Abstract
Notch signaling is an evolutionarily conserved cell signaling pathway involved in cell fate during development, stem cell renewal and differentiation in postnatal tissues. Roles for Notch in carcinogenesis, in the biology of cancer stem cells and tumor angiogenesis have been reported. These features identify Notch as a potential therapeutic target in oncology. Based on the molecular structure of Notch receptor, Notch ligands and Notch activators, a set of Notch pathway inhibitors have been developed. Most of these inhibitors had shown anti-tumor effects in preclinical studies. At the same time, the combinatorial effect of these inhibitors with current chemotherapeutical drugs still under study in different clinical trials. In this review, we describe the basics of Notch signaling and the role of Notch in normal and cancer stem cells as a logic way to develop different Notch inhibitors and their current stage of progress for cancer patient’s treatment.
Keywords: Notch signaling, Notch inhibitors, cancer, cancer stem cells
1. Introduction
Targeted therapies have emerged over the last decade as a new and stimulating noveltystrategy for cancer treatment. Nevertheless, to develop and validate a targeted molecule takes some timeHowever, the development and validation of these agents requires significant investment in cancer biology. The first step is to identify a molecular target marker that is crucial for cancer cell proliferation and survival, ideally one that and it is expressed/repressed preferentially or specifically in the malignant cellstissue. The following step is to develop a targeting strategy based on the structure and function of the putative target. This may be relatively simple in the case of a kinase, and more complex in cases like adaptor proteins or non-kinase receptors. generate and develop a therapeutic agent that target specifically or mostly this molecular target. InIn this step, understanding the structure, function and post-translational modifications of the candidate target, including cross-talk with other druggable targets, are essential to develop a practical targeting strategy.protein structure plays a crucial role to identify the domains or fragments of these proteins that are potentially “druggables”. In this context, Notch receptors and other components of the Notch signaling pathway appear to be goodare potentially attractive therapeutic targets. In this review we will describe the modular structure of Notch receptor and Notch ligands, the effects of the their aberrant expression or dysregulated function of them in cancer and cancer stem cells, and the targeted therapeutic tools developed to inhibit block the activation of the Notch signaling pathway based oin our current understanding of their molecular structures and post-translational modifications.
1.1 Notch receptors
1.1.1 Structure
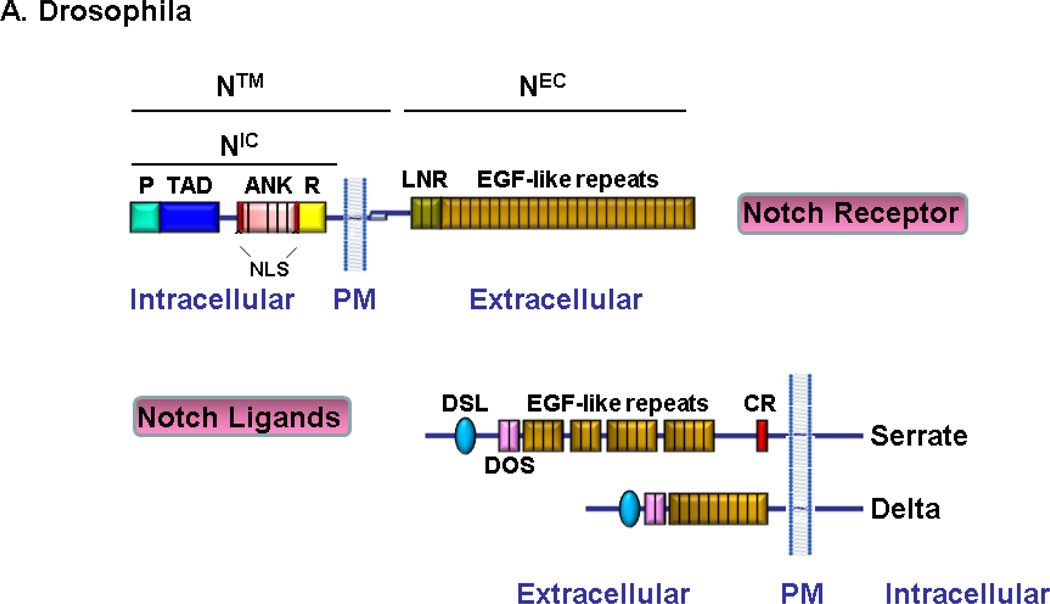
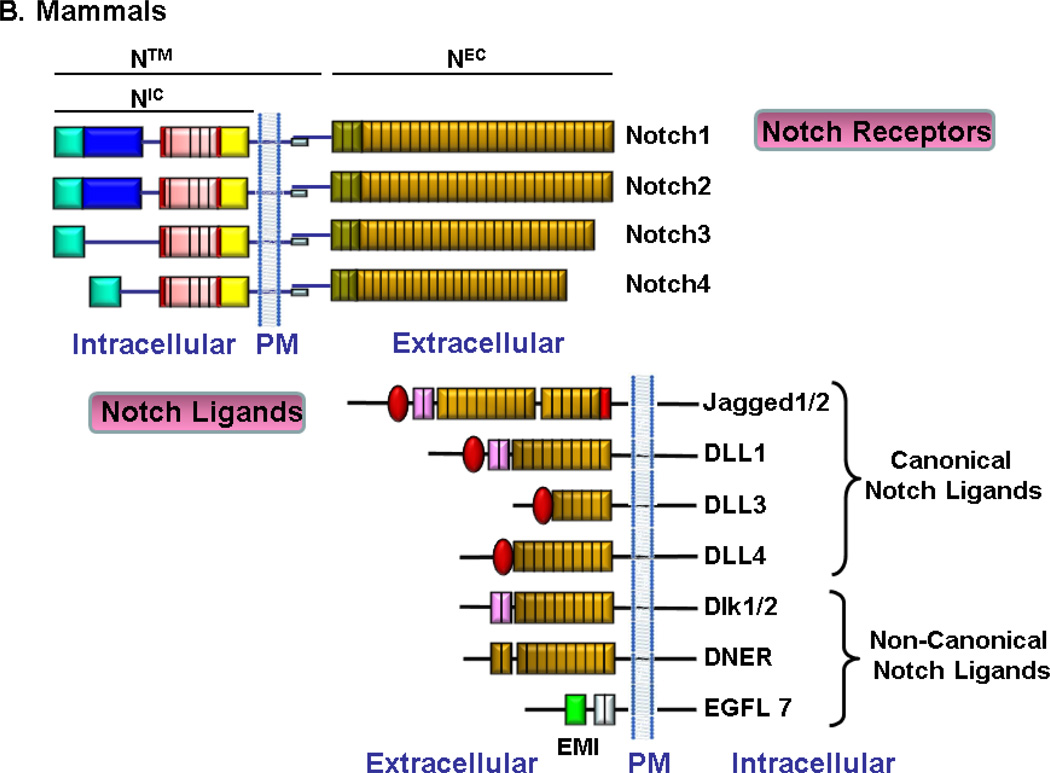
The Notch receptor is a single pass trans-membrane protein evolutionarily conserved from sea urchins to humans. It contains an extracellular domain (NEC), a transmembrane domain (N™) and an intracellular domain (NIC). The extracellular subunit of Notch possesses a multi-modular structure containing multiple Ca2+ binding epidermal growth factor-like repeats (EGF-like) that are required for ligand interaction (Rebay et al., 1991), a negative regulatory region (NRR) which is composed of three cysteine-rich Lin12/Notch repeats (LN) each containing a Ca2+ binding site (Aster et al., 1999; Gordon et al., 2007) and a C-terminal hydrophobic region. The LN repeats stabilize the interaction between subunits by preventing ligand-independent cleavage by metalloproteases (Sanchez-Irizarry et al., 2004). As the name suggests, the NRR holds the mature Notch heterodimer in an auto-inhibited state. The transmembrane subunit includes a short extracellular region containing a pair of conserved cysteines (Kidd et al., 1986; Mumm et al., 2000) thought to participate in heterodimerization (Weinmaster et al., 1992), a Type I transmembrane region and an intracellular region that contains a RBP-jk association module (RAM) that interacts with its transcriptional coactivator RBP-Jk or CSL (CBF-1/Suppressor of Hairless/LAG1) (Tamura et al., 1995). The RAM domain is followed by seven ankyrin (ANK) repeats (Lubman et al., 2004) that interact with CSL and other transcriptional regulators (Nam et al., 2006), two nuclear-localization signals (NLSs) (Lieber et al., 1993), a transactivation domain (TAD) (Kurooka et al., 1998) which ends in a polyglutamine stretch (OPA) (Kurooka et al., 1998) and a C-terminal PEST sequence (a region rich in Proline, Glutamic acid, Serine, and Threonine) that contains multiple phosphorylation sites, which are important for the control of NIC stability and serve as triggers for subsequent ubiquitination and turnover of the receptor (Rechsteiner, 1988).
While Drosophila has only one Notch gene, the mammalian Notch family consists of four members (Notch1, 2, 3, and 4) that are approximately 60% homologous to each other and to Drosophila Notch (Lardelli et al., 1995; Callahan et al., 2001). Although the overall structure of Notch receptors is similar, there are significant differences. The Notch1 and Notch2 receptors contain 36 EGF repeats (Weinmaster et al., 1992; del Amo et al., 1993) in their extracellular domains, similar to Drosophila; whereas Notch3 contain 34 repeats (Lardelli et al., 1994) and Notch4 contains 29 repeats (Uyttendaele et al., 1996). The other difference is in the transactivation domain. Notch1 and Notch2 contain a strong and a weak TAD, respectively (Kurooka et al., 1998), Notch3 has a potent but specific TAD best suited to the activation of the hes5 promoter (Ong et al., 2006). In contrast, Notch4 does not contain a TAD (Fig. 1A–B). These structural differences may offer clues to the functional divergence among mammalian Notch paralogs.
Fig. 1.
Notch receptors and ligands
1.1.2 Post-transcriptional modifications of Notch Receptors
Increasing number of reports have shown that NIC is subject to a variety of post-translational modifications that regulate Notch activity. These modifications include glycosylation, ubiquitylation, phosphorylation, acetylation and hydroxylation.
1.1.2.1 Glycosylation
Glycosylation of Notch receptors by Fringe enzymes (N-acetylglucosaminidyltransferases) affects binding affinities between ligands and specific EGF-repeats (Okajima et al., 2003). Fringe glycosyl transferases initiate elongation of O-linked fucose residues on specific EGF-like repeats of Notch receptors (Bruckner et al., 2000; Moloney, Panin, et al., 2000; Moloney, Shair, et al., 2000). This modification prevents Notch activation by Jagged ligands, but not by Delta-like ligands (Panin et al., 1997). In Drosophila, glycosyltransferase RUMI, also modifies Notch by adding O-glucose to serine residues on particular Notch consensus sequences (Acar et al., 2008) but the importance of this modification in mammals remains to be demonstrated. In mammals three Fringe genes are known, Lunatic Fringe (Lfng), Manic Fringe (Mfng), and Radical Fringe (Rfng) (Cohen et al., 1997). Reduced Lfng expression has been recently demonstrated in basal-like triple-negative breast cancer (TNBC). Importantly, targeted deletion of Lfng in the mouse mammary gland induces TNBC-like mammary cancers with high expression of cleaved Notch receptors. In this model, Lfng blocked mammary stem cells proliferation (Xu et al., 2012).
1.1.2.2 Ubiquitylation
Monoubiquitination has been proposed to result in Notch activation (Gupta-Rossi et al., 2004). Conversely, polyubiquitination can lead to downregulation of Notch signaling. The Ring Finger E3 ubiquitin ligase Deltex along with β-arrestin/Kurtz (Mukherjee et al., 2005), E3 ubiquitin ligases Itch/AIP4 (Atrophin-1 interacting protein 4) (Qiu et al., 2000), NEDD4 (neural precursor cell expressed developmentally down-regulated 4) (Sakata et al., 2004) and Cbl (Casitas B-lineage lymphoma) (Jehn et al., 2002) can poly-ubiquitinate Notch in the cytoplasm and direct Notch receptor via endocytosis towards lysosomal degradation or toward recycling to the plasma membrane (Nichols et al., 2007). Several E3 ubiquitin ligases including Fbw7/Sel-10 (Oberg et al., 2001), Itch (Qiu et al., 2000), c-Cbl (Jehn et al., 2002), and Deltex (Mukherjee et al., 2005) can ubiquitinate active Notch and target it to the proteasome for degradation. Endocytosis can sort Notch to either activation (see above) or degradation pathways. Numb is a cytoplasmic negative regulator of Notch. Numb, in cooperation with the AP2 (adaptor protein-2) component α-adaptin promotes Notch endocytosis (Santolini et al., 2000) followed by proteasome-mediated degradation (McGill et al., 2003). Prolyl isomerase Pin-1 can modify NIC, increasing its intracellular half-life (Rustighi et al., 2009). Pin-1 in turn is regulated by mixed lineage kinases (MLK), potentially placing this pathway upstream of Notch (Rangasamy et al., 2012).
1.1.2.3 Phosphorylation
The NIC is phosphorylated by several kinases at different residues. Phosphorylation of NIC by glycogen synthase kinase 3 β (GSK3β) occurs C-terminally to the ANK repeats and inhibits Notch2 IC-mediated induction of genes such as hairy and enhancer of split 1 (Hes1), but stabilizes NIC (Foltz et al., 2002). Granulocyte colony stimulating factor (Csf) also induces phosphorylation of Notch2IC, leading to its inactivation (Ingles-Esteve et al., 2001). Cyclin C/cyclin-dependent kinase (CDK) 8 phosphorylates NotchIC, and this modification is important for both the activity and turnover of NIC (Fryer et al., 2004).
1.1.2.4 Acetylation
Acetylation controls the stability of NIC (Popko-Scibor et al., 2011; Palermo et al., 2012). The deacetylase Sirtuin 1(SIRT1) has been reported as a key regulator of the endothelial Notch signaling (Guarani et al., 2011).
1.1.2.5 Hydroxylation
It have been described that the asparagine hydroxylase factor-inhibiting HIF1α (FIH1α), which also operates in the cellular hypoxic response, hydroxylates NIC at two residues (N1945 and N2012) (Coleman et al., 2007; Zheng et al., 2008). Interestingly, Notch1IC, 2IC and 3IC, but not Notch4IC, are hydroxylated by FIH1α, and this might contribute to differential signaling. In vitro data suggest that FIH1 negatively regulates Notch signaling, but the biological significance of the FIH1-mediated modifications is not fully understood, and mice targeted for FIH1 do not display an overt Notch gain-of-function phenotype (Zhang et al., 2010)
1.2 Notch ligands
Drosophila has 2 canonical ligands, Delta and Serrate. Mammals express five canonical Notch ligands: three are homologous to Delta and are named Delta-like-1,−3 and −4 (DLL1, DLL3 and DLL4) and two are homologous to Serrate and are named Jagged1 and Jagged2 (Lindsell et al., 1995; Shawber et al., 1996; Dunwoodie et al., 1997; Shutter et al., 2000). These ligands are Type I single-pass transmembrane proteins with an extracellular region consisting of an N-terminal region, a cysteine-rich DSL (an acronym for Delta, Serrate and LAG-2) motif and varying number of EGF-like repeats, similar to the Notch proteins (Kopan et al., 2009). The N-terminal region, the DSL domain and the first two EGF-like repeats are necessary for interaction with EGF repeats 11 and 12 of Notch receptors (Shimizu et al., 1999; Parks et al., 2006). The intracellular regions of DSL ligands are not conserved, but some contain multiple lysine residues and a C-terminal PDZL (PSD-95/Dlg/ZO-1 ligand) motif involved in the ligand signal activity and interactions with the cytoskeleton (Pintar et al., 2007). Notch signaling can also be activated by “non-canonical” ligands other than Delta/Jagged, such as F3/contactin (Hu et al., 2003), DLK1 & 2, DNER, and EGFL7 (Schmidt et al., 2009; D'Souza et al., 2010) (Fig. 1A–B).
The structural variability observed in mammals among the four Notch proteins and their differential contex-dependent functions open the possibility of specific targeting with monoclonal antibodies (mAbs) against the least conserved regions of the proteins.
2. Notch signaling pathway
2.1 Canonical Notch signaling pathways
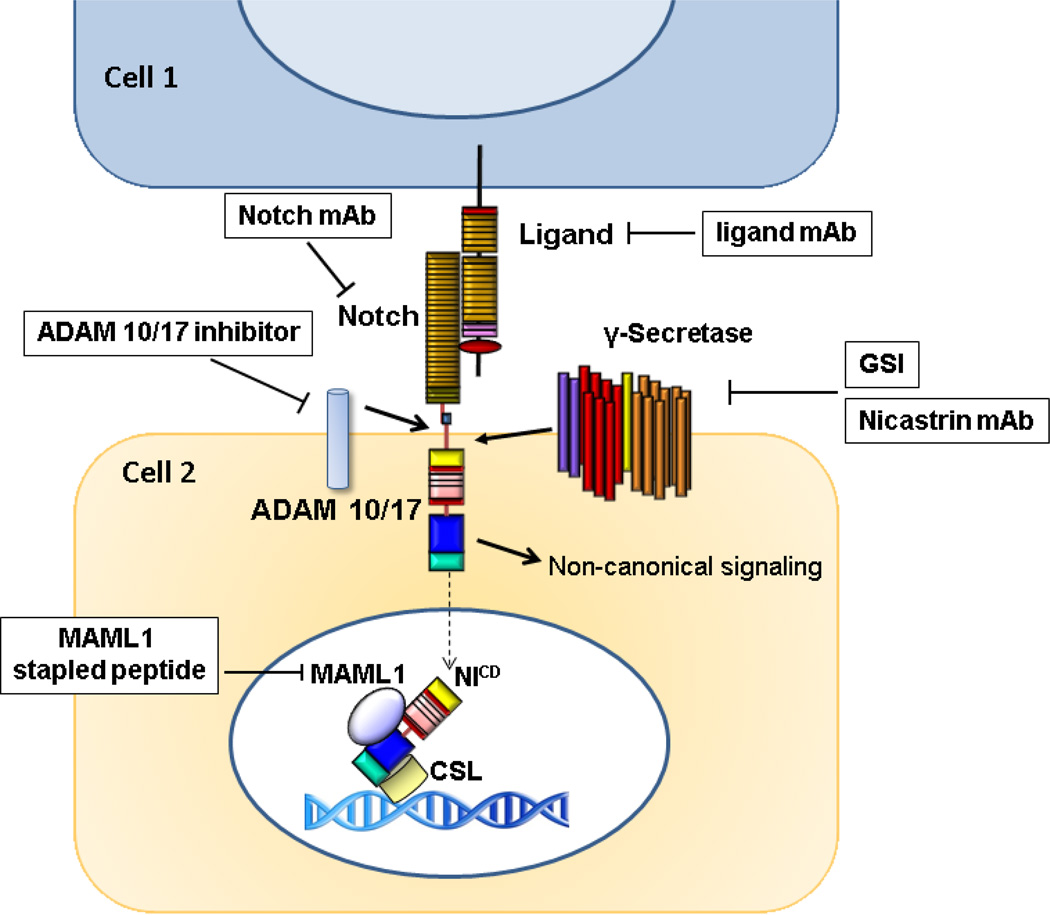
Most of our information on the canonical Notch signaling pathway is derived from studies on Drosophila Notch and its mammalian orthologue Notch1. The Notch precursor protein is produced as a single-chain transmembrane protein in the endoplasmic reticulum where it interacts with O-fucosyltransferase 1 (OFUT1 in Drosophila, POFUT1 in mammals)(Okajima et al., 2005). It is then transported to the Golgi where it is cleaved by a Furin-like convertase at site 1 (S1) (Logeat et al., 1998) and glycosylated by OFUT (Okajima et al., 2002; Shi et al., 2003) and Fringe family N-acetylglucosaminidyl transferases (Haines et al., 2003). Cleaved, glycosylated Notch is transported to the cell surface as a mature heterodimer.
At the plasma membrane, Notch signaling is initiated by a Notch receptor-ligand interaction between two neighboring cells, which induces two successive proteolytic cleavages within the N™ subunit that are required to release the intracellular fragment of Notch (NIC) from the membrane (Mumm et al., 2000). The interaction between Notch and its ligand DSL in trans generates an activating interaction on neighboring cells. In contrast, inhibitory cis interactions between receptor and ligand in the same cell suppress Notch signaling (de Celis et al., 1997; Li et al., 2004; Miller et al., 2009). The cis interaction between Notch and DSL is thought to be bidirectional (Becam et al., 2010; Sprinzak et al., 2010). Ubiquitin ligases Mindbomb (Itoh et al., 2003) or Neuralized (Deblandre et al., 2001; Lai et al., 2001; Pavlopoulos et al., 2001) interact with the intracellular domains of ligands to promote ligand ubiquitination and internalization by transendocytosis to the ligand-expressing cell (Parks et al., 2000). This endocytic process may be required to generate sufficient mechanical force to disrupt the hydrophobic interactions between the N-terminal portion of N™ and the C-terminal portion of NEC (Tiyanont et al., 2011). Subunit dissociation exposes a cleavage site (S2) on N™ on the extracellular side of the membrane for A Disintegrin And Metalloprotease 10 (ADAM10) or 17 (ADAM17) (Brou et al., 2000; Mumm et al., 2000). ADAM cleavage produce a short extracellular truncation fragment, and a clipped transmembrane spanning intermediate called NEXT (Notch Extracellular Truncation) which serves as a substrate for the final proteolytic cleavage (Saxena et al., 2001). The latter occur at site 3 (S3)(Schroeter et al., 1998) and site 4 (S4) within the transmembrane domain and are mediated by the γ-secretase activity of a multi-protein complex consisting of four subunits, presenilin 1 or 2 (the catalytic subunit, an aspartyl protease) (Chen et al., 2006), nicastrin (which maintains complex stability and regulates intracellular protein trafficking) (Zhang et al., 2005), APH1 (anterior pharynx-defective 1; required for the proteolytic activity) (Lee et al., 2004) and PEN2 (presenilin enhancer 2; stabilizes the complex after presenilin proteolysis has generated the activated N-terminal and C-terminal fragments) (Prokop et al., 2004). γ-Secretase cleavage and release of NIC can occur at the cell surface or in an endosomal compartments, but cleavage at the membrane is thought to produce a more stable form of NIC (Tagami et al., 2008; Kopan et al., 2009). After cleavage, NIC translocates to the nucleus where it binds to its downstream transcription factor CSL and drives canonical Notch-mediated gene transcription (Fortini et al., 1994). CSL is thought to be bound to target DNA in a repressive complex that contains histone deacetylases (Lai, 2002), co-repressors SMRT (silencing mediator for retinoid and thyroid receptor)/N-CoR (nuclear receptor co-repressor) (Kao et al., 1998), CIR (CSL interacting repressor) (Hsieh et al., 1999) and SHARP (SMRT/HDAC-1-associated repressor protein)/MINT/SPEN (Oswald et al., 2002; Oswald et al., 2005). NIC competes with the co-repressor complex to bind to CSL and interacts first through its RAM domain (Nam et al., 2006). The ANK domain then associates with CSL to recruit the coactivator Mastermind-like 1 (MAML1, one of three mammalian MAML homologues of Drosophila Mastermind or MAM) (Wu et al., 2000). The Notch-CSL-MAML1 ternary complex in turn recruits other coactivators like histone acetyltransferases CBP/p300 (Wallberg et al., 2002) or PCAF/GCN5 (Kurooka et al., 2000), which convert CSL from a transcriptional repressor to a transcriptional activator. Crystallographic data have shown that the ankyrin domain of NIC and the N-and C-terminals of the Rel homology domain of CSL form a complex with the long, kinked N-terminal helix of MAML1 (Nam et al., 2006). In this complex, the relatively unstructured N-terminal region of the ANK domain, which includes the RAM sequence, folds to form the N-terminal ANK repeat, creating a 7-repeat domain. CSL-binding sites on some Notch promoters exist in pairs in a head-to-head arrangement and could recruit dimeric Notch transcription complexes (Nam et al., 2007) which could increase the strength of the Notch signal. The end result of canonical Notch activation is transcriptional de-repression of a group of genes, many of which are themselves transcription factors or transcriptional repressors. This generates a cascade of gene regulatory events that can modulate virtually every aspect of cell fate decisions depending on cellular context. Recent ChIP-Seq data (Cho et al., 2011; Wang, Zou, et al., 2011; Zhao et al., 2011) have started to shed light on factors contributing to “cellular context”, at least in T- and B-lineage cells. In T-ALL (T-lymphoblastic leukemia) cells, ETS and RUNX family factors are frequently bound to chromatin close to CSL, and appear to cooperate with Notch/CSL, consistent with their known roles in T-cell development and Notch signaling. CREB also appears to cooperate with Notch/CSL at low affinity CSL sites. Zinc finger protein ZNF143 may control the accessibility of CSL to Notch/CSL complexes. ZNF143 sites were predominantly associated with repressive chromatin marks, as were CSL-only sites that contained CSL but not Notch. In proliferating lymphoblastoid cells (LCLs) expressing EBNA2, Wang et al. (Wang, Zou, et al., 2011) found that EBNA2 and CSL bind predominantly at nonpromoter sites. EBF, ETS, RUNX, PU.1, and NF-κB (RELA) sites were found within 500bp of CSL sites. This correlated strongly with actual occupancy data for these transcription factors. Thus, the choice of genomic CSL sites at which Notch activates transcription may depend, at least in part, on the presence of additional transcriptional regulators that can cooperate with or antagonize the Notch-CSL transcriptional complexes. Different cells or different cellular states may have a variety of Notch target sites based on similar mechanisms. “Classical” Notch target genes include among others nuclear basic helix-loop-helix proteins (bHLH) of the Hairy/Enhancer of Split family (HES1–5) (Lecourtois et al., 1995), the Hairy-related family (HRT) (Nakagawa et al., 2000), and the Hairy/Enhancer of Split-related with YRPW motif (HEY) families (Maier et al., 2000). These negatively modulate the expression of genes such as the Achaete-Scute family that induce neuronal differentiation. NIC is also thought to upregulate Deltex (Choi et al., 2002), several members of the NF-kB family, at least in bone marrow hematopoietic cells, (Oswald et al., 1998; Cheng et al., 2001), PPAR family transcription factors (Nickoloff et al., 2002), as well as cell cycle regulators p21WAF1-CIP1 (Rangarajan et al., 2001), cyclin D1 (Ronchini et al., 2001), and c-Myc (Weng et al., 2006). Recently, the Notch1IC nuclear interactome has been characterized in T-ALL cells (Yatim et al., 2012). In these cells, NIC interacts with numerous transcriptional coactivators, and assembles a multifunctional complex containing AF4p12, the PBAF/BRG1 nucleosome remodeling complex, and histone demethylases LSD1 and PHF8 (Yatim et al., 2012). Knockdown experiments showed that these factors regulate the expression of Notch-target genes. Additionally, Notch1IC was shown to interact with multiple proteins involved in trafficking, signal crosstalk, post-translational modifications, DNA repair/replication and RNA processing. The functional significance and context-specificity of these interactions remain to be determined, but some of these may relate to non-canonical Notch signaling.
2.2 Non-canonical Notch signaling pathways
These pathways have been characterized as signals that respond to Notch independently of CSL (Type I), signals that activate Notch independently of S3 cleavage (Type II), or signals that activate CSL-dependent genes without Notch cleavage and NIC release (Type III) (Sanalkumar et al., 2010). Among suggested mechanism of non-canonical Notch signaling are interactions of Notch with non-CSL transcription factors, such as β-catenin (Hayward et al., 2005), HIF-1a (hypoxia-inducible factor-1a) (Gustafsson et al., 2005), NF-κB ((Guan et al., 1996), reviewed in (Osipo, Golde, et al., 2008)), and estrogen receptor ERα (Hao et al., 2010). Non-canonical Notch functions have mostly been identified in stem/progenitor cells or embryonic/primordial cells across species which are capable of expansion and/or differentiation. This suggests that non-canonical Notch signals might play an important role in undifferentiated early cell populations and might interact with conserved cell regulators.
While canonical Notch ligands are responsible for the majority of Notch signaling, a diverse group of structurally unrelated non-canonical ligands has also been identified that activate Notch and likely contribute to the pleiotropic effects of Notch signaling. Thus the final biological effects of Notch targeting are difficult to predict, and depend both on the involved ligand(s) and receptors, and signaling through the canonical intracellular pathway is modulated by non-canonical signaling.
2.3 Crosstalk between Notch and other oncogenic pathways
Numerous oncogenic pathways that cross-talk with Notch signaling have been described. Notch1 is required for the transforming activity of H-Ras (Weijzen et al., 2002) and TGF-α (Miyamoto et al., 2003). Notch activates the PI3K–AKT pathway (Liu et al., 2006; McKenzie et al., 2006; Bedogni et al., 2008; Calzavara et al., 2008; Graziani et al., 2008; Katoh et al., 2009; Yao et al., 2010; Cornejo et al., 2011; Guo, Teng, et al., 2011) while the AKT pathway upregulates Notch1 in response to VEGF (vascular endothelial growth factor) (Liu et al., 2003). Both the AKT and ERK pathways cooperate with Notch4 in transforming breast epithelial cells (Fitzgerald et al., 2000). Glycogen synthase kinase 3 beta (GSK3β), which is negatively regulated by AKT, decreases the half-life of Notch (Foltz et al., 2002). Our group and others have demonstrated that in ERα-positive breast cancer cells estrogen causes accumulation of Notch1 (Soares et al., 2004; Rizzo, Miao, et al., 2008). We also demonstrated that this Notch1 accumulation occur at the plasma membrane (inactive form) and that estrogen also inhibits Notch signaling while estrogen deprivation reactivates this pathway (Rizzo, Miao, et al., 2008). Similarly, Calaf et al. have shown that estrogen can promote malignant transformation of an immortalized human breast epithelial cell line, MCF-10F, increasing the expression of cell adhesion proteins such as Notch (Calaf et al., 2008). We also reported that HER2/neu overexpression inhibits Notch signaling while downregulation of HER2/neu or inhibition of its signaling caused reactivation of Notch signaling (Osipo, Patel, et al., 2008). Recently, Clementz et al. demonstrated that Notch1 and Notch4 are transcriptional targets of PEA3 (Clementz et al., 2011), a transcription factor whose expression has been associated with the malignant phenotype (Trimble et al., 1993; Shepherd et al., 2001) and with HER2/neu expression in breast carcinoma (Benz et al., 1997), and predicted worse overall survival in this malignancy (Kinoshita et al., 2002). Targeting PEA3 may indirectly inhibit Notch pathways, and provide a new therapeutic strategy for triple-negative and possibly other breast cancer subtypes (Clementz et al., 2011) . Finally, we and others (Aguilera et al., 2004; Fernandez-Majada et al., 2007; Song et al., 2008; Hao et al., 2010) have described physical interactions between Notch1IC and the IKK signalosome or nuclear IKKα. These effects are suggested to mediate Notch-induced activation of NF-κB (Song et al., 2008) and ERα (Hao et al., 2010). Notch3IC has also been shown to bind IKKα homodimers, resulting in activation of the NF-κB alternative pathway (Barbarulo et al., 2011). Conversely, nuclear IKKα has been shown to activate Notch-dependent transcription in colon cancer cells (Fernandez-Majada et al., 2007). Recently, Guo and Gonzalez-Perez showed a new Notch, IL-1, and leptin crosstalk. They demonstrated that the leptin proangiogenic effects, via upregulation of VEGF/VEGFR-2, are mediated by leptin-induced Notch expression in breast cancer (Guo and Gonzalez-Perez, 2011)
In summary, Notch is the nexus of a unique and versatile signaling network that regulates and is regulated by a variety of cellular mechanisms highly dependent on cellular context. Thus, therapeutic targeting of the Notch pathway presents both promise and challenges. Successful development of Notch-targeting agents will require a mechanistic understanding of the role of Notch in specific diseases, and ideally, mechanism-based combination regimens. (Fig. 2)
Fig 2.
Notch signaling pathway
2.4 Notch signaling and cancer
Notch was first identified as an oncogene in T-cell acute lymphoblastic leukemia (T-ALL) in which a t(7;9) chromosomal translocation fuses the N-terminal region of the T-cell receptor beta (TCRβ) to the C-terminus of Notch1 (Ellisen et al., 1991). This leads to expression of a truncated Notch1 protein lacking the extracellular subunit and hence constitutively active (Greenwald, 1994). It was later discovered that over 50% of T-ALL have a variety of mutations that activate Notch1 (Weng et al., 2004). These mutations are concentrated in the heterodimerization region, leading to destabilization of the interaction between the two subunits, and/or in the C-terminal PEST region and prolongation of the intracellular half-life of Notch. Further, loss of the E3 ubiquitin ligase Fbw27/Sel-10, or mutations that target the Fbw7-binding pocket can cause Notch pathway activation in T-ALL (Thompson et al., 2007). The intracellular forms of all four Notch proteins are potentially oncogenic and capable of transforming normal cells (Capobianco et al., 1997; Bellavia et al., 2000; Callahan et al., 2001; Kiaris et al., 2004).
Deregulated expression of Notch proteins, ligands, and targets has been described in a multitude of solid tumors, including cervical (Zagouras et al., 1995), head and neck (Leethanakul et al., 2000), endometrial (Suzuki et al., 2000), renal (Rae et al., 2000), lung (Dang et al., 2000), pancreatic (Miyamoto et al., 2003), ovarian (Hopfer et al., 2005), prostate (Santagata et al., 2004; Domingo-Domenech et al., 2012), ovarian (McAuliffe et al., 2012), esophageal (Subramaniam et al., 2012), oral (Liao et al., 2011), hepatocellular (Wang, Xue, et al., 2009), and gastric (Yeh et al., 2009) carcinomas, osteosarcoma, mesothelioma (Bocchetta et al., 2003), melanoma (Balint et al., 2005), gliomas (Purow et al., 2005), medulloblastomas (Fan et al., 2004) and rhabdomyosarcoma (Raimondi et al., 2012). Dysregulation of Notch signaling has been reported in some hematological malignancies other than T-ALL. These include Hodgkin lymphomas, anaplastic large-cell non-Hodgkin lymphomas (Jundt et al., 2002), some acute myeloid leukemias (AML) (Tohda et al., 2001), B-cell chronic lymphoid leukemias (B-CLL) (Hubmann et al., 2002; Hajdu et al., 2010) multiple myeloma (MM) (Houde et al., 2004; Jundt et al., 2004). For a recent review, see (Pancewicz et al., 2011).
Numerous studies have addressed the role of Notch in breast cancer. The first indication of a link between Notch signaling and breast cancer came from a study characterizing a frequent insertion site of the mouse mammary tumor virus (MMTV) in mice (Gallahan et al., 1987) which resulted in the overexpression of truncated Notch4 proteins. These truncated forms of Notch4 contained the transmembrane and intracellular domains, and similar to the truncated Notch1 subsequently discovered in T-ALL, they were constitutively active and caused spontaneous mammary tumors. This was confirmed when truncated Notch4, expressed in transgenic mice under the control of either the MMTV long terminal repeat or the whey acidic protein promoter (Jhappan et al., 1992; Gallahan et al., 1996) led to mammary carcinogenesis. Besides Notch4, there is evidence that constitutive activation of Notch1 and Notch3 in mouse models causes mammary tumors (Hu et al., 2006). Conversely, Notch2 has been associated with better prognosis in breast cancer (Parr et al., 2004).
Immunohistochemical studies reported that high level expression of Notch4 correlates with proliferative marker Ki67, while expression of Notch1 correlates with node status (Yao et al., 2011). Recent findings demonstrated that chromosomal rearrangements lead to the formation of Notch1 and Notch2 fusion transcripts in a subset of TNBC (Robinson et al., 2011). Fusion proteins behave as constitutively active Notch mutants. Tumors carrying such mutations may be sensitive to Notch inhibition. Recent data showing loss of function of Lfng suggest that it may be one of the most common genetic alteration resulting in hyperactive Notch signaling in breast cancer (Xu et al., 2012). Loss of expression of Lfng was seen in a majority of basal-like TNBC. In the same study, expression levels of Notch1 mRNA were shown to be highest in basal-like TNBC, followed by claudin-low TNBC. Expression levels of Notch1 mRNA are strongly correlated with poor survival in TNBC (Pallavi et al., 2012).
Mechanistically, Notch may contribute to carcinogenesis by inhibiting differentiation, inhibiting apoptosis or promoting proliferation. The intracellular forms of Notch induce transformation when it is expressed with oncoproteins that disable the G1-S checkpoint, such as adenovirus E1A, human papillomavirus E6 and E7, Ras, Myc, or SV40 large T-antigen. Depending on context, Notch also can activate the expression of several oncogenic pathways via direct or indirect induction of cyclins D1 (Cohen et al., 2010) and D3 (Joshi et al., 2009), cyclin A (Qi et al., 2003; Rizzo, Osipo, et al., 2008), SKP2 (Sarmento et al., 2005), c-Myc (Liao et al., 2007; Hsu et al., 2008; Allen et al., 2011) or via activation of PI3K–AKT-mTOR, NF-kB and NF-kB2, β-catenin, signal transducers and activators of transcription-3 (STAT3). Notch can also co-operate with oncogenic pathways such as Wnt or Her2/Neu (reviewed in (D'Angelo et al., 2010)) as we describe above. Recent evidence suggests that Notch1 can induce expression of multidrug resistance transporter MRP1 (ABCC1) in breast cancer cells (Cho et al., 2011). Recent genetic studies in Drosophila indicate that transcription factor MEF (Myocyte-Specific Enhancer Factor, the Drosophila homologue of MEF2A, B, C, D in humans) potentiates the oncogenic activity of NotchIC. In a broad range of human malignancies, expression of Notch1 and MEF2C were strongly correlated (Pallavi et al., 2012).
In addition to its cell-autonomous effects on oncogenic pathways, there is strong evidence for a role of Notch in tumor-stroma interactions. Notch signaling can mediate bidirectional tumor–stroma interactions and tumor– endothelium interactions (reviewed in (Gu et al., 2012)). For example, myeloma cells overexpress Jagged-2, activating Notch in stromal cells, which in turn produce IL-6, a growth factor for myeloma cells (Houde et al., 2004). Conversely, stromal cells express Jagged-1, activating Notch in myeloma cells (Jundt et al., 2004). Head and neck squamous cell carcinomas overexpress Jagged-1 which activates Notch in endothelial cells, promoting angiogenesis (Zeng et al., 2005). Productive tumor angiogenesis requires cooperation between VEGF and Notch signaling in the endothelium. Both DLL4 and Jagged-1 ligands participate in this process, with complementary roles (reviewed in (Gu et al., 2012)). Another poorly understood facet of the role of Notch in tumor microenvironment is the well-documented role of Notch signaling in a variety of immune system cells that can affect tumor growth through inflammation, angiogenesis and cytokines (reviewed in (Gu et al., 2012)).
In contrast to its oncogenic role in numerous tissues, Notch has a tumor suppressor effect in the epidermis. Notch1 induces differentiation in murine (Rangarajan et al., 2001) and human (Nickoloff et al., 2002) keratinocytes. This has been confirmed by tissue-specific ablation of Notch1 in conditional knockout mouse models (Nicolas et al., 2003). The mechanism for the tumor suppressor activity of Notch1 is still unclear. Cell-autonomous effects have been described, such as induction of p21 (Rangarajan et al., 2001), calcineurin (Mammucari et al., 2005) and IRF6 (Restivo et al., 2011). Additionally, Notch signaling is essential for epidermal differentiation / barrier formation as Notch1 KO skin loses barrier integrity leading to inflammation and production of cytokines such as TSLP-1 (Dumortier et al., 2010). Chronic inflammation and cytokine production in turn can lead to keratinocyte transformation, as well as systemic effects such as B-lymphocyte proliferative disorder (Demehri et al., 2008) or myeloproliferative syndrome (Dumortier et al., 2010). Recently, Notch1 inactivating mutations have been described in a subset of oropharyngeal squamous carcinomas, suggesting that Notch1 may have a direct or indirect tumor-suppressor role in some of these tumors (Agrawal et al., 2011; Elango et al., 2011). The role of the other 3 Notch paralogs was not investigated. Conversely, increased expression of Notch1 and Jagged-1 has been reported by other groups to be associated with poor prognostic characteristics in Asian head and neck squamous carcinomas (Zhang et al., 2009; Gu et al., 2010; Lin et al., 2010; Yu et al., 2012). This suggests molecular heterogeneity in these tumors. Whether this correlates with HPV status is currently unclear.
2.5 Notch signaling and cancer stem cells
In recent years, Notch activity has been reported in cancer stem-like cells (CSC) (reviewed in (Pannuti et al., 2010)). Notch activity has been implicated in the maintenance of this “cancer stem cell” phenotype in breast cancer (Farnie et al., 2007; Sansone et al., 2007; Harrison et al., 2010), embryonal brain tumors (Fan et al., 2006), glioma (Shih et al., 2006; Fan et al., 2011), hepatocellular carcinoma (Yao et al., 2009), pancreatic (Wang, Azmi, et al., 2009) and prostate carcinomas (Domingo-Domenech et al., 2012).
CSC are thought to constitute a small subset of cancer cells with stem-like phenotype that are a reservoir of self-sustaining cells with the ability to self-renew, presumably leading to recurrence. The stem-like phenotype is also characterized by enhanced resistance to chemo- and radio-therapy (reviewed in (Pannuti et al., 2010) and (Jang et al., 2012). We demonstrated that breast CSCs of different subtypes and in secondary mammospheres from clinical specimens show higher levels of Notch activity compared with the majority of the tumor cells. Blockage of Notch by γ-secretase inhibitors (GSIs) impaired sphere formation, proliferation and anchorage independent growth in soft agar (Grudzien et al., 2010). This data supports a crucial role for Notch signaling in maintenance of breast cancer stem-like cells and suggest that Notch inhibition may have clinical benefits in targeting them. Indeed, evidences showed in a Her2/Neu positive xenograft model (Pandya et al., 2011) indicate that GSIs used in combination with Herceptin do not increase the effects of Herceptin on tumor volume, but completely abrogate tumor recurrence. This strongly suggests an anti-CSC effect.
Thus, the structural differences observed between Notch proteins and its ligands, the ability of Notch receptors to interact with several types of ligands and regulatory or oncogenic proteins, the production of abnormal levels of Notch receptors or ligands offer a variety of potential targeting strategies, some non-selective and some selective for specific Notch receptors or ligands. The cross-talk with other pathways, which must be examined in specific indications, offers the potential for mechanism-based combinations.
3. Strategies to target the Notch signaling pathway
Based on our current understanding of the structure, function and regulation of Notch receptors and ligands, we can identify several steps that can potentially be targeted to inhibit Notch signaling: 1) expression of ligands, 2) ligand ubiquitination and trans-endocytosis, 3) expression of Notch receptors, 4) ligand-receptor binding, 5) heterodimer dissociation during Notch activation, 6) ADAM-mediated cleavage of Notch, 7) subsequent ubiquitination and endocytosis of the γ-secretase substrate, 8) γ-secretase-mediated cleavage of Notch, 9) assembly of the coactivator complex with Notch and CSL, 10) heterodimerization of Notch transcriptional complexes, 11) Notch post-translational modifications and 12) expression of Notch targets. In the next section we will describe currently available Notch inhibitors that target several of these steps and their current development status.
3.1 Neutralizing Notch antibodies
Blocking monoclonal antibodies (mAb) directed against Notch 1, 2 and 3 are under investigation. Two classes of blocking anti-Notch antibodies had been developed. One is directed to the extracellular negative regulator region (NRR) of Notch, blocking the conformational change that allows the ADAM protease cleavage (Aste-Amezaga et al., 2010). A second class consists of ligand-competitors directed against the EGF-repeat region of Notch receptors, blocking the ligand binding domain (LBD) (Aste-Amezaga et al., 2010). Both NRR- and LBD-Notch antibodies induce a strong and specific downregulation of Notch1 signaling, but LBD required higher antibody concentrations to exert the inhibitory effects (Aste-Amezaga et al., 2010). Interesting, Notch 1 NRR (NRR1) antibodies are also capable to bind and inhibit Notch1 carrying the “class I” NRR mutations (single amino acid substitutions or short insertions or deletions in the NRR domain of Notch 1 that cause increased Notch1 activity) in T-All cells (Aste-Amezaga et al., 2010). NRR-specific anti-Notch1 (NRR1), Notch 2 (NRR2) and Notch 3 (NRR3) antibodies that bind to the extracellular binding domain of Notch have been developed and they are in preclinical or in in vitro studies (Li et al., 2008; Aste-Amezaga et al., 2010; Wu et al., 2010). NRR1 also showed anti-angiogenic effects, inhibited blood circulation to the tumor and dramatically inhibited tumor growth (Li et al., 2008; Aste-Amezaga et al., 2010; Wu et al., 2010). Based on the success of in vitro and preclinical studies using blocking Notch antibodies, a dose escalation Phase I clinical trial of OMP-59R5, a humanized mAb that blocks Notch 2 and Notch 3 signaling, has been opened in metastatic or relapsed solid tumor patients who have received prior treatment with standard chemotherapeutic drugs. Some mAbs specific for the negative regulatory region of Notch3 have been shown to inhibit ligand-induced Notch activation by stabilizing the auto-inhibited conformation of the receptor and preventing heterodimer dissociation (Li et al., 2008).
Blocking antibodies against Notch ligands are under development. Anti-Dll4 mAb (Ridgway et al., 2006) and soluble Dll4-Fc fusion proteins (Noguera-Troise et al., 2006; Scehnet et al., 2007) that bind Notch receptors and prevent their activation by endogenous Dll4 have been generated. These biologics inhibited Notch signaling in endothelial cells, caused disorganized angiogenesis and inhibited tumor growth (Ridgway et al., 2006). They are therefore being investigated as anticancer treatments (Thurston et al., 2007; Yan et al., 2007). Studies using the humanized anti-Dll4 mAb OMP-21M18 that blocks the interaction with Notch1 and Notch4, showed anti-tumor activity in patient-derived xenografts independent of any effect on angiogenesis (Reynolds et al., 2011).
Clinical trials using the OMP-21M18 antibody were designed for patients with solid tumors as colorectal cancer, pancreatic cancer, and small cell cancer. Currently, four active clinical trials using OMP-21M18 are ongoing using it as a single agent (NCT00744562) or in combination with chemotherapy (NCT01189968, NCT01189942, NCT01189929) in different solid tumors. The mAb approach has the advantage of potentially exquisite specificity, with the disadvantages of mAbs including limited biodistribution and prolonged half-life. Specificity may decrease toxicity in cases where a specific Notch signaling protein is pathogenetically involved. On the other hand, when multiple Notch paralogs are involved, such as the case of Lfng-negative breast cancer, targeting of individual receptors may not be the most effective approach.
Recently, a novel mAb against the extracellular domain of nicastrin, A5226A, has been generated. This antibody recognizes fully glycosylated mature nicastrin in the active γ-secretase complex on the cell surface, and inhibits γ-secretase activity by competing with substrate binding in vitro. The A5226A antibody abolished the γ-secretase activity-dependent growth of T-ALL cell lines and tumor growth of a T-ALL xenografts mouse model (Hayashi et al., 2012). A different nicastrin mAb has anti-CSC and therapeutic activity in breast cancer models (Lombardo et al., 2012). Nicastrin mAbs would ideally cause γ-secretase inhibition (and potentially pan-Notch inhibition) without the potential off-target effects of small molecules.
3.2 Decoys
Decoys are soluble forms of the extracellular domain of Notch receptors or Notch ligands. Soluble decoys compete with their endogenous cell surface-bound counterparts and abrogate Notch signaling due to the lack of a transmembrane region necessary for receptor activation. A Notch1 decoy that acts as a ligand-dependent Notch antagonist blocks Notch signaling in endothelial cells, affecting tumor neoangiogenesis and growth. It also reduces Notch1 activity and interferes with Dll1, Dll4 and Jagged1 activities, acting as a pan-ligand inhibitor (Funahashi et al., 2008). Soluble forms of the DSL type ligands Dll1 (Varnum-Finney et al., 2000) and Jagged1 (Small et al., 2001) have also been successfully used to inhibit Notch signaling. The presence of endogenous soluble Notch ligands has been reported as a result of endogenous metalloproteases activity (LaVoie et al., 2003; Six et al., 2003). Thus, there is evidence to support the use of soluble Notch ligands as a therapeutic tool. The extracellular domain of Dll1 can exist in a membrane-tethered and in a soluble form (Smas et al., 1997). Another non-canonical Notch ligand is EGF-like domain 7 (EGFL7), a secreted angiogenic factor expressed in endothelial cells. It binds the extracellular domain of the four Notch receptors and inhibits Notch activation induced by Jagged. EGFL7 inhibits neural stem cells renewal (Schmidt et al., 2009) and inhibits Notch activity in post-natal retina and in primary endothelial cells (Nichol et al., 2010). These results suggest that EGFL7 may be used as a Jagged antagonist in cancer cells. The potential efficacy of decoys will depend in large part on their pharmacokinetics and biodistribution. A decoy that achieves better biodistribution than mAbs inside solid tumors may be an attractive therapeutic candidate.
3.3 γ-secretase inhibitors (GSI)
The activation of Notch depends largely on γ-secretase activity (Aster et al., 2008). Thus, γ-secretase is a promising target for Notch inhibition. Non-selective GSIs, often referred to as “Notch inhibitors” in oncology are widely assumed to be equivalent in terms of biological activity and have cytostatic or cytotoxic activities in various cancer cells. GSIs are in clinical trials in a variety of indications (Tammam et al., 2009; Wei et al., 2010; Fouladi et al., 2011; Pandya et al., 2011). Several chemical classes of GSIs have been developed. Most of them are competitive inhibitors of the catalytic activity of presenilins. The dipeptide inhibitor, z-Ile-Leu-CHO (GSI-I) was showed to have Notch1-dependent anti-neoplastic activity in Ras-transformed fibroblasts (Weijzen et al., 2002) and induced apoptosis in melanoma xenografts (Qin et al., 2004). A similar tripeptide inhibited the proliferation of MDA-MB231 cells and tumor growth in MDA-MB231 xenografts. It also inhibited the growth of ERα+ T47D:A18 cells and had a synergistic inhibitory effect with Tamoxifen on ERα+ xenografts (Rizzo, Miao, et al., 2008). These peptides, however, are not candidate human drugs due to poor pharmacokinetics and off-target effects. GSI Compound E inhibited growth and induced apoptosis by increasing the G0/G1 fraction and decreasing the S-phase fraction in T-ALL cell lines (Weng et al., 2003). LY411,575, a GSI that binds to presenilin 1 (PS1) has been widely used in Alzheimer’s disease, where it reduced the accumulation of amyloid-β peptide (Wong et al., 2004; Hyde et al., 2006). In the HER2+ breast cancer cell line BT474, LY 411,575 treatment increased apoptosis and re-sensitized resistant HER2+ cells to trastuzumab (Osipo, Patel, et al., 2008). GSI MRK-003, the parent compound of clinical agent MK-0752, had good preclinical activity in breast cancer and T-ALL (Tammam et al., 2009; Pandya et al., 2011). This compound is more effective than LY411,575 in human mammospheres (Grudzien, 2010) and it completely abrogates recurrence in HER2+ xenografts (Pandya, 2011) in combination with trastuzumab. The clinical compound, MK-0752, also binds to PS1. It is currently in several Phase 1 clinical trial for pediatric and adult oncology treatment (Macy et al., 2008; Zweidler-McKay, 2008; Zhou et al., 2009; Fouladi et al., 2011). We have completed a pilot clinical trial with MK-0752 in combination with endocrine therapy in the preclinical setting (Albain, 2011). This combination was safe and well tolerated, and, importantly, showed molecular evidence of anti-proliferative and pro-apoptotic effects in tumor tissue. GSI RO4929097 appears to differ from other GSIs in that it induces a “less transformed” and slower-growing tumor phenotype without appreciable pro-apoptotic effects (Luistro et al., 2009). Whether this is due to selectivity for specific Notch paralogs is unclear. Currently it is being evaluated in several NCI-sponsored phase 1 clinical trials for treatment of solid tumors and T-ALL. We (Means-Powell JA, 2012) have just completed a phase 1b trial of this agent in combination with exemestane in metastatic, ER+ breast cancer. The combination was well-tolerated and clinical response was observed in a significant number of patients. Unfortunately, the development of RO4929097 has been hampered by its pharmacokinetic liability, due to auto-induction of hepatic metabolism. GSI PF-03084014 has shown an effect in tumor growth and inducing apoptosis in several tumors (Wei et al., 2010) and it is currently in Phase 1 clinical trials for T-All and solid tumors. BMS-708163 is a Notch-selective, second-generation GSI which is in phase 1 (NCT01454115, NCT00860275, NCT01039194, NCT01002079, NCT01079819). In vivo, most GSIs show evidence of anti-angiogenic effects in addition to direct effects on tumor cells. This is most likely due to inhibition of the Notch-VEGF cross-talk essential for angiogenesis (see above). The relative importance of anti-angiogenic versus direct anti-tumor effects in the in vivo mechanism of action of GSIs is still unclear, and may depend on tumor model and class of GSIs. Interestingly, pro-angiogenic cytokines IL6 and IL8 have been reported to cause resistance to GSI RO4929097 (He et al., 2011). The possible role of Notch inhibition in other tumor-stroma components, including T-cells, macrophages, tumor-associated fibroblasts and others is poorly understood.
In summary, it is safe to say that GSIs have shown anti-tumor effects in numerous preclinical models. Anti-angiogenesis and anti-CSC effects are likely to contribute to their mechanism of action in vivo. Due to the broad spectrum of substrates of γ-secretase, GSIs are likely to have multiple off-target effects in vivo. Their toxicity, however, appears to be almost exclusively Notch-mediated. The most serious adverse effect is diarrhea, caused by goblet cell metaplasia of the small intestine which in turn is due to Notch inhibition in intestinal epithelial stem cells. This effect can be dose-limiting and in many cases it requires intermittent administration. The relative lack of specificity of GSIs is not necessarily a therapeutic problem, and may even be an advantage provided that mechanistically relevant pharmacodynamic biomarkers are identified. However, successful development of these agents will require evidence of target inhibition in tumor tissue to guide dose escalation. Molecular biomarkers indicative of Notch inhibition may differ in different tumors and the classical Notch targets (e.g., HES1) may not be the best biomarkers. In our experience, genes responsive to Notch in ER+ breast cancer cell lines have been confirmed to be modulated by GSIs in patient tissue (Means-Powell JA, 2012) more reliably than “classical” Notch target genes described in the literature. Whenever possible, neo-adjuvant clinical trials guided by strong preclinical evidence may be the best approach to development.
3.4 Blocking peptides
Numerous studies on Notch signaling have demonstrated that the activation of Notch and its nuclear access are required to maintain tumor cell growth and survival. Thus, blocking the transcriptional nuclear complex formed by Notch, CSL and coactivators may be another possible therapeutic tool. In 2003, the first dominant negative peptide derived from MAML1 was described. This peptide forms a transcriptionally inert complex with Notch1 and CSL. It has been shown to inhibit the growth of transformed T-ALL cell lines (Weng et al., 2003). Six year later, a new synthetic, cell-permeable, stabilized α-helical, hydrocarbon-stapled peptide derived from MAML1 was generated (SAHM1) (Moellering et al., 2009). Stapled peptides are a new generation of drugs consisting in peptides outfitted with chemical braces or “staples” (Moellering et al., 2009). SAHM1 peptide showed a direct binding to pre-assembled Notch1–CSL complexes and competitive inhibition of the MAML1 co-activator binding. In addition, SAHM1 induced a direct transcriptional repression that resulted in anti-proliferative effects on T-ALL cell lines. SAHM1 treatment also showed an inhibition of leukemic progression through inhibition of Notch signaling in a murine model of T-ALL (Moellering et al., 2009).
The use of stabilized, cell-permeable peptides to interfere with protein complex formation possesses several attractive features; these molecules have relatively small size, they have a high structural compatibility with target proteins, and have the ability to disrupt protein-protein interfaces. Pharmacokinetics will dictate to what extent these molecules can be used therapeutically in humans. Another consideration is the relative role of canonical (nuclear) versus non-canonical Notch signaling. In situations where non-canonical Notch signaling may be responsible for oncogenic activity, such as with Notch4 in the mammary gland (Raafat et al., 2009), inhibition of canonical Notch-CSL-mediated transcription may not be ideal. Having said that a MAML-mimetic may sequester NIC in inactive complexes with CSL, potentially reducing the overall cellular concentration of NIC and also inhibiting non-nuclear Notch signaling.
3.5 Natural compounds
Natural dietary supplements have received much attention, primarily because epidemiological studies have shown that the consumption of fruits, soybean and vegetables is associated with reduced risk of several types of cancers (Lee et al., 2003; Smith-Warner et al., 2003). Such compounds or mixtures of compounds have notoriously pleiotropic activities, but in many cases their biological effects are very promising. As a result, many groups have focused on elucidating molecular mechanisms and identifying targets of these natural products. Several dietary derived compounds target Notch signaling. Isoflavone genistein, found in soy products, inhibits Notch signaling, decreases cell proliferation and induces apoptosis in pancreatic cancer cells via downregulation of NF-κB activity (Wang et al., 2006a). In prostate cancer cells, genistein reduces cell viability and induces apoptosis through downregulation of Notch1, AKT and FoxM1 (Wang, Li, et al., 2011). Sulforaphane, a natural compound derived from cruciferous vegetables such as broccoli, inhibits breast CSCs growth in vitro and in vivo through down-regulation of the Notch and Wnt/beta-catenin pathways, and inhibits growth of CSC-xenografts derived from prostate and pancreatic tumors (Kallifatidis et al., 2011). Quercetin is a major polyphenol and flavonoid commonly found in many fruits and vegetables. It has been reported that quercetin decreases the levels of Notch1 protein and its active fragment in a leukemia cell line with constitutive Notch1 activation (Kawahara et al., 2009) and has a synergistic effect with GSIs on Notch1 activity (Okuhashi et al., 2011). Quercetin also targets CSCs and the epithelial-mesenchimal transition (EMT) phenotype of pancreatic cancer cells (Zhou et al., 2010). Curcumin is an active compound found in Curcuma longa, which is widely used as a flavoring agent in food (e.g., turmeric). It has been shown to have antitumor activity. Curcumin downregulates Notch1 and induces apoptosis through inactivation of NF-κB in pancreatic cancer cells (Wang et al., 2006b) and in oral cancer cells (Liao et al., 2011). Resveratrol, a polyphenolic compound found in grapes, red wine, purple grape juice, peanuts, and some berries, induces apoptosis in part by inhibiting Notch and PI3K/AKT in T-ALL cells (Cecchinato et al., 2007) and in glioblastoma cells (Lin et al., 2011). Recently, it has been reported that resveratrol can also activate Notch2 as a mechanism of apoptosis induction in medullary thyroid cancer (Truong et al., 2011) and in carcinoid (Pinchot et al., 2011). Notch2 may function as a Notch1 antagonist due to its lower transcriptional activity.
Considering the relatively low toxicity of natural products, the idea of such compounds inhibiting Notch in tumor cells or in CSC is potentially attractive. Chronic, partial Notch inhibition by natural products may contribute to chemopreventive activity. Therapeutic uses in established cancers are likely to require combinations with conventional chemotherapeutic agents.
4. Conclusions
In this brief commentary, we attempted to summarize the role of Notch proteins in cancer and current knowledge on Notch-targeting therapeutic tools. Deregulation of Notch proteins has been associated with specific pathologies including cancer development and progression, and with the self-propagation of cancer stem cells. These and other features of Notch signaling, identify Notch as a candidate diagnostic and prognostic biomarker, and an attractive target for cancer therapy. Currently, most Notch-directed therapies involve the use of GSIs, but a variety of biopharmaceuticals and natural products deserve further investigation. As is the case for most embryonic/CSC pathway inhibitors, the development of Notch inhibitors will need to be guided by biology. Biomarkers indicative of Notch activity (and of its inhibition by investigational drugs) will have to be identified and validated in each indication. Additionally, mechanism-based combinations will play a key role. We have demonstrated that combinations with endocrine therapy and trastuzumab can have remarkable therapeutic activity in breast cancer models compared to single agent treatment. Importantly, in the case of Her2-positive breast cancer the effect of Notch inhibition was to prevent recurrence rather than to decrease tumor volume (Pandya et al., 2011). This implies that tumor volume may not be the most informative endpoint in clinical trials of Notch-targeting agents. Recurrence-free survival and/or good surrogate endpoints predictive of survival (e.g., circulating tumor cells, mammosphere-forming cells) are likely to be more informative. These challenges do not diminish the tremendous therapeutic opportunity offered by a pathway that is essential for CSC maintenance, angiogenesis and in many cases proliferation and survival of cancer cells.
Table I.
Notch inhibitors and their current development stage
| Agent | Notch pathway target |
Compound | Condition | Development Phase |
|---|---|---|---|---|
|
Neutralizing antibodies |
Interference with ligand-induced Notch subunit separation and Notch ligands. Specific for Notch 1, 2, 3; DLL1, 4 |
OMP-59R5 anti-Notch2/3 mAb (OncoMed Pharmaceuticals) |
Solid tumors | Phase 1 NCT01277146 |
| NRR1 anti-Notch1 mAb (Genentech and Exelixis; Merck) |
Breast cancer Colon Cancer Anaplastic carcinoma T-cell leukemia T-ALL cell line (Li et al., 2008; Aste-Amezaga et al., 2010; Wu et al., 2010) |
Preclinical and In vitro studies |
||
| NRR2 anti-Notch2 mAb (Genentech and Exelixis) |
Breast cancer Colon Cancer Anaplastic carcinoma HEK293T cell line (Wu et al., 2010) |
Preclinical studies |
||
| NRR3 anti-Notch3 mAb (Genentech) |
HEK293T cell line (Li et al., 2008) | In vitro studies | ||
| OMP-21M18 anti-DLL4 mAb (OncoMed Pharmaceuticals) |
Colorectal cancer Small cell lung cancer Pancreatic cancer Solid tumors |
Phase 1 NCT01189929 NCT01189942 NCT01189968 NCT00744562 |
||
| DLL1-Fc and JAG1-Fc Anti-Delta-like1 and Jagged 1 Fc chimeric mAbs |
Autoimmune encephalomyelitis (Fischer et al., 2011; Reynolds et al., 2011) |
In vitro studies | ||
| A5622A Anti-nicastrin mAb |
T-cell leukemia T-ALL tumor (Hayashi et al., 2012) |
T-cell leukemia T-ALL tumor (Hayashi, I, 2011) |
||
| Decoys | Interference with ligand-receptor interaction |
Soluble forms of Notch1, Dll1 and Jagged 1 |
Endothelial cells (Varnum-Finney et al., 2000; Small et al., 2001; Funahashi et al., 2008) |
Preclinical studies |
| γ-Secretase Inhibitor (GSI) | Notch 1, 2, 3, 4; Notch ligands |
RO4929097 (Roche) |
Breast cancer Brain tumors Colorectal cancer Melanoma Solid tumors T-cell leukemia |
Phase 1 NCT01088763 NCT01198535 NCT01149356 NCT01141569 NCT01196416 NCT01218620 NCT01217411 NCT01270438 NCT01238133 NCT01208441 |
| MRK-003 (Merck) |
Breast cancer T-cell leukemia (Tammam et al., 2009; Pandya et al., 2011) |
Preclinical studies |
||
| MRK-0752 (Merck) |
Breast cancer Brain tumors Neoplasms Pancreatic cancer T-cell leukemia |
Phase 1 NCT00756717 NCT00803894 NCT01295632 NCT01098344 NCT00645333 NCT01243762 NCT00572182 NCT00106145 NCT00100152 |
||
| PF-03084014 (Pfizer) |
Neoplasms Solid tumors Lymphoid leukemia T-cell leukemia |
Phase 1 NCT00878189 |
||
| MRK-0752 (Merck) |
Breast cancer Brain tumors Neoplasms Pancreatic cancer T-cell leukemia |
Phase 1 NCT00756717 NCT00803894 NCT01295632 NCT01098344 NCT00645333 NCT01243762 NCT00572182 NCT00106145 NCT00100152 |
||
| PF-03084014 (Pfizer) |
Neoplasms Solid tumors Lymphoid leukemia T-cell leukemia |
Phase 1 NCT00878189 |
||
| Blocking peptide | Interference with Notch nuclear co-activator MAML1 |
MAM peptide antagonist SAHM1 (Aileron Therapeutics) |
T-cell leukemia T-ALL tumors (Weng et al., 2003; Moellering et al., 2009) |
Preclinical studies |
| Natural compounds | Downregulation of Notch activity and Notch pathway |
Genistein Sulforaphane Quercetin Curcumin Resveratrol |
Pancreatic cancer Prostate cancer Thyroid cancer Carcinoid T-ALL cells Glioblastoma cells Oral cancer cells (Wang et al., 2006; Cecchinato et al., 2007; Kawahara et al., 2009; Kallifatidis et al., 2011; Liao et al., 2011; Okuhashi et al., 2011; Pinchot et al., 2011; Truong et al., 2011; Wang et al., 2011) |
Preclinical and In vitro studies |
Acknowledgments
This work was financially support by NIH PO1AG025531. We thank Dr. Christian R. Gomez for his critical review and comments on the manuscript.
Abbreviations
- ADAM10/17
A Disintegrin And Metalloprotease 10/17
- AIP4
Atrophin-1 interacting protein 4
- Akt
Protein Kinase B
- ANK
Ankyrin repeats
- AP2
Adaptor protein 2
- APH1
Anterior Pharynx-defective 1
- B-CLL
B-cell chronic lymphoid leukemias
- bHLH
basic helix-loop-helix
- Ca2+
Calcium
- Cbl
Casitas B-linage lymphoma
- CBP/p300
CREB binding protein (CBP) and p300
- CDK
Cyclin-dependent kinase
- ChIP-Seq
chromatin immunoprecipitation and sequencing
- CSC
Cancer Stem Cells
- CSL
CBF-1, Suppressor of Hairless/LAG1
- Csf
Granulocyte colony stimulating factor
- CIR
CSL Interacting Repressor
- DLL 1, 3, 4
Delta-like-1, −3, −4
- DLK1/2
delta-like 1/2 homolog (Drosophila)
- DNER
Delta and Notch-like epidermal growth factor repeat
- DSL
Delta/Serrate/Lag-2
- EBF
Early B cell factor
- EBNA2
Epstein-Barr virus nuclear antigen 2
- EGF
Epidermal Growth Factor
- EGFL7
EGF-like domain-containing protein 7
- ERK
Extracellular signal-regulated kinases
- ERα
Estrogen receptor alpha
- ETS
E-twenty six
- Fbw7
F-box and WD40 repeat domain–containing 7
- GCN5
General control of amino acid synthesis protein 5
- GSK3β
Glycogen Synthase Kinase 3 beta
- GSIs
Gamma Secretase Inhibitors
- HD
Heterodimerization domain
- HDAC
Histone deacetylase 1
- HER2
Human Epidermal Growth Factor Receptor 2
- HES1–5
Hairy/Enhancer of Split family 1
- HRT
Hairy-related family
- IKKα
IkappaB kinase alpha
- IRF6
Interferon regulatory factor 6
- Lfng
Lunatic fringe
- LSD1
Lysine-specific demethylase 1
- LBD
Ligand binding domain
- LN
Cystein rich-LIN12/Notch repeats
- mAbs
monoclonal antibodies
- MAML1
Mastermind-like 1
- MEF
Myocyte Specific Enhancer Factor
- Mfng
Manic fringe
- MLK
Mixed lineage kinases
- MMTV
Mouse mammary tumor virus
- NIC
Notch intracellular domain
- NEC
Notch extracellular domain
- N™
Notch transmembrane domain
- N-CoR
Nuclear receptor co-Represor
- NEDD4
Neural precursor cell expressed Developmentally Down-regulated 4
- NEXT
Notch Extracellular Truncation
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- NLS
Nuclear localization signal
- NRR
Negative regulatory region
- LCLs
Lymphoblastoids cells
- LN
Cysteine-rich Lin12/Notch repeats
- OFUT1
O-Fucosyltransferase 1
- OPA
Polyglutamine stretch
- PCAF
P300/CBP-associated factor
- PHF8
PHD finger protein 8
- PEN2
Presenilin Enhancer 2
- PEST
Proline, Glutamic acid, Serine and Threonine
- PDZL
Post synaptic density protein (PSD95)/Drosophila disc large tumor suppressor (Dlg1)/Zonula occludens-1 protein (zo-1) Ligand
- PEST
Proline, Glutamic acid, Serine and Threonine
- PS1
Presenilin 1
- PPAR
Peroxisome proliferators-activated receptors
- RAM
RBP-jk association module
- RBP-jk
Recombining binding protein suppressor of hairless
- Rfng
Radical fringe
- RUNX
Runt-related transcription factor
- SHARP
SMRT/HDAC-1-associated repressor protein
- SKP2
S-phase kinase-associated protein 2
- SMRT
Silencing Mediator for Retinoid and Thyroid receptor
- STAT3
Signal transducer and activator of transcription 3
- T-ALL
T-cell Acute Lymphoblastic Leukemia
- TAD
Transactivation domain
- TCRβ
T-cell receptor beta
- TGFα
Transforming growth factor alpha
- TNBC
Triple negative breast cancer
- TSLP-1
thymic stromal lymphopoietin
- VEGF
Vascular endothelial growth factor
- ZNF143
zinc finger protein 143
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. Conflict of interest/disclosures
No conflict of interest.
References
- Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, et al. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell. 2008;132(2):247–258. doi: 10.1016/j.cell.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera C, Hoya-Arias R, Haegeman G, Espinosa L, Bigas A. Recruitment of IkappaBalpha to the hes1 promoter is associated with transcriptional repression. Proc Natl Acad Sci U S A. 2004;101(47):16537–16542. doi: 10.1073/pnas.0404429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albain K, Czerlanis C, Zlobin A, Covington KR, Rajan P, Godellas C, et al. Modulation of Cancer Stem Cell Biomarkers by the Notch Inhibitor MK0752 Added to Endocrine Therapy for Early Stage ER+ Breast Cancer. Cancer Res. 2011;71(24 Suppl):97s. [Google Scholar]
- Allen TD, Rodriguez EM, Jones KD, Bishop JM. Activated Notch1 induces lung adenomas in mice and cooperates with Myc in the generation of lung adenocarcinoma. Cancer Res. 2011;71(18):6010–6018. doi: 10.1158/0008-5472.CAN-11-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aste-Amezaga M, Zhang N, Lineberger JE, Arnold BA, Toner TJ, Gu M, et al. Characterization of Notch1 antibodies that inhibit signaling of both normal and mutated Notch1 receptors. PLoS One. 2010;5(2):e9094. doi: 10.1371/journal.pone.0009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu Rev Pathol. 2008;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aster JC, Simms WB, Zavala-Ruiz Z, Patriub V, North CL, Blacklow SC. The folding and structural integrity of the first LIN-12 module of human Notch1 are calcium-dependent. Biochemistry. 1999;38(15):4736–4742. doi: 10.1021/bi982713o. [DOI] [PubMed] [Google Scholar]
- Balint K, Xiao M, Pinnix CC, Soma A, Veres I, Juhasz I, et al. Activation of Notch1 signaling is required for beta-catenin-mediated human primary melanoma progression. J Clin Invest. 2005;115(11):3166–3176. doi: 10.1172/JCI25001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbarulo A, Grazioli P, Campese AF, Bellavia D, Di Mario G, Pelullo M, et al. Notch3 and canonical NF-kappaB signaling pathways cooperatively regulate Foxp3 transcription. J Immunol. 2011;186(11):6199–6206. doi: 10.4049/jimmunol.1002136. [DOI] [PubMed] [Google Scholar]
- Becam I, Fiuza UM, Arias AM, Milan M. A role of receptor Notch in ligand cis-inhibition in Drosophila. Curr Biol. 2010;20(6):554–560. doi: 10.1016/j.cub.2010.01.058. [DOI] [PubMed] [Google Scholar]
- Bedogni B, Warneke JA, Nickoloff BJ, Giaccia AJ, Powell MB. Notch1 is an effector of Akt and hypoxia in melanoma development. J Clin Invest. 2008;118(11):3660–3670. doi: 10.1172/JCI36157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellavia D, Campese AF, Alesse E, Vacca A, Felli MP, Balestri A, et al. Constitutive activation of NF-kappaB and T-cell leukemia/lymphoma in Notch3 transgenic mice. EMBO J. 2000;19(13):3337–3348. doi: 10.1093/emboj/19.13.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz CC, O'Hagan RC, Richter B, Scott GK, Chang CH, Xiong X, et al. HER2/Neu and the Ets transcription activator PEA3 are coordinately upregulated in human breast cancer. Oncogene. 1997;15(13):1513–1525. doi: 10.1038/sj.onc.1201331. [DOI] [PubMed] [Google Scholar]
- Bocchetta M, Miele L, Pass HI, Carbone M. Notch-1 induction, a novel activity of SV40 required for growth of SV40-transformed human mesothelial cells. Oncogene. 2003;22(1):81–89. doi: 10.1038/sj.onc.1206097. [DOI] [PubMed] [Google Scholar]
- Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, et al. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5(2):207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- Bruckner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 2000;406(6794):411–415. doi: 10.1038/35019075. [DOI] [PubMed] [Google Scholar]
- Calaf GM, Roy D. Cell adhesion proteins altered by 17beta estradiol and parathion in breast epithelial cells. Oncol Rep. 2008;19(1):165–169. [PubMed] [Google Scholar]
- Callahan R, Raafat A. Notch signaling in mammary gland tumorigenesis. J Mammary Gland Biol Neoplasia. 2001;6(1):23–36. doi: 10.1023/a:1009512414430. [DOI] [PubMed] [Google Scholar]
- Calzavara E, Chiaramonte R, Cesana D, Basile A, Sherbet GV, Comi P. Reciprocal regulation of Notch and PI3K/Akt signalling in T-ALL cells in vitro. J Cell Biochem. 2008;103(5):1405–1412. doi: 10.1002/jcb.21527. [DOI] [PubMed] [Google Scholar]
- Capobianco AJ, Zagouras P, Blaumueller CM, Artavanis-Tsakonas S, Bishop JM. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol Cell Biol. 1997;17(11):6265–6273. doi: 10.1128/mcb.17.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchinato V, Chiaramonte R, Nizzardo M, Cristofaro B, Basile A, Sherbet GV, et al. Resveratrol-induced apoptosis in human T-cell acute lymphoblastic leukaemia MOLT-4 cells. Biochem Pharmacol. 2007;74(11):1568–1574. doi: 10.1016/j.bcp.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Chen F, Hasegawa H, Schmitt-Ulms G, Kawarai T, Bohm C, Katayama T, et al. TMP21 is a presenilin complex component that modulates gamma-secretase but not epsilon-secretase activity. Nature. 2006;440(7088):1208–1212. doi: 10.1038/nature04667. [DOI] [PubMed] [Google Scholar]
- Cheng P, Zlobin A, Volgina V, Gottipati S, Osborne B, Simel EJ, et al. Notch-1 regulates NF-kappaB activity in hemopoietic progenitor cells. J Immunol. 2001;167(8):4458–4467. doi: 10.4049/jimmunol.167.8.4458. [DOI] [PubMed] [Google Scholar]
- Cho S, Lu M, He X, Ee PL, Bhat U, Schneider E, et al. Notch1 regulates the expression of the multidrug resistance gene ABCC1/MRP1 in cultured cancer cells. Proc Natl Acad Sci U S A. 2011;108(51):20778–20783. doi: 10.1073/pnas.1019452108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JW, Pampeno C, Vukmanovic S, Meruelo D. Characterization of the transcriptional expression of Notch-1 signaling pathway members, Deltex and HES-1, in developing mouse thymocytes. Dev Comp Immunol. 2002;26(6):575–588. doi: 10.1016/s0145-305x(01)00095-7. [DOI] [PubMed] [Google Scholar]
- Clementz AG, Rogowski A, Pandya K, Miele L, Osipo C. NOTCH-1 and NOTCH-4 are novel gene targets of PEA3 in breast cancer: novel therapeutic implications. Breast Cancer Res. 2011;13(3):R63. doi: 10.1186/bcr2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen B, Bashirullah A, Dagnino L, Campbell C, Fisher WW, Leow CC, et al. Fringe boundaries coincide with Notch-dependent patterning centres in mammals and alter Notch-dependent development in Drosophila. Nat Genet. 1997;16(3):283–288. doi: 10.1038/ng0797-283. [DOI] [PubMed] [Google Scholar]
- Cohen B, Shimizu M, Izrailit J, Ng NF, Buchman Y, Pan JG, et al. Cyclin D1 is a direct target of JAG1-mediated Notch signaling in breast cancer. Breast Cancer Res Treat. 2010;123(1):113–124. doi: 10.1007/s10549-009-0621-9. [DOI] [PubMed] [Google Scholar]
- Coleman ML, McDonough MA, Hewitson KS, Coles C, Mecinovic J, Edelmann M, et al. Asparaginyl hydroxylation of the Notch ankyrin repeat domain by factor inhibiting hypoxia-inducible factor. J Biol Chem. 2007;282(33):24027–24038. doi: 10.1074/jbc.M704102200. [DOI] [PubMed] [Google Scholar]
- Cornejo MG, Mabialah V, Sykes SM, Khandan T, Lo Celso C, Lopez CK, et al. Crosstalk between NOTCH and AKT signaling during murine megakaryocyte lineage specification. Blood. 2011;118(5):1264–1273. doi: 10.1182/blood-2011-01-328567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo RC, Wicha MS. Stem cells in normal development and cancer. Prog Mol Biol Transl Sci. 2010;95:113–158. doi: 10.1016/B978-0-12-385071-3.00006-X. [DOI] [PubMed] [Google Scholar]
- D'Souza B, Meloty-Kapella L, Weinmaster G. Canonical and non-canonical Notch ligands. Curr Top Dev Biol. 2010;92:73–129. doi: 10.1016/S0070-2153(10)92003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang TP, Gazdar AF, Virmani AK, Sepetavec T, Hande KR, Minna JD, et al. Chromosome 19 translocation, overexpression of Notch3, and human lung cancer. J Natl Cancer Inst. 2000;92(16):1355–1357. doi: 10.1093/jnci/92.16.1355. [DOI] [PubMed] [Google Scholar]
- de Celis JF, Bray S. Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development. 1997;124(17):3241–3251. doi: 10.1242/dev.124.17.3241. [DOI] [PubMed] [Google Scholar]
- Deblandre GA, Lai EC, Kintner C. Xenopus neuralized is a ubiquitin ligase that interacts with XDelta1 and regulates Notch signaling. Dev Cell. 2001;1(6):795–806. doi: 10.1016/s1534-5807(01)00091-0. [DOI] [PubMed] [Google Scholar]
- del Amo FF, Gendron-Maguire M, Swiatek PJ, Jenkins NA, Copeland NG, Gridley T. Cloning, analysis, and chromosomal localization of Notch-1, a mouse homolog of Drosophila Notch. Genomics. 1993;15(2):259–264. doi: 10.1006/geno.1993.1055. [DOI] [PubMed] [Google Scholar]
- Demehri S, Liu Z, Lee J, Lin MH, Crosby SD, Roberts CJ, et al. Notch-deficient skin induces a lethal systemic B-lymphoproliferative disorder by secreting TSLP, a sentinel for epidermal integrity. PLoS Biol. 2008;6(5):e123. doi: 10.1371/journal.pbio.0060123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo-Domenech J, Vidal SJ, Rodriguez-Bravo V, Castillo-Martin M, Quinn SA, Rodriguez-Barrueco R, et al. Suppression of acquired docetaxel resistance in prostate cancer through depletion of notch- and hedgehog-dependent tumor-initiating cells. Cancer Cell. 2012;22(3):373–388. doi: 10.1016/j.ccr.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumortier A, Durham AD, Di Piazza M, Vauclair S, Koch U, Ferrand G, et al. Atopic dermatitis-like disease and associated lethal myeloproliferative disorder arise from loss of Notch signaling in the murine skin. PLoS One. 2010;5(2):e9258. doi: 10.1371/journal.pone.0009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwoodie SL, Henrique D, Harrison SM, Beddington RS. Mouse Dll3: a novel divergent Delta gene which may complement the function of other Delta homologues during early pattern formation in the mouse embryo. Development. 1997;124(16):3065–3076. doi: 10.1242/dev.124.16.3065. [DOI] [PubMed] [Google Scholar]
- Elango KJ, Suresh A, Erode EM, Subhadradevi L, Ravindran HK, Iyer SK, et al. Role of human papilloma virus in oral tongue squamous cell carcinoma. Asian Pac J Cancer Prev. 2011;12(4):889–896. [PubMed] [Google Scholar]
- Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66(4):649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- Fan L, Liu Y, Ying H, Xue Y, Zhang Z, Wang P, et al. Increasing of blood-tumor barrier permeability through paracellular pathway by low-frequency ultrasound irradiation in vitro. J Mol Neurosci. 2011;43(3):541–548. doi: 10.1007/s12031-010-9479-x. [DOI] [PubMed] [Google Scholar]
- Fan X, Matsui W, Khaki L, Stearns D, Chun J, Li YM, et al. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 2006;66(15):7445–7452. doi: 10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- Fan X, Mikolaenko I, Elhassan I, Ni X, Wang Y, Ball D, et al. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 2004;64(21):7787–7793. doi: 10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- Farnie G, Clarke RB. Mammary stem cells and breast cancer--role of Notch signalling. Stem Cell Rev. 2007;3(2):169–175. doi: 10.1007/s12015-007-0023-5. [DOI] [PubMed] [Google Scholar]
- Fernandez-Majada V, Aguilera C, Villanueva A, Vilardell F, Robert-Moreno A, Aytes A, et al. Nuclear IKK activity leads to dysregulated notch-dependent gene expression in colorectal cancer. Proc Natl Acad Sci U S A. 2007;104(1):276–281. doi: 10.1073/pnas.0606476104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K, Harrington A, Leder P. Ras pathway signals are required for notch-mediated oncogenesis. Oncogene. 2000;19(37):4191–4198. doi: 10.1038/sj.onc.1203766. [DOI] [PubMed] [Google Scholar]
- Foltz DR, Santiago MC, Berechid BE, Nye JS. Glycogen synthase kinase-3beta modulates notch signaling and stability. Curr Biol. 2002;12(12):1006–1011. doi: 10.1016/s0960-9822(02)00888-6. [DOI] [PubMed] [Google Scholar]
- Fortini ME, Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994;79(2):273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- Fouladi M, Stewart CF, Olson J, Wagner LM, Onar-Thomas A, Kocak M, et al. Phase I trial of MK-0752 in children with refractory CNS malignancies: a pediatric brain tumor consortium study. J Clin Oncol. 2011;29(26):3529–3534. doi: 10.1200/JCO.2011.35.7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer CJ, White JB, Jones KA. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol Cell. 2004;16(4):509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Funahashi Y, Hernandez SL, Das I, Ahn A, Huang J, Vorontchikhina M, et al. A notch1 ectodomain construct inhibits endothelial notch signaling, tumor growth, and angiogenesis. Cancer Res. 2008;68(12):4727–4735. doi: 10.1158/0008-5472.CAN-07-6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallahan D, Callahan R. Mammary tumorigenesis in feral mice: identification of a new int locus in mouse mammary tumor virus (Czech II)-induced mammary tumors. J Virol. 1987;61(1):66–74. doi: 10.1128/jvi.61.1.66-74.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallahan D, Jhappan C, Robinson G, Hennighausen L, Sharp R, Kordon E, et al. Expression of a truncated Int3 gene in developing secretory mammary epithelium specifically retards lobular differentiation resulting in tumorigenesis. Cancer Res. 1996;56(8):1775–1785. [PubMed] [Google Scholar]
- Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14(4):295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- Graziani I, Eliasz S, De Marco MA, Chen Y, Pass HI, De May RM, et al. Opposite effects of Notch-1 and Notch-2 on mesothelioma cell survival under hypoxia are exerted through the Akt pathway. Cancer Res. 2008;68(23):9678–9685. doi: 10.1158/0008-5472.CAN-08-0969. [DOI] [PubMed] [Google Scholar]
- Greenwald I. Structure/function studies of lin-12/Notch proteins. Curr Opin Genet Dev. 1994;4(4):556–562. doi: 10.1016/0959-437x(94)90072-b. [DOI] [PubMed] [Google Scholar]
- Grudzien P, Lo S, Albain KS, Robinson P, Rajan P, Strack PR, et al. Inhibition of Notch signaling reduces the stem-like population of breast cancer cells and prevents mammosphere formation. Anticancer Res. 2010;30(10):3853–3867. [PubMed] [Google Scholar]
- Gu F, Ma Y, Zhang Z, Zhao J, Kobayashi H, Zhang L, et al. Expression of Stat3 and Notch1 is associated with cisplatin resistance in head and neck squamous cell carcinoma. Oncol Rep. 2010;23(3):671–676. doi: 10.3892/or_00000683. [DOI] [PubMed] [Google Scholar]
- Gu JW, Rizzo P, Pannuti A, Golde T, Osborne B, Miele L. Notch signals in the endothelium and cancer "stem-like" cells: opportunities for cancer therapy. Vasc Cell. 2012;4:7. doi: 10.1186/2045-824X-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan E, Wang J, Laborda J, Norcross M, Baeuerle PA, Hoffman T. T cell leukemia-associated human Notch/translocation-associated Notch homologue has I kappa B-like activity and physically interacts with nuclear factor-kappa B proteins in T cells. J Exp Med. 1996;183(5):2025–2032. doi: 10.1084/jem.183.5.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarani V, Deflorian G, Franco CA, Kruger M, Phng LK, Bentley K, et al. Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature. 2011;473(7346):234–238. doi: 10.1038/nature09917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Teng Q, Ji C. NOTCH and phosphatidylinositide 3-kinase/phosphatase and tensin homolog deleted on chromosome ten/AKT/mammalian target of rapamycin (mTOR) signaling in T-cell development and T-cell acute lymphoblastic leukemia. Leuk Lymphoma. 2011;52(7):1200–1210. doi: 10.3109/10428194.2011.564696. [DOI] [PubMed] [Google Scholar]
- Guo S, Gonzalez-Perez RR. Notch, IL-1 and leptin crosstalk outcome (NILCO) is critical for leptin-induced proliferation, migration and VEGF/VEGFR-2 expression in breast cancer. PLoS One. 2011;6(6):e21467. doi: 10.1371/journal.pone.0021467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta-Rossi N, Six E, LeBail O, Logeat F, Chastagner P, Olry A, et al. Monoubiquitination and endocytosis direct gamma-secretase cleavage of activated Notch receptor. J Cell Biol. 2004;166(1):73–83. doi: 10.1083/jcb.200310098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MV, Zheng X, Pereira T, Gradin K, Jin S, Lundkvist J, et al. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell. 2005;9(5):617–628. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Haines N, Irvine KD. Glycosylation regulates Notch signalling. Nat Rev Mol Cell Biol. 2003;4(10):786–797. doi: 10.1038/nrm1228. [DOI] [PubMed] [Google Scholar]
- Hajdu M, Kopper L, Sebestyen A. Notch-regulation upon Dll4-stimulation of TGFb-induced apoptosis and gene expression in human B-cell non-Hodgkin lymphomas. Scand J Immunol. 2010;71(1):29–37. doi: 10.1111/j.1365-3083.2009.02346.x. [DOI] [PubMed] [Google Scholar]
- Hao L, Rizzo P, Osipo C, Pannuti A, Wyatt D, Cheung LW, et al. Notch-1 activates estrogen receptor-alpha-dependent transcription via IKKalpha in breast cancer cells. Oncogene. 2010;29(2):201–213. doi: 10.1038/onc.2009.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison H, Farnie G, Howell SJ, Rock RE, Stylianou S, Brennan KR, et al. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res. 2010;70(2):709–718. doi: 10.1158/0008-5472.CAN-09-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi I, Takatori S, Urano Y, Miyake Y, Takagi J, Sakata-Yanagimoto M, et al. Neutralization of the gamma-secretase activity by monoclonal antibody against extracellular domain of nicastrin. Oncogene. 2012;31(6):787–798. doi: 10.1038/onc.2011.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward P, Brennan K, Sanders P, Balayo T, DasGupta R, Perrimon N, et al. Notch modulates Wnt signalling by associating with Armadillo/beta-catenin and regulating its transcriptional activity. Development. 2005;132(8):1819–1830. doi: 10.1242/dev.01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Luistro L, Carvajal D, Smith M, Nevins T, Yin X, et al. High tumor levels of IL6 and IL8 abrogate preclinical efficacy of the gamma-secretase inhibitor, RO4929097. Mol Oncol. 2011;5(3):292–301. doi: 10.1016/j.molonc.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfer O, Zwahlen D, Fey MF, Aebi S. The Notch pathway in ovarian carcinomas and adenomas. Br J Cancer. 2005;93(6):709–718. doi: 10.1038/sj.bjc.6602719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houde C, Li Y, Song L, Barton K, Zhang Q, Godwin J, et al. Overexpression of the NOTCH ligand JAG2 in malignant plasma cells from multiple myeloma patients and cell lines. Blood. 2004;104(12):3697–3704. doi: 10.1182/blood-2003-12-4114. [DOI] [PubMed] [Google Scholar]
- Hsieh JJ, Zhou S, Chen L, Young DB, Hayward SD. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc Natl Acad Sci U S A. 1999;96(1):23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu KW, Hsieh RH, Lee YH, Chao CH, Wu KJ, Tseng MJ, et al. The activated Notch1 receptor cooperates with alpha-enolase and MBP-1 in modulating c-myc activity. Mol Cell Biol. 2008;28(15):4829–4842. doi: 10.1128/MCB.00175-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Dievart A, Lupien M, Calvo E, Tremblay G, Jolicoeur P. Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. Am J Pathol. 2006;168(3):973–990. doi: 10.2353/ajpath.2006.050416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu QD, Ang BT, Karsak M, Hu WP, Cui XY, Duka T, et al. F3/contactin acts as a functional ligand for Notch during oligodendrocyte maturation. Cell. 2003;115(2):163–175. doi: 10.1016/s0092-8674(03)00810-9. [DOI] [PubMed] [Google Scholar]
- Hubmann R, Schwarzmeier JD, Shehata M, Hilgarth M, Duechler M, Dettke M, et al. Notch2 is involved in the overexpression of CD23 in B-cell chronic lymphocytic leukemia. Blood. 2002;99(10):3742–3747. doi: 10.1182/blood.v99.10.3742. [DOI] [PubMed] [Google Scholar]
- Hyde LA, McHugh NA, Chen J, Zhang Q, Manfra D, Nomeir AA, et al. Studies to investigate the in vivo therapeutic window of the gamma-secretase inhibitor N2-[(2S)-2-(3,5-difluorophenyl)-2-hydroxyethanoyl]-N1-[(7S)-5-methyl-6-oxo-6,7-di hydro-5H-dibenzo[b,d]azepin-7-yl]-L-alaninamide (LY411,575) in the CRND8 mouse. J Pharmacol Exp Ther. 2006;319(3):1133–1143. doi: 10.1124/jpet.106.111716. [DOI] [PubMed] [Google Scholar]
- Ingles-Esteve J, Espinosa L, Milner LA, Caelles C, Bigas A. Phosphorylation of Ser2078 modulates the Notch2 function in 32D cell differentiation. J Biol Chem. 2001;276(48):44873–44880. doi: 10.1074/jbc.M104703200. [DOI] [PubMed] [Google Scholar]
- Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, et al. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4(1):67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- Jang JY, Kim MK, Jeon YK, Joung YK, Park KD, Kim CW. Adenovirus adenine nucleotide translocator-2 shRNA effectively induces apoptosis and enhances chemosensitivity by the down-regulation of ABCG2 in breast cancer stem-like cells. Exp Mol Med. 2012;44(4):251–259. doi: 10.3858/emm.2012.44.4.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehn BM, Dittert I, Beyer S, von der Mark K, Bielke W. c-Cbl binding and ubiquitin-dependent lysosomal degradation of membrane-associated Notch1. J Biol Chem. 2002;277(10):8033–8040. doi: 10.1074/jbc.M108552200. [DOI] [PubMed] [Google Scholar]
- Jhappan C, Gallahan D, Stahle C, Chu E, Smith GH, Merlino G, et al. Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands. Genes Dev. 1992;6(3):345–355. doi: 10.1101/gad.6.3.345. [DOI] [PubMed] [Google Scholar]
- Joshi I, Minter LM, Telfer J, Demarest RM, Capobianco AJ, Aster JC, et al. Notch signaling mediates G1/S cell-cycle progression in T cells via cyclin D3 and its dependent kinases. Blood. 2009;113(8):1689–1698. doi: 10.1182/blood-2008-03-147967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jundt F, Anagnostopoulos I, Forster R, Mathas S, Stein H, Dorken B. Activated Notch1 signaling promotes tumor cell proliferation and survival in Hodgkin and anaplastic large cell lymphoma. Blood. 2002;99(9):3398–3403. doi: 10.1182/blood.v99.9.3398. [DOI] [PubMed] [Google Scholar]
- Jundt F, Probsting KS, Anagnostopoulos I, Muehlinghaus G, Chatterjee M, Mathas S, et al. Jagged1-induced Notch signaling drives proliferation of multiple myeloma cells. Blood. 2004;103(9):3511–3515. doi: 10.1182/blood-2003-07-2254. [DOI] [PubMed] [Google Scholar]
- Kallifatidis G, Labsch S, Rausch V, Mattern J, Gladkich J, Moldenhauer G, et al. Sulforaphane increases drug-mediated cytotoxicity toward cancer stem-like cells of pancreas and prostate. Mol Ther. 2011;19(1):188–195. doi: 10.1038/mt.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, et al. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12(15):2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y, Katoh M. Integrative genomic analyses on GLI1: positive regulation of GLI1 by Hedgehog-GLI, TGFbeta-Smads, and RTK-PI3K-AKT signals, and negative regulation of GLI1 by Notch-CSL-HES/HEY, and GPCR-Gs-PKA signals. Int J Oncol. 2009;35(1):187–192. doi: 10.3892/ijo_00000328. [DOI] [PubMed] [Google Scholar]
- Kawahara T, Kawaguchi-Ihara N, Okuhashi Y, Itoh M, Nara N, Tohda S. Cyclopamine and quercetin suppress the growth of leukemia and lymphoma cells. Anticancer Res. 2009;29(11):4629–4632. [PubMed] [Google Scholar]
- Kiaris H, Politi K, Grimm LM, Szabolcs M, Fisher P, Efstratiadis A, et al. Modulation of notch signaling elicits signature tumors and inhibits hras1-induced oncogenesis in the mouse mammary epithelium. Am J Pathol. 2004;165(2):695–705. doi: 10.1016/S0002-9440(10)63333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd S, Young MW. Transposon-dependent mutant phenotypes at the Notch locus of Drosophila. Nature. 1986;323(6083):89–91. doi: 10.1038/323089a0. [DOI] [PubMed] [Google Scholar]
- Kinoshita J, Kitamura K, Tanaka S, Sugimachi K, Ishida M, Saeki H. Clinical significance of PEA3 in human breast cancer. Surgery. 2002;131(1 Suppl):S222–S225. doi: 10.1067/msy.2002.119792. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurooka H, Honjo T. Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J Biol Chem. 2000;275(22):17211–17220. doi: 10.1074/jbc.M000909200. [DOI] [PubMed] [Google Scholar]
- Kurooka H, Kuroda K, Honjo T. Roles of the ankyrin repeats and C-terminal region of the mouse notch1 intracellular region. Nucleic Acids Res. 1998;26(23):5448–5455. doi: 10.1093/nar/26.23.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC. Keeping a good pathway down: transcriptional repression of Notch pathway target genes by CSL proteins. EMBO Rep. 2002;3(9):840–845. doi: 10.1093/embo-reports/kvf170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Deblandre GA, Kintner C, Rubin GM. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of delta. Dev Cell. 2001;1(6):783–794. doi: 10.1016/s1534-5807(01)00092-2. [DOI] [PubMed] [Google Scholar]
- Lardelli M, Dahlstrand J, Lendahl U. The novel Notch homologue mouse Notch 3 lacks specific epidermal growth factor-repeats and is expressed in proliferating neuroepithelium. Mech Dev. 1994;46(2):123–136. doi: 10.1016/0925-4773(94)90081-7. [DOI] [PubMed] [Google Scholar]
- Lardelli M, Williams R, Lendahl U. Notch-related genes in animal development. Int J Dev Biol. 1995;39(5):769–780. [PubMed] [Google Scholar]
- LaVoie MJ, Fraering PC, Ostaszewski BL, Ye W, Kimberly WT, Wolfe MS, et al. Assembly of the gamma-secretase complex involves early formation of an intermediate subcomplex of Aph-1 and nicastrin. J Biol Chem. 2003;278(39):37213–37222. doi: 10.1074/jbc.M303941200. [DOI] [PubMed] [Google Scholar]
- Lecourtois M, Schweisguth F. The neurogenic suppressor of hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev. 1995;9(21):2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- Lee MM, Gomez SL, Chang JS, Wey M, Wang RT, Hsing AW. Soy and isoflavone consumption in relation to prostate cancer risk in China. Cancer Epidemiol Biomarkers Prev. 2003;12(7):665–668. [PubMed] [Google Scholar]
- Lee SF, Shah S, Yu C, Wigley WC, Li H, Lim M, et al. A conserved GXXXG motif in APH-1 is critical for assembly and activity of the gamma-secretase complex. J Biol Chem. 2004;279(6):4144–4152. doi: 10.1074/jbc.M309745200. [DOI] [PubMed] [Google Scholar]
- Leethanakul C, Patel V, Gillespie J, Pallente M, Ensley JF, Koontongkaew S, et al. Distinct pattern of expression of differentiation and growth-related genes in squamous cell carcinomas of the head and neck revealed by the use of laser capture microdissection and cDNA arrays. Oncogene. 2000;19(28):3220–3224. doi: 10.1038/sj.onc.1203703. [DOI] [PubMed] [Google Scholar]
- Li K, Li Y, Wu W, Gordon WR, Chang DW, Lu M, et al. Modulation of Notch signaling by antibodies specific for the extracellular negative regulatory region of NOTCH3. J Biol Chem. 2008;283(12):8046–8054. doi: 10.1074/jbc.M800170200. [DOI] [PubMed] [Google Scholar]
- Li Y, Baker NE. The roles of cis-inactivation by Notch ligands and of neuralized during eye and bristle patterning in Drosophila. BMC Dev Biol. 2004;4:5. doi: 10.1186/1471-213X-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Xia J, Chen Z, Zhang S, Ahmad A, Miele L, et al. Inhibitory effect of curcumin on oral carcinoma CAL-27 cells via suppression of Notch-1 and NF-kappaB signaling pathways. J Cell Biochem. 2011;112(4):1055–1065. doi: 10.1002/jcb.23019. [DOI] [PubMed] [Google Scholar]
- Liao WR, Hsieh RH, Hsu KW, Wu MZ, Tseng MJ, Mai RT, et al. The CBF1-independent Notch1 signal pathway activates human c-myc expression partially via transcription factor YY1. Carcinogenesis. 2007;28(9):1867–1876. doi: 10.1093/carcin/bgm092. [DOI] [PubMed] [Google Scholar]
- Lieber T, Kidd S, Alcamo E, Corbin V, Young MW. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 1993;7(10):1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- Lin H, Xiong W, Zhang X, Liu B, Zhang W, Zhang Y, et al. Notch-1 activation-dependent p53 restoration contributes to resveratrol-induced apoptosis in glioblastoma cells. Oncol Rep. 2011;26(4):925–930. doi: 10.3892/or.2011.1380. [DOI] [PubMed] [Google Scholar]
- Lin JT, Chen MK, Yeh KT, Chang CS, Chang TH, Lin CY, et al. Association of high levels of Jagged-1 and Notch-1 expression with poor prognosis in head and neck cancer. Ann Surg Oncol. 2010;17(11):2976–2983. doi: 10.1245/s10434-010-1118-9. [DOI] [PubMed] [Google Scholar]
- Lindsell CE, Shawber CJ, Boulter J, Weinmaster G. Jagged: a mammalian ligand that activates Notch1. Cell. 1995;80(6):909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- Liu ZJ, Shirakawa T, Li Y, Soma A, Oka M, Dotto GP, et al. Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis. Mol Cell Biol. 2003;23(1):14–25. doi: 10.1128/MCB.23.1.14-25.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZJ, Xiao M, Balint K, Smalley KS, Brafford P, Qiu R, et al. Notch1 signaling promotes primary melanoma progression by activating mitogen-activated protein kinase/phosphatidylinositol 3-kinase-Akt pathways and up-regulating N-cadherin expression. Cancer Res. 2006;66(8):4182–4190. doi: 10.1158/0008-5472.CAN-05-3589. [DOI] [PubMed] [Google Scholar]
- Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah NG, et al. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci U S A. 1998;95(14):8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo Y, Filipovic A, Molyneux G, Periyasamy M, Giamas G, Hu Y, et al. Nicastrin regulates breast cancer stem cell properties and tumor growth in vitro and in vivo. Proc Natl Acad Sci U S A. 2012;109(41):16558–16563. doi: 10.1073/pnas.1206268109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubman OY, Korolev SV, Kopan R. Anchoring notch genetics and biochemistry; structural analysis of the ankyrin domain sheds light on existing data. Mol Cell. 2004;13(5):619–626. doi: 10.1016/s1097-2765(04)00120-0. [DOI] [PubMed] [Google Scholar]
- Luistro L, He W, Smith M, Packman K, Vilenchik M, Carvajal D, et al. Preclinical profile of a potent gamma-secretase inhibitor targeting notch signaling with in vivo efficacy and pharmacodynamic properties. Cancer Res. 2009;69(19):7672–7680. doi: 10.1158/0008-5472.CAN-09-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macy ME, Sawczyn KK, Garrington TP, Graham DK, Gore L. Pediatric developmental therapies: interesting new drugs now in early-stage clinical trials. Curr Oncol Rep. 2008;10(6):477–490. doi: 10.1007/s11912-008-0073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier MM, Gessler M. Comparative analysis of the human and mouse Hey1 promoter: Hey genes are new Notch target genes. Biochem Biophys Res Commun. 2000;275(2):652–660. doi: 10.1006/bbrc.2000.3354. [DOI] [PubMed] [Google Scholar]
- Mammucari C, Tommasi di Vignano A, Sharov AA, Neilson J, Havrda MC, Roop DR, et al. Integration of Notch 1 and calcineurin/NFAT signaling pathways in keratinocyte growth and differentiation control. Dev Cell. 2005;8(5):665–676. doi: 10.1016/j.devcel.2005.02.016. [DOI] [PubMed] [Google Scholar]
- McAuliffe SM, Morgan SL, Wyant GA, Tran LT, Muto KW, Chen YS, et al. Targeting Notch, a key pathway for ovarian cancer stem cells, sensitizes tumors to platinum therapy. Proc Natl Acad Sci U S A. 2012;109(43):E2939–E2948. doi: 10.1073/pnas.1206400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem. 2003;278(25):23196–23203. doi: 10.1074/jbc.M302827200. [DOI] [PubMed] [Google Scholar]
- McKenzie G, Ward G, Stallwood Y, Briend E, Papadia S, Lennard A, et al. Cellular Notch responsiveness is defined by phosphoinositide 3-kinase-dependent signals. BMC Cell Biol. 2006;7:10. doi: 10.1186/1471-2121-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means-Powell JA MS, Mayer IA, Abramson VG, Ismail-Khan R, Arteaga CL, Ayers DA, Sanders MS, Lush RM, Miele L. A Phase Ib Dose Escalation Trial of RO4929097 (a γ-secretase inhibitor) in Combination with Exemestane in Patients with ER + Metastatic Breast Cancer. Cancer Res. 2012;72(24 Suppl):280s. doi: 10.1016/j.clbc.2021.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AC, Lyons EL, Herman TG. cis-Inhibition of Notch by endogenous Delta biases the outcome of lateral inhibition. Curr Biol. 2009;19(16):1378–1383. doi: 10.1016/j.cub.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, Maitra A, Ghosh B, Zechner U, Argani P, Iacobuzio-Donahue CA, et al. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3(6):565–576. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462(7270):182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, et al. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406(6794):369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- Moloney DJ, Shair LH, Lu FM, Xia J, Locke R, Matta KL, et al. Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. J Biol Chem. 2000;275(13):9604–9611. doi: 10.1074/jbc.275.13.9604. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Veraksa A, Bauer A, Rosse C, Camonis J, Artavanis-Tsakonas S. Regulation of Notch signalling by non-visual beta-arrestin. Nat Cell Biol. 2005;7(12):1191–1201. doi: 10.1038/ncb1327. [DOI] [PubMed] [Google Scholar]
- Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5(2):197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- Nakagawa O, McFadden DG, Nakagawa M, Yanagisawa H, Hu T, Srivastava D, et al. Members of the HRT family of basic helix-loop-helix proteins act as transcriptional repressors downstream of Notch signaling. Proc Natl Acad Sci U S A. 2000;97(25):13655–13660. doi: 10.1073/pnas.250485597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Pear WS, Aster JC, Blacklow SC. Cooperative assembly of higher-order Notch complexes functions as a switch to induce transcription. Proc Natl Acad Sci U S A. 2007;104(7):2103–2108. doi: 10.1073/pnas.0611092104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124(5):973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- Nichol D, Shawber C, Fitch MJ, Bambino K, Sharma A, Kitajewski J, et al. Impaired angiogenesis and altered Notch signaling in mice overexpressing endothelial Egfl7. Blood. 2010;116(26):6133–6143. doi: 10.1182/blood-2010-03-274860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols JT, Miyamoto A, Weinmaster G. Notch signaling--constantly on the move. Traffic. 2007;8(8):959–969. doi: 10.1111/j.1600-0854.2007.00592.x. [DOI] [PubMed] [Google Scholar]
- Nickoloff BJ, Qin JZ, Chaturvedi V, Denning MF, Bonish B, Miele L. Jagged-1 mediated activation of notch signaling induces complete maturation of human keratinocytes through NF-kappaB and PPARgamma. Cell Death Differ. 2002;9(8):842–855. doi: 10.1038/sj.cdd.4401036. [DOI] [PubMed] [Google Scholar]
- Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33(3):416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- Noguera-Troise I, Daly C, Papadopoulos NJ, Coetzee S, Boland P, Gale NW, et al. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006;444(7122):1032–1037. doi: 10.1038/nature05355. [DOI] [PubMed] [Google Scholar]
- Oberg C, Li J, Pauley A, Wolf E, Gurney M, Lendahl U. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J Biol Chem. 2001;276(38):35847–35853. doi: 10.1074/jbc.M103992200. [DOI] [PubMed] [Google Scholar]
- Okajima T, Irvine KD. Regulation of notch signaling by o-linked fucose. Cell. 2002;111(6):893–904. doi: 10.1016/s0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- Okajima T, Xu A, Irvine KD. Modulation of notch-ligand binding by protein O-fucosyltransferase 1 and fringe. J Biol Chem. 2003;278(43):42340–42345. doi: 10.1074/jbc.M308687200. [DOI] [PubMed] [Google Scholar]
- Okajima T, Xu A, Lei L, Irvine KD. Chaperone activity of protein O-fucosyltransferase 1 promotes notch receptor folding. Science. 2005;307(5715):1599–1603. doi: 10.1126/science.1108995. [DOI] [PubMed] [Google Scholar]
- Okuhashi Y, Itoh M, Nara N, Tohda S. Effects of combination of notch inhibitor plus hedgehog inhibitor or Wnt inhibitor on growth of leukemia cells. Anticancer Res. 2011;31(3):893–896. [PubMed] [Google Scholar]
- Ong CT, Cheng HT, Chang LW, Ohtsuka T, Kageyama R, Stormo GD, et al. Target selectivity of vertebrate notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J Biol Chem. 2006;281(8):5106–5119. doi: 10.1074/jbc.M506108200. [DOI] [PubMed] [Google Scholar]
- Osipo C, Golde TE, Osborne BA, Miele LA. Off the beaten pathway: the complex cross talk between Notch and NF-kappaB. Lab Invest. 2008;88(1):11–17. doi: 10.1038/labinvest.3700700. [DOI] [PubMed] [Google Scholar]
- Osipo C, Patel P, Rizzo P, Clementz AG, Hao L, Golde TE, et al. ErbB-2 inhibition activates Notch-1 and sensitizes breast cancer cells to a gamma-secretase inhibitor. Oncogene. 2008;27(37):5019–5032. doi: 10.1038/onc.2008.149. [DOI] [PubMed] [Google Scholar]
- Oswald F, Kostezka U, Astrahantseff K, Bourteele S, Dillinger K, Zechner U, et al. SHARP is a novel component of the Notch/RBP-Jkappa signalling pathway. EMBO J. 2002;21(20):5417–5426. doi: 10.1093/emboj/cdf549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F, Liptay S, Adler G, Schmid RM. NF-kappaB2 is a putative target gene of activated Notch-1 via RBP-Jkappa. Mol Cell Biol. 1998;18(4):2077–2088. doi: 10.1128/mcb.18.4.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F, Winkler M, Cao Y, Astrahantseff K, Bourteele S, Knochel W, et al. RBP-Jkappa/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol Cell Biol. 2005;25(23):10379–10390. doi: 10.1128/MCB.25.23.10379-10390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo R, Checquolo S, Giovenco A, Grazioli P, Kumar V, Campese AF, et al. Acetylation controls Notch3 stability and function in T-cell leukemia. Oncogene. 2012;31(33):3807–3817. doi: 10.1038/onc.2011.533. [DOI] [PubMed] [Google Scholar]
- Pallavi SK, Ho DM, Hicks C, Miele L, Artavanis-Tsakonas S. Notch and Mef2 synergize to promote proliferation and metastasis through JNK signal activation in Drosophila. EMBO J. 2012;31(13):2895–2907. doi: 10.1038/emboj.2012.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancewicz J, Nicot C. Current views on the role of Notch signaling and the pathogenesis of human leukemia. BMC Cancer. 2011;11:502. doi: 10.1186/1471-2407-11-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya K, Meeke K, Clementz AG, Rogowski A, Roberts J, Miele L, et al. Targeting both Notch and ErbB-2 signalling pathways is required for prevention of ErbB-2-positive breast tumour recurrence. Br J Cancer. 2011;105(6):796–806. doi: 10.1038/bjc.2011.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panin VM, Papayannopoulos V, Wilson R, Irvine KD. Fringe modulates Notch-ligand interactions. Nature. 1997;387(6636):908–912. doi: 10.1038/43191. [DOI] [PubMed] [Google Scholar]
- Pannuti A, Foreman K, Rizzo P, Osipo C, Golde T, Osborne B, et al. Targeting Notch to target cancer stem cells. Clin Cancer Res. 2010;16(12):3141–3152. doi: 10.1158/1078-0432.CCR-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AL, Klueg KM, Stout JR, Muskavitch MA. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development. 2000;127(7):1373–1385. doi: 10.1242/dev.127.7.1373. [DOI] [PubMed] [Google Scholar]
- Parks AL, Stout JR, Shepard SB, Klueg KM, Dos Santos AA, Parody TR, et al. Structure-function analysis of delta trafficking, receptor binding and signaling in Drosophila. Genetics. 2006;174(4):1947–1961. doi: 10.1534/genetics.106.061630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr C, Watkins G, Jiang WG. The possible correlation of Notch-1 and Notch-2 with clinical outcome and tumour clinicopathological parameters in human breast cancer. Int J Mol Med. 2004;14(5):779–786. doi: 10.3892/ijmm.14.5.779. [DOI] [PubMed] [Google Scholar]
- Pavlopoulos E, Pitsouli C, Klueg KM, Muskavitch MA, Moschonas NK, Delidakis C. neuralized Encodes a peripheral membrane protein involved in delta signaling and endocytosis. Dev Cell. 2001;1(6):807–816. doi: 10.1016/s1534-5807(01)00093-4. [DOI] [PubMed] [Google Scholar]
- Pinchot SN, Jaskula-Sztul R, Ning L, Peters NR, Cook MR, Kunnimalaiyaan M, et al. Identification and validation of Notch pathway activating compounds through a novel high-throughput screening method. Cancer. 2011;117(7):1386–1398. doi: 10.1002/cncr.25652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintar A, De Biasio A, Popovic M, Ivanova N, Pongor S. The intracellular region of Notch ligands: does the tail make the difference? Biol Direct. 2007;2:19. doi: 10.1186/1745-6150-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popko-Scibor AE, Lindberg MJ, Hansson ML, Holmlund T, Wallberg AE. Ubiquitination of Notch1 is regulated by MAML1-mediated p300 acetylation of Notch1. Biochem Biophys Res Commun. 2011;416(3–4):300–306. doi: 10.1016/j.bbrc.2011.11.030. [DOI] [PubMed] [Google Scholar]
- Prokop S, Shirotani K, Edbauer D, Haass C, Steiner H. Requirement of PEN-2 for stabilization of the presenilin N-/C-terminal fragment heterodimer within the gamma-secretase complex. J Biol Chem. 2004;279(22):23255–23261. doi: 10.1074/jbc.M401789200. [DOI] [PubMed] [Google Scholar]
- Purow BW, Haque RM, Noel MW, Su Q, Burdick MJ, Lee J, et al. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res. 2005;65(6):2353–2363. doi: 10.1158/0008-5472.CAN-04-1890. [DOI] [PubMed] [Google Scholar]
- Qi R, An H, Yu Y, Zhang M, Liu S, Xu H, et al. Notch1 signaling inhibits growth of human hepatocellular carcinoma through induction of cell cycle arrest and apoptosis. Cancer Res. 2003;63(23):8323–8329. [PubMed] [Google Scholar]
- Qin H, Wang J, Liang Y, Taniguchi Y, Tanigaki K, Han H. RING1 inhibits transactivation of RBP-J by Notch through interaction with LIM protein KyoT2. Nucleic Acids Res. 2004;32(4):1492–1501. doi: 10.1093/nar/gkh295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L, Joazeiro C, Fang N, Wang HY, Elly C, Altman Y, et al. Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J Biol Chem. 2000;275(46):35734–35737. doi: 10.1074/jbc.M007300200. [DOI] [PubMed] [Google Scholar]
- Raafat A, Lawson S, Bargo S, Klauzinska M, Strizzi L, Goldhar AS, et al. Rbpj conditional knockout reveals distinct functions of Notch4/Int3 in mammary gland development and tumorigenesis. Oncogene. 2009;28(2):219–230. doi: 10.1038/onc.2008.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae FK, Stephenson SA, Nicol DL, Clements JA. Novel association of a diverse range of genes with renal cell carcinoma as identified by differential display. Int J Cancer. 2000;88(5):726–732. doi: 10.1002/1097-0215(20001201)88:5<726::aid-ijc7>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Raimondi L, Ciarapica R, De Salvo M, Verginelli F, Gueguen M, Martini C, et al. Inhibition of Notch3 signalling induces rhabdomyosarcoma cell differentiation promoting p38 phosphorylation and p21(Cip1) expression and hampers tumour cell growth in vitro and in vivo. Cell Death Differ. 2012;19(5):871–881. doi: 10.1038/cdd.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan A, Talora C, Okuyama R, Nicolas M, Mammucari C, Oh H, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20(13):3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangasamy V, Mishra R, Sondarva G, Das S, Lee TH, Bakowska JC, et al. Mixed-lineage kinase 3 phosphorylates prolyl-isomerase Pin1 to regulate its nuclear translocation and cellular function. Proc Natl Acad Sci U S A. 2012;109(21):8149–8154. doi: 10.1073/pnas.1200804109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell. 1991;67(4):687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M. Regulation of enzyme levels by proteolysis: the role of pest regions. Adv Enzyme Regul. 1988;27:135–151. doi: 10.1016/0065-2571(88)90014-3. [DOI] [PubMed] [Google Scholar]
- Restivo G, Nguyen BC, Dziunycz P, Ristorcelli E, Ryan RJ, Ozuysal OY, et al. IRF6 is a mediator of Notch pro-differentiation and tumour suppressive function in keratinocytes. EMBO J. 2011;30(22):4571–4585. doi: 10.1038/emboj.2011.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds ND, Lukacs NW, Long N, Karpus WJ. Delta-like ligand 4 regulates central nervous system T cell accumulation during experimental autoimmune encephalomyelitis. J Immunol. 2011;187(5):2803–2813. doi: 10.4049/jimmunol.1100160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway J, Zhang G, Wu Y, Stawicki S, Liang WC, Chanthery Y, et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444(7122):1083–1087. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- Rizzo P, Miao H, D’Souza G, Osipo C, Song LL, Yun J, et al. Cross-talk between notch and the estrogen receptor in breast cancer suggests novel therapeutic approaches. Cancer Res. 2008;68(13):5226–5235. doi: 10.1158/0008-5472.CAN-07-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo P, Osipo C, Foreman K, Golde T, Osborne B, Miele L. Rational targeting of Notch signaling in cancer. Oncogene. 2008;27(38):5124–5131. doi: 10.1038/onc.2008.226. [DOI] [PubMed] [Google Scholar]
- Robinson DR, Kalyana-Sundaram S, Wu YM, Shankar S, Cao X, Ateeq B, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med. 2011;17(12):1646–1651. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchini C, Capobianco AJ. Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): implication for cell cycle disruption in transformation by Notch(ic) Mol Cell Biol. 2001;21(17):5925–5934. doi: 10.1128/MCB.21.17.5925-5934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustighi A, Tiberi L, Soldano A, Napoli M, Nuciforo P, Rosato A, et al. The prolyl-isomerase Pin1 is a Notch1 target that enhances Notch1 activation in cancer. Nat Cell Biol. 2009;11(2):133–142. doi: 10.1038/ncb1822. [DOI] [PubMed] [Google Scholar]
- Sakata T, Sakaguchi H, Tsuda L, Higashitani A, Aigaki T, Matsuno K, et al. Drosophila Nedd4 regulates endocytosis of notch and suppresses its ligand-independent activation. Curr Biol. 2004;14(24):2228–2236. doi: 10.1016/j.cub.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Sanalkumar R, Dhanesh SB, James J. Non-canonical activation of Notch signaling/target genes in vertebrates. Cell Mol Life Sci. 2010;67(17):2957–2968. doi: 10.1007/s00018-010-0391-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Irizarry C, Carpenter AC, Weng AP, Pear WS, Aster JC, Blacklow SC. Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol Cell Biol. 2004;24(21):9265–9273. doi: 10.1128/MCB.24.21.9265-9273.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone P, Storci G, Giovannini C, Pandolfi S, Pianetti S, Taffurelli M, et al. p66Shc/Notch-3 interplay controls self-renewal and hypoxia survival in human stem/progenitor cells of the mammary gland expanded in vitro as mammospheres. Stem Cells. 2007;25(3):807–815. doi: 10.1634/stemcells.2006-0442. [DOI] [PubMed] [Google Scholar]
- Santagata S, Demichelis F, Riva A, Varambally S, Hofer MD, Kutok JL, et al. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 2004;64(19):6854–6857. doi: 10.1158/0008-5472.CAN-04-2500. [DOI] [PubMed] [Google Scholar]
- Santolini E, Puri C, Salcini AE, Gagliani MC, Pelicci PG, Tacchetti C, et al. Numb is an endocytic protein. J Cell Biol. 2000;151(6):1345–1352. doi: 10.1083/jcb.151.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmento LM, Huang H, Limon A, Gordon W, Fernandes J, Tavares MJ, et al. Notch1 modulates timing of G1-S progression by inducing SKP2 transcription and p27 Kip1 degradation. J Exp Med. 2005;202(1):157–168. doi: 10.1084/jem.20050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena MT, Schroeter EH, Mumm JS, Kopan R. Murine notch homologs (N1–4) undergo presenilin-dependent proteolysis. J Biol Chem. 2001;276(43):40268–40273. doi: 10.1074/jbc.M107234200. [DOI] [PubMed] [Google Scholar]
- Scehnet JS, Jiang W, Kumar SR, Krasnoperov V, Trindade A, Benedito R, et al. Inhibition of Dll4-mediated signaling induces proliferation of immature vessels and results in poor tissue perfusion. Blood. 2007;109(11):4753–4760. doi: 10.1182/blood-2006-12-063933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MH, Bicker F, Nikolic I, Meister J, Babuke T, Picuric S, et al. Epidermal growth factor-like domain 7 (EGFL7) modulates Notch signalling and affects neural stem cell renewal. Nat Cell Biol. 2009;11(7):873–880. doi: 10.1038/ncb1896. [DOI] [PubMed] [Google Scholar]
- Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393(6683):382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- Shawber C, Boulter J, Lindsell CE, Weinmaster G. Jagged2: a serrate-like gene expressed during rat embryogenesis. Dev Biol. 1996;180(1):370–376. doi: 10.1006/dbio.1996.0310. [DOI] [PubMed] [Google Scholar]
- Shepherd TG, Kockeritz L, Szrajber MR, Muller WJ, Hassell JA. The pea3 subfamily ets genes are required for HER2/Neu-mediated mammary oncogenesis. Curr Biol. 2001;11(22):1739–1748. doi: 10.1016/s0960-9822(01)00536-x. [DOI] [PubMed] [Google Scholar]
- Shi S, Stanley P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc Natl Acad Sci U S A. 2003;100(9):5234–5239. doi: 10.1073/pnas.0831126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih AH, Holland EC. Notch signaling enhances nestin expression in gliomas. Neoplasia. 2006;8(12):1072–1082. doi: 10.1593/neo.06526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Chiba S, Kumano K, Hosoya N, Takahashi T, Kanda Y, et al. Mouse jagged1 physically interacts with notch2 and other notch receptors. Assessment by quantitative methods. J Biol Chem. 1999;274(46):32961–32969. doi: 10.1074/jbc.274.46.32961. [DOI] [PubMed] [Google Scholar]
- Shutter JR, Scully S, Fan W, Richards WG, Kitajewski J, Deblandre GA, et al. Dll4, a novel Notch ligand expressed in arterial endothelium. Genes Dev. 2000;14(11):1313–1318. [PMC free article] [PubMed] [Google Scholar]
- Six E, Ndiaye D, Laabi Y, Brou C, Gupta-Rossi N, Israel A, et al. The Notch ligand Delta1 is sequentially cleaved by an ADAM protease and gamma-secretase. Proc Natl Acad Sci U S A. 2003;100(13):7638–7643. doi: 10.1073/pnas.1230693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small D, Kovalenko D, Kacer D, Liaw L, Landriscina M, Di Serio C, et al. Soluble Jagged 1 represses the function of its transmembrane form to induce the formation of the Src-dependent chordlike phenotype. J Biol Chem. 2001;276(34):32022–32030. doi: 10.1074/jbc.M100933200. [DOI] [PubMed] [Google Scholar]
- Smas CM, Chen L, Sul HS. Cleavage of membrane-associated pref-1 generates a soluble inhibitor of adipocyte differentiation. Mol Cell Biol. 1997;17(2):977–988. doi: 10.1128/mcb.17.2.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Warner SA, Spiegelman D, Yaun SS, Albanes D, Beeson WL, van den Brandt PA, et al. Fruits, vegetables and lung cancer: a pooled analysis of cohort studies. Int J Cancer. 2003;107(6):1001–1011. doi: 10.1002/ijc.11490. [DOI] [PubMed] [Google Scholar]
- Soares R, Balogh G, Guo S, Gartner F, Russo J, Schmitt F. Evidence for the notch signaling pathway on the role of estrogen in angiogenesis. Mol Endocrinol. 2004;18(9):2333–2343. doi: 10.1210/me.2003-0362. [DOI] [PubMed] [Google Scholar]
- Song LL, Peng Y, Yun J, Rizzo P, Chaturvedi V, Weijzen S, et al. Notch-1 associates with IKKalpha and regulates IKK activity in cervical cancer cells. Oncogene. 2008;27(44):5833–5844. doi: 10.1038/onc.2008.190. [DOI] [PubMed] [Google Scholar]
- Sprinzak D, Lakhanpal A, Lebon L, Santat LA, Fontes ME, Anderson GA, et al. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature. 2010;465(7294):86–90. doi: 10.1038/nature08959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam D, Ponnurangam S, Ramamoorthy P, Standing D, Battafarano RJ, Anant S, et al. Curcumin Induces Cell Death in Esophageal Cancer Cells through Modulating Notch Signaling. PLoS One. 2012;7(2):e30590. doi: 10.1371/journal.pone.0030590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Aoki D, Susumu N, Udagawa Y, Nozawa S. Imbalanced expression of TAN-1 and human Notch4 in endometrial cancers. Int J Oncol. 2000;17(6):1131–1139. doi: 10.3892/ijo.17.6.1131. [DOI] [PubMed] [Google Scholar]
- Tagami S, Okochi M, Yanagida K, Ikuta A, Fukumori A, Matsumoto N, et al. Regulation of Notch signaling by dynamic changes in the precision of S3 cleavage of Notch-1. Mol Cell Biol. 2008;28(1):165–176. doi: 10.1128/MCB.00863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammam J, Ware C, Efferson C, O’Neil J, Rao S, Qu X, et al. Down-regulation of the Notch pathway mediated by a gamma-secretase inhibitor induces anti-tumour effects in mouse models of T-cell leukaemia. Br J Pharmacol. 2009;158(5):1183–1195. doi: 10.1111/j.1476-5381.2009.00389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, et al. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H) Curr Biol. 1995;5(12):1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204(8):1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston G, Noguera-Troise I, Yancopoulos GD. The Delta paradox: DLL4 blockade leads to more tumour vessels but less tumour growth. Nat Rev Cancer. 2007;7(5):327–331. doi: 10.1038/nrc2130. [DOI] [PubMed] [Google Scholar]
- Tiyanont K, Wales TE, Aste-Amezaga M, Aster JC, Engen JR, Blacklow SC. Evidence for increased exposure of the Notch1 metalloprotease cleavage site upon conversion to an activated conformation. Structure. 2011;19(4):546–554. doi: 10.1016/j.str.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohda S, Nara N. Expression of Notch1 and Jagged1 proteins in acute myeloid leukemia cells. Leuk Lymphoma. 2001;42(3):467–472. doi: 10.3109/10428190109064603. [DOI] [PubMed] [Google Scholar]
- Trimble MS, Xin JH, Guy CT, Muller WJ, Hassell JA. PEA3 is overexpressed in mouse metastatic mammary adenocarcinomas. Oncogene. 1993;8(11):3037–3042. [PubMed] [Google Scholar]
- Truong M, Cook MR, Pinchot SN, Kunnimalaiyaan M, Chen H. Resveratrol induces Notch2-mediated apoptosis and suppression of neuroendocrine markers in medullary thyroid cancer. Ann Surg Oncol. 2011;18(5):1506–1511. doi: 10.1245/s10434-010-1488-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyttendaele H, Marazzi G, Wu G, Yan Q, Sassoon D, Kitajewski J. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development. 1996;122(7):2251–2259. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- Varnum-Finney B, Wu L, Yu M, Brashem-Stein C, Staats S, Flowers D, et al. Immobilization of Notch ligand, Delta-1, is required for induction of notch signaling. J Cell Sci 113 Pt. 2000;23:4313–4318. doi: 10.1242/jcs.113.23.4313. [DOI] [PubMed] [Google Scholar]
- Wallberg AE, Pedersen K, Lendahl U, Roeder RG. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol Cell Biol. 2002;22(22):7812–7819. doi: 10.1128/MCB.22.22.7812-7819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zou J, Zhao B, Johannsen E, Ashworth T, Wong H, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proc Natl Acad Sci U S A. 2011;108(36):14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Xue L, Cao Q, Lin Y, Ding Y, Yang P, et al. Expression of Notch1, Jagged1 and beta-catenin and their clinicopathological significance in hepatocellular carcinoma. Neoplasma. 2009;56(6):533–541. doi: 10.4149/neo_2009_06_533. [DOI] [PubMed] [Google Scholar]
- Wang Z, Azmi AS, Ahmad A, Banerjee S, Wang S, Sarkar FH, et al. TW-37, a small-molecule inhibitor of Bcl-2, inhibits cell growth and induces apoptosis in pancreatic cancer: involvement of Notch-1 signaling pathway. Cancer Res. 2009;69(7):2757–2765. doi: 10.1158/0008-5472.CAN-08-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li Y, Ahmad A, Banerjee S, Azmi AS, Kong D, et al. Down-regulation of Notch-1 is associated with Akt and FoxM1 in inducing cell growth inhibition and apoptosis in prostate cancer cells. J Cell Biochem. 2011;112(1):78–88. doi: 10.1002/jcb.22770. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang Z, Zhang Y, Banerjee S, Li Y, Sarkar FH. Inhibition of nuclear factor kappab activity by genistein is mediated via Notch-1 signaling pathway in pancreatic cancer cells. Int J Cancer. 2006a;118(8):1930–1936. doi: 10.1002/ijc.21589. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang Y, Banerjee S, Li Y, Sarkar FH. Notch-1 down-regulation by curcumin is associated with the inhibition of cell growth and the induction of apoptosis in pancreatic cancer cells. Cancer. 2006b;106(11):2503–2513. doi: 10.1002/cncr.21904. [DOI] [PubMed] [Google Scholar]
- Wei P, Walls M, Qiu M, Ding R, Denlinger RH, Wong A, et al. Evaluation of selective gamma-secretase inhibitor PF-03084014 for its antitumor efficacy and gastrointestinal safety to guide optimal clinical trial design. Mol Cancer Ther. 2010;9(6):1618–1628. doi: 10.1158/1535-7163.MCT-10-0034. [DOI] [PubMed] [Google Scholar]
- Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat Med. 2002;8(9):979–986. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- Weinmaster G, Roberts VJ, Lemke G. Notch2: a second mammalian Notch gene. Development. 1992;116(4):931–941. doi: 10.1242/dev.116.4.931. [DOI] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20(15):2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng AP, Nam Y, Wolfe MS, Pear WS, Griffin JD, Blacklow SC, et al. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol Cell Biol. 2003;23(2):655–664. doi: 10.1128/MCB.23.2.655-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong GT, Manfra D, Poulet FM, Zhang Q, Josien H, Bara T, et al. Chronic treatment with the gamma-secretase inhibitor LY-411,575 inhibits beta-amyloid peptide production and alters lymphopoiesis and intestinal cell differentiation. J Biol Chem. 2004;279(13):12876–12882. doi: 10.1074/jbc.M311652200. [DOI] [PubMed] [Google Scholar]
- Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD. MAML1, a human homologue of Drosophila mastermind, is a transcriptional coactivator for NOTCH receptors. Nat Genet. 2000;26(4):484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464(7291):1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- Xu K, Usary J, Kousis PC, Prat A, Wang DY, Adams JR, et al. Lunatic Fringe Deficiency Cooperates with the Met/Caveolin Gene Amplicon to Induce Basal-like Breast Cancer. Cancer Cell. 2012;21(5):626–641. doi: 10.1016/j.ccr.2012.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Plowman GD. Delta-like 4/Notch signaling and its therapeutic implications. Clin Cancer Res. 2007;13(24):7243–7246. doi: 10.1158/1078-0432.CCR-07-1393. [DOI] [PubMed] [Google Scholar]
- Yao J, Qian C. Inhibition of Notch3 enhances sensitivity to gemcitabine in pancreatic cancer through an inactivation of PI3K/Akt-dependent pathway. Med Oncol. 2010;27(3):1017–1022. doi: 10.1007/s12032-009-9326-5. [DOI] [PubMed] [Google Scholar]
- Yao K, Rizzo P, Rajan P, Albain K, Rychlik K, Shah S, et al. Notch-1 and notch-4 receptors as prognostic markers in breast cancer. Int J Surg Pathol. 2011;19(5):607–613. doi: 10.1177/1066896910362080. [DOI] [PubMed] [Google Scholar]
- Yao Z, Mishra L. Cancer stem cells and hepatocellular carcinoma. Cancer Biol Ther. 2009;8(18):1691–1698. doi: 10.4161/cbt.8.18.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatim A, Benne C, Sobhian B, Laurent-Chabalier S, Deas O, Judde JG, et al. NOTCH1 Nuclear Interactome Reveals Key Regulators of Its Transcriptional Activity and Oncogenic Function. Mol Cell. 2012;48(3):445–458. doi: 10.1016/j.molcel.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh TS, Wu CW, Hsu KW, Liao WJ, Yang MC, Li AF, et al. The activated Notch1 signal pathway is associated with gastric cancer progression through cyclooxygenase-2. Cancer Res. 2009;69(12):5039–5048. doi: 10.1158/0008-5472.CAN-08-4021. [DOI] [PubMed] [Google Scholar]
- Yu B, Wei J, Qian X, Lei D, Ma Q, Liu Y. Notch1 signaling pathway participates in cancer invasion by regulating MMPs in lingual squamous cell carcinoma. Oncol Rep. 2012;27(2):547–552. doi: 10.3892/or.2011.1492. [DOI] [PubMed] [Google Scholar]
- Zagouras P, Stifani S, Blaumueller CM, Carcangiu ML, Artavanis-Tsakonas S. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc Natl Acad Sci U S A. 1995;92(14):6414–6418. doi: 10.1073/pnas.92.14.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q, Li S, Chepeha DB, Giordano TJ, Li J, Zhang H, et al. Crosstalk between tumor and endothelial cells promotes tumor angiogenesis by MAPK activation of Notch signaling. Cancer Cell. 2005;8(1):13–23. doi: 10.1016/j.ccr.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Zhang N, Fu Z, Linke S, Chicher J, Gorman JJ, Visk D, et al. The asparaginyl hydroxylase factor inhibiting HIF-1alpha is an essential regulator of metabolism. Cell Metab. 2010;11(5):364–378. doi: 10.1016/j.cmet.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Luo WJ, Wang H, Lin P, Vetrivel KS, Liao F, et al. Nicastrin is critical for stability and trafficking but not association of other presenilin/gamma-secretase components. J Biol Chem. 2005;280(17):17020–17026. doi: 10.1074/jbc.M409467200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZP, Sun YL, Fu L, Gu F, Zhang L, Hao XS. Correlation of Notch1 expression and activation to cisplatin-sensitivity of head and neck squamous cell carcinoma. Ai Zheng. 2009;28(2):100–103. [PubMed] [Google Scholar]
- Zhao B, Zou J, Wang H, Johannsen E, Peng CW, Quackenbush J, et al. Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc Natl Acad Sci U S A. 2011;108(36):14902–14907. doi: 10.1073/pnas.1108892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Linke S, Dias JM, Gradin K, Wallis TP, Hamilton BR, et al. Interaction with factor inhibiting HIF-1 defines an additional mode of cross-coupling between the Notch and hypoxia signaling pathways. Proc Natl Acad Sci U S A. 2008;105(9):3368–3373. doi: 10.1073/pnas.0711591105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BB, Zhang H, Damelin M, Geles KG, Grindley JC, Dirks PB. Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat Rev Drug Discov. 2009;8(10):806–823. doi: 10.1038/nrd2137. [DOI] [PubMed] [Google Scholar]
- Zhou W, Kallifatidis G, Baumann B, Rausch V, Mattern J, Gladkich J, et al. Dietary polyphenol quercetin targets pancreatic cancer stem cells. Int J Oncol. 2010;37(3):551–561. doi: 10.3892/ijo_00000704. [DOI] [PubMed] [Google Scholar]
- Zweidler-McKay PA. Notch signaling in pediatric malignancies. Curr Oncol Rep. 2008;10(6):459–468. doi: 10.1007/s11912-008-0071-2. [DOI] [PubMed] [Google Scholar]