Abstract
The human microbiome represents a vastly complex ecosystem that is tightly linked to our development, physiology, and health. Our increased capacity to generate multiple channels of omic data from this system, brought about by recent advances in high throughput molecular technologies, calls for the development of systems-level methods and models that take into account not only the composition of genes and species in a microbiome but also the interactions between these components. Such models should aim to study the microbiome as a community of species whose metabolisms are tightly intertwined with each other and with that of the host, and should be developed with a view towards an integrated, comprehensive, and predictive modeling framework. Here, we review recent work specifically in metabolic modeling of the human microbiome, highlighting both novel methodologies and pressing challenges. We discuss various modeling approaches that lay the foundation for a full-scale predictive model, focusing on models of interactions between microbial species, metagenome-scale models of community-level metabolism, and models of the interaction between the microbiome and the host. Continued development of such models and of their integration into a multi-scale model of the microbiome will lead to a deeper mechanistic understanding of how variation in the microbiome impacts the host, and will promote the discovery of clinically- and ecologically-relevant insights from the rich trove of data now available.
Introduction
Our ability to study microbial communities in their natural environments has improved dramatically over the past few years thanks to exciting advances in high-throughput molecular methods and biological assays. Specifically, the adaptation of –omic technologies from traditional clonal biology to the study of mixed microbial consortia has bypassed the need to isolate and to culture individual species and has removed many of the obstacles that commonly hindered research in microbial ecology. Such meta’omic technologies [1], including, first and foremost, metagenomics [2], but also metatranscriptomics, metaproteomics and metametabolomics [3], can provide valuable information about the diversity, composition, function, and metabolic capacity of a given microbial ecosystem. Shotgun metagenomic sequences, for example, can be aligned to known genomes or mapped to known gene orthology groups to determine the taxonomic or functional composition of the community.
Using these technologies, researchers are now starting to explore the communities found in various habitats, ranging from soil and marine environments [4–6] to the intestines of mammals [7]. The human microbiome, in particular, has been a major focus of study, due to its massive impact on human health [1]. The human microbiome comprises 10 times the number of cells as the human host [8] and more than 150 times as many genes [9]. Recent studies have extensively cataloged the composition of the healthy human microbiome, determining the core taxa and genes present in the microbiome and the range of variation [9–11]. Additional studies have tracked individual subjects across time to explore the dynamics of the microbiome in infants [12] and in healthy adults [13], and the response of the microbiome to perturbations [14]. Most importantly, comparative metagenomic studies have demonstrated significant associations between the taxonomic and genomic composition of the microbiome and several complex diseases such as obesity [15,16], diabetes [17], and inflammatory bowel disease [18].
Clearly, a comprehensive understanding of the microbiome, its activity, and its impact on the human host cannot be gained by cataloging species and gene composition alone. Systems-level interactions between species, across pathways, and with the host all contribute to the assembly, function, dynamics, and resilience of the microbiome and must be taken into account [19–22]. Microbiome research would therefore benefit tremendously from going beyond statistical and comparative studies and from applying systems-biology approaches to study the microbiome across multiple levels [23–25]. Multiple such approaches have been proposed, drawing on established modeling concepts from a variety of disciplines, including graph theory-based modeling, dynamical modeling, game-theoretical approaches, and agent-based modeling [26–29]. Here, we focus specifically on network-based and stoichiometric frameworks, which have proved successful in elucidating causal mechanisms underlying the behavior of single species [30–32], their ecology [33], and their evolution [34,35]. These frameworks can easily be applied on a large-scale and utilize single time-point data. Ultimately, however, microbiome research should aim to generate a comprehensive systems-level model of the microbiome, capable of predicting function and dynamics from detailed data on molecular, genomic, and species composition. Such a model will not only serve as a touchstone for a profound understanding of the microbiome but will allow clinicians to design and offer individualized microbiome-based therapies [19].
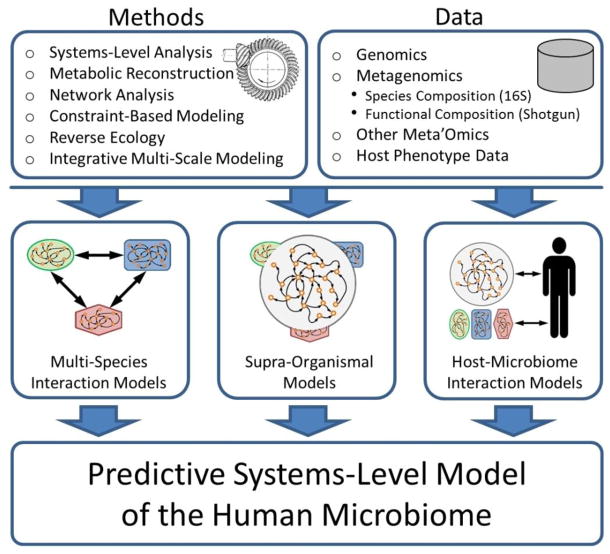
Modeling the human microbiome – a complex and still largely uncharted and poorly understood biological system – is clearly a daunting task. The development of a predictive systems-level model of the microbiome represents a major leap forward and may remain out of reach for many years to come. In this review, we discuss preliminary efforts to tackle this challenge. While our primary motivation and emphasis is the human microbiome (and specifically, the human gut microbiome), many of the studies discussed below focus on other microbial communities or on the development of more generic modeling frameworks, as such studies face similar challenges and often introduce broadly applicable solutions. We focus mostly on metabolic modeling, although ultimately, other processes such as regulation and signaling should be integrated into such models. In the sections below we discuss models that highlight various aspects of microbial communities, including species interactions within the community, community-wide metabolism, and interactions between the community and the host. We believe that these aspects are all crucial for the construction of a comprehensive model of the microbiome and that it is only by integrating such models across multiple scales that a predictive understanding of the microbiome can be gained (Fig 1).
Figure 1.
An overview of microbiome modeling. Genomic, metagenomic, and other meta’omic data are combined with various computational methodologies to model different aspects of the microbiome. These models can then be integrated to obtain a comprehensive, systems-level, predictive model.
A Microbial Tangled Bank
In the concluding paragraph of On the Origin of Species, Darwin marvels about the complexity of the living world. He contemplates “a tangled bank”, inhabited by many different species of plants, birds, insects, and worms, which are all “dependent upon each other in so complex a manner”. Nowadays, researchers express similar marvel at the microbial world. Just like Darwin’s tangled bank, the complexity of microbial ecosystems stems not only from the surprisingly high number of species comprising many microbial communities, but also from the myriad ways these species interact. An organism may compete resources away from one neighbor, while supplying essential nutrients to another [36,37]. Highly organized syntrophic consortia are common and often form multicellular structures with close physical proximity between partners [38]. Antibiotic production and resistance may partition the community into distinct social units [39,40]. Community members can communicate through a variety of signaling molecules and may even disrupt signaling pathways of competitors [41]. As a result of these mutual dependencies, microbes often depend completely on the community for survival and resist isolation and culturing attempts [42].
Due to the prevalence and complexity of interactions among microbes, much effort has been invested in mapping such interactions and in characterizing the impact one species may have on the growth of another. One straightforward approach to obtain insights about potential interactions relies on co-culture growth assays. For example, growth assays in a synthetic oral system revealed a web of interacting commensal organisms responsible for colonizing the enamel, developing biofilm, and ultimately causing disease [43–46]. A similar approach was used to evaluate the productivity of many simple two-species communities, demonstrating that the majority of combinations have a negative effect and suggesting that adaptation typically results in competitive, rather than cooperative, interaction [47]. Chemostat co-cultures of several gut bacteria and a statistical analysis of the observed temporal abundance data were used to examine colonization patterns and species interactions in a simplified model of the intestinal tract [21].
More recently, with the availability of metagenomic-based species composition data, co-occurrence analysis has become a common practice for inferring potential dependencies between species [48]. Such analyses are often performed on a very large scale, identifying for example, thousands of co-occurrence and co-exclusion relationships within the human microbiome [9,49] or throughout the environment [50]. Similar approaches have revealed a web of horizontal gene transfers among species that share similar ecologies [51]. Integrating such co-occurrence studies with phylogenetic measures further demonstrated that intestinal communities are structured by non-neutral processes and that observed species abundance patterns are driven by deterministic interactions between species and with the environment [52].
Notably, however, the methods discussed above for studying species interactions all take a phenomenological approach; Evidence of interaction is inferred from association in abundance across samples, which can be largely effected by sampling depth, or from some deviation observed in a co-culture from the growth obtained in a mono-culture. Exact mechanisms of interaction are not integrated into such methods, nor can they be directly inferred by them. In contrast, the systems biology approaches described below offer a unique suite of tools to model microbial organisms, to examine potential channels of interaction, and to obtain a mechanistic understanding of the way species interact.
Network-Based Modeling of Species Interaction
One approach for predicting metabolic interactions between species relies on simple connectivity-based network models of microbial metabolism coupled with a careful analysis of their structure and topology. In such studies, the metabolic network of each species is reconstructed based on the set of enzymatic genes encoded in its genome. As an organism evolves and adapts to its environment, the set of metabolic reactions it can catalyze and the overall organization of its metabolic network reflect its interactions with the environment and with other species [53,54]. Analysis of the metabolic network topology can therefore be used to detect such adaptive signatures and to obtain insights into the organism’s habitat and ecology. This ‘reverse-ecology’ research approach has proved successful in elucidating the relationship between microbial species and their environment on a large scale [33,55–57].
Specifically, at the core of several studies of species interactions are two methods for characterizing a species’ habitat and metabolic capacity: First, the seed set framework [56] applies a graph theory-based algorithm to analyze the topology of a metabolic network and to detect the set of metabolites acquired from the environment. Such metabolites represent the biochemical habitat of an organism. Second, the network expansion algorithm [58] identifies the set of metabolites an organism can synthesize from a given set of precursors. These two methods have been recently combined to predict metabolic competition between microbial species and the deleterious effect that the presence of one species may have on the metabolic capacity of the other [59]. When applied to clusters of literature-derived co-occurring species, it was shown that higher competition within a cluster is associated with lower mean growth. The seed set framework and the expansion algorithm have similarly been used to investigate ecological strategies to cope with competitive interactions across hundreds of species [60]. This analysis demonstrated that competitive interactions force species to adopt one of two ecological strategies: adapt to a narrow set of environments with lower competition, or to a wide set of environments while maintaining high growth rates. Additional approaches, specifically those that apply such methods on a large scale to study complex communities and that compare predicted interactions to metagenomic-derived species co-occurrence have great potential to reveal the forces that act on these communities and shape their characteristic structure.
Clearly, however, competition is only one of many ways microbial species interact. Cooperative metabolic interactions can lead to increased growth or to the activation of novel pathways. An extension of the network expansion algorithm, termed metabolic synergy, was introduced to quantify the increase in metabolic capacity a pair of organisms achieves through cooperation [61]. By determining the metabolic synergy of a large set of pairs of organisms, it was shown that particular structural features and reactions are more important for synergy than overall dissimilarity between the cooperating networks. This suggests that metabolic synergy is most effective when the cooperating organisms are neither too similar nor too dissimilar. Topological approaches have also been used to investigate specific exchanges between organisms known to cooperate. As an example, Cottret et al. applied the seed set framework to a host-microbe system consisting of the sharpshooter Homalodisca coagulata and the endocytobionts Baumannia cicadellinicola and Sulcia muelleri [62]. B. ciccadellinocola and S. muelleri are known to acquire some metabolites from their insect host, and to exchange others amongst themselves. Notably, it was shown that all seeds common to both endocytobionts are synthesized by the host, highlighting their shared dependency on these compounds. An additional network-based method (PITUFO; [63]) was then used to characterize the metabolic exchanges between the two endocytobionts and to implicate specific pathways in their cooperative growth.
Constraint-Based Modeling of Species Interaction
Constraint-based approaches [30] aim to model, characterize, and quantify metabolic processes by defining a set of simple stoichiometric and thermodynamic constraints that control the metabolic fluxes in the cell. The flux through each reaction and the overall metabolic activity of the cell in a given environment can then be determined by optimizing some cellular objective with respect to these constraints. For example, Flux Balance Analysis (FBA) assumes that maximal growth is governing the metabolic function of the cell and accordingly uses optimal growth as an objective function [64]. Alternatively, different objectives have been proposed for modeling knockout strains that have not evolved toward an optimal growth in a mutant state [65,66].
Although constraint-based methods have been shown to appropriately predict cellular metabolism [67,68] and proved useful for biomedical and industrial applications [69], constraint-based modeling of microbial communities has grown slowly. This is partly because high-quality constraint-based models require a meticulous reconstruction protocol [70], and to date, such models are available only for a small number of the many human microbiome species recently characterized and sequenced (but see also Refs [71,72]). Previous efforts to model microbial communities using constraint-based methods have therefore focused mainly on simple two-species communities, and set out to elucidate specific aspects of species interactions. Stolyar et al. [73] constructed the first such model to examine syntrophic interactions between Desulfovibrio vulgaris and Methanococcus maripaludis. Freilich et al. [74] identified possible cooperative, competitive, and neutral interactions between pairs of species, interestingly finding that predicted cooperative interactions are typically unidirectional. Wintermute and Silver [75] modeled pairs of E. coli auxotroph mutants to qualitatively predict metabolic mutualism.
While these simple models demonstrate the tremendous potential of constraint-based models for studying species interaction and inferring inter-species metabolic transfer, they also highlight two important challenges facing researchers that follow this modeling approach. First, the choice of an objective function is a crucial step and is not at all clear in the context of a community model. Stolyar et al. [73] used as an objective function a simple fixed combination of biomass from both organisms, favoring the growth of D. vulgaris. Another straightforward definition of an objective function is overall community growth [74,75]. Notably, however, such an objective inherently assumes that community members cooperate and act for the common good of the community. This may, for example, lead to predictions where one species barely grows (although nutrients are available) to enable the growth of another species. The development of a biologically meaningful and broadly applicable method to integrate many different species’ objectives within a community context is a challenging task and one for which no clear consensus is currently available (see also Ref. [76]). The second challenge concerns the design of model compartments (e.g. representing species or organelles) and the partitioning of the reactions and the metabolites between the different compartments. The choice of a compartmentalization scheme may depend on the scale of the study, the data available, and the specific question one may wish to address [77]. It can, however, markedly impact the predicted fluxes and should therefore be carefully designed [78].
Recently, more involved methods have been introduced to partly address these challenges. For example one study introduced temporal dynamics to account for the competition between Geobacter sulfurreducens and Rhodoferax ferrireducens and to predict the relative proportion of the community members at loci with different environmental conditions [79]. Taking a different perspective, Klitgord and Segrè [80] developed a novel algorithm to explore environmental conditions that may induce symbiosis between two species, providing exciting insights into the evolution of microbial communities. Extending such models to more than two species has also proved challenging, and only very few studies have gone beyond a two-species interaction model. Taffs et al. explored several methods for applying elementary mode analysis to a community of three distinct microbial guilds: oxygenic phototrophs, filamentous anoxygenic phototrophs, and sulfate-reducing bacteria [77]. More recently, a multi-level optimization scheme was employed to study trade-offs between individual and community level fitness, making community growth a primary objective and individual growth a secondary (and potentially suboptimal) objective [81]. This method was used to elucidate the extent and direction of inter-species metabolic transfer in both two- and three-species communities. Future work in this area will be of immense value to the field.
Modeling Community-Level Metabolism
While the study of species interactions within a microbiome is crucial for understanding community function and dynamics, it is only one of the components required for the development of a comprehensive model of the community. Multi-species models that focus on the interactions between species may fail to explain, for example, how variations in gene or species composition affect the overall metabolic activity of the microbiome or how the microbiome as a whole impacts the host. Such questions call for an alternative approach to model the microbiome, wherein the entire community is regarded as a single biological entity, ignoring both the boundaries between species and the species of origin of each gene. In this supra-organismal approach [82], metabolic pathways are assumed to function at the community-level, eliminating species-level compartmentalization and allowing metabolites to transfer freely between species and to interact directly with the microbiome environment.
Microbiome-wide profiling of enzymatic gene content and community-level characterization of metabolic potential are common in comparative metagenomics. Using a variety of statistical approaches, the prevalence of certain metabolic pathways in the metagenome have been shown to correlate with various host states [9,16,17] and environmental factors [83,84]. A number of tools have been developed to analyze and to visualize metagenomic data and to assess microbiome-wide presence and abundance of various metabolic pathways [85–87]. Notably, some tools go beyond simple tallies of gene components and utilize information about metabolic pathway topology to infer the presence of missing enzymes or to correct the abundance of others. For example, HUMAnN [88], the primary tool used for metabolic analysis of the Human Microbiome Project data [11], applies such methods to translate the relative abundance of sets of enzymatic genes within metagenomic samples to pathway coverage. Similarly, MetaPath [89] overlays metagenome-wide abundance data onto global metabolic pathway structures to heuristically detect differentially abundant pathways across samples.
Such pathway-based analyses are not only powerful and effective tools for studying the functional composition of the microbiome but are also an important first step towards the reconstruction of full-scale models of community-level metabolism. Yet, relatively few studies go beyond characterization of pathway abundances and directly take into account the relationships between the various pathways or the overall organization of the metabolic network. The most extensive effort to date to generate and analyze a microbiome-wide metabolic model focused on the gut microbiome and its impact on obesity and on IBD [22]. Specifically, shotgun metagenomic data was used to reconstruct community-level metabolic networks and to examine the position of disease-associated enzymatic genes in the network, as well as the topology of the network as a whole. This metagenomic systems biology framework revealed that enzymes whose abundances in the metagenome are associated with host health tend to occupy positions at the perimeter of the network. This finding suggests that disease-associated microbiomes differ in the way they interact with the host environment rather than in core metabolic processes. Moreover, by constructing host state-specific community-level metabolic networks, it was further shown that networks derived from obese individuals are significantly less modular than those derived from lean individuals, suggesting that clinically-relevant differences in the microbiome may be associated with distinct systems-level organization. Such network-based approaches are also gaining traction in studying community-level metabolism beyond the human microbiome and across environmental microbiomes. For example, a recent study [90] used a network-based method to infer the turnover of metabolites from marine metagenomic samples from the Western English Channel and demonstrated an association between predicted relative metabolic turnover and seasonal changes of various environmental parameters. Integration with pathway based tools such as those mentioned above may further improve community-level models, addressing the sampling and annotation errors inherent to all metagenomic studies. Ultimately, such modeling efforts, taking a systems-based approach to study the microbiome as a whole, provide valuable insights into the contribution of functional elements to microbiome potential and into the functioning of the microbiome within the context of an ecosystem [23,24].
Modeling Host-Microbiome Metabolic Interaction
Modeling the human microbiome, and for that matter modeling any host-associated microbiome, is further complicated by the tight commensal relationship between the microbiome and its host and by the dynamic nature of a host-derived environment [91]. Host-microbiome interactions play a key role in host metabolism [92], immune response [93,94], development [95], and drug response [96]. Moreover, gut microbes are crucial for processing otherwise inaccessible nutrients and for harvesting energy from the host diet [97,98]. Clearly, some metabolic, energetic, temporal, and spatial considerations may differ significantly between microbe-microbe models and host-microbe models. These considerations may affect the choice of compartmentalization scheme or of other design elements used when constructing these models. Yet, at its heart, the approach taken for modeling host-microbe metabolic interactions is similar to the one described above for modeling microbial interspecific interactions, and usually involves some integration between a microbial metabolic model and a model of the host metabolism. Fortunately, metabolic models of the human host have already been introduced [99], as well as tissue-specific models of human metabolism [100,101].
Several studies have already used this approach to study the interaction between the host and specific host-associated microbial species. Comparing the topology of the host metabolic network with that of a symbiont or a parasite can reveal novel relationships or complementarities in metabolic functions. For example, gaps identified in some essential pathways of an insect host were found to be compensated by the capacity of an endosymbiont [102]. More complex topological methods for finding metabolic complementarities have made use of graph-based algorithms to predict the extent of nutritional support between the metabolic networks of a host and a parasite [103]. Predictions based on this framework have been shown to closely parallel inter-species relationships, providing new insight into evolutionary and ecological trends. A related approach was used to obtained a comprehensive view of the metabolic exchanges between two endocytobionts residing in the same insect host (see also above) by determining the set of exogenously acquired nutrients needed to synthesize the metabolites involved in symbiotic interactions [62]. Constraint-based approaches have also been applied to the study of host-microbe interactions. For example, integrating two genome-scale network reconstructions, adding interfacial constraints, and altering the objective function to reflect a pathogenic lifestyle, a recent study was able to accurately depict different types of M. tuberculosis infection in a human host [104]. Another study captured cross-feeding and competition between a mouse host and a commensal microbe via a combined constraint-based model with the addition of a joint external compartment facilitating metabolite exchange [105]. These studies, again, speak to the challenge of designing an appropriate compartmentalization scheme that most accurately reflects inter-species dynamics. For an in depth review of constraint-based methods in the context of host-microbe interactions, see Ref [106].
Alternatively, one can model the interaction between the host and the microbiome as a whole, following the supra-organism paradigm discussed above. Though such efforts are currently scarce, the rapidly accelerating pace of studies generating microbiome-wide genomic, transcriptomic, and metabolomic data may provide the foundation for continued development in this area. A preliminary attempt to model the human-microbiome interactome was recently introduced, in which a microbiome-wide metabolic network was used to identify sets of bacteria-specific metabolites known to interact with human protein complexes [107]. Interestingly, more than half of these interactions involved complexes associated with disease and many of the metabolites identified were highly similar to known small-molecule drugs, suggesting a significant role for the microbiome in both host metabolic activity and in maintenance of host health. Integrating such community-level models with tissue-specific models of human metabolism [100,101] will allow us to further study metabolic dependencies between the microbiome and the host and to better predict the impact of the microbiome on human health.
Putting it All Together: Future Directions and Challenges
The modeling frameworks described above provide valuable insights into the capacity of the microbiome and illuminate various facets of the human microbiome system. These studies show great promise and highlight some of the potential, as well as the challenges, in modeling different aspects of the microbiome’s metabolic processes. Clearly, however, a fully comprehensive model of the microbiome, encompassing its activity, dynamics, and impact on the host, must not only utilize these various modeling approaches but also integrate them across temporal and organizational scales. Such an integrative model should account, for example, for the way species interactions affect the abundances of the various species, and ultimately the functional composition of the microbial supra-organism, over time. Similarly, the activity of the microbiome as a whole and its interaction with the host in turn affect the biochemical environment of the gut and the context in which microbiome species function. Finally, the host diet, metabolism, and immune response exert strong selective pressures on the microbiome, further affecting its species composition. At longer time scales, the above pressures and interactions may additionally induce genomic adaptation of resident species, resulting in strain-level variation and altering community-wide abundances of specific genes [108]. Modeling and integrating such evolutionary dynamics and their potential functional effects on the metabolic system remains an outstanding challenge. Ultimately though, a predictive and clinically relevant model of the microbiome can only be achieved by considering such complex dependencies and their impact.
Recently introduced multi-scale models of microbe-microbe interactions are a promising first step in this direction. Some models, for example, apply a multi-level and multi-objective optimization scheme [81] or temporal dynamics [79,109] to account for the trade-off between individual species and the community and for the changes in the relative abundances of the interacting species. An additional constraint-based model has recently been constructed to study the interaction between a host and a representative gut microbe [105]. This model was used to characterize diet-dependent changes in uptake and secretion rates and was shown to successfully predict growth dependencies and cross feeding. Multi-scale models of human metabolism, integrating cellular scale models with whole-body physiology, have also been introduced [110]. Still, a comprehensive framework that integrates microbe-microbe and host-microbe interactions, that accounts for the interaction of the microbiome as a whole with the host, and that can be scaled up to effectively model the vast number of species comprising the human microbiome, is lacking.
Moreover, it should be noted that our focus in this review is centered primarily on models of metabolism. This is potentially the most clinically-relevant process, especially in understanding the capacity of the gut microbiome, and the one for which modeling frameworks are relatively well established. Other cellular processes, however, clearly contribute as well to the activity of the microbiome and to its overall influence on the host. Modeling such processes and integrating them with metabolic models is a challenging task. For individual species, a few multi-scale models that integrate metabolism with macromolecular synthesis [111] or with regulation [112] are already available. Most notably, a remarkable effort to computationally model the entire life cycle of the human pathogen Mycoplasma genitalium was recently introduced [113]. This multi-scale model integrates 28 different cellular processes, capturing every facet of the biology of this species and provides the first whole-cell model of a living organism. Taking a similar approach to model all the species in the human microbiome and their interactions is clearly a mammoth task. Many gut dwelling microbes have not yet been identified, let alone sequenced or studied extensively. Yet, such complete single-species models provide a promising glimpse into the potential of in silico modeling of complex biological systems.
Conclusions and Opportunities
Clearly, there is still much work ahead to achieve a comprehensive multi-scale model of the human microbiome. The works summarized above, however, indicate definite progress, with research already moving beyond the consideration of genes or species in isolation and towards a clearer focus on various systems-level aspects of the microbiome. Looking ahead, the implications of such efforts are tremendous. The ability to predict the specific effect of nutritional additives [114,115], drug treatments [116], or microbial supplements [117,118] on the metabolic activity of the microbiome will enable both better diagnostics and intervention in cases of medical importance. Initial promise stemmed from reports of disease elimination through complete microbiome transfer [119], but recently more targeted therapies have been introduced as well [120–122]. A more informed, integrated view of microbial interactions would make such procedures safer, more cost-efficient, and ultimately more effective. Malnourishment could be allayed by the administration of a cocktail of probiotic species carefully designed with in silico support for their sustained growth and activity in the human gut [123]. Similarly, early biomarkers for a digestive disorder could consist of disrupted cross-talk between microbial species or genes, revealed only by explicitly modeling their expected interactions.
Many of the resources required for continued progress are already here and the foundations have been laid. Multiple channels of data from the human microbiome are available, can be easily accessed via specialized databases [10,85,87], and analyzed by multiple tools already in place [88]. Similarly, thousands of annotated microbial genomes are now available [124], opening the door to the reconstruction of numerous genome-scale models. Integration of transcriptomic, proteomic, or regulatory information may further improve the reconstruction and validation of such models [125]. Network-based and constraint-based modeling frameworks have been used extensively to study individual microbial species, and additional frameworks and analytical techniques are constantly being developed. Future research on community modeling will undoubtedly make use of this vast array of systems and pathway-based tools originally designed for single organisms.
The work presented above represents significant progress in just a few short years. We hope that highlighting current work in this sphere will call attention to this promising mode of research and inspire further innovation in this direction. Ultimately, continued research in this field will bring us closer to a principled understanding of the microbiome and will facilitate informed manipulation of this complex system.
Highlights.
Multiple high-throughput methods for studying the human microbiome are available
System-level models are needed to gain a predictive understanding of the microbiome
Preliminary modeling efforts show promise but many challenges remain
Interactions between species, across the community, and with a host can be modeled
Integration of multiple approaches is required to construct a comprehensive model
Acknowledgments
R.L. is supported by an NSF Graduate Research Fellowship under grant no. DGE-0718124. E.B. is an Alfred P. Sloan Research Fellow. This work was supported in part by a New Innovator Award DP2 AT 007802-01 to E.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Morgan XC, Segata N, Huttenhower C. Biodiversity and functional genomics in the human microbiome. Trends in Genetics. 2013;29:51–58. doi: 10.1016/j.tig.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schloss PD, Handelsman J. Metagenomics for studying unculturable microorganisms: cutting the Gordian knot. Genome biology. 2005;6:229. doi: 10.1186/gb-2005-6-8-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, Gordon JI. An invitation to the marriage of metagenomics and metabolomics. Cell. 2008;134:708–13. doi: 10.1016/j.cell.2008.08.025. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert JA, Meyer F, Antonopoulos D, Balaji P, Brown CT, Brown CT, Desai N, Eisen JA, Evers D, Field D, et al. Meeting Report: The Terabase Metagenomics Workshop and the Vision of an Earth Microbiome Project. Standards in genomic sciences. 2010;3:243–248. doi: 10.4056/sigs.1433550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knight R, Jansson J, Field D, Fierer N, Desai N, Fuhrman JA, Hugenholtz P, van der Lelie D, Meyer F, Stevens R, et al. Unlocking the potential of metagenomics through replicated experimental design. Nature Biotechnology. 2012;30:513–20. doi: 10.1038/nbt.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen Ja, Hoffman JM, Remington K, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS biology. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, et al. Evolution of mammals and their gut microbes. Science (New York, NY) 2008;320:1647–51. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wooley JC, Godzik A, Friedberg I. A primer on metagenomics. PLoS Computational Biology. 2010;6:e1000667. doi: 10.1371/journal.pcbi.1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9•.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. This large-scale study of the gut microbiomes of 124 European individuals established a non-redundant collection of over 3.3 million microbial genes present in the human intestinal tract and identified a core set of functions present in the human gut microbiome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Methé Ba, Nelson KE, Pop M, Creasy HH, Giglio MG, Huttenhower C, Gevers D, Petrosino JF, Abubucker S, Badger JH, et al. A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, Fitzgerald MG, Fulton RS, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. This paper details a large-scale study of the microbiomes of 18 body sites across 242 healthy American adults as part of the Human Microbiome Project. The community composition and metabolic function of the healthy Western microbiome is characterized. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12•.Sharon I, Morowitz MJ, Thomas BC, Costello EK, Relman DA, Banfield JF. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome research. 2012;23:111–120. doi: 10.1101/gr.142315.112. This study analyzes time-series metagenomic samples of a single infant’s gut microbiome, providing an exciting example of individualized metagenomic data. Full genomes of prevalent bacterial species are reconstructed and the variations in species’ strains are tracked over time. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, Knights D, Gajer P, Ravel J, Fierer N, et al. Moving pictures of the human microbiome. Genome biology. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proceedings of the National Academy of Sciences. 2011;108 (Suppl):4554–61. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11070–5. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 18.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, Leleiko N, Snapper SB, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome biology. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19•.Lemon KP, Armitage GC, Relman DA, Fischbach MA. Microbiota-targeted therapies: an ecological perspective. Science translational medicine. 2012;4:137rv5. doi: 10.1126/scitranslmed.3004183. An extensive review on the progress and potential of microbiota-targeted therapies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hood L. Tackling the microbiome. Science. 2012;336:1209. doi: 10.1126/science.1225475. [DOI] [PubMed] [Google Scholar]
- 21.Trosvik P, Rudi K, Straetkvern KO, Jakobsen KS, Naes T, Stenseth NC. Web of ecological interactions in an experimental gut microbiota. Environmental microbiology. 2010;12:2677–87. doi: 10.1111/j.1462-2920.2010.02236.x. [DOI] [PubMed] [Google Scholar]
- 22••.Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proceedings of the National Academy of Sciences. 2012;109:594–599. doi: 10.1073/pnas.1116053109. This study introduces the Metagenomic Systems Biology framework, reconstructing and analyzing microbiome-wide metabolic networks based on metagenomic data from 124 human gut microbiome samples. Overlaying information about the differential abundance of enzymes across samples and across host states onto these networks reveals several topological characteristics that typify disease states. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raes J, Bork P. Molecular eco-systems biology: towards an understanding of community function. Nature reviews Microbiology. 2008;6:693–9. doi: 10.1038/nrmicro1935. [DOI] [PubMed] [Google Scholar]
- 24.Röling WF, Ferrer M, Golyshin PN. Systems approaches to microbial communities and their functioning. Current opinion in biotechnology. 2010;21:532–538. doi: 10.1016/j.copbio.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 25.Borenstein E. Computational systems biology and in silico modeling of the human microbiome. Briefings in Bioinformatics. 2012;13:769–780. doi: 10.1093/bib/bbs022. [DOI] [PubMed] [Google Scholar]
- 26.Ruppin E, Papin Ja, de Figueiredo LF, Schuster S. Metabolic reconstruction, constraint-based analysis and game theory to probe genome-scale metabolic networks. Current opinion in biotechnology. 2010 doi: 10.1016/j.copbio.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Hellweger FL, Bucci V. A bunch of tiny individuals—Individual-based modeling for microbes. Ecological Modelling. 2009;220:8–22. [Google Scholar]
- 28.Mitri S, Xavier JB, Foster KR. Social evolution in multispecies biofilms. Proceedings of the National Academy of Sciences of the United States of America. 2011;108 (Suppl ):10839–10846. doi: 10.1073/pnas.1100292108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Momeni B, Brileya Ka, Fields MW, Shou W. Strong inter-population cooperation leads to partner intermixing in microbial communities. eLife. 2013;2:e00230. doi: 10.7554/eLife.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reed JL, Palsson BØ. Thirteen years of building constraint-based in silico models of Escherichia coli. Journal Of Bacteriology. 2003;185:2692–2699. doi: 10.1128/JB.185.9.2692-2699.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durot M, Bourguignon PY, Schachter V. Genome-scale models of bacterial metabolism: reconstruction and applications. FEMS microbiology reviews. 2009;33:164–90. doi: 10.1111/j.1574-6976.2008.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cloots L, Marchal K. Network-based functional modeling of genomics, transcriptomics and metabolism in bacteria. Current Opinion in Microbiology. 2011;14:599–607. doi: 10.1016/j.mib.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Levy R, Borenstein E. Reverse Ecology: From Systems to Environments and Back. Advances in Experimental Medicine and Biology. 2012;751:329–345. doi: 10.1007/978-1-4614-3567-9_15. [DOI] [PubMed] [Google Scholar]
- 34.Papp B, Pál C, Hurst LD. Metabolic network analysis of the causes and evolution of enzyme dispensability in yeast. Nature. 2004;429:661–4. doi: 10.1038/nature02636. [DOI] [PubMed] [Google Scholar]
- 35.Pál C, Papp B, Lercher MJ, Csermely P, Oliver SG, Hurst LD. Chance and necessity in the evolution of minimal metabolic networks. Nature. 2006;440:667–70. doi: 10.1038/nature04568. [DOI] [PubMed] [Google Scholar]
- 36.Schink B. Synergistic interactions in the microbial world. 2002;81:257–261. doi: 10.1023/a:1020579004534. [DOI] [PubMed] [Google Scholar]
- 37.Little AEF, Robinson CJ, Peterson SB, Raffa KF, Handelsman J. Rules of engagement: interspecies interactions that regulate microbial communities. Annual review of microbiology. 2008;62:375–401. doi: 10.1146/annurev.micro.030608.101423. [DOI] [PubMed] [Google Scholar]
- 38.Stams AJM, Plugge CM. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nature Reviews Microbiology. 2009;7:568–577. doi: 10.1038/nrmicro2166. [DOI] [PubMed] [Google Scholar]
- 39.Lee HH, Molla MN, Cantor CR, Collins JJ. Bacterial charity work leads to population-wide resistance. Nature. 2010;467:82–85. doi: 10.1038/nature09354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cordero OX, Wildschutte H, Kirkup B, Proehl S, Ngo L, Hussain F, Le Roux F, Mincer T, Polz MF. Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance. Science (New York, NY) 2012;337:1228–31. doi: 10.1126/science.1219385. [DOI] [PubMed] [Google Scholar]
- 41.Uroz S, Chhabra SR, Cámara M, Williams P, Oger P, Dessaux Y. N-Acylhomoserine lactone quorum-sensing molecules are modified and degraded by Rhodococcus erythropolis W2 by both amidolytic and novel oxidoreductase activities. Microbiology. 2005;151:3313–3322. doi: 10.1099/mic.0.27961-0. [DOI] [PubMed] [Google Scholar]
- 42.Vartoukian SR, Palmer RM, Wade WG. Strategies for culture of “unculturable” bacteria. FEMS Microbiology Letters. 2010;309:1–7. doi: 10.1111/j.1574-6968.2010.02000.x. [DOI] [PubMed] [Google Scholar]
- 43.Periasamy S, Kolenbrander PE. Aggregatibacter actinomycetemcomitans builds mutualistic biofilm communities with Fusobacterium nucleatum and Veillonella species in saliva. Infection and immunity. 2009;77:3542–51. doi: 10.1128/IAI.00345-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Periasamy S, Kolenbrander PE. Mutualistic biofilm communities develop with Porphyromonas gingivalis and initial, early, and late colonizers of enamel. Journal of bacteriology. 2009;191:6804. doi: 10.1128/JB.01006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Periasamy S, Chalmers NI, Du-Thumm L, Kolenbrander PE. Fusobacterium nucleatum ATCC 10953 requires Actinomyces naeslundii ATCC 43146 for growth on saliva in a three-species community that includes Streptococcus oralis 34. Applied and environmental microbiology. 2009;75:3250–7. doi: 10.1128/AEM.02901-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolenbrander PE. Multispecies communities: interspecies interactions influence growth on saliva as sole nutritional source. International journal of oral science. 2011;3:49–54. doi: 10.4248/IJOS11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foster K, Bell T. Competition, not Cooperation, Dominates Interactions among Culturable Microbial Species. Current Biology. 2012;22:1845–1850. doi: 10.1016/j.cub.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 48.Faust K, Raes J. Microbial interactions: from networks to models. Nature Reviews Microbiology. 2012;10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 49.Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C. Microbial Co-occurrence Relationships in the Human Microbiome. PLoS Computational Biology. 2012;8:e1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chaffron S, Rehrauer H, Pernthaler J, von Mering C. A global network of coexisting microbes from environmental and whole-genome sequence data. Genome research. 2010;20:947–59. doi: 10.1101/gr.104521.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480:241–4. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- 52.Jeraldo P, Sipos M, Chia N, Goldenfeld N. Quantification of the relative roles of niche and neutral processes in structuring gastrointestinal microbiomes. Proceedings of the National Academy of Sciences. 2012;109:9692–9698. doi: 10.1073/pnas.1206721109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu D, Daugherty SC, Van Aken SE, Pai GH, Watkins KL, Khouri H, Tallon LJ, Zaborsky JM, Dunbar HE, Tran PL, et al. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS biology. 2006;4:e188. doi: 10.1371/journal.pbio.0040188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCutcheon JP, Moran Na. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19392–7. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Janga SC, Babu MM. Network-based approaches for linking metabolism with environment. Genome biology. 2008;9:239. doi: 10.1186/gb-2008-9-11-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56•.Borenstein E, Kupiec M, Feldman MW, Ruppin E. Large-scale reconstruction and phylogenetic analysis of metabolic environments. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14482–14487. doi: 10.1073/pnas.0806162105. This paper introduces the seed set framework and applies it to 478 microbial species. Seed set composition is shown to successfully predict known biochemical dependencies and provides insights into species’ lifestyles and environments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carr R, Borenstein E. NetSeed: A network-based reverse-ecology tool for calculating the metabolic interface of an organism with its environment. Bioinformatics. 2012 doi: 10.1093/bioinformatics/btr721. no volume. [DOI] [PubMed] [Google Scholar]
- 58.Handorf T, Ebenhöh O, Heinrich R. Expanding metabolic networks: scopes of compounds, robustness, and evolution. Journal of molecular evolution. 2005;61:498–512. doi: 10.1007/s00239-005-0027-1. [DOI] [PubMed] [Google Scholar]
- 59•.Freilich S, Kreimer A, Meilijson I, Gophna U, Sharan R, Ruppin E. The large-scale organization of the bacterial network of ecological co-occurrence interactions. Nucleic Acids Research. 2010;38:3857–3868. doi: 10.1093/nar/gkq118. This study combines the seed-set framework with the network expansion algorithms, introducing the Effective Metabolic Overlap score for species competition. Clusters of co-occurring species are partitioned into low-competition/high-growth and high-competition/low-growth, supporting the r/K selection theory. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Freilich S, Kreimer A, Borenstein E, Yosef N, Sharan R, Gophna U, Ruppin E. Metabolic-network-driven analysis of bacterial ecological strategies. Genome biology. 2009;10:R61. doi: 10.1186/gb-2009-10-6-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Christian N, Handorf T, Ebenhöh O. Metabolic synergy: increasing biosynthetic capabilities by network cooperation. Genome informatics International Conference on Genome Informatics. 2007;18:320–329. [PubMed] [Google Scholar]
- 62•.Cottret L, Milreu PV, Acuña V, Marchetti-Spaccamela A, Stougie L, Charles H, Sagot M-F. Graph-Based Analysis of the Metabolic Exchanges between Two Co-Resident Intracellular Symbionts, Baumannia cicadellinicola and Sulcia muelleri, with Their Insect Host, Homalodisca coagulata. PLoS Computational Biology. 2010;6:13. doi: 10.1371/journal.pcbi.1000904. Network-based modeling is combined with graph theory-based algorithms to study to a complex, 3 species symbiosis. While comparative genomics of the involved endosymbionts provided clues into their inherent complementarity, the methods presented revealed specific pathways used for metabolic exchange, as well as compounds neither was capable to provide to its partner. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cottret L, Milreu PV, Acu V. Enumerating Precursor Sets of Target Metabolites in a Metabolic Network. Algorithms in Bioinformatics. 2008;5251:233–244. [Google Scholar]
- 64.Orth JD, Thiele I, Palsson BØ. What is flux balance analysis? Nature Biotechnology. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Segrè D, Vitkup D, Church GM. Analysis of optimality in natural and perturbed metabolic networks. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15112–7. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shlomi T, Berkman O, Ruppin E. Regulatory on/off minimization of metabolic flux changes after genetic perturbations. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7695–700. doi: 10.1073/pnas.0406346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Varma A, Palsson BO. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Applied and environmental microbiology. 1994;60:3724–31. doi: 10.1128/aem.60.10.3724-3731.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ibarra R, Edwards J. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature. 2002;420:20–23. doi: 10.1038/nature01149. [DOI] [PubMed] [Google Scholar]
- 69.Milne CB, Kim P-J, Eddy JA, Price ND. Accomplishments in Genome-Scale In Silico Modeling for Industrial and Medical Biotechnology. Biotechnology Journal. 2009;4:1653–1670. doi: 10.1002/biot.200900234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70•.Thiele I, Palsson BØ. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nature Protocols. 2010;5:93–121. doi: 10.1038/nprot.2009.203. A step by step protocol for manual construction of FBA models. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71•.Henry CS, DeJongh M, Best Aa, Frybarger PM, Linsay B, Stevens RL. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nature Biotechnology. 2010;28:969–974. doi: 10.1038/nbt.1672. The automatic SEED pipeline for constraint-based model reconstruction from sequenced genomes. [DOI] [PubMed] [Google Scholar]
- 72.Feng X, Xu Y, Chen Y, Tang YJ. MicrobesFlux: a web platform for drafting metabolic models from the KEGG database. BMC systems biology. 2012;6:94. doi: 10.1186/1752-0509-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73••.Stolyar S, Van Dien S, Hillesland KL, Pinel N, Lie TJ, Leigh JA, Stahl DA. Metabolic modeling of a mutualistic microbial community. Molecular Systems Biology. 2007;3:92. doi: 10.1038/msb4100131. The first attempt to model metabolic interactions between two organisms with a simplified constraint-based model that contains a limited number of pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74•.Freilich S, Zarecki R, Eilam O, Segal ES, Henry CS, Kupiec M, Gophna U, Sharan R, Ruppin E. Competitive and cooperative metabolic interactions in bacterial communities. Nature Communications. 2011;2:589. doi: 10.1038/ncomms1597. This study utilizes automatically reconstructed models to assess competitive and cooperative potential among numerous bacterial pairs. Analysis of the predicted interactions coupled with ecological data demonstrates the role of competition in community assembly and the unidirectional nature of cooperative interactions. [DOI] [PubMed] [Google Scholar]
- 75•.Wintermute EH, Silver PA. Emergent cooperation in microbial metabolism. Molecular Systems Biology. 2010;6:1–7. doi: 10.1038/msb.2010.66. A computational and experimental study of metabolic cross-feeding between E. coli auxotrophs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zengler K, Palsson BO. A road map for the development of community systems (CoSy) biology. Nature reviews Microbiology. 2012;10:366–72. doi: 10.1038/nrmicro2763. [DOI] [PubMed] [Google Scholar]
- 77••.Taffs R, Aston JE, Brileya K, Jay Z, Klatt CG, McGlynn S, Mallette N, Montross S, Gerlach R, Inskeep WP, et al. In silico approaches to study mass and energy flows in microbial consortia: a syntrophic case study. BMC systems biology. 2009;3:114. doi: 10.1186/1752-0509-3-114. A theoretical study that presents three fundamentally different schemes for microbial consortia modeling, including a compartmentalized modeling approach, a pooled modeling approach and a nested modeling approach. These methods are examined using data from thermophilic, phototrophic mat communities, highlighting various considerations for choosing an appropriate modeling scheme. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klitgord N, Segrè D. The importance of compartmentalization in metabolic flux models: yeast as an ecosystem of organelles. Genome informatics International Conference on Genome Informatics. 2010;22:41–55. [PubMed] [Google Scholar]
- 79.Zhuang K, Izallalen M, Mouser P, Richter H, Risso C, Mahadevan R, Lovley DR. Genome-scale dynamic modeling of the competition between Rhodoferax and Geobacter in anoxic subsurface environments. The ISME journal. 2011;5:305–316. doi: 10.1038/ismej.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Klitgord N, Segrè D. Environments that induce synthetic microbial ecosystems. PLoS computational biology. 2010;6:e1001002. doi: 10.1371/journal.pcbi.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zomorrodi AR, Maranas CD. OptCom: A Multi-Level Optimization Framework for the Metabolic Modeling and Analysis of Microbial Communities. PLoS computational biology. 2012;8:e1002363. doi: 10.1371/journal.pcbi.1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gordon JI, Klaenhammer TR. Colloquium Paper: A rendezvous with our microbes. Proceedings of the National Academy of Sciences. 2011;108:4513–4515. doi: 10.1073/pnas.1101958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gianoulis TA, Raes J, Patel PV, Bjornson R, Korbel JO, Letunic I, Yamada T, Paccanaro A, Jensen LJ, Snyder M, et al. Quantifying environmental adaptation of metabolic pathways in metagenomics. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1374–1379. doi: 10.1073/pnas.0808022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dinsdale Ea, Edwards Ra, Hall D, Angly F, Breitbart M, Brulc JM, Furlan M, Desnues C, Haynes M, Li L, et al. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–32. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- 85.Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yamada T, Letunic I, Okuda S, Kanehisa M, Bork P. iPath2. 0: interactive pathway explorer. Nucleic acids research. 2011;39:W412–5. doi: 10.1093/nar/gkr313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Markowitz VM, Chen I-min A, Chu K, Szeto E, Palaniappan K, Grechkin Y, Ratner A, Jacob B, Pati A, Huntemann M, et al. IMG / M : the integrated metagenome data management and comparative analysis system. 2012;40:123–129. doi: 10.1093/nar/gkr975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88••.Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, Rodriguez-Mueller B, Zucker J, Thiagarajan M, Henrissat B, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Computational Biology. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. The HUMaN software pipeline maps metagenomic data to modules and pathways and calculates scores of abundance and evenness. Procedures like gap-filling and smoothing facilitate a focus on functionally-oriented subsystems within the microbiome, allowing one to infer the presence or absence of genes by placing them in a functional context. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89•.Liu B, Pop M. MetaPath : identifying differentially abundant metabolic pathways in metagenomic datasets. BMC Proceedings. 2011;5:S9. doi: 10.1186/1753-6561-5-S2-S9. MetaPath applies a greedy heuristic algorithm to enzyme abundances overlaid on KEGG global metabolism pathways to identify subnetworks that are differentially abundant among groups of metagenomic samples. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90•.Larsen PE, Collart FR, Field D, Meyer F, Keegan KP, Henry CS, McGrath J, Quinn J, Gilbert JA. Predicted Relative Metabolomic Turnover (PRMT): determining metabolic turnover from a coastal marine metagenomic dataset. Microbial informatics and experimentation. 2011;1:4. doi: 10.1186/2042-5783-1-4. A methodology is described to quantify differential metabolite consumption and production in the context of community-wide abundance of enzymatic reactions. A matrix linking annotated enzymes in a metagenomic sample to associated substrates and products is multiplied by normalized enzyme abundances to generate a turnover score for each metabolite. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hooper LV. Commensal Host-Bacterial Relationships in the Gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 92.Fischbach Ma, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell host & microbe. 2011;10:336–47. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nature Reviews Immunology. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Heijtz RD, Wang S, Anuar F, Qian Y, Björkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S. Normal gut microbiota modulates brain development and behavior. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3047–52. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wilson ID, Nicholson JK. The role of gut microbiota in drug response. Current Pharmaceutical Design. 2009;15:1519–1523. doi: 10.2174/138161209788168173. [DOI] [PubMed] [Google Scholar]
- 97.Turnbaugh PJ, Ley RE, Mahowald Ma, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 98.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science (New York, NY) 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 99.Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson BØ. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang Y, Eddy JA, Price ND. Reconstruction of genome-scale metabolic models for 126 human tissues using mCADRE. BMC Systems Biology. 2012;6:153. doi: 10.1186/1752-0509-6-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jerby L, Shlomi T, Ruppin E. Computational reconstruction of tissue-specific metabolic models: application to human liver metabolism. Molecular Systems Biology. 2010;6:401. doi: 10.1038/msb.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103•.Borenstein E, Feldman MW. Topological signatures of species interactions in metabolic networks. Journal of computational biology : a journal of computational molecular cell biology. 2009;16:191–200. doi: 10.1089/cmb.2008.06TT. This study extends the seed set framework and presents the Biosynthetic Support Score for inferring the level of nutritional support between a host and a parasite. This score is shown to successfully predict host-parasite interactions on a large scale. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bordbar A, Lewis NE, Schellenberger J, Palsson BØ, Jamshidi N. Insight into human alveolar macrophage and M. tuberculosis interactions via metabolic reconstructions. Molecular Systems Biology. 2010;6:422. doi: 10.1038/msb.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105••.Heinken A, Sahoo S, Fleming RMT, Thiele I. Systems-level characterization of a host-microbe metabolic symbiosis in the mammalian gut. Gut microbes. 2013;4:1–13. doi: 10.4161/gmic.22370. A joined FBA model of host-microbe metabolism, characterizing syntrophic and competitive interactions between Mouse and a representative gut microbe. The authors examine changes of uptake and secretion rates in the host with and without the existence of the gut microbe, under different diets, and with varying ratios of host-microbe growth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thiele I, Heinken A, Fleming RM. A systems biology approach to studying the role of microbes in human health. Current opinion in biotechnology. doi: 10.1016/j.copbio.2012.10.001. date unknown. [DOI] [PubMed] [Google Scholar]
- 107.Jacobsen UP, Nielsen HB, Hildebrand F, Raes J, Sicheritz-Ponten T, Kouskoumvekaki I, Panagiotou G. The chemical interactome space between the human host and the genetically defined gut metabotypes. The ISME journal. 2012 doi: 10.1038/ismej.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schloissnig S, Arumugam M, Sunagawa S, Mitreva M, Tap J, Zhu A, Waller A, Mende DR, Kultima JR, Martin J, et al. Genomic variation landscape of the human gut microbiome. Nature. 2013;493:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hanly TJ, Henson Ma. Dynamic flux balance modeling of microbial co-cultures for efficient batch fermentation of glucose and xylose mixtures. Biotechnology and bioengineering. 2011;108:376–85. doi: 10.1002/bit.22954. [DOI] [PubMed] [Google Scholar]
- 110.Krauss M, Schaller S, Borchers S, Findeisen R, Lippert J, Kuepfer L. Integrating Cellular Metabolism into a Multiscale Whole-Body Model. PLoS Computational Biology. 2012;8:e1002750. doi: 10.1371/journal.pcbi.1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Thiele I, Fleming RMT, Que R, Bordbar A, Diep D, Palsson BO. Multiscale Modeling of Metabolism and Macromolecular Synthesis in E. coli and Its Application to the Evolution of Codon Usage. PloS one. 2012;7:e45635. doi: 10.1371/journal.pone.0045635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Covert MW. Computational Systems Biology. Academic Press; 2005. Integrated regulatory and metabolic models; pp. 191–204. [Google Scholar]
- 113••.Karr JR, Sanghvi JC, Macklin DN, Gutschow MV, Jacobs JM, Bolival B, Assad-Garcia N, Glass JI, Covert MW. A Whole-Cell Computational Model Predicts Phenotype from Genotype. Cell. 2012;150:389–401. doi: 10.1016/j.cell.2012.05.044. The most extensive effort to date to reconstruct a whole-cell model. This multi-scale model of Mycoplasma genitalium integrates 28 cellular processes that are modeled on a short timescale to update global cellular variables, which can then affect each of the processes. A large array of data is used to validate the predictions of the model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Martin F-PJ, Wang Y, Sprenger N, Yap IKS, Rezzi S, Ramadan Z, Peré-Trepat E, Rochat F, Cherbut C, van Bladeren P, et al. Top-down systems biology integration of conditional prebiotic modulated transgenomic interactions in a humanized microbiome mouse model. Molecular systems biology. 2008;4:205. doi: 10.1038/msb.2008.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science (New York, NY) 2011;333:101–4. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proceedings of the National Academy of Sciences. 2009;106:14728–33. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Science translational medicine. 2011;3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bron PA, van Baarlen P, Kleerebezem M. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nature Reviews Microbiology. 2012;10:66–78. doi: 10.1038/nrmicro2690. [DOI] [PubMed] [Google Scholar]
- 119.Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. Journal of Clinical Gastroenterology. 2010;44:354–360. doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- 120.Holmes E, Kinross J, Gibson GR, Burcelin R, Jia W, Pettersson S, Nicholson JK. Therapeutic modulation of microbiota-host metabolic interactions. Science translational medicine. 2012;4:137rv6. doi: 10.1126/scitranslmed.3004244. [DOI] [PubMed] [Google Scholar]
- 121.Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, Goulding D, Rad R, Schreiber F, Brandt C, et al. Targeted Restoration of the Intestinal Microbiota with a Simple, Defined Bacteriotherapy Resolves Relapsing Clostridium difficile Disease in Mice. PLoS Pathog. 2012;8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Relman DA. Restoration of the gut microbial habitat as a disease therapy. Nature Biotechnology. 2013;31:35–37. doi: 10.1038/nbt.2475. [DOI] [PubMed] [Google Scholar]
- 123.Gordon JI, Dewey KG, Mills DA, Medzhitov RM. The human gut microbiota and undernutrition. Science translational medicine. 2012;4:137ps12. doi: 10.1126/scitranslmed.3004347. [DOI] [PubMed] [Google Scholar]
- 124.Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, et al. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Research. 2012;40:D115–22. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bordbar A, Mo ML, Nakayasu ES, Schrimpe-Rutledge AC, Kim Y-M, Metz TO, Jones MB, Frank BC, Smith RD, Peterson SN, et al. Model-driven multi-omic data analysis elucidates metabolic immunomodulators of macrophage activation. Molecular Systems Biology. 2012;8:1–12. doi: 10.1038/msb.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]