Abstract
KRAS mutations define a clinically-distinct subgroup of lung adenocarcinoma patients, characterized by smoking history, resistance to EGFR-targeted therapies, and adverse prognosis. Whether KRAS- mutated lung adenocarcinomas also have distinct histopathologic features is not well established. We tested 180 resected lung adenocarcinomas for KRAS and EGFR mutations by high-sensitivity mass spectrometry-based genotyping (Sequenom) and PCR-based sizing assays. All tumors were assessed for the proportion of standard histologic patterns (lepidic, acinar, papillary, micropapillary, solid and mucinous), several other histologic and clinical parameters, and TTF-1 expression by immunohistochemistry. Among 180 carcinomas, 63 (35%) had KRAS mutations (KRAS+), 35 (19%) had EGFR mutations (EGFR+), and 82 (46%) had neither mutation (KRAS-/EGFR-). Solid growth pattern was significantly over-represented in KRAS+ carcinomas: the mean ± standard deviation for the amount of solid pattern in KRAS+ carcinomas was 27 ± 34% compared to 3 ± 10% in EGFR+ (P<0.001) and 15 ± 27% in KRAS-/EGFR- (P=0.033) tumors. Furthermore, at least focal (>20%) solid component was more common in KRAS+ (28/63; 44%) compared to EGFR+ (2/35; 6%; P<0.001) and KRAS-/EGFR- (21/82; 26%; P=0.012) carcinomas. KRAS mutations were also over-represented in mucinous carcinomas, and were significantly associated with the presence of tumor-infiltrating leukocytes and heavier smoking history. EGFR mutations were associated with non-mucinous non-solid patterns, particularly lepidic and papillary, lack of necrosis, lack of cytologic atypia, hobnail cytology, TTF-1 expression, and never/light smoking history. In conclusion, extended molecular and clinicopathologic analysis of lung adenocarcinomas reveals a novel association of KRAS mutations with solid histology and tumor-infiltrating inflammatory cells, and expands on several previously recognized morphologic and clinical associations of KRAS and EGFR mutations. Solid growth pattern was recently shown to be a strong predictor of aggressive behavior in lung adenocarcinomas, which may underlie the unfavorable prognosis associated with KRAS mutations in these tumors.
Keywords: KRAS, EGFR, lung, adenocarcinoma, TTF-1
INTRODUCTION
Kristen rat sarcoma viral oncogene (KRAS) mutations are one of the most common oncogenic events in human carcinomas of endodermal origin, occurring at high frequency in adenocarcinomas of lung, pancreatic, and colorectal origin.1, 2 KRAS is an “old oncogene” in lung cancer, having been first described in these tumors in 1984,3 but recent years have witnessed a revamped interest in the role of KRAS in lung adenocarcinoma because of the rapid advances in molecularly-targeted therapies. Although the efforts to therapeutically target mutant KRAS have thus far proven unsuccessful, KRAS has emerged as a useful negative predictive marker because it occurs in a mutually-exclusive fashion with several recently-identified targetable mutations, including Epidermal Growth Factor Receptor (EGFR) – the molecular target of EGFR tyrosine kinase inhibitors erlotinib and gefitinib. Thus, routine predictive molecular testing of lung adenocarcinomas now commonly combines screening for KRAS together with EGFR mutations.4, 5
Clinically, KRAS and EGFR mutations define two distinct and contrasting subgroups of lung adenocarcinoma patients. While KRAS mutations are more common in western than East Asian patients (25-35% vs 5-10%, respectively), EGFR mutations have an inverse prevalence in these ethnic groups (10-20% vs >50%, respectively).6 In addition, KRAS mutations are more common in smokers, whereas EGFR mutations – in never or light smokers.6 Although the data on prognostic significance of KRAS and EGFR mutations has been conflicting across studies, the adverse prognostic impact of KRAS mutations and the favorable impact of EGFR mutations have been demonstrated in several studies over the years7-10 and in recent studies from our institution.11, 12 In addition, several studies also suggested that KRAS mutations may be markers of resistance not only to EGFR tyrosine kinase inhibitors,4, 5 but also to conventional cisplatin-based chemotherapy.13-15
Histologically, it is well-established that EGFR mutations occur preferentially in non-mucinous adenocarcinomas with lepidic/bronchioloalveolar and papillary patterns (reviewed in Travis et al16). In contrast, KRAS mutations are over-represented in mucinous adenocarcinomas.17-20 However, mucinous carcinomas account for only a minority of lung adenocarcinomas with KRAS mutations in western populations,18, 20, 21 and therefore this association is unlikely to explain the distinct clinical characteristics imparted by KRAS mutations. Several prior studies also suggested that KRAS mutations are associated with poor differentiation,22-24 but this finding has been inconsistent across publications. Furthermore, because grading of lung carcinomas is not well-established, it is not known which morphologic features (growth pattern, cytologic features, necrosis, etc) may have imparted this association.
The goal of this study was therefore to re-examine potential histopathologic correlates of KRAS mutations, particularly in non-mucinous adenocarcinomas. In addition to recent clarification regarding adverse prognostic significance of KRAS mutations, this re-examination was also prompted by advances in mutation testing methodology, with emergence of methods like mass spectrometry-based genotyping (Sequenom platform), which detect a wide spectrum of KRAS mutations with higher analytical sensitivity than standard Sanger sequencing. The use of a higher-sensitivity method to detect KRAS mutations can be anticipated to yield a more robust molecular baseline for the study of histologic and other clinicopathologic correlates of mutations.
With these considerations in mind, we performed a detailed histologic and clinicopathologic analysis of 180 lung adenocarcinomas annotated for KRAS and EGFR mutations by mass spectrometry-based genotyping and sensitive PCR-based assays with the main goal to re-examine potential histopathologic characteristics associated with KRAS mutations.
MATERIALS AND METHODS
Study design
One hundred and eighty surgical resections of primary lung adenocarcinomas, which had undergone routine genotyping for EGFR and KRAS mutations as part of prospective reflex protocol in 2009-2010, were randomly selected from the archives of Memorial Sloan-Kettering Cancer Center, New York. Only conventional invasive adenocarcinomas were included, whereas adenocarcinomas in situ (formerly pure bronchioloalveolar carcinoma) and minimally-invasive adenocarcinomas16 were excluded. All tumors were reviewed by two thoracic pathologists (NR, AM). The study was performed with the approval of Institutional Review Board of Memorial Sloan-Kettering Cancer Center.
Histologic and immunohistochemical analysis
Using a modification of histologic scoring system proposed by IASLC/ATS/ERS,16 each tumor was scored semi-quantitatively for the percentage (0-100%) of 7 composite histologic patterns, including 5 standard non-mucinous patterns (lepidic/bronchioloalveolar, acinar, papillary, micropapially and solid) plus “complex glandular” and mucinous patterns. “Complex glandular” pattern was defined as either 1) cribriform morphology (resembling mammary ductal carcinoma in situ) or 2) complex arborizing intra-glandular proliferations and/or formation of slit-like multilumina (resembling mammary usual duct hyperplasia). Detailed description of morphology and clinicopathologic characteristics of complex glandular patterns will be reported separately (Moreira et al, in preparation). Solid pattern was defined as pavement-like sheets of cells with no glandular lumina (i.e. cells with circumferential attachment to other cells) with or without focal cytoplasmic mucin. In cases where solid pattern had abundant pink cytoplasm with sharp cell borders, imparting a “squamoid” appearance, the distinction from a true squamous component was made on the basis of positive TTF-1 and/or negative p40 (ΔNp63) immunostains (data not shown). For the purposes of this study, all mucinous carcinomas were analyzed as a single group, which included tumors with non-solid histology with prominent cytoplasmic mucin, including mucinous carcinomas with lepidic growth pattern (former “mucinous bronchioloalveolar carcinoma”25/“invasive mucinous adenocarcinoma”16), colloid carcinomas, and carcinomas with mucinous features, not otherwise specified. Carcinomas were classified as “mucinous” if mucinous component represented >20% of the tumor. Similarly, for the purposes of data analysis, non-mucinous patterns were defined as “present” if they represented >20% of the tumor relative to other patterns (based on prior data that clinically-significant amount of patterns, as measured by differences in propensities to metastasize, is above 10%26). All patterns were recorded in 5% increments.
In addition, all tumors were also scored for the following histologic parameters:
- Necrosis: scored as 2+/extensive= involving >20% of the tumor, 1+/ focal = involving <20% of the tumor and 0/absent.
- Cytologic atypia: defined as anisonucleosis, nucleomegaly, irregular nuclear membranes and/or macronucleoli; scored as 2+ = marked and diffuse; 1+ = moderate or focal; 0 = minimal.
- Hobnail cytology: defined as cell outlines individually projecting into luminal spaces (as opposed to forming a smooth luminal border), thus resembling type II pneumocytes or Clara cells, analogous to what has been described as a defining features of terminal respiratory unit-type histology by Yatabe et al;27 scored as 2+ = diffuse; 1+ = focal; 0 = none.
- Tumor-infiltrating leukocytes: defined as lymphocytes and/or other inflammatory cells involving intra- and peri-tumoral stroma and/or infiltrating in-between tumor cells; scored as 2+/marked = prominent at low power (4X objective), 1+/moderate = easily noticeable at low power, and 0/none or minimal = inconspicuous at low power.
A representative whole tissue section from each tumor was analyzed for TTF-1 expression by immunohistochemistry, as previously described.28 Presence of any TTF-1 reactivity was scored as positive. In addition, percentage of immunoreactive cells (0-100%) and intensity of staining (1+, 2+ or 3+) were recorded, and H scores were calculated by multiplying the percentage by intensity score (0- 300).
Clinicopathologic analysis
The following clinicopathologic parameters were recorded: age, gender, smoking status (never vs current/former smoker), pack-year smoking history (defined as packs of cigarettes per day multiplied by years of smoking), tumor size, and tumor stage (American Joint Committee on Cancer 7th edition). Smoking history was collected based on a prospectively-administered questionnaire. Never smokers were defined as patients who smoked <100 cigarettes in a lifetime.
Molecular analysis
KRAS and EGFR point mutations were tested by Sequenom Mass ARRAY system (Sequenom Inc) – a mass spectrometry-based multiplex genotyping platform – which detects 22 non-synonymous KRAS mutations in codons 12, 13 and 61 and 20 EGFR mutations, as previously described.30 Based on prior studies, Sequenom has analytical sensitivity for a mutated allele of ~5% (i.e. required minimal tumor cell content is ~10%).29 EGFR Exon 19 deletions were identified by length analysis of fluorescently-labeled PCR products, as previously described.30
Statistical analysis
Comparison of categorical variables was performed by Fisher’s exact or Chi-square test, and comparison of continuous variable was performed by a Mann-Whitney test. P values of <0.05 were considered statistically significant.
RESULTS
Clinicopathologic, molecular and histologic characteristics
The clinical characteristics of 180 patients with lung adenocarcinoma were as follows: age median (range) 67 (34-87), female gender n=107 (59%), never smoker n=31 (17%), and smoking pack-years median (range) 30 (0-200). Tumor stage was as follows: stage I n=120 (67%), stage II n=35 (19%), and stage III/IV n=20 (11%). Surgical procedures included wedge resection n=60, segmentectomy n=2, bronchial tumor resection n=1, lobectomy n=116 and pneumonectomy n=1.
As shown in Table 1, mutation analysis revealed that among 180 adenocarcinomas, 63 (35%) had KRAS mutations (KRAS+), 35 (19%) had EGFR mutations (EGFR+), and 82 (46%) had neither mutation (KRAS-/EGFR-). KRAS and EGFR mutations were mutually exclusive, with no tumor containing both mutations. KRAS mutations were distributed in codons 12 (n=59), 13 (n=2) and 61 (n=2).
Table 1.
Summary of mutations in 180 lung adenocarcinomas.
| Mutation | N (%) |
|---|---|
|
KRAS G12A G12C G12D G12F G12R G12V G13C G13D Q61H |
63 (35%) 7 24 15 2 1 10 1 1 2 |
|
EGFR L858R Exon 19 Δ S768V |
35 (19%) 21 13 1 |
| No EGFR or KRAS mutations | 82 (46%) |
The distribution of 7 histologic patterns (lepidic, acinar, papillary, micropapillary, complex glandular, solid and mucinous) in 180 adenocarcinomas is shown in Table 2. The majority (162/180; 90%) of adenocarcinomas were highly heterogeneous, consisting of a mixture of 2-6 patterns. The number of mixed patterns per tumor was 2 in 45 cases, 3-4 in 99 cases, and 5-6 in 19 cases (mean number of patters per tumor = 3). At least focal (>20%) acinar, papillary, solid, complex glandular, micropapillary and lepidic patterns were present in 59%, 48%, 28%, 27%, 22% and 19% of cases, respectively. Mucinous patterns were rare in our unselected patient population, occurring in >20% amount in only 17 (9%) of cases, which included mucinous bronchioloalveolar carcinoma/”invasive mucinous adenocarcinoma” (n=6), colloid carcinoma (n=1), and carcinoma with mucinous features, not otherwise specified (n=10). We also attempted to classify carcinomas based on a single predominant pattern, as recently recommended;16 however, 29% (53/180) of cases had two or more patterns in a similar co-dominant amount, precluding objective assignment of a single predominant pattern.
Table 2.
Distribution of histologic patterns in 180 lung adenocarcinomas.
| N (%)# of cases with indicated amount of pattern: | Pattern amount: mean ± SD (range) |
||||
|---|---|---|---|---|---|
| ≥5% | ≥20% | ≥50% | 100% | ||
| Lepidic/bronchioloalveolar | 53 (29) | 35 (19) | 7 (4) | 0* | 7 ± 14 (0-70) |
| Acinar | 131 (73) | 106 (59) | 49 (27) | 0 | 28 ± 25 (0-90) |
| Papillary | 96 (53) | 86 (48) | 19 (11) | 4 (2) | 19 ± 24 (0-100) |
| Micropapillary | 73 (41) | 40 (22) | 5 (3) | 0 | 8 ± 14 (0-80) |
| Complex glandular | 91 (51) | 48 (27) | 11 (6) | 1 (1) | 12 ± 18 (0-90) |
| Solid | 78 (43) | 51 (28) | 31 (17) | 1 (1) | 17 ± 29 (0-100) |
| Mucinous | 19 (11) | 17 (9) | 16 (9) | 12 (7) | 9 ± 27 (0-100) |
The denominator for shown percentages is the total number of cases (n=180).
Entirely lepidic carcinomas (pure bronchioloalveolar carcinomas/“adenocarcinomas in situ”) were excluded from this study. SD standard deviation
Association of mutations and histologic patterns
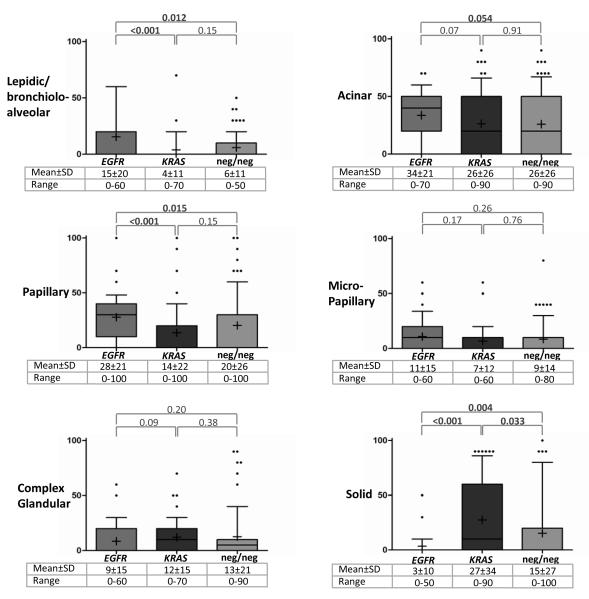
Association of mutations and histologic patterns is shown in Table 3, where patterns were analyzed as categorical variables (i.e. pattern absent vs present), and Figure 1, where patterns were analyzed as continuous variables (i.e. by comparing the mean amount of a pattern according to mutation). Overall, no pattern was invariably present or absent in any molecular group, except for the exclusion of mucinous histology in EGFR+ carcinomas (Table 3). The only pattern that was significantly over-represented in KRAS+ carcinomas compared to EGFR+ and KRAS-/EGFR- groups was solid: the mean ± standard deviation for the amount of solid pattern in KRAS+ carcinomas was 27 ± 34% compared to 3 ± 10% in EGFR+ (P<0.001) and 15 ± 27% in KRAS-/EGFR- (P=0.033) tumors (Figure 1). Conversely, presence of at least focal (>20%) solid component was significantly more frequent in KRAS+ carcinomas (28/63; 44%) compared to EGFR+ (2/35; 6%, P<0.001) and KRAS-/EGFR- (21/82; 26%; P=0.022) groups (Table 3). The rate of KRAS mutations in carcinomas with at least focal solid component was 55% (28/63 cases).
Table 3.
Distribution of histologic patterns according to mutation.
| Total N=180 |
Mutation | P value | |||||
|---|---|---|---|---|---|---|---|
|
EGFR+ n=35 |
KRAS+ n=63 |
neg/neg n=82 |
KRAS+ vs EGFR+ |
KRAS+ vs neg/neg |
EGFR+ vs neg/neg |
||
| Lepidic/bronchioloalveolar | 35 (19) | 15 (43) | 7 (11) | 13 (16) | <0.001 | 0.47 | 0.004 |
| Acinar | 106 (59) | 28 (80) | 34 (54) | 44 (54) | 0.016 | 1.00 | 0.008 |
| Papillary | 86 (48) | 26 (74) | 22 (35) | 38 (46) | <0.001 | 0.18 | 0.008 |
| Micropapillary | 40 (22) | 9 (26) | 12 (19) | 19 (23) | 0.45 | 0.69 | 0.82 |
| Complex glandular | 48 (27) | 9 (26) | 21 (33) | 18 (22) | 0.49 | 0.14 | 0.64 |
| Solid | 51 (28) | 2 (6) | 28 (44) | 21 (26) | <0.001 | 0.022 | 0.012 |
| Mucinous | 17 (9) | 0 | 7 (11) | 10 (12) | 0.048 | 1.00 | 0.032 |
The denominator for shown (percentages) is the total number of cases with each mutation. Pattern was defined as “present” using a ≥20% threshold (see Materials and Methods). neg/neg = cases negative for KRAS and EGFR mutations.
FIGURE 1.
Distribution of patterns according to mutation: the box plots. Y axis indicates the amount of pattern per tumor (0-100%). Upper and lower box borders = 25th and 75th percentiles, whisker = 10th and 90th percentiles, horizontal line = median, plus sign = mean, and dots = outliers. If box borders or a median are not visible – their value is 0. Bolded P values (Mann-Whitney test) are statistically significant. Mucinous pattern was excluded from this analysis due to the overall low number of cases with this pattern. SD standard deviation
Table 3 also shows that of 63 KRAS+ adenocarcinomas, only 7 (11%) cases were mucinous, whereas the rest (89%) of KRAS mutations occurred in non-mucinous carcinomas. The rate of KRAS mutations in mucinous carcinomas overall was 41% (7/17 cases) and the rate of KRAS mutations specifically in mucinous bronchioloalveolar/”invasive mucinous” carcinoma subset was 67% (4/6 cases); statistical analysis of these associations was limited by the overall rarity of mucinous carcinomas in our unselected patient population. The association of KRAS mutations and solid pattern was a property of non-mucinous carcinomas, since solid component was rare in mucinous tumors (present in only 1/17 cases).
The patterns significantly associated with EGFR mutations compared to KRAS+ and KRAS- /EGFR- groups were non-solid and non-mucinous patterns, specifically lepidic, papillary, and to a lesser degree acinar (Table 3; Figure 1).
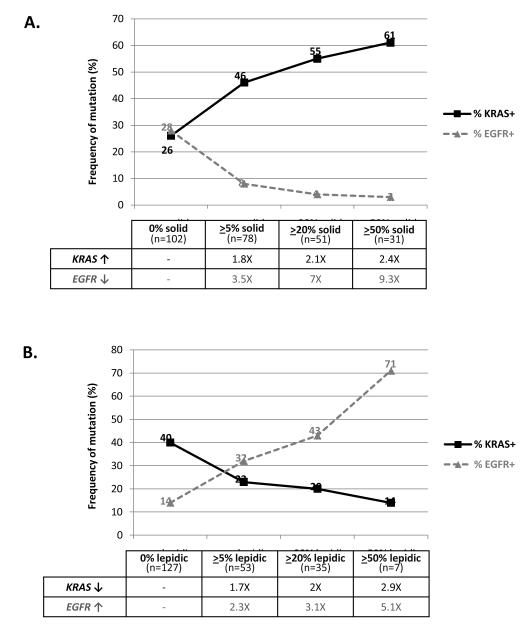
Notably, the amount of solid growth pattern had a graduated effect on the frequency of KRAS and EGFR mutations (Figure 2A). The incremental increase in the amount of solid pattern from 0% → >5% → >20% →>50% lead to the enrichment of KRAS mutations from 26% → 46% → 55% → 61% (2.4X), respectively, while the rate of EGFR mutations had a pronounced 9.3X decrease. In contrast, the amount of lepidic pattern exerted the opposite graduated effect on the likelihood of KRAS and EGFR mutations (Figure 2B). None of the other patterns showed a similar graduated effect on either KRAS or EGFR mutations.
FIGURE 2.
An inverse graduated effects of the amount of solid and lepidic patterns on the likelihood of KRAS and EGFR mutations. The denominator for shown percentages is the number of cases with indicated amount of pattern.
Association of mutations with other histologic characteristics and TTF-1 expression
In addition to the distribution of patterns, we also analyzed the association of mutations with several other histologic features (Table 4). This revealed that KRAS mutations were significantly associated with the presence of tumor-infiltrating leukocytes: 86% of KRAS+ carcinomas featured moderate-marked tumor-associated inflammation compared to 66% of EGFR+ (P=0.038) and 67% of KRAS-/EGFR- (P=0.012) tumors. Furthermore, marked (2+) inflammation was uncommon in EGFR+ carcinomas relative to other groups, but without reaching statistical significance. Similar to solid pattern, association of inflammation with KRAS mutations was only seen in non-mucinous but not in mucinous carcinomas (data not shown). KRAS+ carcinomas also had more necrosis and cytologic atypia relative to EGFR+ and KRAS-/EGFR- groups, although the differences with KRAS-/EGFR- group were not statistically significant. Adenocarcinomas with EGFR mutations had less necrosis and cytologic atypia than the two other molecular groups, but the strongest histologic association for EGFR mutations relative to KRAS+ and KRAS-/EGFR- mutations was with hobnail cytology.
Table 4.
Distribution of other histologic features and TTF-1 expression according to mutation.
| Total N=180 |
Mutation | P value† | |||||
|---|---|---|---|---|---|---|---|
|
EGFR+ n=35 |
KRAS+ n=63 |
neg/neg n=82 |
KRAS vs
EGFR |
KRAS vs neg/neg |
EGFR vs neg/neg |
||
|
Necrosis: N (%) 2+ 1+ 0 |
24 (13) 36 (20) 120 (67) |
1 (3) 4 (11) 30 (86) |
11 (18) 16 (25) 36 (57) |
12 (14) 16 (20) 54 (66) |
0.004 | 0.31 | 0.042 |
|
Cytologic atypia: N (%) 2+ 1+ 0 |
14 (8) 34 (19) 132 (73) |
1 (3) 3 (9) 31 (89) |
7 (11) 16 (25) 40 (63) |
6 (7) 15 (18) 61 (74) |
0.009 | 0.20 | 0.14 |
|
Hobnail cytology: N (%) 2+ 1+ 0 |
48 (27) 24 (13) 108 (60) |
21 (60) 5 (14) 9 (26) |
12 (19) 11 (17) 40 (64) |
15 (18) 8 (10) 59 (72) |
<0.001 | 0.29 | <0.001 |
|
Tumor infiltrating leukocytes: N (%) 2+ 1+ 0 |
24 (13) 108 (60) 48 (27) |
2 (6) 21 (60) 12 (34) |
11 (18) 43 (68) 9 (14) |
11 (13) 44 (54) 27 (33) |
0.038 | 0.012 | 1.00 |
|
TTF-1: N (%) Positive Negative |
162 (90) 18 (10) |
35 (100) 0 |
56 (89) 7 (11) |
71 (87) 11 (13) |
0.048 | 0.80 | 0.032 |
|
TTF-1 in non-mucinous carcinomas: N (%) Positive Negative |
N=163 152 (94) 9 (6) |
n=35 35 (100) 0 |
n=56 53 (95) 3 (5) |
n=74 64 (89) 8 (11) |
0.52 | 0.29 | 0.09 |
P values were analyzed for 2 groups – feature present (1-2+) vs absent (0).
While it is well established that adenocarcinomas with EGFR mutations are almost invariably TTF-1-positive, TTF-1 status in KRAS+ carcinomas is not well established. We therefore analyzed TTF-1 expression by immunohistochemistry in each molecular group (Table 4). Overall, 162 of 180 (90%) adenocarcinomas were positive for TTF-1. Among molecular subgroups, 100% of EGFR+ carcinomas were TTF-1-positive compared to 89% (56/63) of KRAS+ (P=0.048) and 87% (71/82) of KRAS-/EGFR- (P=0.032) carcinomas. Notably, the lack of TTF-1 expression was rare in KRAS+ non-mucinous carcinomas (3/56; 5%) but was seen in the majority of KRAS+ mucinous carcinomas (4/7; 57%); P=0.002. Among non-mucinous carcinomas overall, there was no statistical difference in the number of TTF-1-positive tumors between KRAS+ (53/56; 95%) and EGFR+ (35/35; 100%) tumors; P=0.52. Furthermore, the extent of TTF-1 reactivity in non-mucinous carcinomas was similar in KRAS+ vs EGFR+ groups, which showed mean (range) for TTF-1 H scores of 258 (0-300) vs 281 (60-300), respectively (P=0.29).
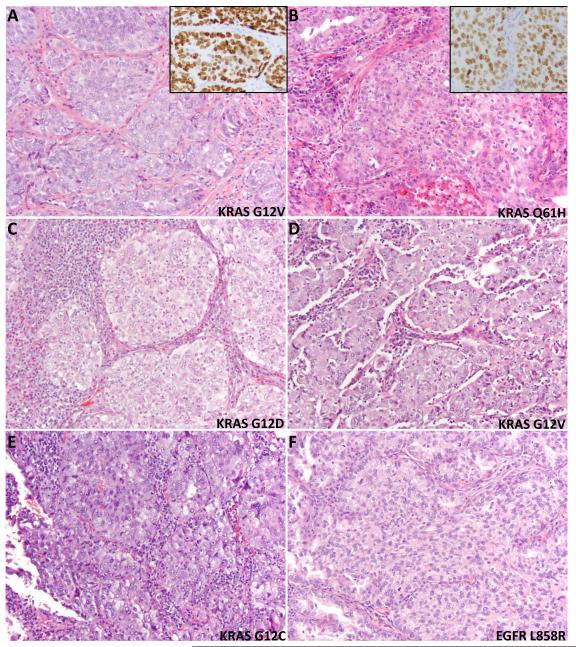
Examples of histologic findings in KRAS+ adenocarcinomas are illustrated in Figure 3.
FIGURE 3.
Examples of solid component in adenocarcinomas with KRAS mutations. At least focal (>20%) solid component was present in 55% of KRAS+ carcinomas (A-E) compared to 4% of EGFR+ carcinomas (F). These solid areas, some of which have “squamoid” appearance, were distinguished from a true squamous component by immunohistochemistry for TTF-1 (inset in A and B), and/or negative p40/ΔNp63 (not shown). Images also illustrate a spectrum of cytologic atypia from minimal (A, B) to moderate (C, D) to marked (E), and spectrum of tumor-associated leukocytes from minimal (A,B) to moderate (D, E) to marked (C) in KRAS+ carcinomas.
Association of mutations and patient characteristics
Distribution of patient characteristics according to mutation is shown in Table 5. Consistent with prior studies, EGFR mutations were strongly associated with never-smoker status and lower pack-year smoking history. In contrast, KRAS mutations were associated with a greater mean pack-year smoking history than EGFR+ (42 vs 13, P<0.001) and KRAS-/EGFR- (42 vs 35; P=0.041) groups. Finally, women were under-represented in KRAS-/EGFR- group. There were no differences in the distribution of age, tumor size and stage. The length of clinical follow-up was too short for survival analysis.
Table 5.
Distribution of clinicopathologic features according to mutation.
| Total N=180 |
Mutation | P value | |||||
|---|---|---|---|---|---|---|---|
|
EGFR+ N=35 |
KRAS+ N=63 |
neg/neg N=82 |
KRAS+ vs EGFR+ |
KRAS+ vs neg/neg |
EGFR+ vs neg/neg |
||
| Age: mean (range) | 66 (38-87) | 65 (51-80) | 66 (38-85) | 67 (34-87) | 0.68 | 0.38 | 0.29 |
|
Gender: N (%) Female Male |
107 (59) 73 (41) |
27 (77) 8 (23) |
41 (65) 22 (35) |
39 (48) 43 (52) |
0.26 | 0.044 | 0.004 |
|
Smoking status: N (%) Never Current/former |
31 (17) 149 (83) |
17 (49) 18 (51) |
4 (6) 59 (64) |
10 (12) 72 (88) |
<0.001 | 0.27 | <0.001 |
|
Smoking pack years: mean (range) |
33 (0-200) | 13 (0-60) | 42 (0-120) | 35 (0-200) | <0.001 | 0.041 | <0.001 |
|
Tumor size, cm: mean (range) |
2.3 (0-9.4) | 2 (0.7-5) | 2.3 (0.5-7.8) | 2.5 (0.6-9.4) | 0.29 | 0.32 | 0.08 |
|
Stage: N (%) I II-IV |
121 (67) 59 (33) |
26 (74) 9 (26) |
44 (70) 19 (30) |
51 (62) 31 (38) |
0.82 | 0.38 | 0.29 |
Comparison between different types of KRAS and EGFR mutations
We next examined whether histologic associations were linked to specific types of KRAS and EGFR mutations. We thus compared tumors with KRAS mutations known to be smoking associated (i.e. transversion mutations involving purine ↔ pyrimidine substitutions, n=47) vs mutations unrelated to smoking (i.e. transition mutations involving purine ↔ purine or pyrimidine ↔ pyrimidine substitutions, n=16), and tumors with EGFR Exon 19 deletions (n=13) vs L858R mutations (n=21) for the distribution of 7 histologic patterns, necrosis, cytologic atypia, hobnail cytology, tumor-infiltrating leukocytes, and TTF-1 expression. No significant differences between molecular subgroups were identified (data not shown), but this analysis was limited by relatively small number of cases in each subgroup.
DISCUSSION
In the present study, by combining detailed histopathologic analysis with high-sensitivity mutation detection method, we identified a novel association of KRAS mutations with solid growth pattern and tumor-infiltrating leukocytes in non-mucinous lung adenocarcinomas. In addition, we expanded on several previously-described histologic and clinical associations of KRAS and EGFR mutations.
While this is the first study to identify a propensity of KRAS+ lung adenocarcinomas for solid growth pattern, several prior studies did hint at this association. First, several studies showed an association of KRAS mutations with poor differentiation.22-24 Although there is currently no standardized grading system for lung adenocarcinomas, solid growth pattern is the central parameter in grading of adenocarcinomas system-wide, and it is likely that the presence of solid growth pattern, at least in part, explains the association of KRAS mutations and poor differentiation in those studies. Furthermore, an association of KRAS mutations with a gene expression profile correlating with solid histology was noted in a study by Motoi et al.21 Lastly, an association of KRAS mutations and “tumor islands”, which in turn were associated with solid growth pattern, was recently reported by Onozato et al.31
Three potential factors could have contributed to the differences in the reported histologic associations of KRAS mutations in lung adenocarcinomas across studies:
One potential factor is under-detection of KRAS mutations by assays with suboptimal sensitivity, such as Sanger sequencing. The relevance of method sensitivity is particularly supported by our finding that lung carcinomas harboring KRAS mutations are enriched with inflammatory cells. Standard macrodissection of such tumors may fail to enrich for tumor cells due to their intimate association with inflammation, and consequently extracted DNA may be diluted by DNA contributed by inflammatory cells. Thus, KRAS-mutated carcinomas may be particularly prone to false-negative results by standard Sanger sequencing, which has a notoriously low analytical sensitivity, requiring high tumor cell content (40-50%).29 In contrast, Sequenom platform, used in this study, requires ~10% tumor cell content.29 The possibility that KRAS mutations may be under-detected by Sanger sequencing is indirectly supported by the data in colorectal carcinomas, where in a matched comparison, Sanger sequencing was found to under-estimate the frequency of exon 2 KRAS mutations by 9% compared to more sensitive methods.29
The second factor potentially contributing to the variability in molecular/histologic correlation results in individual studies could be the variation in the designation of histologic patterns. This is illustrated in a recent inter-observer reproducibility study, which showed significant variability in designation of histologic patterns in lung adenocarcinomas among pathologists.32 Although solid pattern showed one of the highest concordances, a potential source of variability comes from the lack of agreement on the designation of complex glandular patterns (such as cribriform), which are currently variably classified as acinar or solid.32 These patterns were annotated as a distinct category in this study, and, while over-represented in KRAS+ carcinomas, they did not reach a statistical association with any molecular group. Another potential confounder is recently-recommended classification based solely on a single histologic pattern, judged to be predominant relative to other patterns,16 which we found to be difficult to assign objectively in a fair number (29%) of cases due to two or more patterns being present in a similar co-dominant amount. In addition, this annotation may exclude tumors in which a pattern is present in a minor amount but is still biologically significant, as illustrated by our finding that both solid and lepidic patterns have a significant effect on the frequency of KRAS and EGFR mutations even when present as a minor component of a tumor.
Lastly, genotype/phenotype associations could be influenced by ethnic factors. In particular, a potential confounding factor is still a largely unexplained significant difference in the rate of KRAS (and EGFR) mutations in lung adenocarcinomas between western and East Asian populations. Specifically, the baseline rate of KRAS mutations is low in East Asian populations (5-10%), with a substantial (40-60%) proportion of mutations occurring in mucinous carcinomas.19, 33 In contrast, KRAS mutations occur in 25-35% of lung adenocarcinomas in western patients, with the majority (89% in this series) of mutations occurring in non-mucinous carcinomas. Thus, the rarity of KRAS+ non-mucinous carcinomas in East Asian populations could have contributed to the difficulty in detecting histologic associations in this tumor subset in the Asian studies.19 Analogously, the lack of association of KRAS mutations with smoking in some East Asian studies could be a result of concentration of mutations in mucinous adenocarcinomas, which have an inconsistent link with smoking history.19
Overall, KRAS mutations appear to have a dual histologic association in lung adenocarcinomas – one with non-mucinous carcinomas with a solid component, which we found to have KRAS mutations in more than 50% of cases, and the other with mucinous carcinomas formerly designated mucinous bronchioloalveolar carcinoma25 (“invasive mucinous adenocarcinoma”16), which are reported to harbor KRAS mutations in 30% to >80% of cases.17-20 In this study, KRAS mutations were also over-represented in the latter tumors, occurring in 67% (4/6) of cases; although statistical analysis of this association was limited by overall rarity of this tumor type in our unselected patient population. In addition to the dual role of KRAS in invasive adenocarcinomas, paradoxical over-representation of KRAS mutations in pre-invasive glandular lesions – pure bronchioloalveolar carcinomas/adenocarcinomas in situ – has been reported;34 these lesions were excluded from the present study to focus the analysis on conventional invasive adenocarcinomas. From the perspective of lung cancer pathogenesis, these pleotropic histologic associations may hint at the complex role of KRAS mutations in stem cells. One hypothesis is that KRAS mutations may arise in distinct stem cells, giving rise to neoplasms with divergent histology. Alternatively, KRAS mutations may arise in a common pleuripotent stem cell with a broad differentiation potential. These possibilities are in line with pre-clinical data that KRAS-mediated tumorigenesis is significantly influenced by the cellular context.35
Our finding that KRAS mutations are associated with solid histology and tendency for greater necrosis and cytologic atypia may represent the underlying link between KRAS+ genotype and aggressive clinical behavior in lung adenocarcinomas. Several recent studies have demonstrated that solid growth pattern is a strong predictor of adverse clinical outcome, whereas lepidic pattern – associated with EGFR mutations – is a predictor of indolent behavior in lung adenocarcinomas.26, 36 Thus, the distinct association of KRAS and EGFR mutations with aggressive vs indolent histologies, respectively, parallels the differences in prognosis. Because the follow-up available for patients in this series was too short for survival analysis, future studies with survival data and multivariate analysis will be needed to determine whether KRAS and EGFR mutations exert their prognostic effects via a link to histologic subsets, or whether these effects are histology-independent.
Of interest, the association of KRAS mutations and solid histology in lung adenocarcinomas ties in with our recent description of a high frequency (40%) of KRAS mutations in large cell (undifferentiated) carcinomas showing glandular immunophenotype.37 We proposed that these clinically-aggressive tumors represent a spectrum of adenocarcinomas with an extreme amount of solid growth pattern. The high frequency of KRAS mutations in conventional adenocarcinomas with partial solid histology reported in this study is in line with that proposal, as is the low frequency EGFR mutations in adenocarcinomas with solid component vs large cell carcinomas with glandular immunoprofile. The propensity for solid growth/poor differentiation of KRAS-mutant tumors is also consistent with the finding of a high rate (38%) of KRAS mutations in sarcomatoid/pleomorphic lung carcinomas.38
The finding that KRAS-mutated carcinomas are associated with tumor-infiltrating leukocytes, in addition to representing a potential confounder in molecular testing, may itself have biological and clinical significance. Presence of inflammatory cells has been implicated as both favorable and unfavorable prognostic indicator in several malignances, consistent with the capacity of immunity to exert both anti-tumor and pro-tumor effects depending on both tumor and host factors.39, 40 Non-small cell lung carcinomas are frequently associated with prominent tumor-infiltrating lymphocytes and other inflammatory cells, but their significance remains controversial. Both adverse41, 42 and favorable43 prognostic effects having been reported, which may be related to different subsets of inflammatory cells, scoring criteria, and patient populations.44 In this study, only the overall extent of inflammatory infiltrate was analyzed, and further study will be needed to evaluate specific leukocyte subsets. While association of inflammation and KRAS mutations is a novel observation, Dacic et al noted that high level of tumor-infiltrating lymphocytes is uncommon in adenocarcinomas with EGFR mutations;20 this trend for pan-inflammatory infiltrate was also seen in the present study. We cannot exclude that the degree of inflammation in adenocarcinomas with KRAS vs EGFR mutations reflects tissue response to tumors with more vs less aggressive histology, respectively. Nevertheless, these data raise the possibility that patients with KRAS-mutated lung adenocarcinomas may be an especially attractive subset for clinical trials of immunomodulatory agents aimed at enhancing the anti-tumor activity of tumor-infiltrating lymphocytes, such as therapeutic antibodies to PD-1 and PD-L1.45, 46
In addition to describing novel histologic associations of KRAS mutations, this study also expanded on the previously-recognized histologic and clinical association of EGFR mutations. Prior studies consistently reported that EGFR mutations are associated with non-mucinous non-solid histology, as also seen in this study. The association of EGFR mutations with individual histologic patterns has significant variability in the literature, and includes lepidic, papillary, micropapillary, and in some studies acinar.16 In this study, individual patterns associated with EGFR mutations were lepidic, papillary and to a lesser degree acinar. However, the strongest association of EGFR mutations was with hobnail cytologic features, which were typically seen in carcinomas with lepidic component and/or characteristic serrated intra-glandular infoldings, which could be variably described as displaying papillary/micropapillary/acinar patterns (data not shown). Hobnail cytology is proposed as a defining feature of terminal respiratory unit-type adenocarcinomas,27 and it is possible that several architectural patterns emerging as associated with EGFR mutations in individual studies represent variable annotation of architectural manifestations of this type of adenocarcinoma.
It has been suggested that TTF-1 expression represents a feature of terminal respiratory unit-type adenocarcinomas, and that while EGFR+ adenocarcinomas are uniformly TTF-1-positive, carcinomas with KRAS mutations tend to be TTF-1-negative.47 Here we clarify that the lack of TTF-1 expression in KRAS+ carcinomas applies primarily to mucinous carcinomas, of which 57% in this study were TTF-1-negative, whereas the lack of TTF-1 expression in KRAS+ non-mucinous carcinomas is rare (5% in this series). Similarly, we clarify that despite propensity for solid growth, a subset of KRAS+ non-mucinous carcinomas displays hobnail cytologic features in better differentiated areas, suggesting that these tumors do not always belong to a non-terminal respiratory unit lineage, consistent with prior observations.48
A notable observation in this study is that KRAS+ adenocarcinomas have a greater propensity for solid growth pattern compared not only to EGFR+ but also KRAS-/EGFR- carcinomas. It is worth noting, however, that KRAS-/EGFR- is not a molecularly-homogenous group, but rather a mixture of carcinomas with various low-frequency molecular alterations, including ALK, BRAF, HER2, ROS1, and RET (frequency of each ranging from <1%-5%), as well as tumors with yet unidentified molecular events.49, 50 Despite its heterogeneous nature, the pooled clinical outcome for this group was found to be favorable compared to KRAS+ tumors and inferior compared to EGFR+ tumors in clinical studies,11, 12 which parallels the different propensities of these groups for solid growth pattern identified in this study, although at least some molecular subsets within KRAS-/EGFR- group – namely ALK-rearranged carcinomas – are also known to also show a propensity for solid histology (in addition to the classic association with signet ring cells)51 and aggressive behavior.52 Greater pack-year smoking history in patient with KRAS+ carcinomas compared to both EGFR+ as well as KRAS-/EGFR- group in this series is in line with a link to never-smokers of EGFR+ group and several known molecular subsets within KRAS- /EGFR- group, including ALK, ROS1, and RET.
From a practical perspective on predictive molecular testing, our data support prior conclusion that while both EGFR and KRAS mutations are associated with propensities for distinct histologic and clinicopathologic characteristics, none of these associations have sufficient predictive value to allow triage of cases for molecular studies, and therefore all lung adenocarcinomas should undergo molecular testing irrespective of histologic and clinical features (with possible exception being the exclusion of mucinous carcinomas from testing for EGFR mutations).53 On the other hand, estimation of pre-test probability of mutations may have value in some clinical settings. For such situations, nomograms, based on clinical +/- histologic features, have been recently developed to predict the likelihood of EGFR mutations.54, 55 The findings in this study, particularly the predictive effect of solid histology on the likelihood of KRAS and EGFR mutations, may be of value for refinement of such nomograms. Although KRAS mutations have thus far evaded therapeutic targeting, and the current value of testing for these mutations in lung carcinomas is to serve as negative predictors for other targetable mutations, it is hoped that effective targeted therapies for mutant KRAS will emerge in the near future.56
In summary, here we have described a novel association of KRAS mutations with propensity for solid histology in non-mucinous lung adenocarcinomas, which may explain the adverse clinical outcome portended by KRAS mutations. We also describe an association of KRAS mutations with tumor-infiltrating leukocytes, which raises the possibility that patients with KRAS-mutated adenocarcinomas may benefit from novel immunomodulatory agents.
ACKNOWLEDGEMENTS
This work was supported in part by funding from NIH P01 CA129243 grant.
Footnotes
DISCLOSURE/CONFLICT OF INTEREST: none to disclose
REFERENCES
- 1.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–65. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 2.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–9. [PubMed] [Google Scholar]
- 3.Santos E, Martin-Zanca D, Reddy EP, et al. Malignant activation of a K-ras oncogene in lung carcinoma but not in normal tissue of the same patient. Science. 1984;223:661–4. doi: 10.1126/science.6695174. [DOI] [PubMed] [Google Scholar]
- 4.Riely GJ, Ladanyi M. KRAS mutations: an old oncogene becomes a new predictive biomarker. J Mol Diagn. 2008;10:493–5. doi: 10.2353/jmoldx.2008.080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riely GJ, Marks J, Pao W. KRAS mutations in non-small cell lung cancer. Proc Am Thorac Soc. 2009;6:201–5. doi: 10.1513/pats.200809-107LC. [DOI] [PubMed] [Google Scholar]
- 6.Suda K, Tomizawa K, Mitsudomi T. Biological and clinical significance of KRAS mutations in lung cancer: an oncogenic driver that contrasts with EGFR mutation. Cancer Metastasis rev. 2010;29:49–60. doi: 10.1007/s10555-010-9209-4. [DOI] [PubMed] [Google Scholar]
- 7.Slebos RJ, Kibbelaar RE, Dalesio O, et al. K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med. 1990;323:561–5. doi: 10.1056/NEJM199008303230902. [DOI] [PubMed] [Google Scholar]
- 8.Mitsudomi T, Steinberg SM, Oie HK, et al. ras gene mutations in non-small cell lung cancers are associated with shortened survival irrespective of treatment intent. Cancer Res. 1991;51:4999–5002. [PubMed] [Google Scholar]
- 9.Woo T, Okudela K, Yazawa T, et al. Prognostic value of KRAS mutations and Ki-67 expression in stage I lung adenocarcinomas. Lung Cancer. 2009;65:355–62. doi: 10.1016/j.lungcan.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 10.Guan JL, Zhong WZ, An SJ, et al. KRAS Mutation in Patients with Lung Cancer: A Predictor for Poor Prognosis but Not for EGFR-TKIs or Chemotherapy. Ann Surg Oncol. 2012 doi: 10.1245/s10434-012-2754-z. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 11.Marks JL, Broderick S, Zhou Q, et al. Prognostic and therapeutic implications of EGFR and KRAS mutations in resected lung adenocarcinoma. J Thorac Oncol. 2008;3:111–6. doi: 10.1097/JTO.0b013e318160c607. [DOI] [PubMed] [Google Scholar]
- 12.Johnson ML, Sima CS, Chaft J, et al. Association of KRAS and EGFR mutations with survival in patients with advanced lung adenocarcinomas. Cancer. 2013;8:123–5. doi: 10.1002/cncr.27730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winton T, Livingston R, Johnson D, et al. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med. 2005;352:2589–97. doi: 10.1056/NEJMoa043623. [DOI] [PubMed] [Google Scholar]
- 14.Loriot Y, Mordant P, Deutsch E, Olaussen KA, Soria JC. Are RAS mutations predictive markers of resistance to standard chemotherapy? Nat Rev Clin Oncol. 2009;6:528–34. doi: 10.1038/nrclinonc.2009.106. [DOI] [PubMed] [Google Scholar]
- 15.Sklar MD. Increased resistance to cis-diamminedichloroplatinum(II) in NIH 3T3 cells transformed by ras oncogenes. Cancer Res. 1988;48:793–7. [PubMed] [Google Scholar]
- 16.Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–85. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finberg KE, Sequist LV, Joshi VA, et al. Mucinous differentiation correlates with absence of EGFR mutation and presence of KRAS mutation in lung adenocarcinomas with bronchioloalveolar features. J Mol Diagn. 2007;9:320–6. doi: 10.2353/jmoldx.2007.060182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchetti A, Martella C, Felicioni L, et al. EGFR mutations in non-small-cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol. 2005;23:857–65. doi: 10.1200/JCO.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 19.Kakegawa S, Shimizu K, Sugano M, et al. Clinicopathological features of lung adenocarcinoma with KRAS mutations. Cancer. 2011;117:4257–66. doi: 10.1002/cncr.26010. [DOI] [PubMed] [Google Scholar]
- 20.Dacic S, Shuai Y, Yousem S, Ohori P, Nikiforova M. Clinicopathological predictors of EGFR/KRAS mutational status in primary lung adenocarcinomas. Mod Pathol. 2010;23:159–68. doi: 10.1038/modpathol.2009.154. [DOI] [PubMed] [Google Scholar]
- 21.Motoi N, Szoke J, Riely GJ, et al. Lung adenocarcinoma: modification of the 2004 WHO mixed subtype to include the major histologic subtype suggests correlations between papillary and micropapillary adenocarcinoma subtypes, EGFR mutations and gene expression analysis. Am J Surg Pathol. 2008;32:810–27. doi: 10.1097/PAS.0b013e31815cb162. [DOI] [PubMed] [Google Scholar]
- 22.Rodenhuis S, Slebos RJ. Clinical significance of ras oncogene activation in human lung cancer. Cancer Res. 1992;52:2665s–9s. [PubMed] [Google Scholar]
- 23.Tam IY, Chung LP, Suen WS, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res. 2006;12:1647–53. doi: 10.1158/1078-0432.CCR-05-1981. [DOI] [PubMed] [Google Scholar]
- 24.Marotti JD, Schwab MC, McNulty NJ, et al. Cytomorphologic features of advanced lung adenocarcinomas tested for EGFR and KRAS mutations: A retrospective review of 50 cases. Diagn Cytopathol. 2013;41:15–21. doi: 10.1002/dc.21749. [DOI] [PubMed] [Google Scholar]
- 25.Colby TV, Nobuchi M, Henschke C, et al. Adenocarcinoma. In: Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC, editors. Pathology & Genetics of Tumours of the Lung, Pleura, Thymus and Heart. World Health Organization Classification of Tumours. IARC Press; Lyon: 2004. pp. 35–44. [Google Scholar]
- 26.Sica G, Yoshizawa A, Sima CS, et al. A grading system of lung adenocarcinomas based on histologic pattern is predictive of disease recurrence in stage I tumors. Am J Surg Pathol. 2010;34:1155–62. doi: 10.1097/PAS.0b013e3181e4ee32. [DOI] [PubMed] [Google Scholar]
- 27.Yatabe Y, Kosaka T, Takahashi T, Mitsudomi T. EGFR mutation is specific for terminal respiratory unit type adenocarcinoma. Am J Surg Pathol. 2005;29:633–9. doi: 10.1097/01.pas.0000157935.28066.35. [DOI] [PubMed] [Google Scholar]
- 28.Rekhtman N, Ang DC, Sima CS, Travis WD, Moreira AL. Immunohistochemical algorithm for differentiation of lung adenocarcinoma and squamous cell carcinoma based on large series of whole-tissue sections with validation in small specimens. Mod Pathol. 2011;24:1348–59. doi: 10.1038/modpathol.2011.92. [DOI] [PubMed] [Google Scholar]
- 29.Arcila M, Lau C, Nafa K, Ladanyi M. Detection of KRAS and BRAF mutations in colorectal carcinoma roles for high-sensitivity locked nucleic acid-PCR sequencing and broad-spectrum mass spectrometry genotyping. J Mol Diagn. 2011;13:64–73. doi: 10.1016/j.jmoldx.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rekhtman N, Paik PK, Arcila ME, et al. Clarifying the spectrum of driver oncogene mutations in biomarker-verified squamous carcinoma of lung: lack of EGFR/KRAS and presence of PIK3CA/AKT1 mutations. Clin Cancer Res. 2012;18:1167–76. doi: 10.1158/1078-0432.CCR-11-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onozato ML, Kovach AE, Yeap BY, et al. Tumor Islands in Resected Early-stage Lung Adenocarcinomas are Associated With Unique Clinicopathologic and Molecular Characteristics and Worse Prognosis. Am J Surg Pathol. 2013;37:287–94. doi: 10.1097/PAS.0b013e31826885fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thunnissen E, Beasley MB, Borczuk AC, et al. Reproducibility of histopathological subtypes and invasion in pulmonary adenocarcinoma. An international interobserver study. Mod Pathol. 2012;25:1574–83. doi: 10.1038/modpathol.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakuma Y, Matsukuma S, Yoshihara M, et al. Distinctive evaluation of nonmucinous and mucinous subtypes of bronchioloalveolar carcinomas in EGFR and K-ras gene-mutation analyses for Japanese lung adenocarcinomas: confirmation of the correlations with histologic subtypes and gene mutations. Am J Clin Pathol. 2007;128:100–8. doi: 10.1309/WVXFGAFLAUX48DU6. [DOI] [PubMed] [Google Scholar]
- 34.Sakamoto H, Shimizu J, Horio Y, et al. Disproportionate representation of KRAS gene mutation in atypical adenomatous hyperplasia, but even distribution of EGFR gene mutation from preinvasive to invasive adenocarcinomas. J Pathol. 2007;212:287–94. doi: 10.1002/path.2165. [DOI] [PubMed] [Google Scholar]
- 35.Ray KC, Bell KM, Yan J, et al. Epithelial tissues have varying degrees of susceptibility to Kras(G12D)-initiated tumorigenesis in a mouse model. PLoS One. 2011;6:e16786. doi: 10.1371/journal.pone.0016786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshizawa A, Motoi N, Riely GJ, et al. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod Pathol. 2011;24:653–64. doi: 10.1038/modpathol.2010.232. [DOI] [PubMed] [Google Scholar]
- 37.Rekhtman N, Tafe LJ, Chaft JE, et al. Distinct profile of driver mutations and clinical features in immunomarker-defined subsets of pulmonary large-cell carcinoma. Mod Pathol. 2012 doi: 10.1038/modpathol.2012.195. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Italiano A, Cortot AB, Ilie M, et al. EGFR and KRAS status of primary sarcomatoid carcinomas of the lung: implications for anti-EGFR treatment of a rare lung malignancy. Int J Cancer. 2009;125:2479–82. doi: 10.1002/ijc.24610. [DOI] [PubMed] [Google Scholar]
- 39.Hanahan D, Lanzavecchia A, Mihich E. Fourteenth Annual Pezcoller Symposium: the novel dichotomy of immune interactions with tumors. Cancer Res. 2003;63:3005–8. [PubMed] [Google Scholar]
- 40.Lanca T, Silva-Santos B. The split nature of tumor-infiltrating leukocytes: Implications for cancer surveillance and immunotherapy. Oncoimmunology. 2012;1:717–25. doi: 10.4161/onci.20068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petersen RP, Campa MJ, Sperlazza J, et al. Tumor infiltrating Foxp3+ regulatory T-cells are associated with recurrence in pathologic stage I NSCLC patients. Cancer. 2006;107:2866–72. doi: 10.1002/cncr.22282. [DOI] [PubMed] [Google Scholar]
- 42.Shimizu K, Nakata M, Hirami Y, et al. Tumor-infiltrating Foxp3+ regulatory T cells are correlated with cyclooxygenase-2 expression and are associated with recurrence in resected non-small cell lung cancer. J Thorac Oncol. 2010;5:585–90. doi: 10.1097/JTO.0b013e3181d60fd7. [DOI] [PubMed] [Google Scholar]
- 43.Horne ZD, Jack R, Gray ZT, et al. Increased levels of tumor-infiltrating lymphocytes are associated with improved recurrence-free survival in stage 1A non-small-cell lung cancer. J Surg Res. 2011;171:1–5. doi: 10.1016/j.jss.2011.03.068. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki K, Kachala SS, Kadota K, et al. Prognostic immune markers in non-small cell lung cancer. Clin Cancer Res. 2011;17:5247–56. doi: 10.1158/1078-0432.CCR-10-2805. [DOI] [PubMed] [Google Scholar]
- 45.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–65. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hurbin A, Wislez M, Busser B, et al. Insulin-like growth factor-1 receptor inhibition overcomes gefitinib resistance in mucinous lung adenocarcinoma. J Pathol. 2011;225:83–95. doi: 10.1002/path.2897. [DOI] [PubMed] [Google Scholar]
- 48.Takeuchi T, Tomida S, Yatabe Y, et al. Expression profile-defined classification of lung adenocarcinoma shows close relationship with underlying major genetic changes and clinicopathologic behaviors. J Clin Oncol. 2006;24:1679–88. doi: 10.1200/JCO.2005.03.8224. [DOI] [PubMed] [Google Scholar]
- 49.Pao W, Hutchinson KE. Chipping away at the lung cancer genome. Nat Med. 2012;18:349–51. doi: 10.1038/nm.2697. [DOI] [PubMed] [Google Scholar]
- 50.Cheng L, Alexander RE, Maclennan GT, et al. Molecular pathology of lung cancer: key to personalized medicine. Mod Pathol. 2012;25:347–69. doi: 10.1038/modpathol.2011.215. [DOI] [PubMed] [Google Scholar]
- 51.Yoshida A, Tsuta K, Nakamura H, et al. Comprehensive Histologic Analysis of ALK-Rearranged Lung Carcinomas. Am J Surg Pathol. 2011;35:1226–34. doi: 10.1097/PAS.0b013e3182233e06. [DOI] [PubMed] [Google Scholar]
- 52.Yang P, Kulig K, Boland JM, et al. Worse disease-free survival in never-smokers with ALK+ lung adenocarcinoma. J Thorac Oncol. 2012;7:90–7. doi: 10.1097/JTO.0b013e31823c5c32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.NCCN Guidelines: Non-Small Cell Lung Cancer. National Comprehensive Cancer Network FW; PA: Version 2.2012. [Google Scholar]
- 54.Girard N, Sima CS, Jackman DM, et al. Nomogram to predict the presence of EGFR activating mutation in lung adenocarcinoma. Eur Respir J. 2012;39:366–72. doi: 10.1183/09031936.00010111. [DOI] [PubMed] [Google Scholar]
- 55.Dogan S, Shen R, Ang DC, et al. Molecular Epidemiology of EGFR and KRAS Mutations in 3,026 Lung Adenocarcinomas: Higher Susceptibility of Women to Smoking-Related KRAS-Mutant Cancers. Clin Cancer Res. 2012;18:6169–77. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Janne PA, Shaw AT, Pereira JR, et al. Phase II double-blind, randomized study of selumetinib (SEL) plus docetaxel (DOC) versus DOC plus placebo as second-line treatment for advanced KRAS mutant non-small cell lung cancer (NSCLC) J Clin Oncol. 2012;30 abstr 7503. [Google Scholar]