Abstract
Genome sequence data can be used to analyze genome plasticity by whole genome PCR scanning. Small sized chromosomes can indeed be fully amplified by long-range PCR with a set of primers designed using a reference strain and applied to several other strains. Analysis of the resulting patterns can reveal the genome plasticity. To facilitate such analysis, we have developed GenoFrag, a software package for the design of primers optimized for whole genome scanning by long-range PCR. GenoFrag was developed for the analysis of Staphylococcus aureus genome plasticity by whole genome amplification in ∼10 kb-long fragments. A set of primers was generated from the genome sequence of S.aureus N315, employed here as a reference strain. Two subsets of primers were successfully used to amplify two portions of the N315 chromosome. This experimental validation demonstrates that GenoFrag is a robust and reliable tool for primer design and that whole genome PCR scanning can be envisaged for the analysis of genome diversity in S.aureus, one of the major public health concerns worldwide.
INTRODUCTION
Genomes, and consequently phenotypes, can be highly heterogeneous in bacterial strains, even if they belong to the same species. The reasons for this plasticity are punctual mutations, chromosomal rearrangements such as deletions or inversion and/or the acquisition of mobile genetic elements such as plasmids, bacteriophages and transposons, etc. A clearer understanding of bacterial heterogeneity is of considerable interest for epidemiological and evolution studies or strain identification. Until recently, genome variability analysis in bacteria relied on approaches taking account of the entire DNA content, e.g. analysis of restriction enzyme digestion patterns (1) or arbitrarily primed PCR (2,3). These techniques rely on uncharacterized genomic differences between strains in a given bacterial species and have proved more or less discriminatory, depending on the species. The development of sequencing projects has allowed approaches to genome diversity through methods using mobile genetic elements such as molecular markers (4,5). Other investigations have focused on a few genes, e.g. multilocus sequence typing (6–8), or took account of the entire gene content of a genome, e.g. microarrays (9,10). These approaches can efficiently discriminate between closely related strains and, to some extent, can highlight genomic differences from one strain to another. However, they only partly reflect genomic diversity and do not allow the identification of genetic changes (e.g. chromosomal rearrangement, local small insertions or deletions, etc.). Increasing numbers of complete genome sequences for prokaryotic organisms are now available. More than 121 bacterial species have been completely sequenced (Genome Online Database, http://wit.integratedgenomics.com/GOLD/, August 2003), and several strains have been sequenced in some species. This wealth of data allows comparisons between whole genomes, a powerful and accurate approach to genome diversity. For example, the genome of Buchneria aphidicola, an endosymbiotic bacterium, was demonstrated as being very stable, with no rearrangements or gene acquisition having occurred over the past 50 million years (11). In contrast, the genomes of Escherichia coli and Salmonella spp. are >2000-fold more labile in content and gene order (11). Similarly, strains of Staphylococcus aureus, a Gram-positive pathogenic bacterium, are reportedly genomically and phenotypically highly heterogeneous (9). Staphylococcus aureus is the causative agent of a wide range of diseases in warm-blooded animals. In particular, it is a major cause of nosocomial disease worldwide (12) and is also often involved in food-poisoning outbreaks (13). The sequences of seven S.aureus strains are now available. Some are complete and published: strains N315 and Mu50 (14) and strain MW2 (15). Others are under annotation: COL (TIGR), NCTC8325 (University of Oklahoma) and MRSA252, MSSA476 (Sanger Institute). We initiated the whole genome scanning of S.aureus strains using long-range polymerase chain reaction (LR-PCR). This approach is based on comparative analysis of the whole genome structure of different strains of the same species, as determined by whole genome amplification using the LR-PCR technique. Recently, it was successfully used to study genome diversity in enterohemorrhagic E.coli O157 (16) and Bacillus licheniformis ATCC 14580 (17). However, these authors did not use any specific bioinformatics tool to design their set of primers. Several bioinformatics tools are available to design and test the robustness of primers, e.g. Primer3 (18), PRIDE (19) or PRIMO (20). However, no software exists that can process a whole genome sequence to design primers taking account of the specific requirements of a whole genome PCR scanning project, i.e. the design of primers according to parameters fixed by the users, the selection of primer pairs covering the whole genome and allowing segmentation of the genome into fragments whose length and overlap can be set by the users. We describe herewith GenoFrag, a software package that automatically designs primers optimized for this purpose. A set of primers was generated by GenoFrag from the complete genome sequence of S.aureus N315. Two subsets of primers corresponding to two portions of the N315 chromosome (112 and 107 kb long, respectively) were selected and successfully used for LR-PCR amplification experiments.
MATERIALS AND METHODS
Physical parameters
Specificity and thermodynamic stability. Both the length and G+C content of primers are pre-established in GenoFrag by default, but the values can be modified by users. Thermodynamic stability of the primer–template duplex has to be fixed according to the physical conditions required for LR-PCR. A primer length of 25mer is fixed as a default value [the 25mer size was chosen according to Rychlik (21)]. Thermodynamic stability has to be identical for all primers designed by GenoFrag in order to facilitate large-scale PCR. Here, a 12 G+C content is pre-fixed (corresponding to 48% G+C in the 25mers compared with the 33% G+C content in the S.aureus genome). This default value gives a Tm of 58°C using the rules devised by Suggs et al. (22).
To increase specificity, GenoFrag favors primers that exhibit a higher degree of free energy (ΔG) in the 5′ primer extremity than in the 3′ extremity. The nearest-neighbor method was used to calculate the thermodynamic stability of hybrids (23). Five bases at each extremity are considered: GC clamp in 5′ and lower stability in 3′ are favored since PCR yields dramatically decreased with high ΔG in the 3′ extremity (21).
Words of identical bases. To avoid non-specific annealing of primers, GenoFrag eliminates sequences that contain words of five or more identical and consecutive bases (21).
Secondary structures. Putative hairpin formation is checked and GenoFrag rejects primers when they are likely to form hairpin structures with a stem of 4 nt (minimum) and a loop of 4 nt (minimum). These default values were fixed in line with the suggestion made by Blommers et al. (24).
Self-complementarity. A first selection step eliminates primers with regard to their overall self-complementarity. A second step focuses on the 3′ extremity and further eliminates primers that present self-complementarity but with more discriminative values. Each step requires the implementation of computation involving the nearest neighbor method (23).
Secondary binding sites. One critical point in the analysis of whole genome scanning PCR results is amplicon size. A 10 kb size is set by default but can be modified by the user. This amplicon size allows routine LR-PCR experiments without major difficulties, and size variations around 10 kb can easily be visualized. It is important to avoid any secondary binding sites within a range of length that could be PCR amplified under LR-PCR conditions and thus give rise to non-specific PCR products (additional bands that are smaller or slightly longer than those expected). This criterion is particularly important during the early cycles of PCR amplification because the non-specific PCR fragment can become the predominant template for the remainder of the reaction. The methods employed here to eliminate candidates with secondary binding sites involved the use of alignment algorithms: a candidate is rejected when a putative annealing site is found with minimum values (set by default) of 17 matches with one gap allowed. Secondary binding sites are searched within a limited range of the template sequence corresponding to twice the amplicon size. Indeed, the parameters used for LR-PCR allow amplification of fragments shorter or slightly longer than those expected (e.g. for an expected amplicon of 10 kb, a range of 0 to ∼15 kb could be amplified in the reaction in the event of non-specific annealing).
Inter-primer complementarity. Partial complementarity between two primers for a given pair may interfere with annealing. If complementarity occurs at the 3′ end of the primers, primer dimer formation may take place and will prevent formation of the desired product (i.e. hybridization of the primers with the template) via competition. The methods used here to evaluate interprimer complementarity are similar to that used to calculate self-complementarity.
Operating environment and availability of GenoFrag
GenoFrag can be run under UNIX, Windows or LINUX environments. The source code has been written in C (25), Perl (with Bioperl modules) (26) and CAML (27) languages. In practice, two programs need to be executed sequentially. The first one generates primers, while the second searches for an optimum set of amplicons. Each program takes as its parameter a file describing the different physical values. Both versions are available on request from the authors. An online version can be tested on the West Genopole bioinformatics server: http://genouest.no-ip.org/Services/GenoFrag/.
Databases and analysis of genome sequence. The S.aureus genome sequence used in this study was that of S.aureus N315 (14), which was updated in June 2001 and is available under NCBI accession number NC_002745.
Bacterial strain and isolation of chromosomal DNA. Staphylococcus aureus strain N315 (kindly provided by Dr T. Ito, Department of Bacteriology, Juntendo University, Tokyo, Japan) was used as the reference strain for primer design and the experimental validation of GenoFrag. An overnight culture of the N315 strain grown on TSB (tryptone soy broth, AES, Combourg, France) at 37°C under shaking was used for the isolation of chromosomal DNA, essentially as previously described for B.licheniformis (17) except that lysostaphin digestion was performed prior to cell lysis. Briefly, 50 ml of the overnight culture was pelleted and the cells were washed in 5 ml of TES (Tris 10 mM pH 8, 5 mM EDTA, 0.5 M saccharose), resuspended in 5 ml of TES-lysostaphin (TES; 100 µg/ml lysostaphin) and incubated for 3 h at 37°C. Subsequent steps were performed according to the method described by Lapidus et al. (17). The DNA pellet was resuspended in 1.5 ml distilled water, aliquoted and kept frozen at –20°C.
Long-range PCR. PCRs were performed using 0.1 µg of genomic DNA as the template, and the long-range PCR kit was used as recommended by the supplier (GeneAmpXL PCR kit; Applied BioSystem, Foster City, CA). The final volume of the PCR mix was 50 µl and cycling conditions were as follows: 95°C, 4 min; 30 cycles of 30 s melting at 95°C and 12 min annealing–polymerization–repair at 68°C, increasing the extension time by 15 s at each cycle; at the end of the 30 cycles, 15 min at 72°C. PCR products were analyzed using 0.5% agarose gel electrophoresis.
RESULTS AND DISCUSSION
GenoFrag algorithm
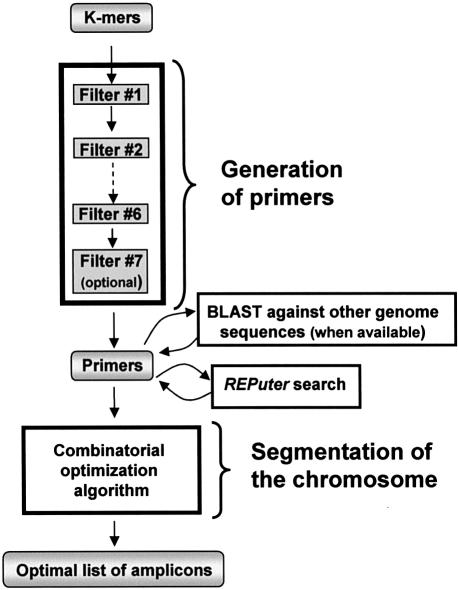
An overview of the GenoFrag software package is shown in Figure 1. It has two principal parts: the generation of primers and segmentation of the genome.
Figure 1.
Synoptic overview of GenoFrag software. Successive filters: filter 1, G+C content; filter 2, number of repeats; filter 3, hairpin; filter 4, stability; filter 5, autocomplementarity; filter 6, similarity; filter 7 (optional), sequence divergence.
Generation of primers. This software program identifies all primers suitable for LR-PCR. It acts as a sequential pipeline of seven filters. All potential K-mers (default value, 25) are considered and enter filter 1. Each filter yields only those oligonucleotides satisfying specific constraints set by the user. In order to limit the computation time, filters with the highest selectivity are the first to be activated. Filter 1 selects oligonucleotides according to their G+C content. Filter 2 removes oligonucleotides with N consecutive identical nucleotides. Filter 3 removes oligonucleotides with hairpin loops. Filter 4 selects oligonucleotides according to thermodynamic stability constraints. Filter 5 tests both overall and 3′ extremity self-complementarity of oligonucleotides. Filter 6 checks that no other binding sites exist in the neighboring sequence. Filter 7 (optional) compares the primer list with other genome sequences available. If sequences other than that of the reference strain are available, the user can ask GenoFrag to perform a BLASTn search (28) with these genome sequences against the primer list. This option allows GenoFrag to reject primers that may not produce PCR amplification because of sequence divergence.
All oligonucleotides passing successfully through the filters are proposed as primers for LR-PCR. In addition, they are labeled with their position in the genome and with their ability to start or end an amplicon.
Segmentation of the genome. This second software program aims to provide a list of amplicons ensuring optimum coverage of the whole genome, or part of the genome, from the set of primers previously generated. Constraints are the minimum and maximum length of amplicons, and the minimum and maximum overlaps allowed. If, for the sake of simplicity, we assume that a solution is made up of a list of N amplicons, and that each of the amplicons can occupy only P different locations, then the number of possibilities is equal to PN. Finding the best option when N is large is clearly a combinatorial problem; computing of all the possibilities is untenable. We have developed computational optimization methods to solve this problem within a reasonable time interval. Two solutions to this problem have been implemented: Shortest Path Problem (SPP) and Single Traverse Algorithm (SITA). SPP looks for an optimum list of amplicons whose sizes are as close as possible to an ideal length. Under the second solution, the ideal length is not required a priori, but is computed by the SITA in such a way that the best segmentation is that which provides homogeneous amplicon sizes and minimizes the difference between the shortest and the longest amplicons. For both solutions we have (i) formulated a suitable combinatorial optimization model and (ii) programmed a dedicated graph algorithm to solve these models [the mathematical analysis described elsewhere (29)]. Both programs provide a list of primer pairs as output. The computation time for processing a sequence of 1 Mb ranges from 1 to 2 min on a 1.5 GHz PC, depending on the number of primers selected during the first step. Of course, the higher the number of primers, the longer is the computation time.
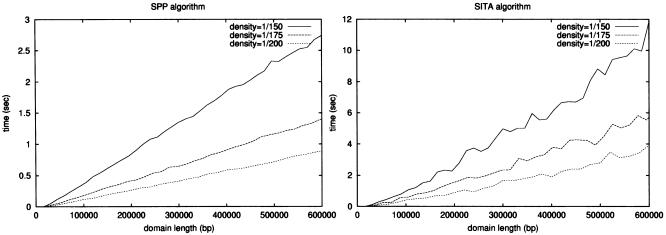
GenoFrag performance. The execution time for the ‘generation of primers’ program to produce a list of all potential primers is around 40 s in the case of a genome 2.8 Mb long (corresponding to the size of the S.aureus chromosome) and using the default parameters (detailed above). This time may vary slightly and linearly, depending on the stringency of the parameters when tested on a 1.6 GHz Linux machine. In order to evaluate and compare the performance of the SPP and SITA algorithms, both were run on randomly generated genomes of increasing length (where primers are uniformly distributed over the segment) but of a fixed primer density for each curve. Computational experiments were performed on a Pentium 4 (1.6 GHz) machine under Linux. Each point on the curves was the average of 10 runs. As shown in Figure 2, these curves exhibited a linear behavior compared with the genome length.
Figure 2.
GenoFrag performance. Software performance was determined on several virtual chromosomal sequences, which were randomly generated and ranged from 10 to 600 kb.
Application to the whole genome PCR scanning analysis of S.aureus. GenoFrag can be used for primer design on any bacterial sequence. We are particularly interested in S.aureus since it is one of the major public health concerns worldwide, and it is reported to be genomically heterogeneous (9). This latter characteristic renders S.aureus a good candidate for whole genome PCR scanning analysis. The complete genome sequence of S.aureus N315 was used to generate a set of primers using GenoFrag. Once the parameters for primer design are set up, GenoFrag automatically generates a set of primers corresponding to optimum coverage of the chromosome.
Sequence treatment before submission to GenoFrag
Genome sequence can be used to perform a search of maximum exact and degenerate repeats using REPuter (30). Using REPuter outputs, users can precisely localize redundant sequences with or without biological significance and their coordinates. This preliminary analysis allows GenoFrag to define genome regions that will not be taken into account for primer design, so as to avoid ambiguous PCR results. These regions are hereinafter referred to as forbidden regions: these include numerous repetitive sequences without known biological significance that are automatically revealed by the REPuter search. Other forbidden regions include short and redundant mobile genetic elements such as tranposons (Tn554, ∼6.7 kb, five copies in the N315 chromosome) or insertion sequences (IS1181, ∼1.5 kb, eight copies in the N315 chromosome). These short mobile genetic elements are included in amplified regions. This is particularly interesting since these mobile genetic elements are often involved in genome plasticity (4,5). Sequences of ribosomal RNA (five copies for 23S and 16S rRNAs, six copies for 5S rRNA) and STAR sequences (STaphylococcus Aureus Repeats) were not used for primer design but retained in amplified regions (14). On the other hand, mobile genetic elements too large to be included in an amplicon (e.g. prophages or pathogenicity islands) are usually unique or present at low-copy levels in the chromosome. They may be absent from strains other than the reference strain, or present elsewhere on the chromosome. They may also be subject to internal rearrangements from one strain to another, thus participating in genome plasticity (31,32). GenoFrag uses these sequences to generate primer pairs. It should be noted that users can also submit these large genetic elements independently to GenoFrag for the design of specific primer sets (e.g. giving smaller amplicons) to focus on their intrinsic plasticity. Altogether, these sequence treatments defined about 100 forbidden regions, which were identified by their coordinates on the N315 chromosome. These data allow a precise location of any mobile genetic element in the reference pattern of amplicons. Users can thus carry out an initial PCR screen, after which it is possible to return to regions with negative/polymorphic PCR to resolve the genomic structure by further PCR and/or directed sequencing. These data were submitted to GenoFrag for primer design.
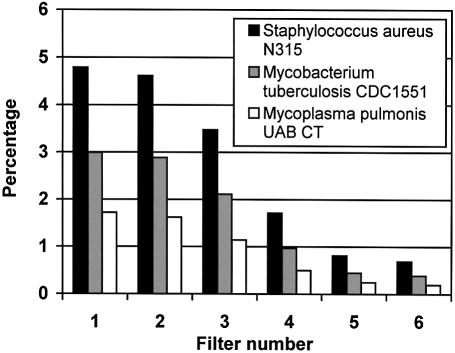
Primer design and primer pair assembly on the S.aureus chromosome. Stepwise design of primers. GenoFrag allows evaluation of the selectivity of different filters in primer design by giving the number of putative primers before and after each filter. Filter selectivity is a function of the parameters set by users. The default parameters used for primer design are summarized in Table 1. Users can modify the set of parameters when they are too stringent, i.e. if the number of primers does not allow optimum coverage of the chromosome. Users can also modify parameters with regard to sequence properties. For example, the efficiency of filter 1 has been evaluated on bacterial genomes with a low or high G+C content. Three genomes were successively tested with the same default parameters: S.aureus and Mycoplasma pulmonis (33) as low-G+C content genomes (32.8 and 26.6%, respectively), and Mycobacterium tuberculosis CDC1551 (34) as a high-G+C content genome (65.6%). Because the G+C content filter was set for a 12 G+C proportion with a 25 nt primer (i.e. 48%), and because the G+C content of the genomes was distant from the default value, the effect of filter 1 was drastic (as shown in Fig. 3, for M.tuberculosis and M.pulmonis in particular). For these two bacteria, the number of primers remaining at the end of the first program was insufficient to enable complete coverage of the genome in the second program. Filter 1 was therefore the most selective filter, eliminating 95, 97 and 98% of primers for S.aureus, M.tuberculosis and M.pulmonis, respectively. The selectivity of subsequent filters did not vary significantly between the three bacteria.
Table 1. Parameters set by default for primer design under GenoFrag.
| Parameter | Value | Significance |
|---|---|---|
| Length of primer | 25 nt | General information |
| G+C content | 12 | |
| Genome type | Circular | |
| Maximum number of repeats | 4 nt | Search for word with identical bases |
| Maximum size of hairpin stem | 3 bp | Hairpin search |
| Maximum size of hairpin loop | 4 nt | |
| Maximum ΔG value at 5′ extremity (first 5 bp) | –8.5 kcal mol–1 | Evaluation of thermodynamic stability |
| Minimum ΔG value at 3′ extremity (last 5 bp) | –10.0 kcal mol–1 | |
| Maximum ΔG value at 3′ extremity (last 5 bp) | –4.0 kcal mol–1 | |
| Minimum size of amplicon | 9000 nt | |
| Maximum size of amplicon | 11 000 nt | |
| Maximum number of matches between a primer and a limited range of the template sequence corresponding to twice the maximum amplicon size | 17 | Secondary binding site search |
| Maximum number of authorized gaps | 0 | |
| Maximum number of matches between a primer and itself | 17 | Autocomplementarity evaluation (as for the intercomplementary evaluation) |
| Maximum ΔG value of an internal dimer with L > 4 bp | –7.0 kcal mol–1 | |
| Maximum ΔG value of an internal dimer at the 3′ extremity (last 10 bp) with L > 3 bp | –5.0 kcal mol–1 |
Figure 3.
Proportion of conserved primer candidates after processing of the chromosomal sequence through each filter. The solution input in filter 1 corresponded to 100%. Histograms give the percentage of primer solutions remaining after each filter for three bacterial genomes. Histogram numbers correspond to filters: 1, selection of oligonucleotides according to their G+C content; 2, elimination of oligonucleotides with N consecutive identical nucleotides; 3, elimination of oligonucleotides with hairpin loops; 4, selection of oligonucleotides according to thermodynamic stability constraints; 5, selection of oligonucleotides according to both overall and 3′ extremity self-complementarity; 6, elimination of oligonucleotides with other binding sites in the neighboring sequence.
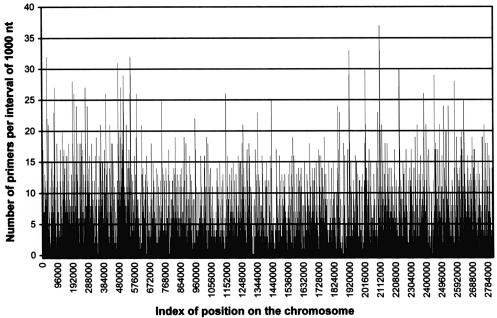
Homogenous repartition of primers on the chromosome. Using the default parameters, we analyzed the repartition of primers on the N315 chromosome in order to check that primer pairs could uniformly cover the whole chromosome. As shown in Figure 4, start and end primers were homogeneously distributed throughout the chromosome. An average of 3.38 start primers and 3.47 end primers were found within 1000-nt intervals. A few 1-kb intervals were exempt of any solution for primer design. Four domains of three to four consecutive 1-kb intervals did not contain any primer solutions. However, these ‘primerless’ domains were smaller than the 10 kb-size of the desired LR-PCR amplicons, and thus could readily be included in one of the possible amplicons during chromosomal segmentation by GenoFrag.
Figure 4.
Homogeneous repartition of primers selected on S.aureus N315 chromosome. The repartition of primers selected by GenoFrag was homogeneous throughout the N315 chromosome. An average of 7.67 primers was found every 1 kb. Only a few 1-kb intervals were exempt of any solution for primer design. However, these primerless intervals were distributed on the chromosome and could easily be integrated into 10 kb amplicons when GenoFrag designed the optimum chromosomal segmentation.
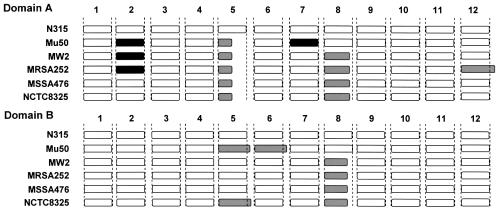
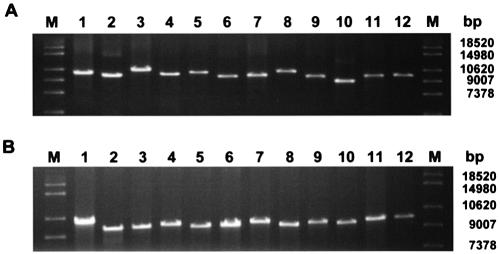
Experimental validation of primers by LR-PCR amplification of a S.aureus chromosome fragment. In order to validate the performance of GenoFrag, we focused on two domains of the S.aureus N315 chromosome, hereinafter referred to as domains A and B. Domain A is 112.5 kb long, and ranges from nucleotide 829 205 to nucleotide 941 785 on the N315 chromosome. Domain B is 107.5 kb long and ranges from nucleotide 1 691 413 to nucleotide 1 799 004. Each domain contains several repeated sequences (STAR sequences) and mobile genetic elements (Tn554b1 and IS1181–3 in Domain A and IS1181–5 in Domain B) that were not to be taken into account for primer design. Primers were generated by GenoFrag without using the optional filter 7. GenoFrag generated a set of primers for 12 amplicons in Domain A and 12 amplicons in Domain B. Primer sequences for Domains A and B, and the expected sizes of amplicons in each domain are shown in Table 2. We first of all performed an in silico validation of GenoFrag using the 48 primers generated for domains A and B in virtual LR-PCR on six different S.aureus strains. The sequence of Mu50, a strain closely related to N315 (14), and sequences of the non-related strains MW2 (15), MRSA272, MSSA476 (Sanger Institute) and NCTC8325 (University of Oklahoma) were used to check that primers designed using N315 were highly likely to generate amplicons during LR-PCR experiments (hybridization and secondary biding sites were checked; data not shown). Results of the virtual LR-PCR experiments are summarized in Figure 5. In most cases, the amplicon patterns for strains other than the N315 reference strain were similar to those seen in N315. In 17 cases, an amplicon shorter or slightly longer than expected was obtained (see Fig. 5 legend for details). This was due to the integration or excision of short mobile genetic elements. In two cases, no PCR product was expected because of the integration of a genetic element which was too large to allow amplification. Only one of the 48 primers tested caused problems with LR-PCR in only one strain, MRSA (a 4 nt mismatch occurred in the 3′ extremity). MRSA is the most divergent strain when compared with N315 (considering the overall sequence of domains; data not shown). This primer was excluded when filter 7 was used. The two sets of primers for Domains A and B were used for LR-PCR amplification on N315 genomic DNA (Fig. 6). As expected, 12 amplicons were obtained at the expected size for each domain. Taken together, these results demonstrate that the primers generated by GenoFrag could successfully be used in large-scale LR-PCR experiments on DNA isolated from different strains and using the same PCR parameters.
Table 2. Subsets of primer pairs designed by GenoFrag and used for LR-PCR on Staphylococcus aureus N315 chromosome.
| Domain | Amplicon | Forward primer (5′→3′) | Reverse primer (5′→3′) | Length (bp) |
|---|---|---|---|---|
| A | 1 | CACCCAGATTTAGCCCAGTTGACAT | GGCAGTCGCATTATCCAACGAAACA | 10 112 |
| 2 | TGCTGTTGCTACAAACGCTGGTCAA | GCCACACTCGTTTCGCCATTATTAG | 10 045 | |
| 3 | CGAAGTTGTAGGGCACGGTTTACTA | AACGGCTAGTGCTTTTACCACACCA | 9220 | |
| 4 | TCGAAAGGGCAATACGCAAAGAGGT | CTGCCAAGAGAATCCCCTCCTAATT | 9905 | |
| 5 | GCAAGCTAGGGATACCCATAACTTC | CGTTGATTCCTCCCTGTGTTGCTTA | 10 752 | |
| 6 | TAGCCGATTCTGATGACGAAGGAGA | CCACCTGACCATACTGCTGAGTTTA | 9947 | |
| 7 | GCCACATTAGCTTGGAAAGCATTGG | CCCCTAACTAGATTACAGGTCCATG | 9763 | |
| 8 | GCCAAGGTGAGGCACAGTTAAATAG | CAAGGTATCGTTGGGTCATTAGGAG | 10 500 | |
| 9 | CGTTGTATCTTCAGTAGGCATCAGG | CCTGGTACTGTAGTGCTTGGTGTAA | 10 107 | |
| 10 | TGCCACTACCAACGATATGATCGGT | CCGCTTCACCTTGAATTGGTTCTAC | 10 929 | |
| 11 | GGGGTATGGGTTAAAGATCCTGAAG | CCAGGAACACCGATGACACGTTTAA | 9913 | |
| 12 | CAGGGACAATTATACCGTGATGACC | TTTGCGAGTGTCGGCGTTAAACCAA | 10 420 | |
| B | 1 | CCTTTCCCCTAACTCAAATGCTGCT | TTGGAGAACAAACAGCACGGACCAT | 10 036 |
| 2 | CAGCACCAGCTTCCACTTGAGAAAT | AACGCAGCAGCTCGTCAACATGAAA | 10 019 | |
| 3 | GACCGCCTACACCTATTGAAGATTG | GCCATCGCACATATTAAAGCAGGTG | 9809 | |
| 4 | TCGAAGGCTTGTCGTCCCATTTAAG | GTGCGTGGCTTAGGCTATAAATTGG | 10 039 | |
| 5 | GCTTTTGAAGGCGTCGTGCTAAATC | CGCATCCAGAAGGTTATCGAAAAGC | 9777 | |
| 6 | TGCTGTACGAACAACTGCTCTTACG | GCTTGAGCATCTTGTTCGCTGATTG | 10 132 | |
| 7 | CGCTCGCTACTTTGTCGTTTTGACT | GGGCAATACACGCACGTTTACTCAA | 9781 | |
| 8 | AATGCTGTTGACAACGATGGACACG | CGTGGATTACCGAGTGATTTTCCTG | 9775 | |
| 9 | TGCGATATACGAATCCTCATCCCTC | GCGTCGTATGTATGGTCAAACAGAC | 9960 | |
| 10 | GCCAAACACCTAGATACAGAAGACC | CGGTGTTAGATACTTGGTGGATGAC | 9854 | |
| 11 | TGGCTTGCGATCTCTAGTGTAACCA | GCAAAATAGACACTACCGTCGTGGA | 9777 | |
| 12 | CCGCTTCTGCTTGTGCTTCTCTTTT | CGAAGCCGTTGTGGTCAAAGCAATA | 10 550 |
Figure 5.
Virtual LR-PCR using primers for Domains A and B on six different S.aureus strains. Genome sequences of S.aureus strains N315, Mu50, MW2, MRSA476, MSSA252 and NCTC8325 were used to perform virtual PCR. Each of the 12 primer pairs for Domains A and B were checked. White: PCR product similar to that of the reference strain N315. Black: no PCR product expected (i.e. the size of the amplicon exceeded 15 kb or one of the primers exhibited excessive sequence divergence). Gray: PCR products shorter or slightly longer than in N315. Domain A: amplicon 2, in Mu50, expected size above 20 kb (integration of Pathogenicity Island, SaPIm); amplicon 2, in MW2, expected size above 20 kb (integration of a SaPI); amplicon 2, in MRSA252, sequence divergence (leading to four mismatches in the 3′ end of the forward primer); amplicon 5, in Mu50, MW2, MRSA252, MSSA476 and NCTC8325, absence of transposon Tn554b1 (present in N315, only); amplicon 7, in Mu50, expected size above 20 kb (integration of bacteriophage φMu50b, 44.4 kb); amplicon 8, ∼1.5 kb shorter in MW2, MRSA252, MSSA476 and NCTC8325 (IS1181–3 present in N315 and Mu50 only); amplicon 12, 1.92 kb longer in MRSA252 (probable presence of an IS). Domain B: amplicon 5, ∼1.5 kb longer in Mu50 (presence of IS1181m1 in the overlap region with amplicon 6); amplicon 5, ∼2 kb longer in NCTC8325 (probable insertion of an IS); amplicon 8, 1.5 kb shorter in MW2, MRSA252, MSSA476 and NCTC8325 (IS1181–5 present in N315 and Mu50 only).
Figure 6.
LR-PCR on the S.aureus N315 chromosome. Two subsets of primers corresponding to Domains A and B (see text and Table 2) were generated and used in LR-PCR for experimental validation.
CONCLUSION
This work concerned the development of GenoFrag, a software package to design primers optimized for whole genome PCR scanning. GenoFrag was used to generate a set of primers from the sequence of S.aureus strain N315. To achieve experimental validation of the software, two subsets of primers were successfully used to amplify two portions of the N315 chromosome with LR-PCR. This result demonstrates the reliability and robustness of GenoFrag. GenoFrag is versatile because it was designed to allow a broad range of parameter implementations (primer length, G+C content of primers, amplicon length, overlap size etc). It has been successfully tested and used to generate primers on other bacterial genomes with different G+C contents [M.pulmonis, 26% G+C (A.Blanchard, INRA Bordeaux, France, personal communication); Bacillus cereus (A.Sorokin, INRA Jouy en Josas, France, personal communication)]. GenoFrag can also be used for viral genomes, after a simple implementation to convert retroviral sequences. GenoFrag could also be implemented to generate primers to fill in the gaps between contigs in unfinished genome sequencing projects.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Dr S. D. Ehrlich for constructive discussions at the beginning of this work, and Dr M. El Karoui and Prof. A. Blanchard for their kind help and advice. N.B.Z. receives financial support in the form of a PhD grant from the French Ministry of Research and Education.
REFERENCES
- 1.Bannerman T.L., Hancock,G.A., Tenover,F.C. and Miller,J.M. (1995) Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol., 33, 551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hadrys H., Balick,M. and Schierwater,B. (1992) Applications of random amplified polymorphic DNA (RAPD) in molecular ecology. Mol. Ecol., 1, 55–63. [DOI] [PubMed] [Google Scholar]
- 3.Dabrowski W., Czekajlo-Kolodziej,U., Medrala,D. and Giedrys-Kalemba,S. (2003). Optimisation of AP-PCR fingerprinting discriminatory power for clinical isolates of Pseudomonas aeruginosa. FEMS Microbiol. Lett., 218, 51–57. [DOI] [PubMed] [Google Scholar]
- 4.Kreiswirth B.N., Lutwick,S.M., Chapnick,E.K., Gradon,J.D., Lutwick,L.I., Sepkowitz,D.V., Eisner,W. and Levi,M.H. (1995) Tracing the spread of methicillin-resistant Staphylococcus aureus by Southern blot hybridization using gene-specific probes of mec and Tn554. Microb. Drug Resist., 1, 307–313. [DOI] [PubMed] [Google Scholar]
- 5.Schneider D., Duperchy,E., Depeyrot,J., Coursange,E., Lenski,R. and Blot,M. (2002) Genomic comparisons among Escherichia coli strains B, K-12 and O157:H7 using IS elements as molecular markers. BMC Microbiol., 2, 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enright M.C. and Spratt,B.G. (1999) Multilocus sequence typing. Trends Microbiol., 7, 482–487. [DOI] [PubMed] [Google Scholar]
- 7.Enright M.C., Day,N.P., Davies,C.E., Peacock,S.J. and Spratt,B.G. (2000) Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol., 38, 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enright M.C., Robinson,D.A., Randle,G., Feil,E.J., Grundmann,H. and Spratt,B.G. (2002) The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl Acad. Sci. USA, 99, 7687–7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgerald J.R. and Musser,J.M. (2001) Evolutionary genomics of pathogenic bacteria. Trends Microbiol., 9, 547–553. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald J.R., Sturdevant,D.E., Mackie,S.M., Gill,S.R. and Musser,J.M. (2001) Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl Acad. Sci. USA, 98, 8821–8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamas I., Klasson,L., Canback,B., Naslund,A.K., Eriksson,A.S., Wernegreen,J.J., Sandstrom,J.P., Moran,N.A. and Andersson,S.G. (2002) 50 million years of genomic stasis in endosymbiotic bacteria. Science, 296, 2376–2379. [DOI] [PubMed] [Google Scholar]
- 12.Oliveira D.C., Tomasz,A. and de Lencastre,H. (2002) Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis., 2, 180–189. [DOI] [PubMed] [Google Scholar]
- 13.LeLoir Y., Baron,F. and Gautier,M. (2003) Staphylococcus aureus and food poisoning. Genet. Mol. Res., 2, 63–76. [PubMed] [Google Scholar]
- 14.Kuroda M., Ohta,T., Uchiyama,I., Baba,T., Yuzawa,H., Kobayashi,I., Cui,L., Oguchi,A., Aoki,K., Nagai,Y. et al. (2001) Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet, 357, 1225–1240. [DOI] [PubMed] [Google Scholar]
- 15.Baba T., Takeuchi,F., Kuroda,M., Yuzawa,H., Aoki,K., Oguchi,A., Nagai,Y., Iwama,N., Asano,K., Naimi,T. et al. (2002) Genome and virulence determinants of high virulence community-acquired MRSA. Lancet, 359, 1819–1827. [DOI] [PubMed] [Google Scholar]
- 16.Ohnishi M., Terajima,J., Kurokawa,K., Nakayama,K., Murata,T., Tamura,K., Ogura,Y., Watanabe,H. and Hayashi,T. (2002) Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl Acad. Sci. USA, 99, 17043–17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lapidus A., Galleron,N., Andersen,J.T., Jorgensen,P.L., Ehrlich,S.D. and Sorokin,A. (2002) Co-linear scaffold of the Bacillus licheniformis and Bacillus subtilis genomes and its use to compare their competence genes. FEMS Microbiol. Lett., 209, 23–30. [DOI] [PubMed] [Google Scholar]
- 18.Rozen S. and Skaletsky,H. (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol., 132, 365–386. [DOI] [PubMed] [Google Scholar]
- 19.Haas S., Vingron,M., Poutska,A. and Wiemann,S. (1998) Primer design for large-scale sequencing. Nucleic Acids Res., 26, 3006–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li P., Kupfer,K.C., Davies,C.J., Burbee,D., Evans,G.A. and Garner,H.R. (1997) PRIMO: a primer design program that applies base quality statistics for automated large-scale DNA sequencing. Genomics, 40, 476–485. [DOI] [PubMed] [Google Scholar]
- 21.Rychlik W. (1995) Selection of primers for polymerase chain reaction. Mol. Biotechnol., 3, 129–134. [DOI] [PubMed] [Google Scholar]
- 22.Suggs S.V., Hirose,T., Myoke,E.H., Kawashima,M.J., Johnson,K.I. and Wallace,R.B. (1981) In Brown,D.D. (ed.), ICN-UCLA Symposium for Developmental Biology Using Purified Gene, Vol. 23. Academic Press, New York, pp. 683–693. [Google Scholar]
- 23.Breslauer K.J., Frank,R., Blaker,H. and Marky,L.A. (1986) Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA, 83, 3746–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blommers M.J.J., Walters,J.A.L.I., Haasnoot,C.A.G., Aelen,J.M.A., van der Marel,G.A., van Boom,J.H. and Hilbers,C.W. (1989) Effects of base sequence on the loop folding in DNA hairpins. Biochemistry, 28, 7491–7498. [DOI] [PubMed] [Google Scholar]
- 25.Kernighan B.W. and Ritchie,D.M. (1988) The C Programming (ANSI C) Language (2nd Edn). Prentice-Hall, Englewood Cliffs, NJ. [Google Scholar]
- 26.Stajich J.E., Block,D., Boulez,K., Brenner,S.E., Chervitz,S.A., Dagdigian,C., Fuellen,G., Gilbert,J.G.R., Korf,I., Lapp,H. et al. (2002) The Bioperl Toolkit: Perl modules for the life sciences. Genome Res., 12, 1161–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weis P., Aponte,M.V., Laville,A. Mauny,M. and Suarez,A. (1990) The CAML Reference Manual. INRIA Research Report RT-0121, Institut National de Recherche en Informatique et Automatique, France. [Google Scholar]
- 28.Altschul S.F., Gish,W., Miller,W., Myers,W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 29.Andonov R., Yanev,N., Lavenier,D. and Veber,P. (2003) Combinatorial approaches for segmenting bacterial genomes. INRIA Report RR-4853, available online at http://www.inria.fr/rrrt/rr-4853.html. [Google Scholar]
- 30.Kurtz S., Choudhuri,J.V., Ohlebusch,E., Schleiermacher,C., Stoye,J. and Giegerich,R. (2001) REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res., 29, 4633–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yarwood J.M., McCormick,J.K., Paustian,M.L., Orwin,P.M., Kapur,V. and Schlievert,P.M. (2002) Characterization and expression analysis of Staphylococcus aureus pathogenicity island 3. Implications for the evolution of staphylococcal pathogenicity islands. J. Biol. Chem. 277, 13138–13147. [DOI] [PubMed] [Google Scholar]
- 32.Ubeda C., Tormo,M.A., Cucarella,C., Trotonda,P., Foster,T.J., Lasa,I. and Penades,J.R. (2003) Sip, an integrase protein with excision, circularization and integration activities, defines a new family of mobile Staphylococcus aureus pathogenicity islands. Mol. Microbiol., 49, 193–210. [DOI] [PubMed] [Google Scholar]
- 33.Chambaud I., Heilig,R., Ferris,S., Barbe,V., Samson,D., Galisson,F., Moszer,I., Dybvig,K., Wroblewski,H., Viari,A. et al. (2001) The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res., 29, 2145–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fleischmann R.D., Alland,D., Eisen,J.A., Carpenter,L., White,O., Peterson,J., DeBoy,R., Dodson,R., Gwinn,M., Haft,D. et al. (2002) Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol., 184, 5479–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]