Abstract
Purpose
To evaluate the effect of flip angle (FA) on accuracy and within-examination repeatability of hepatic proton-density fat fraction (PDFF) estimation with complex data-based magnetic resonance imaging (MRI).
Materials and Methods
PDFF was estimated at 3T in thirty subjects, using two sets of five MRI sequences with FA from 1° to 5° in each set. One set used 7ms repetition time and acquired 6 echoes (TR7/E6); the other used 14ms and acquired 12 echoes (TR14/E12). For each FA in both sets, the accuracy of MRI-PDFF was assessed relative to MR spectroscopy (MRS)-PDFF using four regression parameters (slope, intercept, average bias, R2). Each subject had four random sequences repeated; within-examination repeatability of MRI-PDFF for each FA was assessed with intraclass correlation coefficient (ICC). Pairwise comparisons were made using bootstrap-based tests.
Results
Most FAs provided high MRI-PDFF estimation accuracy (intercept range -1.25–0.84, slope 0.89–1.06, average bias 0.24–1.65, R2 0.85–0.97). Most comparisons of regression parameters between FAs were not significant. Informally, in the TR7/E6 set, FAs of 2° and 3° provided the highest accuracy, while FAs of 1° and 5° provided the lowest. In the TR14/E12 set, accuracy parameters did not differ consistently between FAs. FAs in both sets provided high within-examination repeatability (ICC range 0.981–0.998).
Conclusion
MRI-PDFF was repeatable and, for most FAs, accurate in both sequence sets. In the TR7/E6 sequence set, FAs of 2° and 3° informally provided the highest accuracy. In the TR14/E12 sequence set, all FAs provided similar accuracy.
Keywords: magnetic resonance imaging, proton-density fat fraction, accuracy, repeatability, flip angle
INTRODUCTION
Complex data-based magnetic resonance imaging (MRI) can estimate the hepatic proton density fat fraction (PDFF), the proportion of mobile protons in liver tissue attributable to triglyceride fat (1), with high accuracy in human subjects using magnetic resonance spectroscopy (MRS) as the reference standard (2–4). This technique (5) uses both magnitude and phase information to permit separation of water and triglyceride fat and calculate the triglyceride fat fraction from 0 to 100%. It also uses a low flip angle (FA) relative to repetition time (TR) to reduce bias from T1 relaxation (5); acquires multiple echoes at different echo times (TE) per excitation to permit estimation of and correction for T2* decay (6); and applies prior knowledge of the fat spectrum to model the spectral complexity of fat (7,8). Additonally, this technique applies magnitude discrimination to reduce noise bias and hybrid complex-magnitude fitting to cancel artifactual phase shifts caused by eddy currents (9).
The choice of FA is critical using this technique: the FA must be sufficiently low relative to TR to keep T1 confounding effects negligible but sufficiently high to maintain adequate signal to noise ratio (SNR). In the low FA range, SNR is proportional to FA. Hence, increasing the FA within this range generates higher SNR at the cost of greater T1 bias, while decreasing the FA reduces T1 bias at the cost of lower SNR. Both T1 bias and low SNR can lead to inaccuracy in the MRI-PDFF estimation: the former by causing systematic overestimation, the latter by increasing estimation variability.
At 1.5T, the technique was accurate in human subjects using a FA of 5° and a TR of 14 ms (4), with MRS-measured PDFF (MRS-PDFF) as the reference standard, suggesting a FA of 5° at a TR of 14 ms is appropriate for MRI-PDFF estimation at 1.5T. However, the relative T1s of fat and water protons depend on field strength (10,11). Thus, while a FA/TR combination of 5° and 14 ms may be appropriate for MRI-PDFF estimation at 1.5T, this combination may provide too much T1 weighting and cause PDFF overestimation at 3T. A FA smaller than 5° is likely to be more appropriate at 3T but the optimal FA is not yet known.
The purpose of this technical report is to evaluate the effect of FA on accuracy and within-examination repeatability of hepatic MRI-PDFF estimation with complex data-based MRI.
MATERIALS AND METHODS
Subjects
This cross-sectional clinical study was compliant with the Health Insurance Portability and Accountability Act and approved by an institutional review board. From a cohort of over 1000 pediatric and adult research participants, 30 subjects spanning a wide PDFF spectrum based on prior MRS examinations were prospectively recruited between October 2009 and May 2010. Inclusion criteria were willingness to participate and ability to hold breath for 20 seconds. Exclusion criteria were pregnancy, trying to become pregnant, contraindications to MRI, and severe claustrophobia. Subjects’ demographic information, weight, height and body mass index (BMI) were recorded. Adult subjects gave informed consent. Pediatric subjects gave informed assent with parental consent.
MR Examinations
Subjects were scanned on a single 3T clinical scanner (Signa HDx, GE Healthcare, Waukesha, WI). Subjects were positioned supine with an 8-channel torso phased array coil centered over the liver. A dielectric pad was placed between the coil and the abdomen.
MRS Technique
Using three-plane localizing images and avoiding major vessels, bile ducts, and artifacts, a single 20×20×20 mm3 voxel was placed in segment V or VIII of the liver, as this location consistently provides reliable MRS measurements. Spatial saturation bands around the voxel were disabled to ensure a uniform spectral response across the frequency range (12). After automatic shimming during free breathing, single-voxel proton MRS was performed in a 21-second breath-hold using stimulated echo acquisition mode (STEAM) (13). The sequence was performed three times with identical parameters. To minimize T1 effects, TR was set at 3500 ms with the lowest allowed mixing time (TM) of 10 ms. Following a single pre-acquisition excitation pulse to balance T1 saturation effects, five STEAM spectra, each with a single average, were collected at equally spaced TEs between 10 and 30 ms in a single acquisition. This range of TEs permits robust T2 correction with minimal confounding from J-coupling (13). The spectra from the individual channels were combined using singular value decomposition (14). An anatomic image showing the MRS voxel location was saved.
MRI Technique
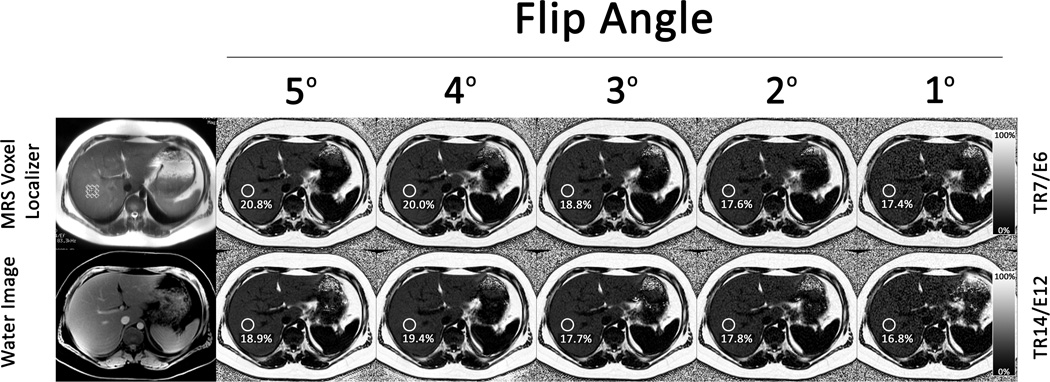
MR images of the liver centered at the level of the MRS voxel were obtained with an investigational version of a chemical shift-based water-fat separation method known as Iterative Decomposition of water and fat with Echo Asymmetry and Least-squares estimation (IDEAL) (15,16), implemented with a multi-echo 3D-spoiled gradient-echo acquisition. Two sets of five sequences, in separate breathholds, were obtained per subject. One set used TR of 7ms and acquired 6 echoes with an echo train length (ETL) of 3 (TR7/E6) with fly-back readouts and with echo spacing chosen to maximize the signal-to-noise ratio performance of the water-fat separation; the other used TR of 14ms and acquired 12 echoes with ETL of 6 (TR14/E12). The 7 ms TR is the lowest TR at 3T that permits acquisiton of six appropriately spaced echoes as done in the recent 1.5T study (4). The 14 ms TR in TR14/E12 matched the TR used in that study. Because the fat-water oscillation frequency at 3T is twice that at 1.5T, 12 appropriately spaced echoes could be obtained using this TR at 3T. Figure 1 shows representative images from each set, along with anatomic images used to co-localize regions of interest (ROIs) to the MRS voxel.
Figure 1.
Representative images from an 18-year old male subject with MRS-PDFF of 19.1% to illustrate co-localization of the region of interest on MRI PDFF maps and MRS voxel location. The MRS localizing image and the anatomic image used to co-localize the ROIs on the MRI-PDFF maps are shown on the left. To the right are shown MRI-PDFF maps acquired with FAs 5° through 1° in the TR7/E6 sequence set (top row) and in the TR14/E12 sequence set (bottom row). For each FA in each sequence set, the MRI-PDFF measured in the region of interest (circle) corresponding to the MRS voxel location is indicated.
To assess FA effects, each sequence within a set was performed using a different FA (5°, 4°, 3°, 2°, 1°), while keeping calibration and all other imaging parameters constant: first echo time = 1 ms, echo spacing = 0.8 ms, bandwidth (BW) = ± 200 kHz, field of view (FOV) = 44×44 cm, slice thickness = 8 mm, and 224×160 matrix. Acquisition time ranged from 18 to 25 seconds. Parallel imaging was not performed.
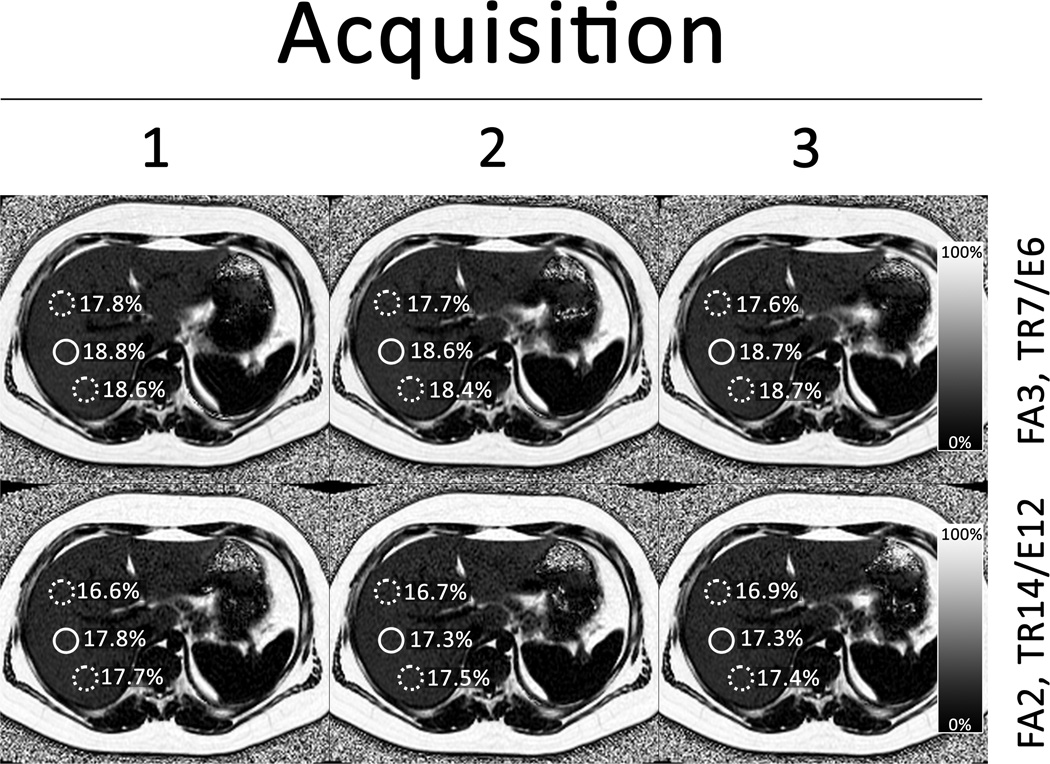
To assess within-examination repeatability, four sequences selected by block randomization were repeated three times in each subject without removing the subject from the scanner. The randomization was designed to repeat each sequence in at least eleven subjects (a reasonable statistical minimum). To avoid excessive burden on research subjects we did not repeat each sequence in each subject three times. A representative set of repeat sequence images is shown in Figure 2.
Figure 2.
Representative images from the same 18-year old male subject shown in Figure 1. The top row of maps are generated from three repeated acquisitions with FA 3° in the TR7/E6 sequence set. The bottom row of maps are generated from three repeated acquisitions with FA 2° in the TR14/E12 sequence set. Note that each map has 3 ROIs drawn to allow greater assessment of repeatability. One ROI (solid circle) was co-localized with the MRS voxel and used in the assessment of accuracy. The other two ROIs (dashed circles) were not co-localized with the MRS voxel.
MR Analysis
MRS Analysis
An MR physicist (--, PhD, 10-years experience) analyzed the MR spectra using Advanced Method for Accurate, Robust, and Efficient Spectral (AMARES) fitting included in Java based Magnetic Resonance User Interface software (http://sermn02.uab.es/mrui) (17,18). Peaks corresponding to water (4.7 ppm) and fat (2.2, 1.3, 0.9 ppm) were modeled as Gaussian resonances and measured. The T2-values of the spectral peaks and the T2-corrected peak areas were calculated by non-linear least-square fitting. Fat peaks at 4.2 and 5.3 ppm are obscured by the water peak and not visible at in-vivo field strengths in human liver; these peak areas were estimated from the visible fat peaks based on the known biochemical structure of human liver triglyceride (19). The MRS-PDFF was calculated separately for each MRS acquisition as the integrated sum of T2-corrected fat peaks divided by the sum of T2-corrected fat and water peaks. The average of the three per-subject co-localized MRS-PDFF values was computed and used as the reference standard. T1 effects were assumed negligible at the acquired TRs and TMs.
MRI Analysis
MR source images were reconstructed using an investigational on-line algorithm to generate water images, fat images, and MRI-PDFF maps. The algorithm generated water and fat images from the complex source data using a region growing approach to avoid water-fat swapping. The algorithm corrected the water and fat images for confounding effects due to T2* decay, noise bias, and eddy currents, and used spectral modeling of fat. MRI-PDFF maps were generated automatically from the corrected water and fat images as (F/(W+F)). Reconstructed images and maps were saved in DICOM format.
A trained research fellow (--, 1-year experience) manually placed one ROI identical in size and co-localized with the MRS voxel on the water images generated by each imaging sequence. (Figures 1 and 2). Co-localization was performed by comparing the water images with the saved anatomic image showing the MRS voxel location (Figure 2). In the repeated sequences, two additional ROIs of the same size were placed on water images elsewhere on the same slice, avoiding blood vessels and artifacts (Figure 2). ROIs then were propagated from the water images to the corresponding MRI-PDFF maps.
Blinding
The MRI analyst and the MR spectroscopist were blinded to each other’s results. Both were blinded to clinical data and prior MRS-PDFF measurements,.
Statistical Analysis
Accuracy Assessments
For both sets of sequences (TR7/E6 and TR14/E12), the average MRS-PDFF was regressed on MRI-PDFF using a single-predictor regression for each FA in each sequence set.
The reason MRS-PDFF was regressed on MRI-PDFF is to adhere to a statistical convention, whereby the predictor (e.g. the test measurement, here MRI-PDFF) is placed on the x-axis and the outcome (e.g. the reference measurement, here MRS-PDFF) on the y-axis. The resulting regression equation describes how to transform the observed MRI-PDFF predictor to estimate the MRS-PDFF outcome.
Four parameters were computed from each regression model:
The intercept of the regression line, which is the predicted value of the MRS-PDFF when the MRI-PDFF is equal to 0. As MRS-PDFF was regressed on MRI-PDFF, a positive intercept indicates that MRI tends to underestimate PDFF, while a negative intercept indicates that MRI tends to overestimate PDFF.
The slope of the regression line. As MRS-PDFF was regressed on MRI-PDFF, a slope greater than 1 indicates that MRI tends to underestimate PDFF, while a slope less than 1 indicates that MRI tends to overestimate PDFF.
The average bias of the fitted regression, defined as the square root of the average squared difference between the regression line and the MRS = MRI line (intercept=0, slope=1). The average bias reflects the magnitude of systematic estimation error, with higher values indicating greater error.
The R2 of the fitted regression line, which corresponds to the proportion of variance explained by the regression. An R2 of 1 would imply MRS-PDFF is perfectly predicted by MRI-PDFF, while lower values of R2 correspond to higher residual variance and lower predictive accuracy.
Parametric 95% confidence intervals (CI) were constructed around the regression intercept and slope, while bootstrap-based 95% CIs were constructed around average bias and R2. Bootstrap-based tests were used to make pairwise comparisons of the performance parameters obtained with different FAs in each sequence set. A Bonferroni correction was used to adjust for multiple comparisons within each sequence set, setting the significance threshold for comparisons within each sequence set at alpha = 0.005.
Repeatability Assessments
For each FA in both sequence sets, intra-class correlation (ICC) was estimated for the three repeated MRI-estimated PDFF values in the subset of subjects with that sequence repeated. To adjust for within-subject dependence, bootstrap-based 95% CIs were constructed around each ICC. Bootstrap-based tests were used to make pairwise comparisons of the ICC obtained with different FAs in each sequence set. P-values below 0.005 were considered significant (Bonferroni correction).
RESULTS
Subject Characteristics
The 30 enrolled subjects (12 female, 18 male) were 15 to 66 years old (mean 38 years), with BMI from 20.2 to 40.4 kg/m2 (mean 29.5 kg/m2). MRS-PDFF ranged from 1% to 25.2% (mean 12.1%): 6 subjects had 0–2% MRS-PDFF, 1 had 3–5%, 5 had 6–10%, 8 had 11–15%, 6 had 16–20%, and 4 had ≥ 21%.
Effect of Flip Angle on PDFF Estimation Accuracy for Each Set of Sequences
Accuracy Assessments
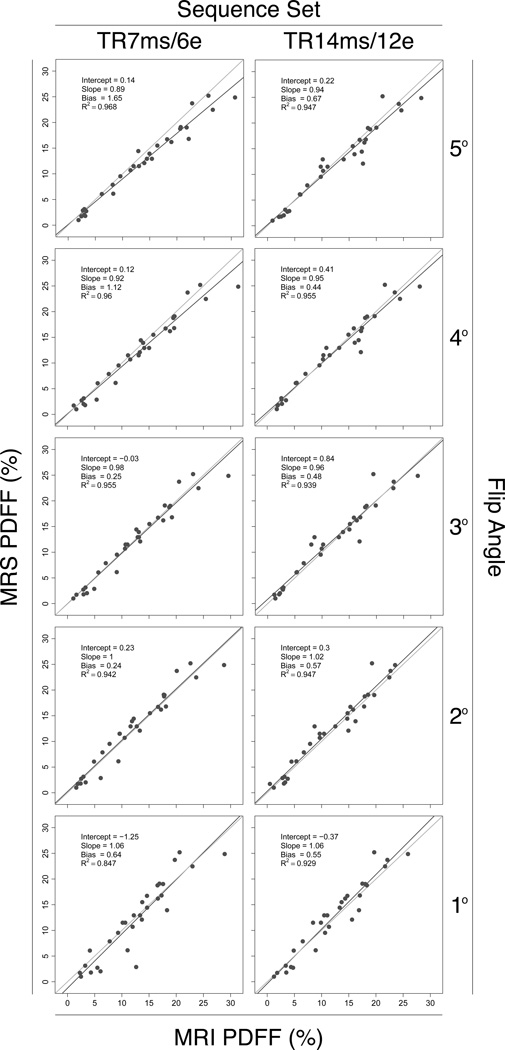
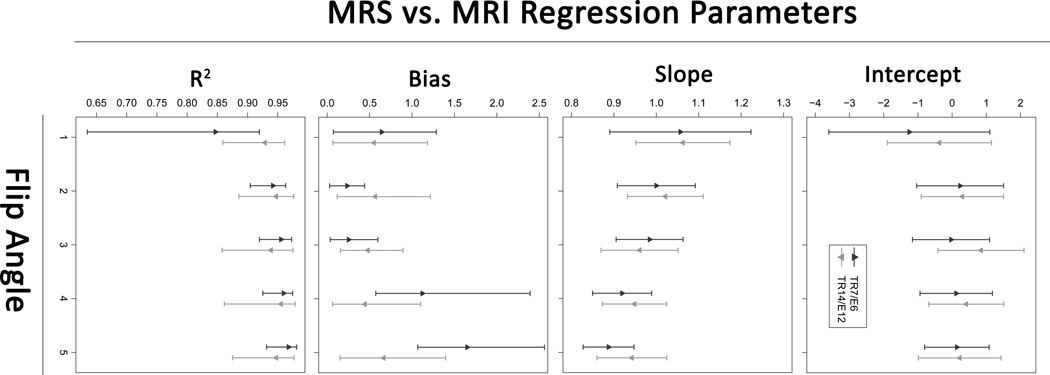
Scatterplots of MRS-PDFF vs. MRI-PDFF for each FA in both sequence sets are shown in Figure 3. The four regression parameters for each FA in both sequence sets (intercept, slope, average bias and R2) are summarized in Table 1 and Figure 4.
Figure 3.
Scatterplots of MRS-PDFF vs. MRI-PDFF for all FAs in both sequence sets. The dark line is the regression line. The light grey line is the equality (MRS-PDFF=MRI-PDFF) line. Regression parameters from a MRS-PDFF vs. MRI-PDFF univariate regression are shown on each plot.
Table 1. MRS-PDFF vs. MRI-PDFF Regression Parameters.
For each FA in both sequence sets the mean and, in brackets, 95% CI are summarized for each of the four regression parameters: intercept, slope, average bias and R2.
| Sequence set |
FA° | Intercept [95%CI] | Slope [95%CI] | Average Bias [95% CI] | R2 [95% CI] |
|---|---|---|---|---|---|
| TR7/E6 | 5 | 0.140 [−0.803, 1.083] | 0.887 [0.828, 0.947] | 1.650 [1.067, 2.561] | 0.968 [0.931, 0.981] |
| 4 | 0.120 [−0.938, 1.179] | 0.919 [0.850, 0.989] | 1.121 [0.573, 2.392] | 0.960 [0.925, 0.975] | |
| 3 | −0.031 [−1.159, 1.098] | 0.984 [0.905, 1.063] | 0.253 [0.035, 0.597] | 0.955 [0.920, 0.973] | |
| 2 | 0.234 [−1.036, 1.505] | 1.000 [0.908, 1.092] | 0.235 [0.028, 0.441] | 0.942 [0.904, 0.963] | |
| 1 | −1.250 [−3.604, 1.104] | 1.057 [0.890, 1.223] | 0.643 [0.073, 1.285] | 0.847 [0.635, 0.920] | |
| TR14/E12 | 5 | 0.220 [−0.987, 1.428] | 0.942 [0.860, 1.025] | 0.666 [0.150, 1.395] | 0.947 [0.875, 0.977] |
| 4 | 0.413 [−0.684, 1.510] | 0.949 [0.873, 1.025] | 0.444 [0.064, 1.101] | 0.955 [0.862, 0.979] | |
| 3 | 0.844 [−0.413, 2.102] | 0.960 [0.870, 1.051] | 0.479 [0.157, 0.894] | 0.939 [0.858, 0.975] | |
| 2 | 0.299 [−0.904, 1.503] | 1.021 [0.932, 1.111] | 0.566 [0.118, 1.217] | 0.947 [0.886, 0.977] | |
| 1 | −0.373 [−1.896, 1.150] | 1.063 [0.952, 1.174] | 0.551 [0.066, 1.182] | 0.929 [0.859, 0.961] | |
Figure 4.
The plots show means and 95% confidence intervals of the four regression parameters (intercept, slope, average bias and R2) for each FA in both sequence sets.
The regression intercepts ranged from −1.25 to 0.84. The 95% CIs for the regression intercepts for all FAs in both sequence sets included 0; hence, no FA in either sequence set achieved an intercept significantly different from 0. Based on the bootstrap tests there were no differences in intercepts between FAs in either sequence.
Regression slopes ranged from 0.89 to 1.06 (with CIs for most sequences covering 1). For both sequence sets, the regression slopes increased monotonically from below 1 (MRI overestimates PDFF) to above 1 (MRI underestimates PDFF) as FA decreased from 5° to 1°. Informally, regression slopes closest to 1 were achieved at FAs of 2° and 3° for both sequence sets. In the TR7/E6 sequence set, the slope achieved with FA 2° was significantly closer to 1 than the slopes achieved with FAs 4° and 5°, and the slope achieved with FA 3° was significantly closer to 1 than the slope achieved with FA 5°. In the TR14/E12 sequence set, no FA provided a slope significantly closer to 1 than any other FA.
Average bias ranged from 0.24% to 1.65%. For both sequence sets, average bias was the highest at FA 5°. In the TR7/E6 sequence set, the average bias at FA 5° was significantly higher than at all other FAs. Informally FAs of 2° and 3° provided the lowest average bias for this sequence set. The average bias at FA 3° was significantly lower than at FA 4°; other pairwise comparisons were not significant for this sequence set. There were no significant differences in average bias between any FAs in the TR14/E12 sequence set.
R2 values ranged from 0.85 to 0.97. For both sequence sets, R2 values were furthest from 1 at FA of 1°. In the TR7/E6 sequence set, R2 achieved with FA 1° was significantly lower than that achieved with all other FAs, and R2 achieved with FA 2° was significantly lower than that achieved with FA 4°, although the difference (0.942 vs. 0.960) between FAs 2° and 4° is probably not meaningful. In the TR14/E12 sequence set, R2 achieved with FA 1° was lower than that achieved with FA 4°, although this difference (0.929 vs. 0.955) is probably also not meaningful.
Repeatability Assessments
All FAs in each sequence set provided high within-examintion repeatability (Table 2). ICC coefficients ranged from 0.981 to 0.998 depending on FA and sequence set. The ICC achieved with FA 1° was significantly lower than that achieved with FA 4° in the TR7/E6 sequence set; this difference (0.982 vs. 0.998) is probably not meaningful.
Table 2. ICC for within-examination repeated MRI-PDFF estimates.
For each FA in both sequence sets the table summarizes number of subjects in whom sequence was repeated, the mean and range of MRS-PDFF values of the subjects, and the mean and, in brackets, 95% CI for the ICC.
| Sequence set |
FA° | Number of Subjects with Repeated Sequences |
MRS-PDFF of Subjects in Each Repeated Sequence |
ICC [95% CI] | |
|---|---|---|---|---|---|
| Mean (% PDFF) | Range (% PDFF) | ||||
| TR7/E6 | 5 | 12 | 13.2% | 2.0% – 25.2% | 0.994 [0.989, 0.998] |
| 4 | 13 | 13.5% | 1.0% – 24.9% | 0.998 [0.996, 0.999] | |
| 3 | 12 | 13.3% | 1.7% – 24.9% | 0.996 [0.994, 0.999] | |
| 2 | 12 | 11.7% | 1.0% – 24.9% | 0.994 [0.986, 0.998] | |
| 1 | 12 | 13.6% | 1.7% – 25.2% | 0.981 [0.962, 0.990] | |
| TR14/E12 | 5 | 11 | 12.5% | 1.8% – 24.9% | 0.987 [0.958, 0.997] |
| 4 | 14 | 11.5% | 1.0% – 25.2% | 0.994 [0.985, 0.998] | |
| 3 | 15 | 12.6% | 1.0% – 24.9% | 0.989 [0.973, 0.997] | |
| 2 | 12 | 13.4% | 2.0% – 25.2% | 0.991 [0.979, 0.997] | |
| 1 | 11 | 14.4% | 1.7% – 24.9% | 0.986 [0.969, 0.995] | |
DISCUSSION
This prospective clinical study assessed the effect of FA on MRI-PDFF estimation accuracy and within-examination repeatability of a complex data-based, T1-independent, T2*-corrected, spectrum-modeled MRI technique at 3T. While this study used an investigational variant of this technique, the technique is now commercially available and is simple to perform and analyze in the clinical setting. MRS-PDFF was the reference standard, as done in prior studies (2–4), because it is considered the most accurate non-invasive method to measure PDFF, and it can be performed contemporaneously to and co-localized with MRI.
Accuracy of MRI-PDFF was assessed by regressing MRS-PDFF on MRI-PDFF for each FA in each sequence set. The four regression parameters (intercept, slope, average bias, and R2) assess two separate aspects of the MRS-MRI relationship: bias and variance. Slope, intercept, and average bias reflect the magnitude of systematic deviations between MRS-PDFF and MRI-PDFF and are all measurements of bias. R2 is a measure of variance; lower R2 corresponds to greater residual variance in the MRI-MRS model.
Most MRI sequences provided high accuracy and all provided high within-examination repeatability for PDFF estimation. The high accuracy of most sequences and the high within-examination repeatability of all sequences confirm the results of other recent studies on PDFF estimation methods (2,20–22).
While most sequences were accurate, some performance differences were observed in the TR7/E6 sequence set. FAs of 1° and 5° provided the worst fit to the MRS-PDFF data, due to variance and bias, respectively. FAs of 2° and 3° provided the best fit overall of MRI-PDFF to MRS-PDFF, with acceptably low bias and acceptably low variance, although only some of the individual performance parameters for these FAs were significantly better than those for other FAs. The observation that FAs of 2° and 3° provided the best fit overall suggests that these FAs provide an optimal compromise in this sequence set between T1 bias, which leads to systematic overestimation, and low SNR, which leads to high variance.
In the TR14/E12 sequence set, no significant differences in performance were observed between FAs, with a single exception (R2 of FA 1° was significantly lower than the R2 of FA 4°, but the difference is probably not meaningful); hence, with our sample size, the performance of the various FAs was similar. The lower dependence of performance on FA observed in this sequence set may in part be due to its longer TR which reduces the degree of T1-weighting, partially offsetting T1 bias.
A limitation of our study was the inability to use internally calibrated parallel imaging with the version of the investigational imaging technique available to us. The use of parallel imaging would have reduced imaging time, albeit at a lower SNR, and the results of this study may not be generalizable to newer versions of the sequence that use internally calibrated parallel imaging. The 14 ms and 7 ms repetition times had different numbers of echoes, 12 and 6, respectively. As a result, this study cannot separate the potential effects of repetition time and the number of echoes.
In conclusion, we found that complex data-based, T1-independent, T2*-corrected, spectrum-modeled magnetic resonance imaging was accurate and repeatable in human subjects at 3T over a range of FAs from 1° to 5°. In the TR7/E6 sequence set, FAs of 2° and 3° may be optimal by providing both acceptably low bias and acceptably low variance. In the TR14/E12 sequence set, all FAs achieved reasonably good performance.
Acknowledgments
Grant support:
This research was supported by the National Institute of Health (grants NIDDK DK075128, NIDDK DK088925, NCMHD EXPORT P60 MD00220), RSNA (RSNA Medical Student Grant Award, Benjamin Johnson), and a research grant from General Electric.
REFERENCES
- 1.Longo R, Pollesello P, Ricci C, et al. Proton MR spectroscopy in quantitative in vivo determination of fat content in human liver steatosis. J Magn Reson Imaging. 1995 May;5:281–285. doi: 10.1002/jmri.1880050311. [DOI] [PubMed] [Google Scholar]
- 2.Yokoo T, Bydder M, Hamilton G, et al. Nonalcoholic fatty liver disease: diagnostic and fat-grading accuracy of low-flip-angle multiecho gradient-recalled-echo MR imaging at 1.5 T. Radiology. 2009;251:67–76. doi: 10.1148/radiol.2511080666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yokoo T, Shiehmorteza M, Hamilton G, et al. Estimation of hepatic proton-density fat fraction by using MR imaging at 3.0 T. Radiology. 2011;258:749–759. doi: 10.1148/radiol.10100659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meisamy S, Hines CD, Hamilton G, et al. Quantification of Hepatic Steatosis with T1-independent, T2*-corrected MR Imaging with Spectral Modeling of Fat: Blinded Comparison with MR Spectroscopy. Radiology. 2011;258:767–775. doi: 10.1148/radiol.10100708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu CY, McKenzie CA, Yu H, Brittain JH, Reeder SB. Fat quantification with IDEAL gradient echo imaging: correction of bias from T(1) and noise. Magn Reson Med. 2007;58:354–364. doi: 10.1002/mrm.21301. [DOI] [PubMed] [Google Scholar]
- 6.Yu H, McKenzie CA, Shimakawa A, et al. Multiecho reconstruction for simultaneous water-fat decomposition and T2* estimation. J Magn Reson Imaging. 2007;26:1153–1161. doi: 10.1002/jmri.21090. [DOI] [PubMed] [Google Scholar]
- 7.Sinha R, Dufour S, Petersen KF, et al. Assessment of skeletal muscle triglyceride content by (1)H nuclear magnetic resonance spectroscopy in lean and obese adolescents: relationships to insulin sensitivity, total body fat central adiposity. Diabetes. 2002;51:1022–1027. doi: 10.2337/diabetes.51.4.1022. [DOI] [PubMed] [Google Scholar]
- 8.Hamilton G, Yokoo T, Bydder M, et al. In vivo characterization of the liver fat (1)H MR spectrum. NMR Biomed. 2011;24:784–790. doi: 10.1002/nbm.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu H, Shimakawa A, Reeder S, McKenzie C, Brittain J. Magnitude Fitting Following Phase Sensitive Water-Fat Separation to Remove Effects of Phase Errors. Paper presented at the17th annual meeting of the ISMRM; Hawai'i. 2009. [Google Scholar]
- 10.Crooks LE, Arakawa M, Hoenninger J, McCarten B, Watts J, Kaufman L. Magnetic resonance imaging: effects of magnetic field strength. Radiology. 1984;151:127–133. doi: 10.1148/radiology.151.1.6701302. [DOI] [PubMed] [Google Scholar]
- 11.Rakow-Penner R, Daniel B, Yu H, Sawyer-Glover A, Glover GH. Relaxation times of breast tissue at 1.5T and 3T measured using IDEAL. J Magn Reson Imaging. 2006;23:87–91. doi: 10.1002/jmri.20469. [DOI] [PubMed] [Google Scholar]
- 12.Held D. Details Held for Review [Google Scholar]
- 13.Hamilton G, Middleton MS, Bydder M, et al. Effect of PRESS and STEAM sequences on magnetic resonance spectroscopic liver fat quantification. J Magn Reson Imaging. 2009;30:145–152. doi: 10.1002/jmri.21809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bydder M, Hamilton G, Yokoo T, Sirlin CB. Optimal phased-array combination for spectroscopy. Magn Reson Imaging. 2008;26:847–850. doi: 10.1016/j.mri.2008.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reeder SB, McKenzie CA, Pineda AR, et al. Water-fat separation with IDEAL gradient-echo imaging. J Magn Reson Imaging. 2007;25:644–652. doi: 10.1002/jmri.20831. [DOI] [PubMed] [Google Scholar]
- 16.Reeder SB, Pineda AR, Wen Z, et al. Iterative decomposition of water and fat with echo asymmetry and least-squares estimation (IDEAL): application with fast spin-echo imaging. Magn Reson Med. 2005;54:636–644. doi: 10.1002/mrm.20624. [DOI] [PubMed] [Google Scholar]
- 17.Naressi A, Couturier C, Castang I, de Beer R, Graveron-Demilly D. Java-based graphical user interface for MRUI, a software package for quantitation of in vivo/medical magnetic resonance spectroscopy signals. Comput Biol Med. 2001;31:269–286. doi: 10.1016/s0010-4825(01)00006-3. [DOI] [PubMed] [Google Scholar]
- 18.Vanhamme L, van den Boogaart A, Van Huffel S. Improved method for accurate and efficient quantification of MRS data with use of prior knowledge. J Magn Reson. 1997;129:35–43. doi: 10.1006/jmre.1997.1244. [DOI] [PubMed] [Google Scholar]
- 19.Hamilton GMM, Yokoo T, Bydder M, Mwangi IW, Schroeder ME, Sirlin CB. In-vivo Determination of the Full H MR Spectrum of Liver Fat. Paper presented at the 19th annual meeting of the ISMRM; Stockholm, Sweden. 2010. [Google Scholar]
- 20.Reeder SB, Robson PM, Yu H, et al. Quantification of hepatic steatosis with MRI: the effects of accurate fat spectral modeling. J Magn Reson Imaging. 2009;29:1332–1339. doi: 10.1002/jmri.21751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hines CD, Yu H, Shimakawa A, McKenzie CA, Brittain JH, Reeder SB. T1 independent, T2* corrected MRI with accurate spectral modeling for quantification of fat: validation in a fat-water-SPIO phantom. J Magn Reson Imaging. 2009;30:1215–1222. doi: 10.1002/jmri.21957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hussain HK, Chenevert TL, Londy FJ, et al. Hepatic fat fraction: MR imaging for quantitative measurement and display--early experience. Radiology. 2005;237:1048–1055. doi: 10.1148/radiol.2373041639. [DOI] [PubMed] [Google Scholar]
- 23.Hines CD, Frydrychowicz A, Hamilton G, et al. T(1) independent, T(2) (*) corrected chemical shift based fat-water separation with multi-peak fat spectral modeling is an accurate and precise measure of hepatic steatosis. J Magn Reson Imaging. 2011;33:873–881. doi: 10.1002/jmri.22514. [DOI] [PMC free article] [PubMed] [Google Scholar]