Abstract
The structural maintenance of chromosomes (SMC) protein encoded by the fission yeast rad18 gene is involved in several DNA repair processes and has an essential function in DNA replication and mitotic control. It has a heterodimeric partner SMC protein, Spr18, with which it forms the core of a multiprotein complex. We have now isolated the human orthologues of rad18 and spr18 and designated them hSMC6 and hSMC5. Both proteins are about 1100 amino acids in length and are 27–28% identical to their fission yeast orthologues, with much greater identity within their N- and C-terminal globular domains. The hSMC6 and hSMC5 proteins interact to form a tight complex analogous to the yeast Rad18/Spr18 heterodimer. In proliferating human cells the proteins are bound to both chromatin and the nucleoskeleton. In addition, we have detected a phosphorylated form of hSMC6 that localizes to interchromatin granule clusters. Both the total level of hSMC6 and its phosphorylated form remain constant through the cell cycle. Both hSMC5 and hSMC6 proteins are expressed at extremely high levels in the testis and associate with the sex chromosomes in the late stages of meiotic prophase, suggesting a possible role for these proteins in meiosis.
INTRODUCTION
Many aspects of eukaryotic cell cycle progression depend on the maintenance and modulation of chromosome architecture. The structural maintenance of chromosomes (SMC) superfamily comprises proteins with important functions in a number of chromosome maintenance activities, including chromosome condensation, sister chromatid cohesion, and DNA repair (reviewed by Hirano, 1998; Hirano, 1999; Strunnikov and Jessberger, 1999; Cobbe and Heck, 2000; Nasmyth et al., 2000). In eukaryotes, six classes of SMC protein have been identified. These are the four classical SMC subfamilies (SMC1-SMC4), named for their Saccharomyces cerevisiae members, along with the somewhat more distantly related Schizosaccharomyces pombe Rad18 (SMC6) and Spr18 (SMC5) groups (Jessberger et al., 1998; Fousteri and Lehmann, 2000). All these SMC proteins share the same characteristic structure, in which two extended coiled-coil domains, separated by a short hinge region, are flanked by highly conserved globular head and tail domains (Figure 1A). The α-helical coiled-coils are involved in protein-protein interactions and the N- and C-terminal domains are thought to mediate ATP binding and hydrolysis (Jessberger et al., 1998). Members of the SMC family form heterodimers, which probably adopt an antiparallel conformation (Melby et al., 1998).
Figure 1.
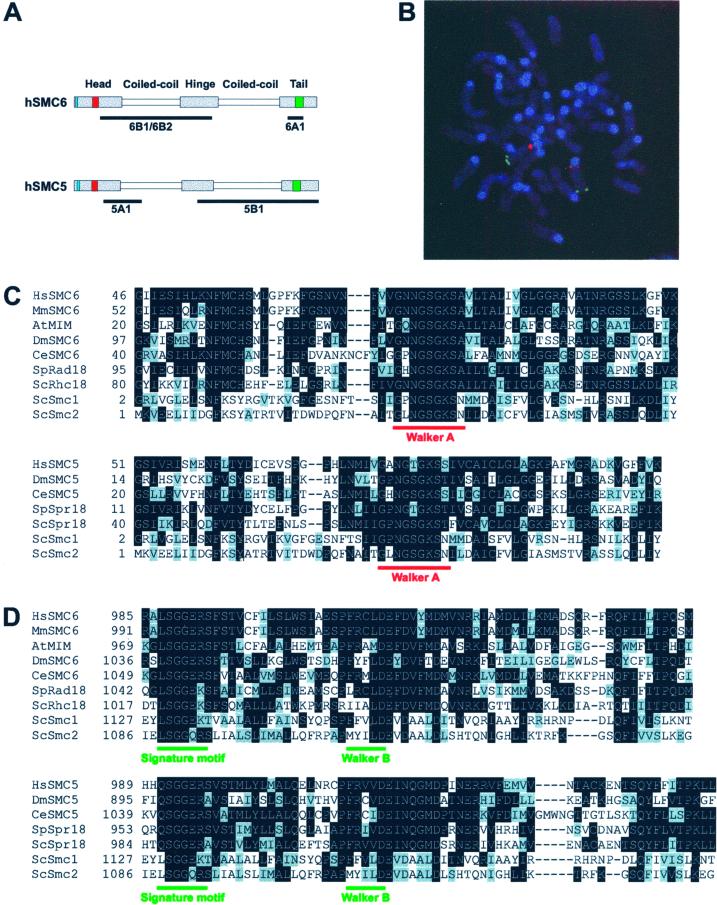
Mammalian SMC5 and SMC6 proteins. (A) Schematic of the hSMC5 and hSMC6 proteins showing the conserved Walker A nucleotide-binding motif and the Walker B motif in red and green, respectively, and the bipartite nuclear localization motif in blue. The underlines represent the regions of protein against which the indicated antibodies were increased. (B) Genomic localization of the mSMC6 gene. Double stain with the use of Texas Red-mSMC6 DNA (red) and an FITC-labeled chromosome 12 telomere-specific probe (green). (C and D) Amino acid sequence alignments between the hSMC6 and hSMC5 N-terminal (C) and C-terminal (D) regions and published SMC protein sequences. Identical amino acids are highlighted in black and conserved residues are indicated in gray.
SMC heterodimers associate with a number of additional proteins to form high molecular mass functional complexes. In budding yeast, the Smc1/Smc3 heterodimer forms the core of the cohesin complex, which is required to hold replicated sister chromatids together until their segregation at anaphase (Guacci et al., 1997; Michaelis et al., 1997). An equivalent cohesin complex has been identified in Xenopus laevis egg extracts (Losada et al., 1998) and human cells (Schmiesing et al., 1998). In bovine cells, bSMC1/bSMC3 have also been shown to form a different complex with DNA ligase III and DNA polymerase ε, which has DNA strand exchange activity in vitro (Jessberger et al., 1996). The Smc2/Smc4 heterodimer is part of the five-member condensin complex, which is required for chromosome condensation at mitosis in S. cerevisiae, S. pombe and X. laevis (Strunnikov et al., 1995; Hirano et al., 1997; Sutani et al., 1999). Human orthologues of Smc2 and Smc4 (hCAP-E and hCAP-C) have been identified and are also found to be part of a complex (Schmiesing et al., 1998; Shimizu et al., 1998). Recently we isolated the third SMC complex from fission yeast, which plays a role in the recombinational repair of DNA strand breaks (Fousteri and Lehmann, 2000). The DNA repair protein Rad18 and its partner Spr18 form the SMC core of this high molecular mass complex, which in addition, contains at least five other uncharacterized components.
Although our understanding of DNA damage responses has made great progress over the last few years, S. pombe rad18 is one of several genes involved in the responses to DNA damage whose functions remain poorly understood. We and others showed in previous studies that the Rad18 protein is required for several different processes, including the repair of DNA double-strand breaks produced by ionizing radiation (Verkade et al., 1999), an alternative excision repair mechanism for removing UV damage (Lehmann et al., 1995), and tolerance of DNA damage during DNA replication (Murray et al., 1997). There is also evidence of a potential role for Rad18 in maintaining checkpoint-dependent cell cycle arrest in response to DNA damage (Verkade et al., 1999). Moreover, analysis of a rad18 null mutant has revealed an essential function, possibly related to DNA replication and mitotic control (Lehmann et al., 1995; Fousteri and Lehmann, 2000). We have speculated that the function of the Rad18/Spr18 complex might be to hold broken DNA molecules in a specific configuration to facilitate repair by recombination. The double-strand DNA damage that is repaired in this manner could be either a double-strand break produced directly by ionizing radiation or a UV photoproduct opposite a gap or stalled replication fork generated during the replication of UV-damaged DNA (Lehmann et al., 1995; Fousteri and Lehmann, 2000). Further evidence of a recombination function for Rad18 has come from recent work with Arabidopsis thaliana, in which a fourfold decrease in intrachromatid recombination was observed after the inactivation of the A. thaliana rad18 orthologue (Mengiste et al., 1999).
In this paper we describe the isolation and characterization of mammalian orthologues of S. pombe Rad18 and Spr18. Confusion surrounds the nomenclature of the yeast rad genes. In particular S. pombe rad18 is unrelated to S. cerevisiae RAD18, and the human homologue of S. cerevisiae RAD18 has already been designated hRad18 (Tateishi et al., 2000). In an attempt to minimize further confusion, we have designated the human homologue of S. pombe rad18 hSMC6 and of its partner spr18 as hSMC5 (c.f., Jessberger et al., 1998).
MATERIALS AND METHODS
Cloning of the hSMC5 and hSMC6 cDNAs
Degenerate oligonucleotide primers, designed to yield a polymerase chain reaction (PCR) product spanning the conserved C-terminal region of the Rad18 (SMC6) family, were used to PCR amplify a fragment from human cDNA. The PCR product was used as a probe to screen a human testis λgt11 cDNA library (Clontech, Palo Alto, CA). Several truncated clones were isolated and a 120-bp fragment from one of these cDNAs was used to rescreen the cDNA library and identify clones containing an additional 5′ sequence. The 5′-end of hSMC6 was PCR amplified from the largest of these clones and was used to assemble the full-length hSMC6 open reading frame (ORF) (DDBJ/EMBL/GenBank accession no. AJ310551). The 3′-end of the mouse SMC6 gene was found in the expressed sequence tag (EST) clone accession no. W62755. We used a 446-bp fragment of this clone to screen a mouse cochlear λZAP cDNA library (gift from G. Richardson) and isolated a single clone that contained the complete mSMC6 ORF (DDBJ/EMBL/GenBank accession no. AJ310552). A partial hSMC5 cDNA was also identified in the EST database (accession no. T10381), and a 380-bp fragment of this clone was used to screen a human testis λgt11 cDNA library (Clontech). Several positive clones were identified, and a fragment from one of the cDNA inserts was used to screen a human thymus 5′-STRETCH PLUS λgt11 cDNA library (Clontech). The 5′-end of the hSMC5 ORF was PCR amplified from a positive clone and was used to assemble the full-length hSMC5 cDNA. We subsequently noted a 45-bp deletion in the sequence derived from EST clone T10381, in comparison with more recently identified hSMC5 ESTs. Having verified the existence of this 45-bp sequence in a fragment of hSMC5 PCR amplified directly from human cDNA, we corrected the deletion in our hSMC5 contig by PCR and subcloning (DDBJ/EMBL/GenBank accession no. AJ310550).
Northern Blot Hybridization
Poly(A)+ mRNA (3 μg) isolated from 1BR.3Neo cells with the QuickPrep micro mRNA purification kit (Pharmacia, Piscataway, NJ) was electrophoresed in formaldehyde-agarose gels, transferred to nitrocellulose membranes, and immobilized by UV cross-linking. Northern blots of poly(A)+ RNA derived from different human tissues (2 μg per lane) were obtained from Clontech. RNA blots were hybridized with [α-32P]dCTP-labeled cDNA probes, prepared with the random primed DNA-labeling kit (Roche, Gipf-Oberfrick, Switzerland).
Antibody Production and Affinity Purification
cDNA fragments corresponding to hSMC6 aa 103–636 and aa 949-1021 and to hSMC5 aa 181–305 and aa 551-1101 were cloned into the vector pQE-30 (Qiagen, Chatsworth, CA) and expressed in Escherichia coli as N-terminal hexahistidine-tagged fusion proteins. Each protein was purified to near homogeneity under denaturing conditions with the use of Ni2+-nitrilotriacetic acid affinity chromatography. Antibodies against each of the recombinant proteins were raised in rabbits. The α-hSMC5 and α-hSMC6 antibodies were affinity purified from crude serum by binding to their respective antigen immobilized on nitrocellulose membrane. Nonspecifically bound protein was removed by extensive washing with phosphate-buffered saline (PBS) before elution of the antibodies with 200 mM glycine, pH 2.5.
Preparation of Cell Extracts and Western Blot Analysis
To prepare whole cell extracts for immunoblotting, cultured cells were trypsinized, washed once with cold buffer A (50 mM Tris-HCl, pH 7.5, 5 mM EDTA, 250 mM NaCl, 1 mM dithiothreitol, 50 mM NaF, 15 mM p-nitrophenyl phosphate, 0.1 mM orthovanadate, 80 mM β-glycerophosphate, and a protease inhibitor cocktail consisting of 10 μg/ml AEBSF, 1 μg/ml trypsin inhibitor, 0.5 μg/ml leupeptin, and 0.5 μg/ml aprotinin), and then resuspended in buffer A to which 1 quarter volume of SDS sample buffer was added. For immunoprecipitations, cells were lysed in cold buffer A containing 0.5% Triton X-100. The lysis mixture was incubated on ice for 10 min and then cleared by centrifugation at 16,000 × g for 10 min. For subcellular fractionation, fibroblasts (1–2 × 107 cells) were resuspended in cold buffer B (20 mM HEPES, pH 7.5, 5 mM potassium acetate, 0.5 mM MgCl2, 0.5 mM dithiothreitol, 20% glycerol, and a protease inhibitor cocktail, as described above, with or without phosphatase inhibitors, 100 mM NaF, 15 mM p-nitrophenyl phosphate, 0.2 mM orthovanadate, and 80 mM β-glycerophosphate) and Triton X-100 was added to 0.02%. The permeabilized cells were briefly homogenized in a Dounce homogenizer (five strokes) and the nuclei were pelleted at 2000 × g for 5 min. The supernatant was cleared by high-speed centrifugation (16,000 × g, 10 min) and designated the cytoplasmic fraction. Nuclei were washed several times in buffer B before being resuspended in extraction buffer. For salt extractions buffer B contained 0.25, 1, or 2 M NaCl. Extractions were performed on ice for 1 h. For DNase I extractions buffer B was supplemented with 5 mM MgCl2 and 150, 250, or 450 U/ml DNase I. DNase I digestion was conducted at 25°C for 1 h. To confirm the digestion, a sample was deproteinized by incubation in 10 mM Tris-HCl, pH 8.0, 10 mM EDTA, 0.5% SDS, 10 μg/ml RNase A, and 100 μg/ml proteinase K for 1 h at 37°C, followed by phenol extraction and electrophoresis on a 0.8% agarose gel. After nuclear extraction the soluble nucleoplasm and insoluble nuclear material were separated by centrifugation (16,000 × g, 5 min) and the nuclear pellet was solubilized by the addition of 1% SDS. Western blot analyses were carried out using standard procedures and immunoreactive bands were visualized by chemiluminescence. Horseradish peroxidase-conjugated goat anti-rabbit antibody (Dako, Carpinteria, CA), horseradish peroxidase-conjugated rabbit anti-mouse antibody (Dako), anti-tubulin antibody (Sigma, St. Louis, MO), anti-proliferating cell nuclear antigen antibody (PC10, Santa Cruz, Santa Cruz, CA), and anti-lamin B antibody (antibody-1, Oncogene Research Products, Cambridge, MA) were used at concentrations recommended by the manufacturers.
Immunoprecipitation
For immunoprecipitations, cell extracts were incubated overnight at 4°C with anti-hSMC5 antibody (5A1), anti-hSMC6 antibody (6A1), or preimmune serum. Protein G-agarose beads were added for 4 h to bind the immune complexes. The beads were washed several times with buffer A and then boiled in SDS sample buffer before SDS-PAGE and Western blot analysis. For in vitro translation of hSMC6 and ΔN-hSMC5, the entire coding sequence of hSMC6 and 810-3306 bp of hSMC5, respectively, were subcloned into the pEPEX vector, behind the T7 promoter. Proteins were synthesized with the TNT Quick Coupled Transcription/Translation System (Promega, Madison, WI). Immunoprecipitations were performed as described above using a 1:1 mixture of in vitro translated proteins in PBS.
Mammalian Cell Culture
1BR.3 primary fibroblasts and 1BR.3Neo transformed cells were grown in MEM supplemented with 15% fetal calf serum. HTC75 osteosarcoma cells (a gift from B. van Steensel) were cultured in DMEM with 10% fetal calf serum. For irradiations, 1BR.3 cells were exposed to 5 Gy γ-radiation. Cells were harvested at 2, 4, 8, 16, and 24 h after irradiation. For cell cycle block and release experiments 1BR.3Neo cells were synchronized at G1/S by growing them for 16 h in the presence of 2.5 mM thymidine. The thymidine was then removed for a period of 13 h before a second cell cycle block (200 μM mimosine) was imposed for 12 h. Human SMC6 was N-terminally tagged with the FLAG epitope tag by subcloning the hSMC6-coding sequence into a modified pCI-neo vector (Promega) harboring the FLAG epitope sequence. For expression in mammalian cells, the construct was transfected into HTC75 cells with the use of polybrene (Smith-Ravin and Jeggo, 1989) and a stable transfectant was isolated.
Immunocytological Methods
Human cells, grown on glass coverslips, were routinely fixed for 30 min at 25°C in PBS containing 3.7% formaldehyde and then permeabilized with 0.5% Triton X-100 in PBS for 2 min. Where indicated, cells were pre-extracted with 0.2% Triton X-100 in PBS for 1 min on ice. For solvent fixation, cells were fixed with 30% methanol/70% acetone for 10 min at −20°C. After fixation the coverslips were washed in PBS and then incubated with primary antibody in bovine serum albumin (BSA) solution (3% BSA in PBS) for 1 h at 25°C. Coverslips were washed in PBS, incubated with fluorescent dye-conjugated secondary antibody in BSA solution (1 h, 25°C), and washed again before being mounted with Glycergel (Dako). Anti-FLAG M5 antibody (Sigma) was used at 20 μg/ml, anti-SC35 antibody (Sigma) was used at a 1:2000 dilution, tetramethylrhodamine B isothiocyanate-conjugated anti-rabbit antibody was used at a 1:150 dilution, and fluorescein isothiocyanate (FITC)-conjugated anti-mouse antibody was used at a 1:50 dilution. The protocol used for detection of hSMC5 and hSMC6 in mouse spermatocyte nuclei has been described (Moens et al., 2000). Chromosome cores were detected with anti-COR1 antibody and a rhodamine-conjugated secondary antibody. The hSMC6 and hSMC5 staining was detected with 6B1 and 5A1, respectively, and an FITC-conjugated secondary antibody.
RESULTS
Isolation of Mammalian Orthologues of Rad18 and Spr18
Using a combination of degenerate PCR and cDNA library screening, we have isolated and assembled the complete ORF (3276 bp) of the human orthologue of S. pombe Rad18. We have designated this ORF hSMC6. With the use of similar procedures, we have also isolated a single clone containing the complete mouse orthologue, mSMC6 (3294 bp). Using fluorescent in situ hybridization with a cDNA clone as a probe, we localized hSMC6 to the short arm of chromosome 2, at 2p23–24. Partial hSMC6 sequences identified in the human EST database form part of the Unigene Hs 34497 cluster, which has been mapped to chromosome 2p22 (Deloukas et al., 1998), confirming our chromosomal localization. Using the mouse cDNA we have isolated several genomic mSMC6 clones (data to be presented elsewhere), one of which was used in fluorescent in situ hybridization studies to localize mSMC6 to chromosome 12A3 (Figure 1B). This chromosome domain is syntenic with human chromosome 2p23.
The hSMC6 and mSMC6 cDNAs encode proteins of 1091 and 1097 aa, respectively, that share the characteristic features of the SMC family, i.e., the N-terminal Walker A and C-terminal Walker B nucleotide-binding motifs, flanking two large coiled-coil domains and a hinge region (Figure 1A). The hSMC6 and mSMC6 proteins are highly related to each other (90% identity) and to the Rad18 orthologues from a variety of species, e.g., S. pombe Rad18 (28% identity; Lehmann et al., 1995), S. cerevisiae Rhc18 (25% identity; Lehmann et al., 1995), Caenorhabditis elegans SMC6 (DDBJ/EMBL/GenBank accession no. CAB01681; 26% identity), Drosophila melanogaster SMC6 (DDBJ/EMBL/GenBank accession no. AAF56254; 26% identity), and A. thaliana Mim (25% identity; Mengiste et al., 1999). The degree of sequence conservation between Rad18 (SMC6) family members is relatively low within the coiled-coil regions which, nevertheless, are predicted to form very similar secondary structures. A much higher level of sequence identity (∼50%) is evident between the globular N- and C-terminal domains of hSMC6 and its orthologues (Figure 1, C and D).
We recently described another SMC protein from fission yeast, Spr18 (Smc5), which is closely related to Rad18 and is in fact its heterodimeric partner (Fousteri and Lehmann, 2000). We were able to identify a human sequence (DDBJ/EMBL/GenBank accession no. T10381) in the EST database whose translation product has significant sequence similarity to the C-terminal two-thirds of S. pombe Spr18. We used this sequence to screen human cDNA libraries and isolated the full-length ORF (3306 bp) of the human orthologue, hSMC5. Further database searching identified several hSMC5 ESTs as part of the Unigene cluster Hs 103283, which has been mapped to chromosome 9q12 (Deloukas et al., 1998). The hSMC5 gene encodes a 1101-residue protein that has all the signature motifs of the SMC family (Figure 1A). Human SMC5 is highly related to the Spr18 orthologues from S. pombe (27% identity; Fousteri and Lehmann, 2000), S. cerevisiae (27% identity; Fousteri and Lehmann, 2000), C. elegans (DDBJ/EMBL/GenBank accession no. AAB00709; 26% identity), and D. melanogaster (DDBJ/EMBL/GenBank accession no. AAF51749; 22% identity), as shown in Figure 1, C and D, which also illustrates the sequence similarity between hSMC5 and hSMC6 (22% identity). Once again the degree of sequence conservation is most pronounced within the globular N- and C-terminal domains of this family of proteins (Figure 1, C and D).
hSMC6 Is Phosphorylated in Vivo
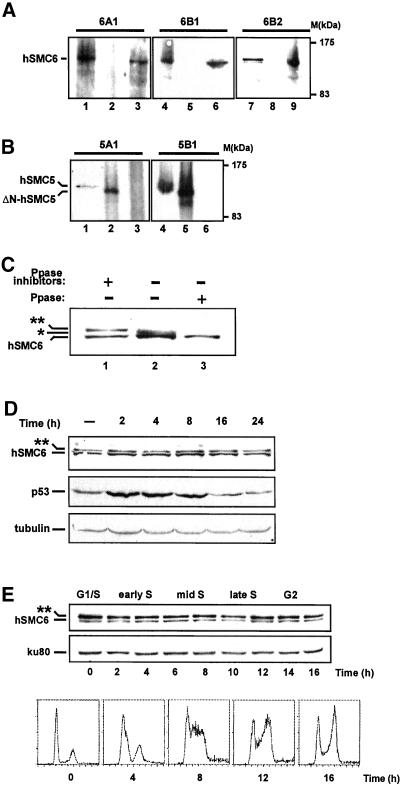
To assist in the characterization of hSMC5 and hSMC6 we raised polyclonal antibodies to each protein. Anti-hSMC6 antibodies were generated against either a C-terminal (aa 949-1021; antibody 6A1) or an N-terminal (aa 103–636; antibodies 6B1 and 6B2) hSMC6 polypeptide (Figure 1A). Anti-SMC5 antibodies were raised against two different fragments of the hSMC5 protein (aa 551-1101 (antibody 5A1) and aa 181–305 (antibody 5B1) (Figure 1A). Western blotting revealed that the anti-hSMC6 antibodies, 6A1 and 6B1, both recognize a band of ∼130 kDa, the predicted molecular mass of hSMC6, in human fibroblast cell extracts (Figure 2A, lanes 1 and 4) as well as full-length hSMC6 protein translated in vitro (Figure 2A, lanes 3 and 6). The anti-hSMC5 antibodies, 5A1 and 5B1, also specifically recognize a 130-kDa protein in human cell extracts (Figure 2B, lanes 1 and 4) and are able to detect a truncated version of hSMC5 translated in vitro (Figure 2B, lanes 2 and 5). Neither of the anti-hSMC6 antibodies is able to recognize in vitro translated hSMC5, and conversely, neither of the anti-hSMC5 antibodies is able to detect in vitro translated hSMC6 (Figure 2, A, lanes 2, 5, and 8, and B, lanes 3 and 6). Together these data indicate that we have generated antibodies with a high degree of specificity for hSMC5 and hSMC6, with no cross-reactivity between the two sets of antibodies despite the structural similarities between hSMC5 and hSMC6.
Figure 2.
Anti-hSMC5 and anti-hSMC6 antibodies and their use in expression studies. (A) Western blots with three different anti-hSMC6 antibodies on human 1BR.3Neo cell extract (lanes 1, 4, and 7), in vitro translated ΔN-hSMC5 (lanes 2, 5, and 8) and in vitro translated hSMC6 (lanes 3, 6, and 9). (B) Western blots with two different anti-hSMC5 antibodies on 1BR.3Neo cell extract (lanes 1 and 4), in vitro translated ΔN-hSMC5 (lanes 2 and 5), and in vitro translated hSMC6 (lanes 3 and 6). (C) 1BR.3Neo cell extracts were prepared in the presence (lane 1) or absence (lane 2) of phosphatase inhibitors or were treated with alkaline phosphatase (lane 3) before immunoblotting with 6B2 anti-hSMC6 antibody. (D) Western blot of 1BR primary fibroblast cell extracts (50 μg per lane) after γ-irradiation (5 Gy). Blots were probed with anti-hSMC6 (6B2), anti-tubulin, and anti-p53 antibodies. (E) Western blot of cell extracts prepared after the release of 1BR.3Neo-transformed fibroblasts from a thymidine-mimosine cell cycle block. Blots were probed with the 6B2 anti-hSMC6 antibody and ku80 antibody (as a loading control).
A third antibody generated with the N-terminal (aa 103–636) hSMC6 polypeptide (antibody 6B2) detects the 130-kDa hSMC6 band but recognizes, in addition, a more slowly migrating species in immunoblots of human cell extract (Figure 2A, lane 7). This additional band has also occasionally been detected with antibody 6B1, albeit at extremely low levels (Taylor and Lehmann, unpublished results). To investigate whether this more slowly migrating band might represent a phosphorylated form of hSMC6, cell extracts were prepared in the presence of a cocktail of phosphatase inhibitors, in the absence of inhibitors, or were treated with calf intestinal alkaline phosphatase before immunoblotting with antibody 6B2. The high molecular mass form is clearly visible as a discrete band when endogenous phosphatase activity is inhibited (Figure 2C, lane 1) but is replaced by a faster migrating species when cells are lysed in the absence of phosphatase inhibitors (Figure 2C, lane 2). This rather indistinct band is further collapsed to the single 130-kDa hSMC6 band upon treatment with phosphatase (Figure 2C, lane 3), indicating that the high molecular mass species recognized by 6B2 is likely to be a phosphorylated form of hSMC6. Because antibody 6B2 does not immunoprecipitate the modified form of hSMC6, we were obliged to use crude extracts for these phosphatase experiments. We are therefore formally unable to exclude the possibility, which we consider unlikely, that the observed shifts in mobility are the result of some other posttranslational modification, which is dependent on the phosphorylation of a different protein. We did not find any evidence for a phosphorylated form of hSMC5.
Because the fission yeast rad18 gene product is required for a number of responses to DNA damage we anticipate that hSMC6 will play a similar role in protecting human cells from genetic damage. To investigate whether the human SMC6 orthologue was regulated in response to genotoxic stress, we performed an immunoblot with the 6B2 antibody to detect hSMC6 after cells were exposed to ionizing radiation (5 Gy). No significant variation in hSMC6 levels was observed (Figure 2D). Moreover, the proportion of hyper- and hypophosphorylated hSMC6 remained unchanged at all time points after irradiation. Similar results were obtained when UV-C (10 J m−2) was used as a damaging agent (Taylor and Lehmann, unpublished results). In contrast, induction of the tumor suppressor p53 is clearly evident under these conditions (Figure 2D). We have also analyzed hSMC6 expression through the mitotic cell cycle and found constant levels of hSMC6 protein and hSMC6 phosphorylation (Figure 2E).
hSMC5 and hSMC6 Form a Complex
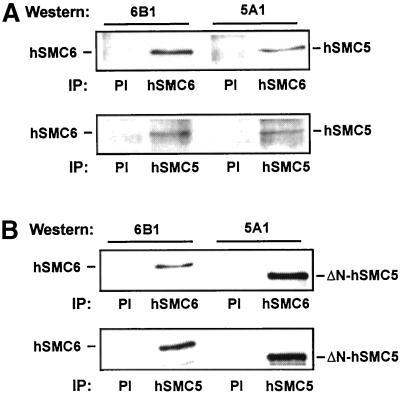
All the members of the SMC family studied thus far in eukaryotes form heterodimeric complexes, and we identified Spr18 as the partner of Rad18 in fission yeast (Fousteri and Lehmann, 2000). To examine whether the mammalian orthologues of Rad18 and Spr18 also interact, we performed immunoprecipitations on human cell extract. Immunoprecipitation with anti-hSMC6 antibody revealed that hSMC5 coimmunoprecipitated with hSMC6 (Figure 3A, top). Similar results were obtained in the reciprocal experiment in which immunoprecipitation was performed with anti-hSMC5 antibody (Figure 3A, bottom). These data demonstrate that hSMC5 and hSMC6 do interact in human cells. To eliminate the possibility that other proteins mediate this interaction we also examined the interaction of hSMC5 and hSMC6 in vitro. For this experiment we mixed in vitro translated hSMC6 with ΔN-hSMC5, an N-terminally deleted form of hSMC5 lacking the first 270 aa. Immunoprecipitations on this mixture with either anti-hSMC6 antibody (Figure 3B, top) or anti-hSMC5 antibody (Figure 3B, bottom) showed that the in vitro translated hSMC6 does indeed coimunoprecipitate with the truncated hSMC5 protein. These data clearly indicate that the interaction between hSMC5 and hSMC6 does not require the presence of additional proteins, nor is it dependent on the extreme N-terminal 270 aa of hSMC5.
Figure 3.
Interaction of hSMC5 and hSMC6. Human cell extracts were immunoprecipitated with preimmune (PI), 6A1, or 5A1 antisera as indicated and immunoblotted with 6B1 anti-hSMC6 (left) or 5A1 anti-hSMC5 (right) antibody. (B) As in A but with in vitro translated hSMC6 or ΔN-hSMC5 in place of human cell extracts.
Intracellular Localization of hSMC5 and hSMC6
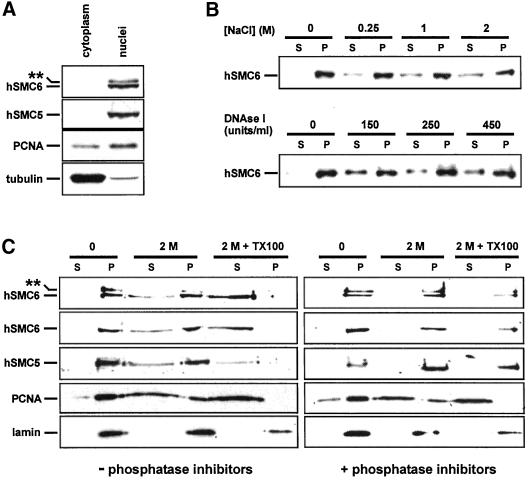
To determine the subcellular distribution of hSMC5 and hSMC6 we examined the expression of both proteins in cytoplasmic, nucleoplasmic, and insoluble nuclear matrix fractions of human fibroblasts. When cytoplasmic and whole nuclear preparations were subjected to Western blot analysis we found that both the hyper- and hypophosphorylated forms of hSMC6 were localized to the nucleus, along with hSMC5 (Figure 4A). This result is in accordance with our observation that both hSMC6 and hSMC5 contain a bipartite nuclear localization sequence at their extreme N-termini (hSMC6 aa 3–19, hSMC5 aa 17–33). Further fractionation of the isolated nuclei, by salt extraction or DNase I digestion, released a significant amount of hSMC6 into the soluble nucleoplasm (Figure 4B), suggesting that this population of hSMC6 is DNA associated. The remaining salt- and DNase-resistant fraction of hSMC6 could be released by the addition of nonionic detergent (2.5% Triton X-100; Figure 4C), implying a hydrophobic interaction between this population of hSMC6 and a component of the nucleoskeleton. A similar extraction profile was seen for hSMC5 (Figure 4C; Moghraby and Lehmann, unpublished results). We next proceeded to examine the distribution of the hyperphosphorylated form of hSMC6 by performing the nuclear extraction procedure in the presence of phosphatase inhibitors. Strikingly, the inclusion of phosphatase inhibitors reduced the extraction of both hSMC5 and hSMC6, even after treatment with Triton X-100 (compare Figure 4C, left and right), suggesting an inverse correlation between phosphorylation status and the extractability of the hSMC5/hSMC6 proteins.
Figure 4.
Subcellular localization of hSMC5 and hSMC6. (A) Nuclear and cytoplasmic extracts of human 1BR.3Neo cells containing 50 μg of protein were electrophoresed and immunoblotted with the indicated antibodies. 6B2 was used for hSMC6 and 5B1 for hSMC5. (B) Nuclei were extracted for 1 h at 4°C with the indicated concentrations of NaCl or for 1 h at 25°C with DNAse I at the indicated units/milliliter. The nuclei were then centrifuged and the amount of hSMC6 in supernatant (S) and pellet (P) were determined by immunoblotting with 6B1 anti-hSMC6. (C) Nuclei were extracted for 1 h at 4°C in the presence or absence of phosphatase inhibitors, without treatment, with 2 M NaCl, or with 2 M NaCl and 2.5% Triton X-100. After centrifugation, the amounts of the indicated proteins in the supernatant (S) and pellet (P) fractions were determined by immunoblotting. Antibodies used were 6B2 (top panel), 6B1 (second panel), 5B1 (third panel), anti-PCNA (fourth panel), and anti-lamin B (fifth panel).
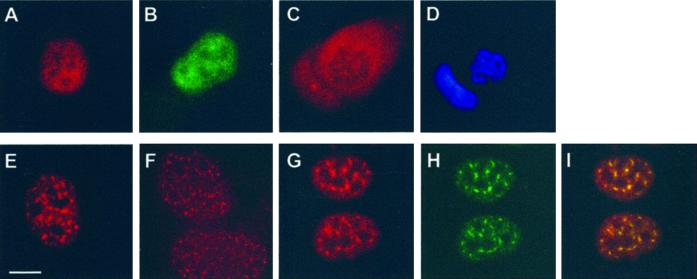
To extend our analysis of the intracellular distribution of hSMC6 we visualized the protein by indirect immunofluorescence. Using affinity-purified 6B1 antibody to stain an asynchronous population of 1BR.3Neo fibroblasts, we observed intense nuclear staining of interphase cells (Figure 5A). The hSMC6 immunofluorescence signal is evenly distributed throughout the nucleus, with the exception of the nucleoli from which it is excluded. Comparable staining has also been obtained with a second anti-hSMC6 antibody, 6A1 (Taylor and Lehmann, unpublished results) and with a monoclonal α-FLAG antibody to examine the localization of FLAG epitope-tagged hSMC6 in a stably transfected cell line (Figure 5B). During mitosis hSMC6 staining undergoes a striking redistribution, because it is excluded from the condensed mitotic chromosomes and is dispersed instead through the rest of the cell volume after nuclear envelope breakdown (Figure 5, C and D).
Figure 5.
Immunofluorescence studies of hSMC6. (A) Human 1BR.3Neo fibroblasts were stained with 6B1 anti-hSMC6. (B) HTC75 cells were transfected with FLAG-tagged hSMC6 and then stained with anti-FLAG M5 antibody. (C and D) A mitotic cell stained with 6B1 (C) or 4′,6-diamidino-2-phenylindole (D). (E–I) Cells fixed with 3.7% formaldehyde (E), 30% methanol/70% acetone (F), or pre-extracted with 0.5% Triton X-100 before formaldehyde fixation (G) were stained with 6B2 anti-hSMC6. The cells shown in G were counterstained with anti-SC35 splicing factor antibody (H). The overlay image of the two antibodies is presented in I. Bar, 10 μm.
Next, we performed immunofluorescence staining of human cells with the 6B2 antibody, which is capable of recognizing a phosphorylated form of hSMC6 in immunoblots. Once again the staining that we observed was localized to cell nuclei, but interestingly the signal was concentrated in distinct foci or nuclear speckles (Figure 5E). This nuclear speckle pattern was observed in a variety of human cell lines, under different fixation conditions, and is highly resistant to detergent extraction (Figure 5, E–G). We had previously noted that 6B2 antisera taken at early stages of the immunization procedure recognized the phosphorylated form of hSMC6 with much greater efficiency than antisera obtained during the later stages of immunization. When we compared the immunofluorescence staining of early and late batches of antisera we found that the nuclear speckle pattern largely correlated with the ability to recognize hyperphosphorylated hSMC6 in immunoblots, whereas the diffuse nuclear staining is more readily apparent with antisera, which preferentially recognizes hypophosphorylated hSMC6. Taken together these data suggest that it is the phosphorylated form of hSMC6 that localizes to the nuclear speckles. The speckled pattern that we observe with 6B2 is highly reminiscent of the speckle domains that have previously been described for pre-mRNA splicing factors (Spector, 1993). To test whether or not hSMC6 speckles correspond to the splicing factor-enriched nuclear domains termed interchromatin granule clusters (IGCs), we performed double immunofluorescence staining with 6B2 and an anti-SC35 splicing factor antibody. Figure 5, G–I, shows that the two sets of nuclear speckles colocalize, suggesting that the phosphorylated form of hSMC6 is indeed associated with IGCs.
Expression of hSMC5 and hSMC6 in Various Tissues
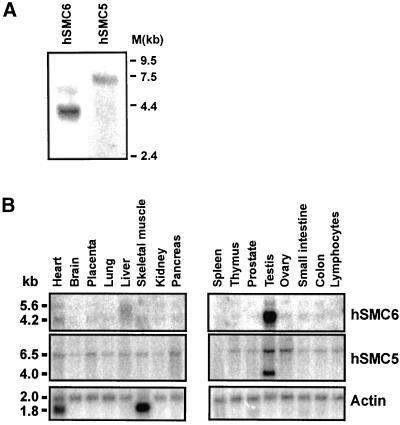
Northern blots on mRNA prepared from SV40-transformed human fibroblasts (1BR.3Neo) revealed that hSMC6 encodes a major transcript of 4.2 kb, as well as a second, minor transcript of 5.6 kb (Figure 6A). The hSMC5 gene encodes a single mRNA species of ∼6.5 kb in length (Figure 6A). We examined the pattern of expression of both hSMC5 and hSMC6 in various human tissues. Northern blotting revealed that the 4.2- and 5.6-kb hSMC6 transcripts are both present at very low levels in all the tissues examined, with the exception of testis, where the 4.2-kb hSMC6 transcript is greatly up-regulated (∼30-fold) (Figure 6B). A similar degree of transcriptional induction was also observed for the mouse SMC6 gene in this tissue (Lees and Lehmann, unpublished results). The 6.5-kb hSMC5 transcript is also expressed ubiquitously and is somewhat elevated in testis (approximately threefold). A novel hSMC5 transcript of ∼4 kb, evident only in testis, constitutes the major hSMC5 transcript in this tissue (Figure 6B). The large increases in hSMC5/hSMC6 mRNA levels that we observe in testis may partly reflect the proliferative nature of this tissue. In A. thaliana it has recently been demonstrated that the MIM gene, which encodes the plant SMC6 homologue, is transcriptionally up-regulated in accordance with the degree of cell division activity (Mengiste et al., 1999). Moreover, our own studies indicate that hSMC6 expression is slightly elevated (approximately threefold) in rapidly proliferating transformed cells in comparison with quiescent primary fibroblasts (Taylor and Lehmann, unpublished results). We do not, however, detect any significant increase in hSMC5 or hSMC6 expression in other rapidly proliferating tissues such as thymus, nor do we see any evidence of the novel 4-kb hSMC5 transcript in a rapidly dividing transformed cell line (Figure 6A). These observations suggest that the elevated expression of hSMC5 and hSMC6 most likely reflect a specific requirement for these gene products in testis, perhaps in meiotic progression.
Figure 6.
Tissue-specific expression of hSMC6 and hSMC5. (A) Northern blots of 3 μg of mRNA prepared from proliferating 1BR.3Neo-transformed human fibroblasts hybridized with hSMC6 and hSMC5 probes. (B) Human tissue-specific Northern blot (Clontech) probed for the indicated mRNAs.
Association of hSMC5 and hSMC6 with Meiotic Chromosomes
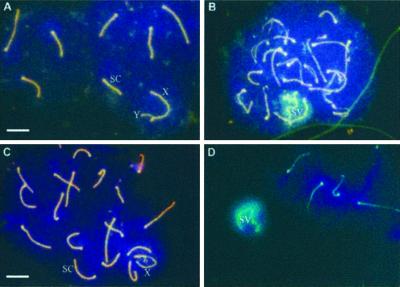
To investigate a possible meiotic role for the mammalian SMC5/SMC6 proteins, we tested whether the mouse SMC5 and SMC6 homologues localize to chromosomes in meiotic prophase. We performed immunofluorescence staining of mouse spermatocytes with anti-hSMC6 together with an antibody against chromosome cores (anti-Cor1; Figure 7, A and B). During early to midmeiotic prophase, as the chromosomes become synapsed and recombination occurs between homologues, we were unable to detect any mSMC6 staining (Figure 7A). During late pachytene/diplotene, however, we observed intense staining of the X-Y chromosome pair within the sex vesicle (Figure 7B). This staining is specific for mSMC6 because no staining of the X-Y bivalent was seen with preimmune sera (Moens, unpublished results). When the experiment was performed with anti-hSMC5 antibody we observed a strikingly similar staining pattern (Figure 7, C and D). The colocalization of mSMC5 and mSMC6 on the X-Y chromosomes during diplotene suggests that these proteins may act together at a late step in meiosis.
Figure 7.
Meiotic prophase localization of hSMC5 and hSMC6. (A) A midpachytene mouse spermatocyte immunostained with antibodies to chromosome cores and with 6B1 anti-hSMC6 antibody. (B) An early diplotene spermatocyte stained with anti-core antibody and 6B1. The sex vesicle and sex chromosome cores are recognized by the anti-hSMC6 antibody. (C and D) Midpachytene (C) and early diplotene (D) spermatocytes stained with anti-COR1 and 5A1 anti-hSMC5 antibody. The X-chromosome (X), Y-chromosome (Y), sex vesicle (SV), and synaptonemal complexes (SC) are indicated. Bar, 10 μm.
DISCUSSION
Heterodimeric Association of hSMC5 and hSMC6
Members of the SMC family of proteins are essential for a variety of chromosomal activities, including chromosome condensation, sister chromatid cohesion, and recombinational repair (Hirano, 1998; Jessberger et al., 1998). In budding and fission yeast three SMC complexes have been identified. SMC1 and SMC3 are components of the cohesin complex, the SMC2/SMC4 heterodimer is part of the condensin complex, and the fission yeast DNA repair protein Rad18 and its partner Spr18 form the core of the third complex (Guacci et al., 1997; Michaelis et al., 1997; Sutani et al., 1999; Fousteri and Lehmann, 2000). Here we have shown that, like the other SMC proteins, Rad18 and Spr18 are conserved throughout eukaryotes with orthologues in C. elegans, D. melanogaster, A. thaliana (Mengiste et al., 1999), mouse, and human. There is ∼25% sequence identity overall between the Rad18 (SMC6) and Spr18 (SMC5) orthologues from different organisms, with much higher levels of conservation between the globular N- and C-terminal domains, which harbor the ATP-binding/hydrolysis motifs. In contrast, the coiled-coil regions of the SMC5/SMC6 proteins are poorly conserved, although each is predicted to form a similar α-helical secondary structure. Interactions between the hydrophobic surfaces of coiled-coil domains are important for the dimerization of these proteins (Lupas, 1996). In bacterial species that have only a single SMC family member, SMC proteins form homodimers (Melby et al., 1998). In contrast, the multiple SMC proteins that have been identified in eukaryotes associate in a heterodimeric manner (Hirano, 1998; Jessberger et al., 1998). Our previous data suggest that Rad18 and Spr18 form a heterodimer in fission yeast (Fousteri and Lehmann, 2000). In this work we have demonstrated a physical interaction between the hSMC5 and hSMC6 proteins, confirming that they are also heterodimeric partners. Preliminary results suggest that the hSMC5/hSMC6 heterodimer, like its S. pombe counterpart, is at the core of a high molecular mass functional complex, and we are currently investigating this complex in more detail.
To date no high-resolution structural studies have been performed on any of the eukaryotic SMC heterodimers. However, an electron microscopy study of bacterial SMC homodimers revealed that the long coiled-coil domains of both the Bacillus subtilis SMC (BsSMC) protein and the E. coli MukB SMC-like protein adopt an antiparallel conformation (Melby et al., 1998). This is somewhat unusual because other coiled-coil proteins are almost all parallel in orientation (Lupas, 1996). As a result of this antiparallel association the N- and C-terminal domains are paired at each end of the dimeric SMC molecule, bringing the Walker A and Walker B motifs into close proximity. The functional association of these ATP-binding motifs has yet to be demonstrated for an SMC dimer, although a recent x-ray crystallographic study has revealed that this is the case for a structurally related ATPase, the DNA double-strand break repair enzyme Rad50 (Hopfner et al., 2000). Although Rad50 is not classified as an SMC protein, it has a very similar structure but lacks a “hinge” domain. Hopfner et al. (2000) cocrystallized the N- and C-terminal globular domains of Rad50 and showed that they formed a tight dimeric complex. In the presence of ATP, two of these N/C dimers associated to form a tetramer with two ATP molecules located in the interface between the two dimers. It is likely that SMC proteins will adopt similar configurations, and this raises several interesting questions. 1) Each heterodimer (e.g., hSMC5/hSMC6) has two dimeric globular domains, one at each end. Is the ATP-binding pocket formed by folding a single heterodimer back on itself at the hinge or by two heterodimers associating with each other? 2) Does the postulated tetramerization on binding ATP affect the association of the SMC protein cores with other proteins in the complex? 3) Can SMC homodimers as well as heterodimers form these structures? We note that, although hSMC5 and hSMC6 show many similar characteristics and clearly form heterodimers, the tissue-specific expression levels of the two mRNAs show some differences (Figure 6B), raising the possibility that they might have individual roles in addition to their joint functions.
Expression of hSMC5 and hSMC6 in Somatic Cells
We have shown that hSMC5 and hSMC6 are nuclear proteins that are, at least in part, associated with the DNA. This observation is consistent with a postulated role for this SMC heterodimer in the recombinational repair of DNA strand breaks. We have, in addition, demonstrated a cell cycle-dependent redistribution of hSMC6, which becomes excluded from the condensed chromosomes during mitosis. This localization pattern, which is remarkably similar to that of the human and mouse SMC cohesin subunits (Schmiesing et al., 1998; Darwiche et al., 1999), is the inverse of the staining that has been described for the condensin SMCs (Hirano and Mitchison, 1994; Schmiesing et al., 1998; Shimizu et al., 1998). This result suggests that hSMC5/hSMC6 function, like that of the cohesins, occurs predominantly during interphase rather than in mitosis.
We anticipate that hSMC6 will, like its orthologues in yeast and A. thaliana, function in response to DNA damage. We did not, however, detect any significant alteration in hSMC6 expression, at either the RNA (Taylor and Lehmann, unpublished results) or protein level, after DNA damage, nor did we find any evidence of posttranslational modification of hSMC6 in response to irradiation. These observations differ from recent data regarding MIM, the A. thaliana orthologue of hSMC6, which is significantly up-regulated after treatment with the DNA-damaging agent methyl methane-sulfonate (Mengiste et al., 1999). The A. thaliana MIM protein is the first eukaryotic SMC protein thus far tested that is not essential for viability but seems to be solely required for the response to genotoxic stress. It will be interesting to see whether hSMC6 is also nonessential or if, like its S. pombe counterpart, hSMC6 is absolutely required for cell viability.
In the course of this study we have identified a phosphorylated form of hSMC6 that can be detected with the use of the 6B2 antibody. This phosphorylated form appears to be constitutive, because its level remained unchanged throughout the cell cycle and after DNA damage. Under certain conditions we find that the hyperphosphorylated form of hSMC6 is more refractory to extraction than hypophosphorylated hSMC6, suggesting an inverse correlation between phosphorylation status and the extractability of hSMC6. Our observation that the addition of excess phosphatase inhibitors significantly reduces the extraction of both hSMC6 and hSMC5 reinforces this correlation. Our immunofluorescence data suggest that the phosphorylated form of hSMC6 localizes to detergent-resistant nuclear speckle domains. Colocalization of this staining with the pre-mRNA splicing factor SC35 identified these nuclear speckles as IGCs, dynamic nuclear organelles that have been shown to serve as pools of pre-mRNA splicing factors (Misteli et al., 1997). It is interesting to note that phosphorylation events seem to play an important role in regulating IGC speckle morphology and in determining the localization of several IGC-associated proteins (Gui et al., 1994; Misteli and Spector, 1996; Mortillaro et al., 1996). The functional significance of hSMC6 localization at IGCs is not immediately apparent. Perhaps IGCs serve as reservoirs for a greater variety of proteins than hitherto suspected. The recently reported biochemical purification of IGCs is likely to prove informative in this regard (Mintz et al., 1999).
hSMC5 and hSMC6 May Have a Role in Meiosis
Both hSMC5 and hSMC6 show extraordinarily high levels of expression in the testis and, in the case of hSMC5, a novel transcript. Although part of this increased transcription is undoubtedly due to an increased proliferation rate, similar increases are not seen in other tissues such as the thymus, which has a comparable proliferation rate to the testis. It is therefore most likely that the increased levels of expression in testis reflect a role for hSMC5/hSMC6 in meiosis. This is supported by our finding of an association of mSMC5 and mSMC6 with the X-Y chromosome pair of meiotic cells. A number of proteins that function in recombination (Rad51, Dmc1, Rad50, Mre11) and DNA damage-induced checkpoint responses (Rad1, ATR) are highly up-regulated in testis (Dolganov et al., 1996; Habu et al., 1996; Keegan et al., 1996; Carney et al., 1998) and have been shown to associate with meiotic chromosomes (Barlow et al., 1997; Moens et al., 1997, 1999, 2000; Freire et al., 1998; Eijpe et al., 2000; Lombard and Guarente, 2000). Rad51/Dmc1 foci formation occurs early in meiotic prophase and is believed to mark the regions of DNA where recombination events occur (Barlow et al., 1997; Moens et al., 1997). The checkpoint proteins Rad1 and ATR stain chromosome cores slightly later in meiotic prophase and, in contrast to Rad51/Dmc1, Rad1, and ATR antigens are abundant on the X-Y chromosome pair in the late stages of meiotic prophase (Freire et al., 1998; Moens et al., 1999). Rad50/Mre11 staining also persists on the X-Y bivalent during late pachytene/diplotene when recombination between homologous chromosomes is complete (Eijpe et al., 2000). This sex vesicle staining is strikingly similar to the localization we have observed for hSMC5 and hSMC6. Eijpe et al. (2000) suggested a possible explanation for the association of Rad50/Mre11 with the X-Y chromosome pair. That is, Rad50/Mre11 could have a role in the processing of DNA double-strand breaks that form in the nonautosomal, asynapsed parts of the X and Y chromosomes. Any such DNA breaks could not be repaired with the use of the homologous chromosome but would persist until the sister chromatids became available for repair. Clearly, the proposed function of the hSMC5/hSMC6 heterodimer, in holding broken DNA molecules together to facilitate strand break repair, could be of importance in this context. It remains to be seen whether this is indeed the case. The precise role of hSMC5/hSMC6 in meiosis and significance of the X-Y association will be explored in future work.
ACKNOWLEDGMENTS
We are indebted to Guy Richardson and Bas van Steensel for materials, to Alastair Waugh for FACS analysis, and to Tony Carr, Penny Jeggo, Bryn Bridges, and Patricia Kannouche for comments on the manuscript. This work was supported in part by European Community contracts F14P-CT95-0010 and FIGH-CT1999-0010.
Abbreviations used:
- BSA
bovine serum albumin
- EST
expressed sequence tag
- FITC
fluorescein isothiocyanate
- IGC
interchromatin granule cluster
- ORF
open reading-frame
- PBS
phosphate-buffered saline
- SMC
structural maintenance of chromosomes
- PCR
polymerase chain reaction
REFERENCES
- Barlow AL, Benson FE, West SC, Hulten MA. Distribution of the Rad51 recombinase in human and mouse spermatocytes. EMBO J. 1997;16:5207–5215. doi: 10.1093/emboj/16.17.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, III, Hays L, Morgan WF, Petrini JHJ. The hMre11/hRad50 protein complex and Nijmegen breakage-syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- Cobbe N, Heck MM. Review: SMCs in the world of chromosome biology: from prokaryotes to higher eukaryotes. J Struct Biol. 2000;129:123–143. doi: 10.1006/jsbi.2000.4255. [DOI] [PubMed] [Google Scholar]
- Darwiche N, Freeman LA, Strunnikov A. Characterization of the components of the putative mammalian sister chromatid cohesion complex. Gene. 1999;233:39–47. doi: 10.1016/s0378-1119(99)00160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloukas P, Schuler GD, Gyapay G, Beasley EM, Soderlund C, Rodriguez-Tome P, Hui L, Matise TC, McKusick KB, Beckmann JS, Bentolila S, Bihoreau M, Birren BB, Browne J, Butler A, Castle AB, Chiannilkulchai N, Clee C, Day PJ, Dehejia A, Dibling T, Drouot N, Duprat S, Fizames C, Bentley DR, et al. A physical map of 30,000 human genes. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- Dolganov GM, Maser RS, Novikov A, Tosto L, Chong S, Bressan DA, Petrini JH. Human Rad50 is physically associated with human Mre11: identification of a conserved multiprotein complex implicated in recombinational DNA repair. Mol Cell Biol. 1996;16:4832–4841. doi: 10.1128/mcb.16.9.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijpe M, Offenberg H, Goedecke W, Heyting C. Localization of RAD50 and MRE11 in spermatocyte nuclei of mouse and rat. Chromosoma. 2000;109:123–132. doi: 10.1007/s004120050420. [DOI] [PubMed] [Google Scholar]
- Fousteri MI, Lehmann AR. A novel SMC protein complex in Schizosaccharomyces pombecontains the Rad18 DNA repair protein. EMBO J. 2000;19:1691–1702. doi: 10.1093/emboj/19.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire R, Murguia JR, Tarsounas M, Lowndes NF, Moens PB, Jackson SP. Human and mouse homologs of Schizosaccharomyces pombe rad1(+) and Saccharomyces cerevisiaeRAD17: linkage to checkpoint control and mammalian meiosis. Genes Dev. 1998;12:2560–2573. doi: 10.1101/gad.12.16.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui JF, Lane WS, Fu XD. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994;369:678–682. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- Habu T, Taki T, West A, Nishimune Y, Morita T. The mouse and human homologs of DMC1, the yeast meiosis-specific homologous recombination gene, have a common unique form of exon-skipped transcript in meiosis. Nucleic Acids Res. 1996;24:470–477. doi: 10.1093/nar/24.3.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. SMC protein complexes and higher-order chromosome dynamics. Curr Opin Cell Biol. 1998;10:317–322. doi: 10.1016/s0955-0674(98)80006-9. [DOI] [PubMed] [Google Scholar]
- Hirano T. SMC-mediated chromosome mechanics: a conserved scheme from bacteria to vertebrates? Genes Dev. 1999;13:11–19. doi: 10.1101/gad.13.1.11. [DOI] [PubMed] [Google Scholar]
- Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophilabarren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- Jessberger R, Frei C, Gasser SM. Chromosome dynamics: the SMC protein family. Curr Opin Gen Dev. 1998;8:254–259. doi: 10.1016/s0959-437x(98)80149-4. [DOI] [PubMed] [Google Scholar]
- Jessberger R, Riwar B, Beachtold H, Akhmedov AT. SMC proteins constitute two subunits of the mammalian recombination complex RC-1. EMBO J. 1996;15:4061–4068. [PMC free article] [PubMed] [Google Scholar]
- Keegan KS, Holtzman DA, Plug AW, Christenson ER, Brainerd EE, Flaggs G, Bentley NJ, Taylor EM, Meyn MS, Moss SB, Carr AM, Ashley T, Hoekstra MF. The Atr and Atm protein-kinases associate with different sites along meiotically pairing chromosomes. Genes Dev. 1996;10:2423–2437. doi: 10.1101/gad.10.19.2423. [DOI] [PubMed] [Google Scholar]
- Lehmann AR, Walicka M, Griffiths DJF, Murray JM, Watts FZ, McCready S, Carr AM. The rad18 gene of Schizosaccharomyces pombedefines a new subgroup of the SMC superfamily involved in DNA repair. Mol Cell Biol. 1995;15:7067–7080. doi: 10.1128/mcb.15.12.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Guarente L. Nijmegen breakage syndrome disease protein and MRE11 at PML nuclear bodies and meiotic telomeres. Cancer Res. 2000;60:2331–2334. [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. Identification of XenopusSMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- Melby TE, Ciampaglio CN, Briscoe G, Erickson HP. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins; long, antiparallel coiled coils, folded at a flexible hinge. J Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste T, Revenkova E, Bechtold N, Paszkowski J. An SMC-like protein is required for efficient homologous recombination in Arabidopsis. EMBO J. 1999;18:4505–4512. doi: 10.1093/emboj/18.16.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Mintz PJ, Patterson SD, Neuwald AF, Spahr CS, Spector DL. Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 1999;18:4308–4320. doi: 10.1093/emboj/18.15.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Misteli T, Spector DL. Serine/threonine phosphatase 1 modulates the subnuclear distribution of pre-mRNA splicing factors. Mol Biol Cell. 1996;7:1559–1572. doi: 10.1091/mbc.7.10.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Chen DJ, Shen Z, Kolas N, Tarsounas M, Heng HHQ, Spyropoulos B. Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma. 1997;106:207–215. doi: 10.1007/s004120050241. [DOI] [PubMed] [Google Scholar]
- Moens PB, Freire R, Tarsounas M, Spyropoulos B, Jackson SP. Expression and nuclear localization of BLM, a chromosome stability protein mutated in Bloom's syndrome, suggest a role in recombination during meiotic prophase. J Cell Sci. 2000;113:663–672. doi: 10.1242/jcs.113.4.663. [DOI] [PubMed] [Google Scholar]
- Moens PB, Tarsounas M, Morita T, Habu T, Rottinghaus ST, Freire R, Jackson SP, Barlow C, Wynshaw-Boris A. The association of ATR protein with mouse meiotic chromosome cores. Chromosoma. 1999;108:95–102. doi: 10.1007/s004120050356. [DOI] [PubMed] [Google Scholar]
- Mortillaro MJ, Blencowe BJ, Wei X, Nakayasu H, Du L, Warren SL, Sharp PA, Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JM, Lindsay HD, Munday CA, Carr AM. Role of Schizosaccharomyces pombeRecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol Cell Biol. 1997;17:6868–6875. doi: 10.1128/mcb.17.12.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Peters JM, Uhlmann F. Splitting the chromosome: cutting the ties that bind sister chromatids. Science. 2000;288:1379–1385. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- Schmiesing JA, Ball AR, Jr, Gregson HC, Alderton JM, Zhou S, Yokomori K. Identification of two distinct human SMC protein complexes involved in mitotic chromosome dynamics. Proc Natl Acad Sci USA. 1998;95:12906–12911. doi: 10.1073/pnas.95.22.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Shirataki H, Honda T, Minami S, Takai Y. Complex formation of SMAP/KAP3, a KIF3A/B ATPase motor-associated protein, with a human chromosome-associated polypeptide. J Biol Chem. 1998;273:6591–6594. doi: 10.1074/jbc.273.12.6591. [DOI] [PubMed] [Google Scholar]
- Smith-Ravin J, Jeggo PA. Use of damaged plasmid to study DNA repair in x-ray sensitive (xrs) strains of Chinese hamster ovary (CHO) cells. Int J Radiat Biol. 1989;56:951–961. doi: 10.1080/09553008914552411. [DOI] [PubMed] [Google Scholar]
- Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Strunnikov AV, Hogan E, Koshland D. SMC2, a Saccharomyces cerevisiaegene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev. 1995;9:587–599. doi: 10.1101/gad.9.5.587. [DOI] [PubMed] [Google Scholar]
- Strunnikov AV, Jessberger R. Structural maintenance of chromosomes (SMC) proteins: conserved molecular properties for multiple biological functions. Eur J Biochem. 1999;263:6–13. doi: 10.1046/j.1432-1327.1999.00509.x. [DOI] [PubMed] [Google Scholar]
- Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K, Yanagida M. Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 1999;13:2271–2283. doi: 10.1101/gad.13.17.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi S, Sakuraba Y, Masuyama S, Inoue H, Yamaizumi M. Dysfunction of human Rad18 results in defective postreplication repair and hypersensitivity to multiple mutagens. Proc Natl Acad Sci USA. 2000;97:7927–7932. doi: 10.1073/pnas.97.14.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkade HM, Bugg SJ, Lindsay HD, Carr AM, O'Connell MJ. Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol Biol Cell. 1999;10:2905–2918. doi: 10.1091/mbc.10.9.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]