Abstract
Fibroblasts are known to eliminate intracellular bacteria, but the lethal hit of the bactericidal mechanism has not been defined. We show that primary embryonic and established fibroblasts can be induced by interferons or by intracellular bacterial infection to express a perforin-like mRNA previously described as macrophage-expressed gene 1 (Mpeg1). The presence and level of the perforin-like mRNA correlate with the ability of primary mouse embryonic fibroblasts (MEF) to eliminate intracellular bacteria. In addition, siRNA knockdown of the perforin-like molecule abolishes bactericidal activity and allows intracellular bacterial replication. Complementation of MEF in which the endogenous perforin-like molecule has been knocked down with a red fluorescent protein-tagged version restores bactericidal activity. The perforin-like molecule has broad bactericidal specificity for pathogenic and non-pathogenic bacteria, including Gram-positive and -negative, and acid fast bacteria. The perforin-like molecule renders previously lysozyme-resistant bacteria sensitive to lysis by lysozyme suggesting physical damage of the outer cell wall by the perforin-like protein. MEF damage cell walls of intracellular bacteria by insertion, polymerization, and pore formation of the perforin-like protein, analogous to pore formers of complement and perforin-1 of cytolytic lymphocytes. We propose the name perforin-2.
Key Words: Bactericidal activity, Immune defense, Innate immunity, Intracellular bacterial replication, Non-phagocytic cells
Introduction
The macrophage-expressed gene 1 (Mpeg1), described in 1995 [1], predicts a perforin-like protein that has been highly conserved throughout evolution (fig. 1). An orthologue of the perforin-like molecule is found already in the earliest metazoan phylum, the sponges (Porifera) that were already present 500 million years ago. In the species Suberites domuncula, Gram-negative bacteria are recognized by a lipopolysaccharide (LPS)-interacting protein inducing the mRNAs for MyD88 and the perforin-like molecule, the latter of which is involved as an ‘executing protein’ in the elimination of Gram-negative but not Gram-positive bacteria [2]. In an evolutionary analysis, perforin of cytotoxic T cells is detected in all Euteleostomi, and may be present in Gnathostomata, but is not found in earlier species suggesting that perforin of cytotoxic lymphocytes and the pore-forming proteins of complement in plasma arose from a primordial perforin gene [3], namely the Mpeg1 gene encoding the perforin-like protein.
Fig. 1.
Phylogenetic conservation of the perforin-like protein. Alignment of predicted protein sequences from several species (Vector NTI, Invitrogen). The MACPF domain is boxed, and the transmembrane domain highlighted in yellow. Color code: red font and yellow highlight = identity in all species; blue highlight = identity in most species; green highlight = identity or conservative replacement in most species listed. Colors refer to the online version only.
The perforin-like protein is predicted to contain a membrane attack complex (MAC)/perforin (MACPF) domain which is also found in the pore-forming proteins of the MAC of complement [4, 5] that kill extracellular bacteria and in the pore-forming protein perforin-1 (P-1) used by cytotoxic T cells and natural killer cells that kill virus-infected cells [6, 7, 8, 9]. The MACPF domain has been identified as the killer domain in perforin responsible for polymerization, membrane insertion, and pore formation [10, 11]. A similar function of the MACPF domain is likely for the perforin-like molecule in macrophages.
Materials and Methods
Plasmid Constructs
The complete coding region of murine Mpeg1 cDNA was constructed from several EST clones and inserted into the pEGFP-N3 plasmid (Clontech). Monomeric red fluorescent protein (RFP; R. Flavell, Yale [12]) was cloned in place of the green fluorescent protein for use in some experiments.
Cells
Murine embryonic fibroblasts (MEF) were isolated as previously described [13]. Thioglycolate-elicited peritoneal macrophages were induced by injecting 1.5 ml of a 3% thioglycolate solution i.p. into C57/B6 mice. Four days later, peritoneal cells were harvested and macrophages were purified by adherence.
Chemicals and Cytokines
MG132, chicken egg white lysozyme, and LPS were purchased from Sigma. Recombinant murine interleukin (IL)-1α, IL-1β, tumor necrosis factor (TNF)-α, interferon (IFN)-γ, IFN-α, IFN-β, and recombinant human IFN-γ and IFN-β were purchased from PeproTech. Recombinant human IFN-α was purchased from R&D systems. Murine IL-1α was supplemented at 10 U/ml where indicated, murine IL-1β at 1 ng/ml, murine TNF-α at 20 ng/ml, and murine IFN-α, IFN-β and IFN-γ were supplemented at 100 U/ml.
Antibodies
Rabbit anti-Mpeg1 polyclonal antibodies were obtained from Abcam and used for Western blot analysis.
qRT-PCR
RNA was extracted from cells following RNeasy (Qiagen) instructions. One microgram of RNA was converted to cDNA using the QuantiTect Reverse Transcription kit (Qiagen) following the supplier's protocol. qRT-PCR was performed using TaqMan® gene expression assays (Applied Biosystems) for murine Mpeg1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), with the latter serving as a housekeeping control gene. All assays were performed on the Applied Biosystems 7300 PCR platform.
Gentamicin Protection Assay for Killing of Intracellular Bacteria
Salmonella typhimurium strain LT2Z (G. Plano, University of Miami), Mycobacterium smegmatis (ATCC), methicillin-resistant Staphylococcus aureus (MRSA; L. Plano, University of Miami), and K12 Escherichia coli were grown from glycerol stocks at 37°C with shaking for 16–18 h in Luria broth (S. typhimurium, MRSA, and E. coli) or 24 h in Middlebrook 7H9 broth (mycobacteria) prior to infection. For S. typhimurium, the culture was then diluted 1:33 in Luria broth and grown for another 3 h to induce the invasive phenotype. MEF were transfected following Lonza's optimized protocol and plated into 12-well plates after transfection. The cells were then stimulated for 14 h with IFN-γ (100 U/ml). Cells were infected at a multiplicity of infection between 10 and 60 for 30 min (S. typhimurium) or 1 h (MRSA, E. coli, and M. smegmatis) at 37°C in a 5% CO2 incubator. After infection, cells were washed twice with ice-cold PBS and fresh media containing gentamicin (50 µg/ml) were added. After 2 h, the media were changed to decrease the concentration of gentamicin (5 µg/ml). At the indicated time points, cells were washed with PBS, lysed using 1% Igepal in double-distilled H2O, diluted and plated in technical triplicates on Luria broth agar plates (S. typhimurium, S. aureus, and E. coli) or Middlebrook 7H11 plates (mycobacteria) and colony-forming units (CFU) were determined after colony growth [14].
RNA Interference
Three Mpeg1-specific chemically synthesized 19-nucleotide siRNA duplexes were obtained from Sigma. Two siRNAs were complementary to the 3′ untranslated region (UTR) of perforin-2 (P-2) and the third complementary to the coding region. The sequences were as follows: CCACCUCACUUUCUAUCAA, GA-GUAUUCUAGGAAACUUU, and CAAUCAAGCUCUUGUGCAC. A scramble siRNA was also generated to serve as a control to the reaction. Transfection of siRNA into all cells was carried out using the Amaxa Nucleofector System (Lonza) according to the manufacturer's instructions. All transfections were carried out using 1–4 × 106 cells and a final concentration of 1 µM siRNA (P-2-specific siRNA were pooled) and 2 µg of plasmid DNA where indicated. Immediately after transfection, cells were plated in antibiotic-free complete medium.
Lysozyme Susceptibility Assay
The assay was carried out as described for the gentamicin protection assay. Following lysis, lysates were divided into 6 equal fractions, treating half to achieve a final concentration 40 µg/ml lysozyme and the other half with equal volume buffer. All fractions were incubated on ice for 30 min prior to plating in technical duplicates for CFU analysis.
Statistical Analyses
GraphPad Prism was used for all statistical analyses. Student's t test was used for estimation of statistical significance as indicated in the figure legends.
Results
Fibroblasts Express an IFN-Inducible Perforin-Like Molecule
It is known that MEF in vitro are able to eliminate bacteria [for a review, see [15]]. Invasion of MEF by S. typhimurium is associated with LC3, a marker of autophagy. Moreover, bacterial replication is increased in autophagy-deficient cells, implying a role in the control of intracellular replication [16, 17, 18]. In vivo, the lethality of intragastric S. typhimurium infection of infant mice was significantly reduced by pretreatment with fibroblast IFN [19]. However, the molecular mediator of the ‘lethal hit’ remains undefined.
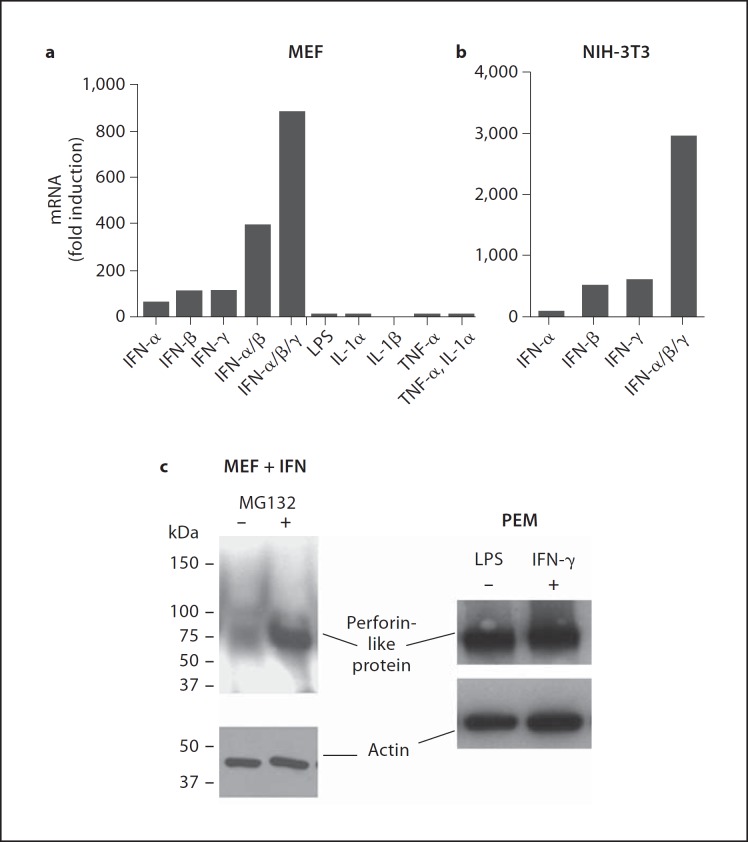
We investigated whether primary MEF express the mRNA for the perforin-like molecule described previously in macrophages [1, 20, 21, 22, 23]. Fresh cultured MEF do not express the perforin-like molecule even after 38 rounds of amplification by real-time TaqMan-PCR (fig. 2a) while the mRNA encoding GAPDH, which was used as positive control, is detected after 16 rounds of amplification (not shown). Eukaryotes respond to microbial invasion with the generation of cytokines as early warning signals that serve to induce immune response genes. We therefore incubated MEF with IFN, LPS, and other cytokines known as early warning signals (fig. 2a). IFN-α, IFN-β, and IFN-γ, when used individually, significantly induced the perforin-like molecule about 100-fold above background (background is 38 cycles). When the IFNs were added together, a synergistic effect of up to 1,000-fold induction is seen (fig. 2a). In contrast, LPS, IL-1α and TNF-α did not induce the perforin-like molecule in MEF. Analysis of induction of the perforin-like molecule by IFN in the established fibroblast NIH-3T3 showed results similar to MEF, including the synergistic activity of the three IFN (fig. 2b).
Fig. 2.
Type I and II IFN induce perforin-like protein mRNA (Mpeg1) in fibroblasts. a Mpeg1 mRNA expression by TaqMan assay in MEF following treatment with IFN-α, IFN-β, IFN-γ, LPS, IL-1α, IL-1β, and TNF-α for 14 h (38 cycles of amplification are considered negative and set to 1 for fold calculation). b Mpeg1 mRNA expression by TaqMan assay in an NIH-3T3 fibroblast cell line following treatment with IFN-α, IFN-β, and IFN-γ for 14 h. Fold induction is relative to untreated control 3T3 cells. c Western blot analysis for perforin-like protein expression. Left panel: MEF cells were stimulated with IFN-γ for 14 h without further treatment (lane 1) or with a 1-hour treatment with the proteasome inhibitor MG132 (25 mM) prior to cell lysis (lane 2). Right panel: thioglycolate-elicted peritoneal macrophages (PEM) were left untreated (lane 1) or were stimulated with LPS and IFN-γ overnight (lane 2). β-Actin expression was used as a loading control for both blots.
Execution of bactericidal functions by the perforin-like molecule requires its translation into a protein. However, multiple attempts failed to detect the perforin-like protein by Western blotting in IFN-induced MEF expressing very high levels of mRNA (fig. 2c). As positive controls for the antibody in Western blots, we used thioglycolate-elicited peritoneal macrophages which, as expected, were positive for the protein even without IFN induction (fig. 2c). It is possible that the perforin-like protein in MEF after IFN induction is rapidly degraded after translation, thereby never reaching a steady-state level sufficient for detection in Western blots. Protein turnover and degradation is a function of the proteasome. Incubation of IFN-induced MEF with the proteasome inhibitor MG132 for 1 h revealed a strong protein band at ∼72 kDa for the perforin-like molecule corresponding to its predicted molecular weight (fig. 2c). The data suggest a rapid turnover of the perforin-like protein in MEF.
Association of Induction of the Perforin-Like Molecule with Bactericidal Activity
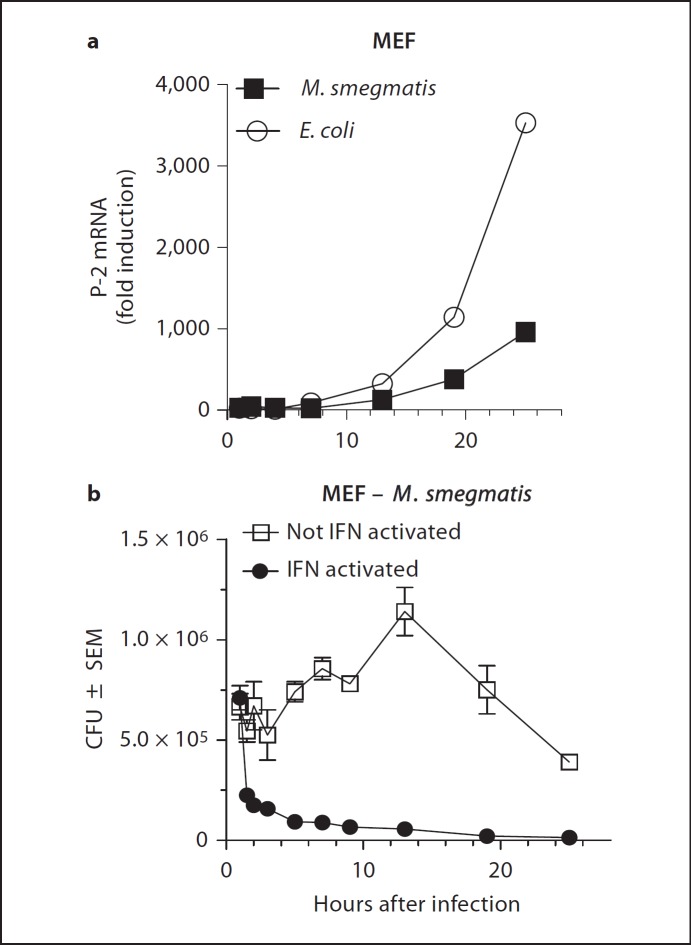
Although LPS did not induce the perforin-like molecule in MEF we determined whether intracellular infection of MEF with M. smegmatis or E. coli was able to do so. In these experiments, IFN-uninduced MEF were incubated for 1 h with bacteria at a multiplicity of infection of 50. After that time, the cells were washed to remove extracellular bacteria and plated again in the presence of gentamicin, a membrane-impermeable antibiotic, to prevent extracellular bacterial growth. Samples were harvested at timed intervals and analyzed for mRNA encoding the perforin-like molecule (fig. 3a). Both E. coli and M. smegmatis rapidly induced the perforin-like molecule in MEF, which was detectable by 8 h and continued to increase over 24 h to very high levels.
Fig. 3.
Perforin-like mRNA expression induced by IFN or by intracellular infection is associated with clearance of intracellular bacteria. a Kinetics of Mpeg1 mRNA expression in MEF following infection with E. coli or M. smegmatis. Fold induction is relative to uninfected control MEF. b MEF were induced or not induced over night with IFN and then infected for 1 h with E. coli or M. smegmatis. Intracellular bacterial killing was determined over the next 24 h in the gentamicin protection assay. The x-axis is presented in two scales to allow representation of early and late kinetics of bacterial survival and killing.
Induction of the perforin-like mRNA may be associated with bactericidal activity in MEF. We determined intracellular survival of bacteria by the gentamicin assay at different times after infection and correlated it to the level of mRNA induced by infection (fig. 3b). During the first 12 h after infection, the level of Mpeg1 mRNA encoding the perforin-like protein is low (fig. 3a) and the number of intracellular M. smegmatis increases, indicating that they replicate (fig. 3b). By 12 h after infection of MEF, the mRNA has reached detectable levels and continues to rise (fig. 3a) while the number of intracellular mycobacteria begins to decline in parallel with increasing mRNA levels (fig. 3b). The association of rising perforin-like mRNA levels with declining intracellular M. smegmatis points to a potential causal relationship between the synthesis of the perforin-like protein, possibly in conjunction with other molecules, and the elimination of intracellular infection.
As a positive control for the presence of the perforin-like molecule we used MEF pre-induced with IFN for 16 h. IFN-induced MEF expressing high levels of the perforin-like mRNA at the time of infection eliminated the majority of intracellular M. smegmatis within the first 2 h after intracellular infection. The kinetics of bactericidal killing by IFN-pre-induced MEF is also consistent with a role of the perforin-like molecule in the execution of bacterial death, possibly in concert with other induced molecules.
Knockdown of the Perforin-Like Molecule in MEF Inhibits Bactericidal Activity
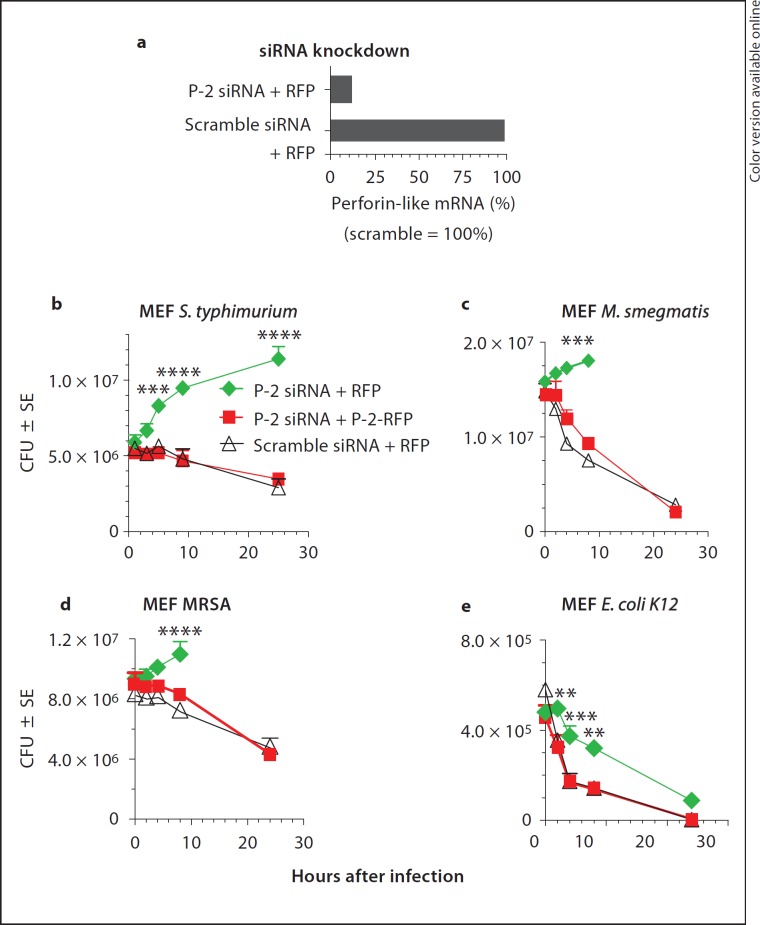
The association of mRNA expression of the perforin-like molecule with bactericidal activity for intracellular bacteria in MEF suggests that the molecule may be necessary for bacterial killing. In that case, the knockdown of the mRNA encoding the perforin-like protein is expected to inhibit bactericidal activity resulting in intracellular bacterial replication. For further validation of the role of the perforin-like protein in the killing of intracellular bacteria, we also conducted complementation experiments reconstituting cells depleted of the perforin-like mRNA and protein by transfection with a C-terminally RFP-tagged perforin-like protein (P-2-RFP). First, we induced the perforin-like mRNA with IFN-γ and concurrently knocked it down again with two siRNAs hybridizing with the 3′ UTR. Twenty hours after siRNA transfection, the knockdown of the perforin-like mRNA was routinely 90–96% (fig. 4a). As non-specific control for specificity of knockdown, we used siRNA in which the same nucleotides were scrambled and which had no suppressive effect on the expression of the perforin-like mRNA (fig. 4a). In a parallel batch, IFN-induced MEF were co-transfected with P-2 siRNA and with P-2-RFP to reconstitute endogenous perforin-like protein. The RFP-tagged form lacks the 3′ UTR of endogenous P-2 and therefore is resistant to siRNA knockdown. Twenty hours after IFN induction and transfection, MEF were ready for use in gentamicin protection assays. For infection, we used a panel of widely different bacteria, namely Gram-negative non-pathogenic E. coli laboratory strain K12 and human pathogenic Salmonella enterica, serovar Typhimurium, clinical isolates of Gram-positive MRSA, and non-pathogenic, acid fast M. smegmatis, in order to determine whether the perforin-like molecule had broad-spectrum activity or was specialized in its bactericidal activity for certain subgroups of bacteria. MEF activated with IFN-γ and transfected with scramble siRNA contain high levels of the perforin-like mRNA and are efficient killers of M. smegmatis, S. typhimurium, MRSA, and E. coli (fig. 4b, c, d, e, triangles). In contrast, P-2 siRNA-transfected MEF, in which the perforin-like molecule is knocked down, cannot kill and bacteria replicate intracellularly (fig. 4b, c, d, diamonds). Intracellular replication of M. smegmatis and MRSA in knockdown MEF results in progressive cell death beyond 8 h (data not shown) causing release of intracellular bacteria that are killed by extracellular gentamicin. The E. coli laboratory strain K12 is also killed at significantly reduced rates upon P-2 siRNA knockdown (fig. 4e, squares) compared with scramble siRNA-treated MEF containing normal levels of P-2 (fig. 4e, triangles). The perforin-like protein thus is a potent killer of E. coli, but other bactericidal factors are also able to kill non-pathogenic laboratory E. coli in fibroblasts. This is in sharp contrast to Salmonella, M. smegmatis, and MRSA, which all required the activity of the perforin-like protein for clearance from the cell.
Fig. 4.
Perforin-like protein is necessary and sufficient for killing of Gram-positive MRSA, Gram-negative S. typhimurium and acid fast M. smegmatis in MEF. a Knockdown of IFN-induced Mpeg1 mRNA by P-2 siRNA but not scramble siRNA. MEF were transfected with siRNA and RFP (as negative control for P-2-RFP) 20 h prior to infection. IFN was added 14 h prior to infection and induced Mpeg1 mRNA in scramble but not P-2 siRNA-transfected cells. b-e IFN induction was the same under all conditions. b Infection and intracellular killing of pathogenic S. typhimurium. c Infection and intracellular killing of M. smegmatis. After 8 h, replicating bacteria kill cells and are released and killed by gentamicin. d Infection and intracellular killing of pathogenic clinical isolates of MRSA. After 8 h, MRSA kill MEF and are released and killed by gentamicin. e Infection and intracellular killing of laboratory strain E. coli K12. ** p < 0.01, *** p < 0.001, **** p < 0.0001 (Student's t test).
Complementation of the perforin-like molecule in P-2-knockdown MEF was employed to further validate the role of the perforin-like protein in bactericidal activity. We tagged the P-2 protein at its C-terminus with RFP to create a P-2-RFP fusion protein. P-2 siRNA transfection of MEF suppresses endogenous P-2, meanwhile the mRNA for P-2-RFP is not suppressed because plasmid-encoded P-2-RFP does not contain the 3′ UTR of the -endogenous perforin-like molecule, which is targeted by the siRNAs. Complementation of knockdown MEF with the RFP-tagged perforin-like molecule (P-2-RFP) restored the level of bactericidal activity to the same level as that seen in control scramble siRNA-transfected MEF (fig. 4b, c, d, e, squares). The data strongly suggest that the perforin-like molecule is required for killing of intracellular bacteria in order to prevent intracellular replication of pathogenic S. typhimurium and MRSA as well as non-pathogenic M. smegmatis. Non-pathogenic laboratory strains of E. coli, on the other hand, are still eliminated, albeit at a less efficient rate, in the absence of the perforin-like molecule.
The Perforin-Like Molecule Enables the Bacteriolytic Activity of Lysozyme
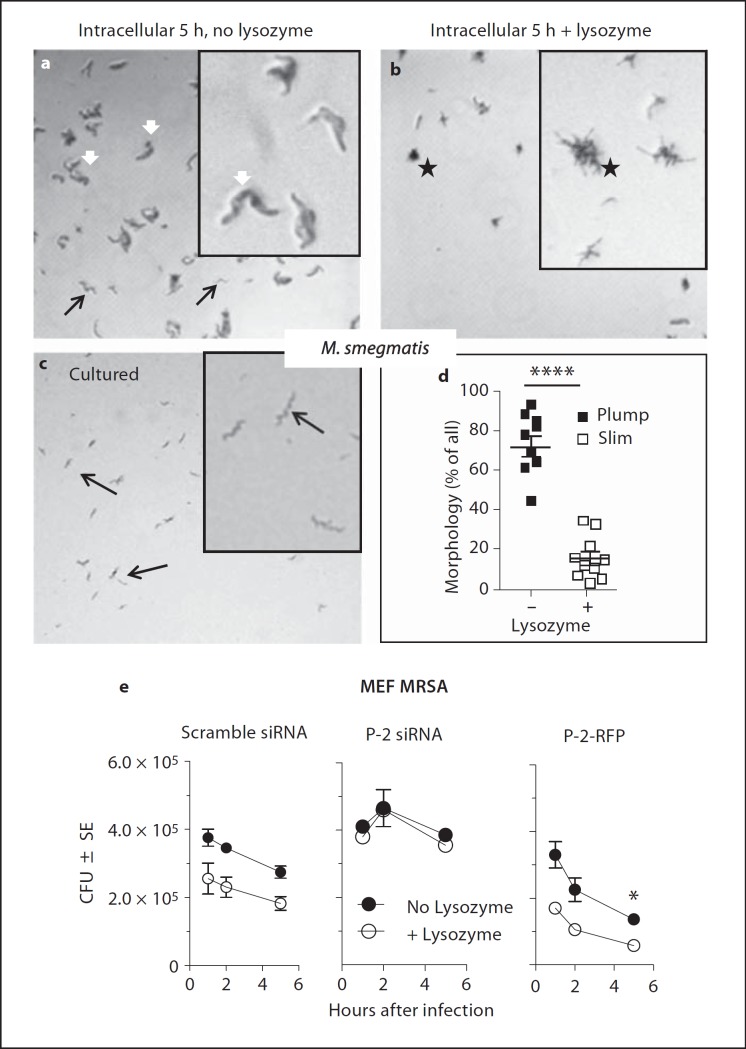
Mycobacteria require 2–3 days to form colonies visible by eye on agar plates. Inspection of plated bacteria with the phase-contrast light microscope allows determination of colony formation within 16 h of plating. Live mycobacteria plated fresh from liquid culture have thin, rod-like morphology (fig. 5c, black arrows) and can form colonies when plated. In contrast, M. smegmatis isolated from host cells 5 h after infection have lost the slim bacillus structure of M. smegmatis and instead exhibited swollen, plump bodies that did not form colonies upon plating, indicating that they were unable to replicate (fig. 5a, white arrows). Swelling to plump bodies suggested loss of the permeability barrier normally provided by the bacterial cell wall. To test this hypothesis, we added lysozyme to mycobacteria isolated from host cells 5 h after infection, incubated the bacteria on ice for 30 min and then plated them on agar over night. Lysozyme caused the disappearance of the plump M. smegmatis bodies seen in the absence of added lysozyme (fig. 5b vs. a) suggesting that they were lysed by lysozyme due to loss of the cell wall permeability barrier and degradation of the peptidoglycan layer. Remaining live bacteria formed small colonies composed of bacteria that had slim, rod-like morphology not affected by lysozyme (fig. 5b, asterisk), consistent with lysozyme resistance of intact M. smegmatis (data not shown). The data are consistent with the assumption that the perforin-like molecule of MEF damages bacterial cell walls rendering the bacteria susceptible to lysis by lysozyme. Quantitation by counting all bacilli and reporting the percentage of plump bodies before and slim bodies after treatment with lysozyme indicates that 5 h after infection 80% of M. smegmatis have plump bodies, are lysozyme sensitive and do not form colonies, and 20% have slim appearance, are lysozyme resistant and can form colonies (fig. 5d).
Fig. 5.
Perforin-like protein of MEF increases susceptibility of intracellular bacteria to lysozyme. a-c Representative images of M. smegmatis were taken by phase light microscopy (magnification: ×50) of plated M. smegmatis on Middlebrook 7H11 agar plates. Insets for each image show a 3-fold electronic zoom. a M. smegmatis isolated from IFN-stimulated MEF 5 h after infection, incubated for 30 min with PBS on ice and then plated for 12 h. White arrows point to plump bodies of single mycobacterial cells. Black arrows show surviving slim mycobacteria. b M. smegmatis isolated from IFN-stimulated MEF 5 h after infection, incubated for 30 min with 35 µg/ml lysozyme in PBS on ice and then plated for 12 h; asterisk shows microcolonies forming from live cells. Plump bodies have disappeared due to lysis by lysozyme. c Freshly cultured M. smegmatis plated prior to incubation with MEF; black arrows point to M. smegmatis slim (healthy) morphology. d Quantification of plump and slim morphology of M. smegmatis isolated from MEF 5 h after infection without and with lysozyme treatment. 1,000 bacteria were counted in each experiment and characterized by morphology with the percentage of plump and slim M. smegmatis reported before and after lysoszyme treatment, respectively. e MEF were transfected 20 h prior to infection with scramble siRNA, P-2-specific siRNA, or P-2-RFP, stimulated with IFN-γ for 14 h, and then infected with MRSA. Bacteria were released by lysis of host cells at the times indicated and tested for lysozyme susceptibility as described in the Materials and Methods section. Graphs shown are representative of 3 independent experiments with 2 replicates (means and SEM), * p < 0.05 (Student's t test).
The role of lysozyme was further studied with MRSA infection of MEF in which the perforin-like molecule was knocked down or in which P-2-RFP was transfected and expressed in addition to the endogenous perforin-like molecule (fig. 5e). In control scramble siRNA-transfected MEF containing normal levels of the perforin-like molecule, the addition of lysozyme to MRSA obtained at different times after infection by detergent lysis of host cells significantly reduced the number of colonies (fig. 5e, left panel). The lytic effect of lysozyme is not detected when the perforin-like molecule is knocked down by siRNA indicating that the perforin-like molecule is required for lysozyme sensitivity (fig. 5e, center panel). When P-2-RFP is overexpressed, the lysozyme effect is somewhat more pronounced consistent with lysozyme sensitivity dependent on the activity of the perforin-like protein (fig. 5e, right panel). Lysozyme sensitivity requires the presence of the perforin-like molecule in the infected cell.
Discussion
This study shows for the first time that fibroblasts can be induced to express the perforin-like protein encoded by Mpeg1. In addition, this is the first demonstration that the perforin-like protein is required for the elimination of intracellular bacteria. The pathogenic and non-pathogenic Gram-negative and -positive as well as the acid fast mycobacteria used in this study are susceptible to killing by the perforin-like protein despite widely different compositions of their outer cell wall. The lack of specificity is consistent with a mechanism of physical attack damaging the outer cell wall of bacteria. The perforin-like protein contains an MACPF domain predicting a pore-forming protein similar to the MAC of complement and perforin of cytotoxic T cells and natural killer cells. In analogy to MAC and perforin, the perforin-like protein of MEF may cause physical attack of bacteria by outer wall damage owing to insertion, polymerization, and pore formation of the perforin-like protein.
We also show that mycobacteria isolated from MEF within 2–4 h after infection have undergone profound morphological changes. The enlarged, plump morphology suggests that the bacteria have lost their normal control over outer cell wall permeability and have taken up large quantities of water resulting in osmotic swelling. Loss of normal permeability control may be due to damage of the outer cell wall by the perforin-like protein. Similar swelling and shape changes have been reported for E. coli attacked by 11 purified complement proteins [24]. Destruction of bacteria attacked by complement required addition of lysozyme.
The proteoglycan layer of bacteria has also been described as the bacterial cytoskeleton, functioning to give bacteria their typical shape. The proteoglycan layer is protected from lysozyme by an impermeable layer of glycolipid that prevents free diffusion of lysozyme and smaller molecules. The mycobacteria and MRSA used in this study are not susceptible to lysozyme up to 40 µg/ml when exposed after culture in nutrient broth. However, lysozyme mediates bacterial lysis when bacteria are isolated from cells between 2 and 5 h after intracellular infection. We conclude that the outer shell of the bacterial cell wall has become permeable to lysozyme due to attack and pore formation of the perforin-like protein resulting in lysozyme entry, degradation of peptidoglycan, and bacterial lysis. This chain of events is analogous to pore formation on bacteria by complement which provides access for lysozyme [24]. Similarly, the pores formed by P-1 on virus-infected or tumor cells provide access for granzymes to enter the cell and mediate lysis. We propose the designation P-2 for the perforin-like molecule expressed by MEF.
This study identifies the perforin-like molecule, P-2, as an innate defense mechanism against replication of intracellular bacteria in MEF. It remains to be seen whether the perforin-like molecule is inducible and used also by other non-phagocytic cells such as epithelial cells (HeLa), which was found in initial results in our laboratory. Furthermore, the mechanisms of triggering pore formation and bactericidal activity require additional studies which are ongoing. Autophagy has been described as an anti-bacterial mechanism in MEF [for a review, see [25]] and it is possible that P-2, which is a membrane protein, participates in the autophagic bactericidal mechanism. Finally, the biological importance of P-2 in comparison to other mechanisms of intracellular bacterial killing is not known and remains to be studied; experiments addressing this are also ongoing.
As is the case for all new discoveries in research, many new questions arise that need to be studied and answered. The one answer that appears clear already is that physical attack of microbes by components of the innate and adaptive immune system is crucial for immune defense. Complement attacks and can kill extracellular bacteria and facilitate phagocytosis. P-1 attacks virus-infected cells. P-2 attacks intracellular bacteria, at least in MEF. The MACPF domain is used by all the proteins involved in microbial attack to create a breach in the outermost defensive layer which is then used by other toxic or lytic components to complete the destruction. Thus, the efficiency of the immune killing mechanisms is derived from the combination of physical and chemical attack.
Acknowledgments
This study was supported by USPHS, NIH/NCI grant: 5R01CA039201-24 and 5PO1CA109094-05.
References
- 1.Spilsbury K, O'Mara MA, Wu WM, Rowe PB, Symonds G, Takayama Y. Isolation of a novel macrophage-specific gene by differential cDNA analysis. Blood. 1995;85:1620–1629. [PubMed] [Google Scholar]
- 2.Wiens M, Korzhev M, Krasko A, Thakur NL, Perovic-Ottstadt S, Breter HJ, Ushijima H, Diehl-Seifert B, Muller IM, Muller WE. Innate immune defense of the sponge suberites domuncula against bacteria involves a myd88-dependent signaling pathway. Induction of a perforin-like molecule. J Biol Chem. 2005;280:27949–27959. doi: 10.1074/jbc.M504049200. [DOI] [PubMed] [Google Scholar]
- 3.D'Angelo ME, Dunstone MA, Whisstock JC, Trapani JA, Bird PI. Perforin evolved from a gene duplication of MPEG1, followed by a complex pattern of gene gain and loss within Euteleostomi. BMC Evol Biol. 2012;12:59. doi: 10.1186/1471-2148-12-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DiScipio RG, Gehring MR, Podack ER, Kan CC, Hugli TE, Fey GH. Nucleotide sequence of cDNA and derived amino acid sequence of human complement component C9. Proc Natl Acad Sci USA. 1984;81:7298–7302. doi: 10.1073/pnas.81.23.7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Podack ER, Tschopp J. Polymerization of the ninth component of complement (C9): formation of poly(C9) with a tubular ultrastructure resembling the membrane attack complex of complement. Proc Natl Acad Sci USA. 1982;79:574–578. doi: 10.1073/pnas.79.2.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kagi D, Ledermann B, Burki K, Seiler P, Odermatt B, Olsen KJ, Podack ER, Zinkernagel RM, Hengartner H. Cytotoxicity mediated by T cells and natural killer cells is greatly impaired in perforin-deficient mice. Nature. 1994;369:31–37. doi: 10.1038/369031a0. [DOI] [PubMed] [Google Scholar]
- 7.Lichtenheld MG, Olsen KJ, Lu P, Lowrey DM, Hameed A, Hengartner H, Podack ER. Structure and function of human perforin. Nature. 1988;335:448–451. doi: 10.1038/335448a0. [DOI] [PubMed] [Google Scholar]
- 8.Shinkai Y, Takio K, Okumura K. Homology of perforin to the ninth component of complement (C9) Nature. 1988;334:525–527. doi: 10.1038/334525a0. [DOI] [PubMed] [Google Scholar]
- 9.Podack ER, Dennert G. Assembly of two types of tubules with putative cytolytic function by cloned natural killer cells. Nature. 1983;302:442–445. doi: 10.1038/302442a0. [DOI] [PubMed] [Google Scholar]
- 10.Baran K, Dunstone M, Chia J, Ciccone A, Browne KA, Clarke CJ, Lukoyanova N, Saibil H, Whisstock JC, Voskoboinik I, Trapani JA. The molecular basis for perforin oligomerization and transmembrane pore assembly. Immunity. 2009;30:684–695. doi: 10.1016/j.immuni.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 11.Law RH, Lukoyanova N, Voskoboinik I, Caradoc-Davies TT, Baran K, Dunstone MA, D'Angelo ME, Orlova EV, Coulibaly F, Verschoor S, Browne KA, Ciccone A, Kuiper MJ, Bird PI, Trapani JA, Saibil HR, Whis-stock JC. The structural basis for membrane binding and pore formation by lymphocyte perforin. Nature. 2010;468:447–451. doi: 10.1038/nature09518. [DOI] [PubMed] [Google Scholar]
- 12.Wan YY, Flavell RA. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proc Natl Acad Sci USA. 2005;102:5126–5131. doi: 10.1073/pnas.0501701102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 14.Elkins KL, Cowley SC, Conlan JW. 2011, vol 14.25.1-14.25.13. Hoboken: Wiley; Mesurement of Macrophage-Mediated Killing of Intracellular Bacteria including Francisella and Mycobacteria. [DOI] [PubMed] [Google Scholar]
- 15.Kuballa P, Nolte WM, Castoreno AB, Xavier RJ. Autophagy and the immune system. Annu Rev Immunol. 2012;30:611–646. doi: 10.1146/annurev-immunol-020711-074948. [DOI] [PubMed] [Google Scholar]
- 16.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 18.Kageyama S, Omori H, Saitoh T, Sone T, Guan JL, Akira S, Imamoto F, Noda T, Yoshimori T. The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Mol Biol Cell. 2011;22:2290–2300. doi: 10.1091/mbc.E10-11-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bukholm G, Berdal BP, Haug C, Degre M. Mouse fibroblast interferon modifies Salmonella typhimurium infection in infant mice. Infect Immun. 1984;45:62–66. doi: 10.1128/iai.45.1.62-66.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He X, Zhang Y, Yu Z. An Mpeg (macrophage expressed gene) from the Pacific oyster Crassostrea gigas: molecular characterization and gene expression. Fish Shellfish Immunol. 2011;30:870–876. doi: 10.1016/j.fsi.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 21.Kemp IK, Coyne VE. Identification and characterisation of the Mpeg1 homologue in the South African abalone, Haliotis midae. Fish Shellfish Immunol. 2011;31:754–764. doi: 10.1016/j.fsi.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Mah SA, Moy GW, Swanson WJ, Vacquier VD. A perforin-like protein from a marine mollusk. Biochem Biophys Res Commun. 2004;316:468–475. doi: 10.1016/j.bbrc.2004.02.073. [DOI] [PubMed] [Google Scholar]
- 23.Wang GD, Zhang KF, Zhang ZP, Zou ZH, Jia XW, Wang SH, Lin P, Wang YL. Molecular cloning and responsive expression of macrophage expressed gene from small abalone Haliotis diversicolor supertexta. Fish Shellfish Immunol. 2008;24:346–359. doi: 10.1016/j.fsi.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Schreiber RD, Morrison DC, Podack ER, Muller-Eberhard HJ. Bactericidal activity of the alternative complement pathway generated from 11 isolated plasma proteins. J Exp Med. 1979;149:870–882. doi: 10.1084/jem.149.4.870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saitoh T, Fujita N, Yoshimori T, Akira S. Autophagy and innate immunity (in Japanese) Tanpakushitsu Kakusan Koso. 2008;53:2279–2285. [PubMed] [Google Scholar]