Abstract
Personal genome resequencing has provided promising lead to personalized medicine. However, due to the limited samples and the lack of case/control design, current interpretation of personal genome sequences has been mainly focused on the identification and functional annotation of the DNA variants that are different from the reference genome. The reference genome was deduced from a collection of DNAs from anonymous individuals, some of whom might be carriers of disease risk alleles. We queried the reference genome against a large high-quality disease-SNP association database and found 3,556 disease-susceptible variants, including 15 rare variants. We assessed the likelihood ratio for risk for the reference genome on 104 diseases and found high risk for type 1 diabetes (T1D) and hypertension. We further demonstrated that the risk of T1D was significantly higher in the reference genome than those in a healthy patient with a whole human genome sequence. We found that the high T1D risk was mainly driven by a R260W mutation in PTPN22 in the reference genome. Therefore, we recommend that the disease-susceptible variants in the reference genome should be taken into consideration and future genome sequences should be interpreted with curated and predicted disease-susceptible loci to assess personal disease risk.
1. Introduction
With the advance of sequencing technology and assembling tools, whole genome sequencing has become a commodity with 10,000 personal genomes being sequenced in the next two years. An urgent question is how to interpret personal genome sequences to comprehensively assess disease risk and optimize personalized treatment. Sixteen personal genomes (1–13) have been fully sequenced and described in the literature, while companies state they are sequencing as many as 500 individuals per month. However, due to the limited samples and lack of case/control design, the current interpretation of these genomes had been mainly focused on the identification and functional annotation of the DNA variants that are different from the reference genome sequence, with an aim to find interesting genomic features. The reference genome was not from a single normal individual; instead, the reference was deduced from a collection of DNAs from anonymous individuals with primarily European origins and assembled into a mosaic haploid genome (14, 15). To our knowledge, the clinical and phenotypic information of the participants had never been published. Although they were very likely to be healthy at the time of study, some of them might be carriers of disease risk alleles. The identification of biologically and clinically important rare and common disease variants in the reference genome and a comprehensive disease risk assessment will improve our understanding of the reference to better assemble and interpret future genome sequences.
We have previously developed a method to assess the risk of a patient for 55 diseases using a quantitative human disease-SNP association database, and showed that we could suggest useful and clinical relevant information using his personal genome sequence (16). Here, we queried the reference genome sequence against our database and identified 3,556 disease-susceptibility variants, including 15 rare variants. We comprehensively assessed the risk of the reference genome for 104 diseases and found high risk for type 1 diabetes (T1D) and hypertension. We further demonstrated that the risk of T1D was also significantly higher in the reference genome than in the genome of the healthy male we previously described (16). Comparing all contributing alleles, we found that the high T1D risk was mainly driven by a R260W mutation in the intracellular tyrosine phosphatase (PTPN22) in the reference genome.
2. Methods
2.1 Identifying the disease susceptible/protective alleles in the reference genome
We downloaded the alleles at 24.5 million SNPs (dbSNP 131 on hg19) of the reference genome from the UCSC genome browser (17, 18), and removed all SNPs that were mapped to multiple locations.
As described previously (16), we manually curated quantitative human disease-SNP associations from the full text, figures, tables, and supplemental materials of 3,333 human genetics papers, and recorded more than 100 features from each paper, including the disease name (e.g. coronary artery disease), specific phenotype (e.g. acute coronary syndrome in coronary artery disease), study population (e.g. Finnish individuals), case and control population (e.g. 2,508 patients with coronary artery disease proven by angiography), gender distribution, genotyping technology, major/minor risk alleles, odds ratio, 95% confidence interval of the odds ratio, published p-value, and genetic model. Studies on similar diseases were categorized and mapped to the Concept Unique Identifiers (CUI) in the Unified Medical Language System (UMLS) (19). For each study, the frequency of each genotype and allele in the case and control populations was recorded.
We queried the reference genome against this disease-SNP database using dbSNP identifiers (17), and identified all disease susceptible or protective alleles in the reference. We then retrieved the Minor Allele Frequency (MAF) from the HapMap II and III projects (20) and identified rare disease-susceptible alleles in the reference that had an MAF<1% in the CEU population.
2.2 Assessing the risk of the reference genome on 104 diseases
We had previously reported the medical assessment of a personal genome sequence from a healthy 40-year-old male by calculating his pre-test probability, likelihood ratio (LR), and post-test probability across 55 diseases (16) using a curated high-quality quantitative human disease-SNP association database. Similarly, for each of 104 diseases, we queried the reference genome sequence against our database, identified all independent disease-associated loci, treated the genotype at each locus as an independent genetic test, and calculated the LR as the increased disease odds from all tests.
For each disease, we identified all SNPs that had been significantly associated with the disease with a p value of ≤10−6 in Genome-Wide Association Studies on more than 5000 individuals, or with a p value of ≤0.01 in candidate gene studies on more than 1000 individuals. We estimated genetic risk using a likelihood ratio for each SNP defined by the relative frequency of the individual’s genotype in the diseased vs. healthy control populations (e.g., given an allele “A”, LR = Pr(A|diseased)/Pr(A|control)). The LR incorporates both the sensitivity and specificity of the test and provides a direct estimate of how much a test result will change the odds of having a disease (21). In addition, the likelihood ratio is taught to medical students and physicians in training(22).
We excluded studies with diseased patients in the control group, and included studies across all ethnicities and genders, because the reference genome was deduced from a mixture of people with different ethnicities and genders. For each allele, we averaged the LRs from multiple studies with a weight of the square root of the sample size to give higher confidence to studies with larger sample size. After removing SNPs in high linkage disequilibrium (R2≥0.8 in HapMap CEU populations), we assumed each locus as an independent genetic test and multiplied LRs to report the combined LR or risk.
2.3 Comparing the disease risk between the reference genome and a healthy patient
We plotted the log(LR) of a 40-year-old healthy male (16) against the log(LR) of the reference genome across 62 shared diseases to identify the diseases where the reference genome had significantly higher risk. All contributing SNPs were plotted for the disease to identify SNPs that drove the observed risk difference between the two genomes. For each SNP, its associated gene was identified using the NCBI Entrez dbSNP (17), and annotated using the UCSC genome browser (18) for its functional type and chromosome location.
3. Results
3.1 Disease susceptible and protective alleles in the reference genome
The reference genome (hg19) contains 21.8 million SNPs, with 17,429 of them known to associate with human disease and other phenotypes, and 12,190 of them known to associate with human diseases (Table 1). It contains slightly more diseases-protective alleles and genotypes (4,052 SNPs for 381 diseases) than disease-susceptible alleles and genotypes (3,556 SNPs for 349 diseases).
Table 1.
Number of disease susceptible and protective alleles in the reference genome
| SNPs | Phenotypes | PubMed count | |
|---|---|---|---|
| Disease/traits# | 17,429 | 1,026 | 3,333 |
| Associated with disease | 12,190 | 561 | 2,695 |
| Susceptibility to disease | 3,556 | 349 | 1,416 |
| Protection from disease | 4,052 | 381 | 1,600 |
Non-disease phenotypes included drug response and clinical measurements
3.2 Rare disease-susceptible variants in the reference genome
The reference genome carries minor alleles at 0.93 million SNPs in the CEU population, and 0.15 million of them were rare variants with MAF<1% in the HapMap II and III projects (20). We found that 15 rare alleles in the reference genome are known to increase the risk of a variety of diseases (Table 2). For example, rs10849033 is close to the 5′ end of C12orf5, a TP53-induced glycolysis and apoptosis regulator. The reference genome has a rare G allele at rs10849033 with an MAF of 0.8%. The G allele had been found to significantly increase the risk of acute lymphoblastic leukemia (ALL) by 2.55 fold, with a p value of 8.5×10−6 in a study on 317 children with ALL and 17,958 non-ALL individuals in a control group (23). This rare ALL-susceptibility variant would likely be missed by recent personal genome resequencing efforts focusing on reporting and studying only those variants different from the reference genome.
Table 2.
Rare disease-susceptible variants (MAF<1%# in Caucasian) in the reference genome
| Disease | Gene | SNP | Allele | Type | PubMed |
|---|---|---|---|---|---|
| Acute lymphoblastic leukemia | C12orf5 | rs10849033 | G | near 5′ | 19684603 |
| Asthma | rs10837012 | G | unknown | 19187332 | |
| rs1335159 | C | unknown | 19187332 | ||
| Breast cancer | RRP1B | rs9306160 | T | missense | 19825179 |
| Coronary artery disease | PON2 | rs7493 | G | missense | 12588779 |
| Focal segmental glomerulosclerosis | WT1 | rs2234591 | T | intron | 15687485 |
| Juvenile idiopathic arthritis | SLC26A2 | rs30832 | T | missense | 17393463 |
| Malaria | FAM53B | rs7076268 | C | intron | 19465909 |
| Obesity | rs7173766 | A | unknown | 19584900 | |
| Parkinson’s disease | ADH1C | rs283413 | A | nonsense | 15642852 |
| NUCKS1 | rs823128 | G | intron | 19915575 | |
| Placental abruption | F5 | rs6025 | T | coding-synon | 18277167 |
| Prostate cancer | GDF15 | rs1058587 | C | missense | 16775185 |
| Schizophrenia | rs4568102 | A | unknown | 18347602 | |
| Type 2 diabetes | ARHGEF11 | rs861086 | G | near 5′ | 17369523 |
| Venous thrombosis | F5 | rs6025 | T | coding-synon | 17284699 |
MAF (minor allele frequency) was retrieved from the HapMap II and III projects
We further found two rare variants in the reference genome increasing the risk of Parkinson’s disease (Table 2). One of them is rs283413, containing an A allele in the reference genome, which leads to the early truncation of ADH1C protein, and has been known to increase the risk of Parkinson’s disease by 3.25 fold (p=0.007) in multiple Swedish and Caucasian studies (24).
A large survey across 17,429 disease SNPs in our database showed that the effect sizes or the odds ratio of disease SNP associations were consistently and negatively associated with the MAF in Caucasian, African, Chinese, and Japanese. This indicated that rare disease-associated SNPs conveyed significantly larger effect size to the observed genetic association across human diseases. With the discovery of several rare alleles known to be associated with disease in the reference genome, we suggest that whole genome resequencing would very likely identify other causal SNPs, possibly explaining some of the currently missing genetic heritability of complex diseases (25). As such, some of the other 0.15 million rare variants in the reference genome could also potentially be associated with disease. Comparing genome sequences against curated disease and rare variants would likely discover many causal variants.
3.3 Risk likelihood ratio of the reference genome on 104 diseases
We analyzed the risk likelihood ratio (LR) of the reference genome on 104 diseases using the independent test likelihood ratio model. We found that the reference genome had an increased risk on 48 diseases (LR>1) and a decreased risk on 56 diseases (LR<1). The LR ranged from 0.14 to 5.14 with a mean LR close to 1.0 (p=0.39, t-test). Strikingly, T1D demonstrated the highest risk with a product LR of 5.14. This LR was calculated from 31 T1D-susceptible alleles and 14 T1D-protective alleles in the reference genome.
The reference genome also had a high likelihood ratio of risk for hypertension with 11 risk and 3 protective alleles. The high risk of hypertension was mainly driven by a G allele at rs3741691 in THAP2 with a LR of 1.26 (26), an A allele at rs2106809 in ACE2 with a LR of 1.26 (27), and an A risk allele at rs3761987 with a LR of 1.21 (26). Table 3 lists the LR and the number of susceptible and protective SNPs on just the 44 diseases with 10 or more SNPs.
Table 3.
Disease risk profile of the reference genome on 44 diseases with ≥10 SNPs
| Disease | LR | Susceptible SNPs | Protective SNPs |
|---|---|---|---|
| Type 1 diabetes | 5.14 | 31 | 14 |
| Hypertension | 2.58 | 10 | 3 |
| Ankylosing spondylitis | 1.90 | 9 | 6 |
| Myocardial infarction | 1.78 | 10 | 3 |
| Prostate cancer | 1.56 | 22 | 19 |
| Breast cancer | 1.28 | 17 | 17 |
| Multiple sclerosis | 1.25 | 10 | 4 |
| Inflammatory bowel disease | 1.21 | 7 | 8 |
| Colorectal cancer | 1.20 | 9 | 12 |
| Lung cancer | 1.03 | 6 | 5 |
| Parkinson’s disease | 1.01 | 14 | 7 |
| Alzheimer’s disease | 0.89 | 10 | 8 |
| Coronary artery disease | 0.86 | 8 | 9 |
| Celiac disease | 0.83 | 9 | 10 |
| Rheumatoid arthritis | 0.76 | 12 | 11 |
| Bipolar disorder | 0.75 | 5 | 5 |
| Schizophrenia | 0.71 | 5 | 10 |
| Ulcerative colitis | 0.70 | 6 | 12 |
| Systemic lupus erythematosus | 0.66 | 26 | 29 |
| Type 2 diabetes | 0.61 | 34 | 37 |
| Crohn’s disease | 0.55 | 12 | 17 |
| Glioma | 0.53 | 4 | 9 |
| Psoriasis | 0.47 | 11 | 10 |
| Obesity | 0.43 | 6 | 14 |
| Basal cell carcinoma | 0.33 | 3 | 8 |
| Melanoma | 0.14 | 4 | 11 |
We then plotted the histogram of log(LR) across all 198 diseases, and observed a symmetric distribution with no significant difference from the mean of zero (p=0.07, t-test). This suggests that our method is unbiased towards overcalling susceptibility or protection across all diseases.
3.4 Disease risk comparison between the reference and a personal genome
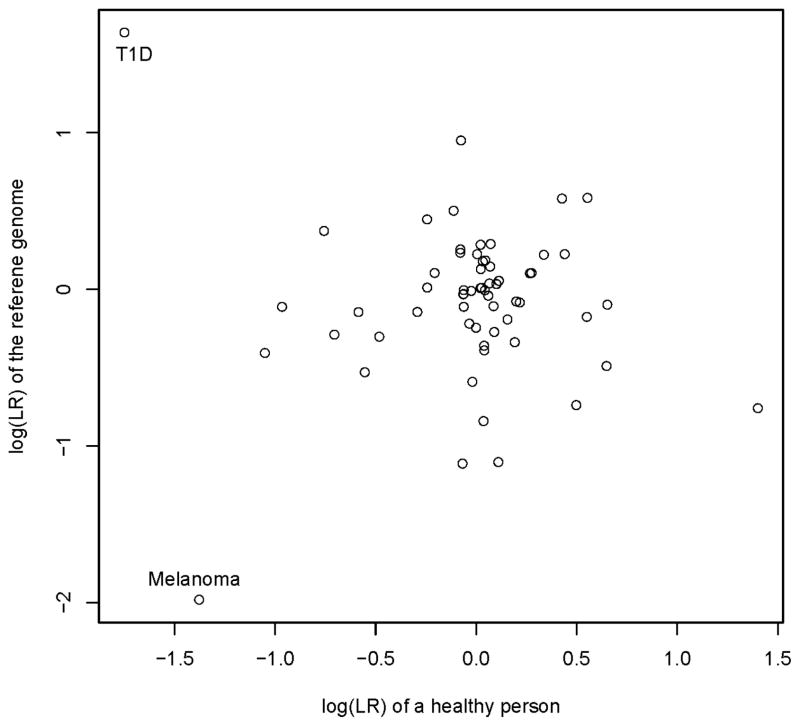
We plotted the log(LR) of a 40-year-old healthy Caucasian male against the log(LR) of the reference genome across 104 shared diseases (Figure 2). Interestingly, the reference genome showed a strikingly increased risk on T1D than the healthy male, and a decreased risk on Melanoma. This indicats that the high T1D risk was likely a result of T1D-susceptible alleles in the reference genome instead of biased T1D-susceptible alleles in the database. Although the reference genome was deduced from a group of healthy persons, some of them might be carriers of T1D-sueceptible alleles. Therefore, the reference genome is not free of predicted disease-risk and these disease-susceptible alleles in the reference genome need to be taken into consideration in interpreting future genome sequences.
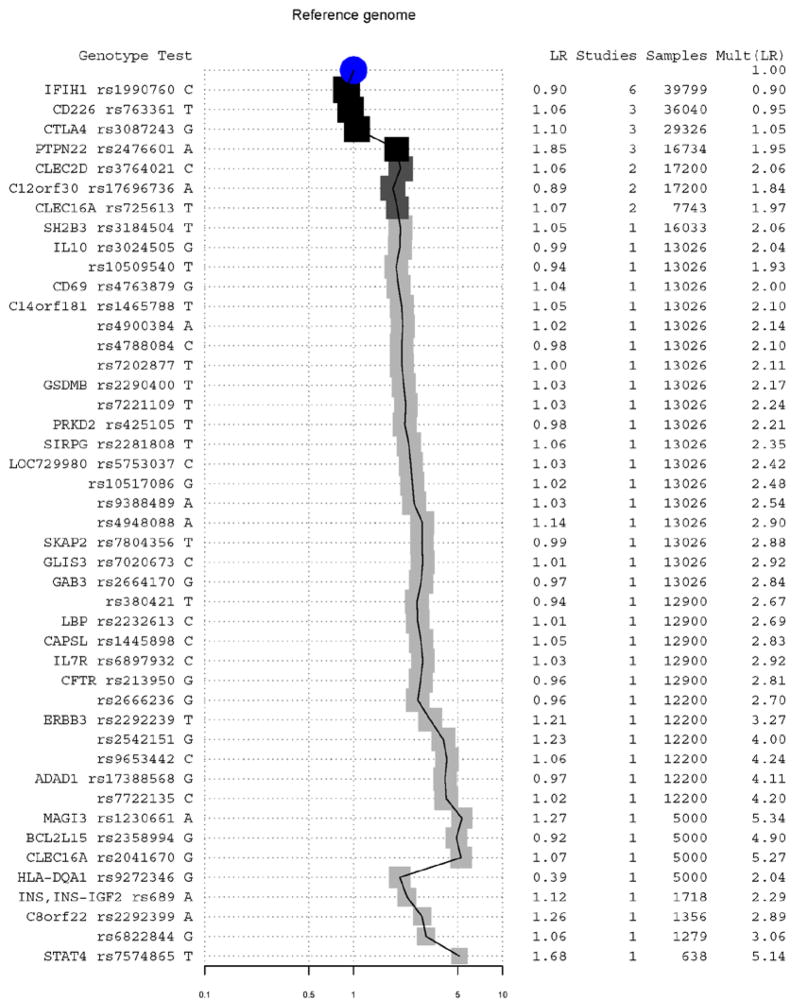
Fig. 2.
Contribution of individual alleles to overall risk LR of T1D of the reference genome. Alleles and their associated genes are listed on the left, ordered from top to bottom by the number of studies in which each was published and the total sum of cohort sizes across those papers. The LR of each independent SNP/allele is listed. A user of this figure could draw a horizontal line at a given threshold of belief, include and exclude alleles, and retrieve the accumulated LR at the right column and shown graphically in the middle. The central graph displays the change in accumulated LR, with darker squares representing more publications and larger squares representing larger sample size.
3.5 T1D-susceptible alleles in the reference genome
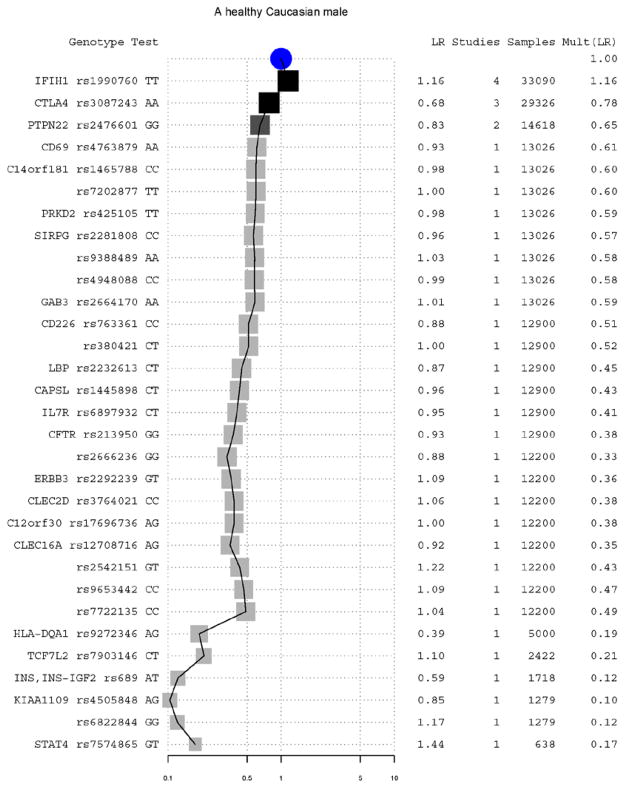
To identity the specific alleles that led to the striking difference on predicted T1D risk between the reference genome and the healthy male, we plotted all contributing T1D susceptible and protective alleles in both the reference genome (Figure 2) and the previously studied 40 year old patient (Figure 3).
Fig. 3.
Contribution of individual genotypes to the overall risk LR of T1D for a previously published 40-year-old healthy Caucasian male. See Figure 3 for details on the graphical elements.
Comparing Figure 2 and 3, we found that the increased T1D risk in the reference genome was mainly due to a highly T1D-susceptible allele A at rs2476601, causing a R260W mutation in the intracellular tyrosine phosphatase (PTPN22). This SNP had been reported to increase the risk of T1D by 2 fold in more than nine studies (28–31). Comparing with the patient, the reference genome also has increased risk of T1D due to the lack of two T1D-protective alleles at rs3087243 in cytotoxic T-lymphocyte-associated protein 4 (CTLA4) (32) and at rs689 in the insulin (INS) (28). These three alleles increased the T1D risk for the reference genome by 6.8 fold comparing with our previously published patient. Interestingly, for rs2476601 in PTPN22, the T1D susceptible allele in the reference genome is the minor allele in most population. The 3,556 known disease-susceptible variants and many unknown ones especially rare variants could be potentially missed if only variants different from the reference were analyzed.
3.6 Disease-susceptible alleles deleted in the reference genome
The reference genome also contains a deletion at 2.7M SNPs with a dbSNP identifier in the dbSNP build 131 (17). We found that 16 SNPs that are known to associate with human diseases at these points of deletion. The clinical relevance of these missing base pairs is not clear.
4. Discussion
We identified 3,556 disease-susceptible variants including 15 rare variants (MAF<1%) in the reference human genome, which provides a useful tool for the annotation of personal genome sequences. Using a curated high-quality quantitative human disease-SNP association database, we assessed the likelihood ratio of increased risk over healthy population on 104 diseases for the reference genome and found the high predictive T1D risk with a R260W mutation in the intracellular tyrosine phosphatase (PTPN22). It reminded us that the reference genome was not from a regular person and was certainly not disease free. Although it had dramatically accelerated personal genome sequencing efforts, focusing on variants different from the reference will likely miss many disease causal variants including rare variants.
With the likely incoming deluge of 10,000 personal genome sequences arriving within the next two years, a method to estimate personal disease risk is urgently needed. Here, we described a method to estimate personal genetic risk using a likelihood ratio for each SNP as the relative frequency of the individual’s genotype in the diseased vs. healthy control populations. We further described a very simple method to treat multiple disease loci outside the linkage disequilibrium as independent genetic test, and estimated their combined effect. We acknowledge that assuming independence of tests is actually a different assumption than assuming that each variant contributes independently to risk. If each measured variant is viewed as an independent test probing disease state, this is arguably closer to our understanding of their use as markers associated with disease instead of actual causal variants (22). We admit that it is likely to be too simple to accurately model the risk of many common diseases, especially those like T1D, which are also influenced by unknown environmental and gene-environmental factors, and we are currently investigating different models to estimate combined effects.
The accurate assessment on personal disease risk is also dependent on the quality and coverage of the genotype/allele frequency in the disease and control population in the literature. We found that many studies, including genome-wide association studies (GWAS) only reported the odds ratio of disease risk between genotypes/alleles, and not their frequencies in the case and control population, which were required for the calculation of the likelihood ratio. For studies reporting both the odds ratio and the minor allele frequency in the control group, we recalculated their allele frequencies. We excluded studies reporting only the odds ratio, and we are investigating the possibility of estimating the genotype/allele frequencies in the control group using the data in the HapMap III project (33). There have been many debates on whether the aggregated genotype frequency data should be published in GWASs (34). Analyses showing association of a single biomarker with disease typically report very detailed characteristic of the populations studied; this is radically different from typical genetic association studies, which often report almost nothing about the subjects (22). Therefore, we strongly recommend the release of the genotype frequency in future GWAS studies as it is critical for us to quantitatively evaluate the disease-SNP association, enabling an accurate personal risk assessment.
We further found that many disease SNPs had been reported as the genotypes in the negative strand without indicating their strand directions. We had identified the strand direction by comparing the major/minor alleles in the study with the major/minor alleles in similar population in the HapMap projects. However, the identification process became difficult when the C/G or A/T alleles share similar frequencies. Therefore, we strongly recommend investigators to report the genotype frequencies in the case and control population and their strand direction in the future GWAS publications. With exponentially increasing personal genome sequences with phenotype information, we will likely to discover more rare causal variants and comprehensively predict personal risk on a variety of diseases.
Fig. 1.
The disease risk comparison between the personal genome of a healthy male and the reference genome. Each circle represents the genetic risk of a disease for the patient and the reference genome.
Acknowledgments
This work was supported by Lucile Packard Foundation for Children’s Health, National Institute of General Medical Sciences (R01 GM079719), National Library of Medicine (R01 LM009719), National Cancer Institute (R01 CA138256), and Howard Hughes Medical Institute. We thank Alex Morgan for R scripts for graphics and suggestions on the maximum likelihood ratio model. We thank Alex Skrenchuk and Boris Oskotsky for computer support, and thank Prajkta Bhide, Priyanka Korde, Anuj Kharnal, Harshal Darokar, and Priyanka Khandelwal from Optra Systems for curating disease-SNP association from the literature under contract.
Contributor Information
RONG CHEN, Department of Pediatrics, Stanford University School of Medicine, Stanford, CA 94305-5479, USA.
ATUL J. BUTTE, Email: abutte@stanford.edu, Department of Pediatrics, Stanford University School of Medicine, Stanford, CA 94305-5479, USA
References
- 1.Lupski JR, et al. N Engl J Med. 2010 Apr 1;362:1181. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pleasance ED, et al. Nature. 2010 Jan 14;463:191. [Google Scholar]
- 3.Drmanac R, et al. Science. 2010 Jan 1;327:78. [Google Scholar]
- 4.Pushkarev D, Neff NF, Quake SR. Nat Biotechnol. 2009 Sep;27:847. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mardis ER, et al. N Engl J Med. 2009 Sep 10;361:1058. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JI, et al. Nature. 2009 Aug 20;460:1011. [Google Scholar]
- 7.McKernan KJ, et al. Genome Res. 2009 Sep;19:1527. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahn SM, et al. Genome Res. 2009 Sep;19:1622. doi: 10.1101/gr.092197.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ley TJ, et al. Nature. 2008 Nov 6;456:66. [Google Scholar]
- 10.Wang J, et al. Nature. 2008 Nov 6;456:60. [Google Scholar]
- 11.Bentley DR, et al. Nature. 2008 Nov 6;456:53. [Google Scholar]
- 12.Wheeler DA, et al. Nature. 2008 Apr 17;452:872. [Google Scholar]
- 13.Levy S, et al. PLoS Biol. 2007 Sep 4;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snyder M, Du J, Gerstein M. Genes Dev. 2010 Mar 1;24:423. doi: 10.1101/gad.1864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lander ES, et al. Nature. 2001 Feb 15;409:860. [Google Scholar]
- 16.Ashley EA, et al. Lancet. 2010 May 1;375:1525. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherry ST, et al. Nucleic acids research. 2001 Jan 1;29:308. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kent WJ, et al. Genome Res. 2002 Jun;12:996. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bodenreider O. Nucleic acids research. 2004 Jan 1;32:D267. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nature. 2003 Dec 18;426:789. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 21.http://www.childrens-mercy.org/stats/definitions/likelihood.htm.
- 22.Morgan AA, Chen R, Butte AJ. Genome Med. 2010;2:30. doi: 10.1186/gm151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trevino LR, et al. Nat Genet. 2009 Sep;41:1001. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buervenich S, et al. Arch Neurol. 2005 Jan;62:74. doi: 10.1001/archneur.62.1.74. [DOI] [PubMed] [Google Scholar]
- 25.Manolio TA, et al. Nature. 2009 Oct 8;461:747. [Google Scholar]
- 26.Kato N, et al. Hum Mol Genet. 2008 Feb 15;17:617. doi: 10.1093/hmg/ddm335. [DOI] [PubMed] [Google Scholar]
- 27.Fan X, et al. Clin Pharmacol Ther. 2007 Aug;82:187. doi: 10.1038/sj.clpt.6100214. [DOI] [PubMed] [Google Scholar]
- 28.Cervin C, et al. Diabetes. 2008 May;57:1433. doi: 10.2337/db07-0299. [DOI] [PubMed] [Google Scholar]
- 29.Smyth DJ, et al. Diabetes. 2008 Jun;57:1730. doi: 10.2337/db07-1131. [DOI] [PubMed] [Google Scholar]
- 30.Kawasaki E, et al. Am J Med Genet A. 2006 Mar 15;140:586. doi: 10.1002/ajmg.a.31124. [DOI] [PubMed] [Google Scholar]
- 31.Criswell LA, et al. Am J Hum Genet. 2005 Apr;76:561. doi: 10.1086/429096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howson JM, et al. Diabetologia. 2007 Apr;50:741. doi: 10.1007/s00125-007-0603-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altshuler DM, et al. Nature. 2010 Sep 2;467:52. [Google Scholar]
- 34.Church G, et al. PLoS Genet. 2009 Oct;5:e1000665. doi: 10.1371/journal.pgen.1000665. [DOI] [PMC free article] [PubMed] [Google Scholar]