguchi K, Yoneyama M, Takemura A, Akira S, Taniguchi T, Namiki H, Fujita T. Viral infections activate types I and III interferon genes through a common mechanism. J Biol Chem. 2007;282:7576–7581. doi: 10.1074/jbc.M608618200. [DOI] [PubMed] [Google Scholar]
- 123.Oshiumi H, Okamoto M, Fujii K, Kawanishi T, Matsumoto M, Koike S, Seya T. The TLR3/TICAM-1 pathway is mandatory for innate immune responses to poliovirus infection. J Immunol. 2011;187:5320–5327. doi: 10.4049/jimmunol.1101503. [DOI] [PubMed] [Google Scholar]
- 124.Osterlund PI, Pietila TE, Veckman V, Kotenko SV, Julkunen I. IFN regulatory factor family members differentially regulate the expression of type III IFN (IFN-lambda) genes. J Immunol. 2007;179:3434–3442. doi: 10.4049/jimmunol.179.6.3434. [DOI] [PubMed] [Google Scholar]
- 125.Otsuka M, Kato N, Moriyama M, Taniguchi H, Wang Y, Dharel N, Kawabe T, Omata M. Interaction between the HCV NS3 protein and the host TBK1 protein leads to inhibition of cellular antiviral responses. Hepatology. 2005;41:1004–1012. doi: 10.1002/hep.20666. [DOI] [PubMed] [Google Scholar]
- 126.Pavio N, Taylor DR, Lai MM. Detection of a novel unglycosylated form of hepatitis C virus E2 envelope protein that is located in the cytosol and interacts with PKR. J Virol. 2002;76:1265–1272. doi: 10.1128/JVI.76.3.1265-1272.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pietschmann T, Zayas M, Meuleman P, Long G, Appel N, Koutsoudakis G, Kallis S, Leroux-Roels G, Lohmann V, Bartenschlager R. Production of infectious genotype 1b virus particles in cell culture and impairment by replication enhancing mutations. PLoS Pathog. 2009;5:e1000475. doi: 10.1371/journal.ppat.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pindel A, Sadler A. The role of protein kinase R in the interferon response. J Interferon Cytokine Res. 2011;31:59–70. doi: 10.1089/jir.2010.0099. [DOI] [PubMed] [Google Scholar]
- 130.Polyak SJ, Khabar KS, Paschal DM, Ezelle HJ, Duverlie G, Barber GN, Levy DE, Mukaida N, Gretch DR. Hepatitis C virus nonstructural 5A protein induces interleukin-8, leading to partial inhibition of the interferon-induced antiviral response. J Virol. 2001;75:6095–6106. doi: 10.1128/JVI.75.13.6095-6106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Polyak SJ, Khabar KS, Rezeiq M, Gretch DR. Elevated levels of interleukin-8 in serum are associated with hepatitis C virus infection and resistance to interferon therapy. J Virol. 2001;75:6209–6211. doi: 10.1128/JVI.75.13.6209-6211.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Preiss S, Thompson A, Chen X, Rodgers S, Markovska V, Desmond P, Visvanathan K, Li K, Locarnini S, Revill P. Characterization of the innate immune signalling pathways in hepatocyte cell lines. J Viral Hepat. 2008;15:888–900. doi: 10.1111/j.1365-2893.2008.01001.x. [DOI] [PubMed] [Google Scholar]
- 133.Randall G, Chen L, Panis M, Fischer AK, Lindenbach BD, Sun J, Heathcote J, Rice CM, Edwards AM, McGilvray ID. Silencing of USP18 potentiates the antiviral activity of interferon against hepatitis C virus infection. Gastroenterology. 2006;131:1584–1591. doi: 10.1053/j.gastro.2006.08.043. [DOI] [PubMed] [Google Scholar]
- 134.Randall G, Chen L, Panis M, Fischer AK, Lindenbach BD, Sun J, Heathcote J, Rice CM, Edwards AM, McGilvray ID. Silencing of USP18 potentiates the antiviral activity of interferon against hepatitis C virus infection. Gastroenterology. 2006;131:1584–1591. doi: 10.1053/j.gastro.2006.08.043. [DOI] [PubMed] [Google Scholar]
- 135.Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, Chien M, Weir DB, Russo JJ, Ju J, Brownstein MJ, Sheridan R, Sander C, Zavolan M, Tuschl T, Rice CM. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci U S A. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Raychoudhuri A, Shrivastava S, Steele R, Kim H, Ray R, Ray RB. ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J Virol. 2011;85:12881–12889. doi: 10.1128/JVI.05633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rehermann B. Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. J Clin Invest. 2009;119:1745–1754. doi: 10.1172/JCI39133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Riordan SM, Skinner NA, Kurtovic J, Locarnini S, McIver CJ, Williams R, Visvanathan K. Toll-like receptor expression in chronic hepatitis C: correlation with pro-inflammatory cytokine levels and liver injury. Inflamm Res. 2006;55:279–285. doi: 10.1007/s00011-006-0082-0. [DOI] [PubMed] [Google Scholar]
- 139.Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005;79:3851–3854. doi: 10.1128/JVI.79.6.3851-3854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rosen HR. Hepatitis C pathogenesis: mechanisms of viral clearance and liver injury. Liver Transpl. 2003;9:S35–43. doi: 10.1053/jlts.2003.50253. [DOI] [PubMed] [Google Scholar]
- 141.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sarasin-Filipowicz M, Oakeley EJ, Duong FH, Christen V, Terracciano L, Filipowicz W, Heim MH. Interferon signaling and treatment outcome in chronic hepatitis C. Proc Natl Acad Sci U S A. 2008;105:7034–7039. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sarasin-Filipowicz M, Wang X, Yan M, Duong FH, Poli V, Hilton DJ, Zhang DE, Heim MH. Alpha interferon induces long-lasting refractoriness of JAK-STAT signaling in the mouse liver through induction of USP18/UBP43. Mol Cell Biol. 2009;29:4841–4851. doi: 10.1128/MCB.00224-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schulz O, Pichlmair A, Rehwinkel J, Rogers NC, Scheuner D, Kato H, Takeuchi O, Akira S, Kaufman RJ, Reis e Sousa C. Protein kinase R contributes to immunity against specific viruses by regulating interferon mRNA integrity. Cell Host Microbe. 2010;7:354–361. doi: 10.1016/j.chom.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Seki E, Brenner DA. Toll-like receptors and adaptor molecules in liver disease: update. Hepatology. 2008;48:322–335. doi: 10.1002/hep.22306. [DOI] [PubMed] [Google Scholar]
- 148.Sen GC. Viruses and interferons. Annu Rev Microbiol. 2001;55:255–281. doi: 10.1146/annurev.micro.55.1.255. [DOI] [PubMed] [Google Scholar]
- 149.Servant MJ, Tenoever B, Lin R. Overlapping and distinct mechanisms regulating IRF-3 and IRF-7 function. J Interferon Cytokine Res. 2002;22:49–58. doi: 10.1089/107999002753452656. [DOI] [PubMed] [Google Scholar]
- 150.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and Characterization of MAVS, a Mitochondrial Antiviral Signaling Protein that Activates NF-kappaB and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 151.Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, Lemon SM. Stabilization of hepatitis C RNA by an Ago2-miR-122 complex. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1112263109. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Shimoike T, McKenna SA, Lindhout DA, Puglisi JD. Translational insensitivity to potent activation of PKR by HCV IRES RNA. Antiviral Res. 2009;83:228–237. doi: 10.1016/j.antiviral.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 153.Silverman RH. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol. 2007;81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sir D, Chen WL, Choi J, Wakita T, Yen TS, Ou JH. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Siren J, Pirhonen J, Julkunen I, Matikainen S. IFN-alpha regulates TLR-dependent gene expression of IFN-alpha, IFN-beta, IL-28, and IL-29. J Immunol. 2005;174:1932–1937. doi: 10.4049/jimmunol.174.4.1932. [DOI] [PubMed] [Google Scholar]
- 156.Smith KR, Suppiah V, O'Connor K, Berg T, Weltman M, Abate ML, Spengler U, Bassendine M, Matthews G, Irving WL, Irving WL, Powell E, Riordan S, Ahlenstiel G, Stewart GJ, Bahlo M, George J, Booth DR. Identification of improved IL28B SNPs and haplotypes for prediction of drug response in treatment of hepatitis C using massively parallel sequencing in a cross-sectional European cohort. Genome Med. 2011;3:57. doi: 10.1186/gm273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Stiffler JD, Nguyen M, Sohn JA, Liu C, Kaplan D, Seeger C. Focal distribution of hepatitis C virus RNA in infected livers. PLoS One. 2009;4:e6661. doi: 10.1371/journal.pone.0006661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Su AI, Pezacki JP, Wodicka L, Brideau AD, Supekova L, Thimme R, Wieland S, Bukh J, Purcell RH, Schultz PG, Chisari FV. Genomic analysis of the host response to hepatitis C virus infection. Proc Natl Acad Sci U S A. 2002;99:15669–15674. doi: 10.1073/pnas.202608199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sumpter R, Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr Regulating Intracellular Antiviral Defense and Permissiveness to Hepatitis C Virus RNA Replication through a Cellular RNA Helicase, RIG-I. J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Syed GH, Amako Y, Siddiqui A. Hepatitis C virus hijacks host lipid metabolism. Trends Endocrinol Metab. 2010;21:33–40. doi: 10.1016/j.tem.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Taguchi T, Nagano-Fujii M, Akutsu M, Kadoya H, Ohgimoto S, Ishido S, Hotta H. Hepatitis C virus NS5A protein interacts with 2′,5′-oligoadenylate synthetase and inhibits antiviral activity of IFN in an IFN sensitivity-determining region-independent manner. J Gen Virol. 2004;85:959–969. doi: 10.1099/vir.0.19513-0. [DOI] [PubMed] [Google Scholar]
- 162.Takahashi K, Asabe S, Wieland S, Garaigorta U, Gastaminza P, Isogawa M, Chisari FV. Plasmacytoid dendritic cells sense hepatitis C virus-infected cells, produce interferon, and inhibit infection. Proc Natl Acad Sci U S A. 2010;107:7431–7436. doi: 10.1073/pnas.1002301107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 164.Tanida I, Fukasawa M, Ueno T, Kominami E, Wakita T, Hanada K. Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy. 2009;5:937–945. doi: 10.4161/auto.5.7.9243. [DOI] [PubMed] [Google Scholar]
- 165.Targett-Adams P, Boulant S, McLauchlan J. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J Virol. 2008;82:2182–2195. doi: 10.1128/JVI.01565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tasaka M, Sakamoto N, Itakura Y, Nakagawa M, Itsui Y, Sekine-Osajima Y, Nishimura-Sakurai Y, Chen CH, Yoneyama M, Fujita T, Wakita T, Maekawa S, Enomoto N, Watanabe M. Hepatitis C virus non-structural proteins responsible for suppression of the RIG-I/Cardif-induced interferon response. J Gen Virol. 2007;88:3323–3333. doi: 10.1099/vir.0.83056-0. [DOI] [PubMed] [Google Scholar]
- 167.Taura M, Eguma A, Suico MA, Shuto T, Koga T, Komatsu K, Komune T, Sato T, Saya H, Li JD, Kai H. p53 regulates Toll-like receptor 3 expression and function in human epithelial cell lines. Mol Cell Biol. 2008;28:6557–6567. doi: 10.1128/MCB.01202-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Taylor DR, Puig M, Darnell ME, Mihalik K, Feinstone SM. New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1. J Virol. 2005;79:6291–6298. doi: 10.1128/JVI.79.10.6291-6298.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Taylor DR, Shi ST, Romano PR, Barber GN, Lai MM. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999:107–110. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- 170.Urban TJ, Thompson AJ, Bradrick SS, Fellay J, Schuppan D, Cronin KD, Hong L, McKenzie A, Patel K, Shianna KV, McHutchison JG, Goldstein DB, Afdhal N. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 2010;52:1888–1896. doi: 10.1002/hep.23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Uze G, Monneron D. IL-28 and IL-29: newcomers to the interferon family. Biochimie. 2007;89:729–734. doi: 10.1016/j.biochi.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 172.Uzri D, Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J Virol. 2009;83:4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, Barber GN. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol. 2007;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- 174.Vestal DJ, Jeyaratnam JA. The guanylate-binding proteins: emerging insights into the biochemical properties and functions of this family of large interferon-induced guanosine triphosphatase. J Interferon Cytokine Res. 2011;31:89–97. doi: 10.1089/jir.2010.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wald O, Weiss ID, Galun E, Peled A. Chemokines in hepatitis C virus infection: pathogenesis, prognosis and therapeutics. Cytokine. 2007;39:50–62. doi: 10.1016/j.cyto.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 177.Wang C, Pflugheber J, Sumpter R, Jr, Sodora DL, Hui D, Sen GC, Gale M., Jr Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J Virol. 2003;77:3898–3912. doi: 10.1128/JVI.77.7.3898-3912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wang N, Liang Y, Devaraj S, Wang J, Lemon SM, Li K. Toll-like receptor 3 mediates establishment of an antiviral state against hepatitis C virus in hepatoma cells. J Virol. 2009;83:9824–9834. doi: 10.1128/JVI.01125-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wang S, Wu X, Pan T, Song W, Wang Y, Zhang F, Yuan Z. Viperin inhibits hepatitis C virus replication by interfering with binding of NS5A to host protein hVAP-33. J Gen Virol. 2012;93:83–92. doi: 10.1099/vir.0.033860-0. [DOI] [PubMed] [Google Scholar]
- 180.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yang Y, Liang Y, Qu L, Chen Z, Yi M, Li K, Lemon SM. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc Natl Acad Sci U S A. 2007;104:7253–7258. doi: 10.1073/pnas.0611506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yao L, Dong H, Zhu H, Nelson D, Liu C, Lambiase L, Li X. Identification of the IFITM3 gene as an inhibitor of hepatitis C viral translation in a stable STAT1 cell line. J Viral Hepat. 2011;18:e523–529. doi: 10.1111/j.1365-2893.2011.01452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yi M, Villanueva RA, Thomas DL, Wakita T, Lemon SM. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc Natl Acad Sci U S A. 2006;103:2310–2315. doi: 10.1073/pnas.0510727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr, Akira S, Yonehara S, Kato A, Fujita T. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 185.Zeremski M, Petrovic LM, Talal AH. The role of chemokines as inflammatory mediators in chronic hepatitis C virus infection. J Viral Hepat. 2007;14:675–687. doi: 10.1111/j.1365-2893.2006.00838.x. [DOI] [PubMed] [Google Scholar]
- 186.Zhang YL, Guo YJ, Bin L, Sun SH. Hepatitis C virus single-strandedRNA induces innate immunity via Toll-like receptor 7. J Hepatol. 2009;51:29–38. doi: 10.1016/j.jhep.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 187.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhou Z, Wang N, Woodson SE, Dong Q, Wang J, Liang Y, Rijnbrand R, Wei L, Nichols JE, Guo JT, Holbrook MR, Lemon SM, Li K. Antiviral activities of ISG20 in positive-strand RNA virus infections. Virology. 2011;409:175–188. doi: 10.1016/j.virol.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhu H, Butera M, Nelson DR, Liu C. Novel type I interferon IL-28A suppresses hepatitis C viral RNA replication. Virol J. 2005;2:80. doi: 10.1186/1743-422X-2-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zhu H, Liu C. Interleukin-1 inhibits hepatitis C virus subgenomic RNA replication by activation of extracellular regulated kinase pathway. J Virol. 2003;77:5493–5498. doi: 10.1128/JVI.77.9.5493-5498.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]