Abstract
While protein-surface interactions have been widely studied, relatively little is understood at this time regarding how protein-surface interaction effects are influenced by protein-protein interactions and how these effects combine with the internal stability of a protein to influence its adsorbed-state structure and bioactivity. The objectives of this study were to develop a method to study these combined effects under widely varying protein-protein interaction conditions using hen egg-white lysozyme (HEWL) adsorbed on silica glass, poly(methyl methacrylate), and polyethylene as our model systems. In order to vary protein-protein interaction effects over a wide range, HEWL was first adsorbed to each surface type under widely varying protein solution concentrations for 2 h to saturate the surface, followed by immersion in pure buffer solution for 15 h to equilibrate the adsorbed protein layers in the absence of additionally adsorbing protein. Periodic measurements were made at selected time points of the areal density of the adsorbed protein layer as an indicator of the level of protein-protein interaction effects within the layer, and these values were then correlated with measurements of the adsorbed protein’s secondary structure and bioactivity. The results from these studies indicate that protein-protein interaction effects help stabilize the structure of HEWL adsorbed on silica glass, have little influence on the structural behavior of HEWL on HDPE, and actually serve to destabilize HEWL’s structure on PMMA. The bioactivity of HEWL on silica glass and HDPE was found to decrease in direct proportion to the degree of adsorption-induce protein unfolding. A direct correlation between bioactivity and the conformational state of adsorbed HEWL was less apparent on PMMA, thus suggesting that other factors influenced HEWL’s bioactivity on this surface, such as the accessibility of HEWL’s bioactive site being blocked by neighboring proteins or the surface itself. The developed methods provide an effective means to characterize the influence of protein-protein interaction effects and provide new molecular-level insights into how protein-protein interaction effects combine with protein-surface interaction and internal protein stability effects to influence the structure and bioactivity of adsorbed protein.
I. INTRODUCTION
The interaction of proteins with material surfaces is of primary importance in many areas of biotechnology and biomedical engineering, including biosensors, enzyme based technologies, tissue engineering and regenerative medicine, implants, and biodefense. The key element in all of these applications is the bioactive state of the protein, which can be strongly influenced by adsorption-induced changes in a protein’s structure on an adsorbent surface. While much work on this topic has already been reported, a fundamental understanding on the role of different material surfaces on the conformational state, packing arrangement, and bioactivity of adsorbed proteins is still not well understood. These limitations are partly due to the complexities introduced by protein-protein interactions on the adsorption responses of proteins with various levels of internal protein stability in combination with protein-surface interactions [1-3].
As previously described by Norde [4-6] and others [7-11], when a material is exposed to a protein-containing solution, proteins rapidly adsorb to its surfaces. Once adsorbed, forces between the protein, surface, and solvent (e.g., electrostatic, hydrogen bonding, hydrophobic, and/or dispersion interactions) can alter the thermodynamic state of the system leading to spontaneous shifts in an adsorbed protein’s structure from its native state and subsequent unfolding and spreading out on the surface. The amount that an adsorbed protein will unfold and spread out on a surface is largely determined by the strength of the protein-surface interactions relative to the internal stability of the protein. The extent to which unfolding will occur is also influenced by whether or not the adjacent areas of the surface are occupied by other adsorbed proteins and subsequent interactions with such neighboring proteins; which, when present, result in protein-protein interactions that tend to sterically block further unfolding and spreading. The degree to which protein-protein interaction effects limit the unfolding or spreading of a protein on a surface can thus be simply controlled by adjusting the concentration of the protein in solution, which influences the rate that neighboring sites are filled.
At surface saturation, the conformational state of the final resulting adsorbed layer of protein will thus be dependent on the combined influences of internal protein stability, protein-surface interaction, and protein-protein interaction effects. Protein-protein interaction effects are the least understood of these types of interactions and can be generally expected to be proportional to the amount of the protein adsorbed on the surface (i.e., areal density of the protein on the surface) [2]. Based on this assumption, the influence of protein-protein interaction effects on the structure of adsorbed protein for a given type of surface should be able to be assessed by adsorbing the protein to the surface under conditions that will provide different degrees of areal density, which can be controlled for a given surface by varying the protein solution concentration from which the protein is adsorbed, with higher solution concentrations generally resulting in higher areal densities at surface saturation [12].
The objective of this research was therefore to study the influence of protein-protein interaction effects on the structural changes and corresponding bioactivity of adsorbed protein on three different surface chemistries, each with the potential to interact with proteins through a distinctly different molecular mechanism. The experimental approach that we designed to address these issues was to first adsorb the protein from varying solution concentrations for a period of time previously determined to be sufficient to saturate the surface (adsorption time) in order to vary the initial areal density of protein on the surface and the subsequent degree of protein-protein interaction effects occurring within the adsorbed layer of protein. We then rinsed the surfaces with pure buffer to remove weakly adsorbed proteins, replaced the protein solution with pure buffer solution to remove the ability of new proteins to adsorb to the surface, and allowed the adsorbed protein layers to equilibrate under pure buffer conditions while monitoring their areal density and conformational structure by measuring the shift in absorbance and circular dichroism (CD), respectively, until they stabilized to an apparent equilibrated state (equilibration time). Following equilibration, bioactivity studies were then finally conducted to quantify the influence of the applied adsorption processes on the bioactive state of the adsorbed protein. Under these experimental conditions, differences in the optical characteristics of the adsorbed HEWL layers with different areal densities on a given surface can be considered to occur under constant internal protein stability and protein-surface interaction conditions, thus isolating the influence of protein-protein interaction effects on the structural and bioactive response of the adsorbed protein.
II. EXPERIMENTAL METHODS
II.a. Protein and Material Surfaces
Hen egg white lysozyme (HEWL) was selected for use in this study as one of the most well characterized protein model systems [12-19]. Being a small (MW 14 kDa) relatively ‘hard’ protein with 4 disulfide bonds stabilizing its structure, HEWL is generally considered to be a protein with relatively high internal stability (i.e., high internal protein stability) [2, 18].
The selected material surfaces included fused silica glass (glass), high density polyethylene (HDPE), and poly(methyl methacrylate) (PMMA). These three materials were chosen to represent some of the most commonly used materials in biotechnological and biomedical engineering applications [19-25]. They were also selected because their chemical compositions provide them with the potential to interact with proteins by three distinctly different mechanisms.
Being composed of a silicon-oxygen network with a high density of hydroxyl groups on the surface, the glass surface has strong potential to form both accepting and donating-type hydrogen bonds with hydrogen bondable groups of a protein as well as ionic groups for electrostatic interactions. Because hydrogen bonds stabilize the secondary structures of a protein (as well as playing a role in tertiary structural stability), this type of surface thus has the potential to substantially destabilize a protein’s secondary and tertiary structures by competing with the hydrogen bonds that serve to stabilize the protein’s internal structure.
In contrast to glass, HDPE is entirely composed of saturated nonpolar alkane chains, thus lacking the ability to interact with a protein via either hydrogen bonding or electrostatic effects, while having the potential to exhibit strong hydrophobic interactions with a protein’s hydrophobic amino acid residues. Given the fact that the tertiary structure of a protein is generally stabilized by hydrophobic interactions, HDPE thus has the potential to strongly induce tertiary unfolding of a protein, which in turn can be expected to potentially destabilize the native secondary structures as well.
Our third surface, PMMA, can be considered to have much lower potential to interact with the secondary structure of proteins compared to the glass surfaces since it has a much lower density of hydrogen bondable groups, with these representing only hydrogen-bond-accepting groups but not hydrogen-bond-donating groups. In addition, because the hydrogen bondable groups that are present in PMMA subsequently reduce the hydrophobicity of the surface, it can be expected to exhibit weaker hydrophobic interactions with proteins compared with HDPE. Therefore, theoretically, PMMA should exhibit lower protein-surface interaction effects than either glass or HDPE, with the greater protein-surface interaction effects from glass and HDPE occurring through distinctly different mechanisms.
II.b. Material Surface Preparation and Characterization
II.b.1. Preparation of Material Surfaces
Custom cut glass slides (0.375″ × 1.625″ × 0.0625″, Chemglass Life Sciences) were procured to fit our custom designed CD cuvettes[12]. HDPE and PMMA surfaces were spin–coated onto glass slides from dodecalin (0.5% (w/w) at 1500 rpm for 60s) and chloroform solutions (1.5% (w/w) at 1000 rpm for 60s), respectively. All chemicals including the HDPE (Mw =125,000, Sigma 181900) and PMMA (Mw=350,000, Sigma 445746) and the solvents such as dodecalin (Sigma 294772) and chloroform (EMD Chemicals, CX 1054) were used as supplied by the manufacturer.
Glass substrates used for adsorption studies were cleaned by sonicating in “piranha” (7:3 (v/v) H2SO4 (EMD Chemicals, SX 1244)/H2O2 (Ricca Chemicals, 3821) and basic solution (1:1:3 (v/v/v) NH4OH (BDH Chemicals, BDH3016)/ H2O2/ H2O) at 50°C for 1 minute. Prior to adsorption studies, all the substrates were rinsed in absolute ethanol, followed by nanopure water and then dried under a steady stream of nitrogen gas.
II.b.2. Characterization of Material Surfaces
Surface characterization was performed to determine the static air–water contact angle, atomic composition, film thickness, and surface roughness of the substrates used. For all the surfaces, the static air–water contact angle values were analyzed using a contact–angle goniometer (Kruss, DSA-20E). Similarly, the atomic compositions were verified via X–ray photoelectron spectroscopy (NESCA/BIO, University of Washington) and the average surface roughness was analyzed using atomic force microscopy (Asylum Research, MFP–3D) over an area of 5μm×5μm. The thicknesses of the polymer films were characterized using variable angle spectroscopic ellipsometry (Sopra Inc., GES–5).
II.c. Protein Adsorption and Equilibration
II.c.1 Protein Adsorption
Stock solutions of 5.0 mg/ml HEWL (Sigma, L6876) were prepared in 10 mM potassium phosphate buffer (PPB) and filtered to remove any insoluble aggregates. The final protein concentrations were verified via biuret method (Thermo scientific, 23225) or absorbance at 205 nm (A205)[14, 26]. PPB was prepared by mixing appropriate amounts of monobasic potassium phosphate (Sigma, P8708) and dibasic potassium phosphate (Sigma, P8508) to a final solution pH of 7.4.
All adsorbent surfaces were first incubated in PPB at room temperature and then the required amount of protein stock solution was pipetted into the buffer to make up to the desired bulk solution concentration, with care taken to ensure that the pipet tip was below the air–water interface during injection to avoid denaturation of the protein at this interface. As previously mentioned, the effects of protein-protein interaction on the adsorption responses of protein on glass, PMMA and HDPE surfaces were then varied by controlling the adsorption-desorption kinetic parameters, namely (i) the protein concentration in solution from which the protein was adsorbed, (ii) the time that the surfaces were exposed to the protein solution, and (iii) the equilibration time following adsorption with the adsorbed protein layers immersed in protein-free buffer solution.
II.c.2. Bulk Concentrations of Protein for Adsorption Studies
Protein adsorption was conducted at room temperature under eight different protein solution concentrations (0.03 mg/ml, 0.05 mg/ml, 0.10 mg/ml, 0.20 mg/ml, 0.40 mg/ml, 0.60 mg/ml, 0.80 mg/ml and 1.00 mg/ml) following the adsorption process described in section S.1 of the supporting information.
II.c.3. Adsorption Time in Protein Solution
For the protein solution concentrations used in this study, an adsorption time of 2 hours was determined to be sufficient to saturate the surface for each material and protein solution concentration (see S.2 of the supporting information), which is consistent with the previous reports on the adsorption time required by HEWL to saturate an adsorbent surface for a wide range of solution concentrations [13, 15].
II.c.4. Equilibration Time in Pure Buffer Solution
Following the adsorption process in the respective protein solution concentrations, the material surfaces were subsequently washed under a steady gentle flow (12 mL/min) of protein-free buffer for five minutes in order to remove the bulk protein solution and to desorb loosely adherent proteins. The surfaces with the adsorbed layer of protein were then immersed in protein-free buffer solutions for 15 hours to allow the adsorbed protein layers to structurally equilibrate.
II.d. Analysis of Adsorbed Proteins Using CD Spectroscopy
The structure of HEWL in solution, the amount of protein adsorbed on each surface, and the subsequent adsorption-induced conformational changes on these proteins on each material surface were determined using CD spectropolarimetry following our standardized methods [12]. The CD spectra (consisting of the ellipticity and absorbance values over wavelengths ranging from 190 nm to 300 nm) were obtained at room temperature using a Jasco J-810 spectropolarimeter.
The solution structure of the proteins was determined in quartz cuvettes (Starna Cells) while the structure of the adsorbed proteins was determined using a custom-designed cuvette, which has been previously described [12]. The CD instrument as well as the path length of the cuvettes used in this study were calibrated to be within the recommend standards [14] prior to these analyses (see section S.3 of the supporting information).
II.d.1. Determination of Molar Extinction Coefficient of Protein at 205 nm and Structure of Protein in Solution
The molar extinction coefficient of the protein (ε205) in solution at 205 nm was determined by recording the background corrected absorbance at this wavelength (A205) for five different solution concentrations (0.20 mg/ml, 0.40 mg/ml, 0.60 mg/ml, 0.80 mg/ml and 1.00 mg/ml) in 0.10 mm pathlength (L) demountable quartz cuvettes (Starna). The solution concentrations (Csoln) were first verified using the biuret method (Thermo scientific, 23225)[26], following which the molar extinction coefficient of protein in solution at 205 nm was obtained from the slope of the absorbance (A205) vs (Csoln*L) plot.
The CD spectrum for protein in solution was then measured in the same 0.10 mm pathlength demountable quartz cuvette (Starna) at 1.00 mg/ml solution concentration using parameters and techniques previously described [12]. Briefly, the background-corrected solution CD spectra were recorded from 190 nm to 300 nm at a scan rate of 50 nm/min with a response time of 0.25 sec using six accumulations, with the CD spectra then analyzed using the methods described in section II.d.3 below.
II.d.2. Determination of the Surface Density of Adsorbed Protein (Qads) and Adsorbed Protein Structure
The slides supporting the material surfaces with the adsorbed protein layers were transferred into custom-designed cuvettes and the CD spectra were recorded before and after protein adsorption. Throughout the study, slides remained hydrate in buffer solution. Following our established methods [12], the absorbance is dependent only on the total mass of protein per unit area that the polarized light beam passes through. The areal surface density of adsorbed protein (Qads) was thus estimated by the following equation:
| (1) |
where A205 is the background-corrected absorbance at 205 nm, and ε205 is the molar extinction coefficient that was determined for the protein solution at a wavelength of 205 nm in section II.d.1.
II.d.3. Quantification of Secondary Structure in Solution and Adsorbed State of Protein
The background corrected CD signals that were obtained as described above in sections II.d.1 and II.d.2 were converted to molar ellipticity (θmol) using equations 2 and 3, respectively:
| (2) |
| (3) |
where θraw is the background corrected raw CD signal, L is the path length of the cuvette (cm), Csoln is the solution concentration of the protein (g/mL), Qads is the surface density of adsorbed protein (g/cm2), and M is the mean residue molecular weight of 115 g/mol.
Once the CD signals were converted to their respective molar ellipticity units, the spectra were then deconvoluted to predict secondary structure using the CONTIN/LL, SELCON3, and CDSStr methods provided with the CDPro package using the SP43 and SP48 protein reference datasets[16]. Each of the deconvoluted spectra was then assessed for quality by analyzing the R-fit using non-linear regression [16]. The final secondary structures represent the averaged structures obtained from all of the reliable outputs (R-fit < 10) resulting from the above described data analysis methods, which are consistent with the data analysis recommendations for CD [14, 17]. For the purposes of this study, we were primarily interested in the percent helical structure of HEWL as a sensitive indicator of adsorption-induced changes in the HEWL’s structure.
II.e. Bioactivity Assays of HEWL
We used a turbidometric assay to measure the enzymatic activity of HEWL, which was carried out using a custom-designed cuvette that was previously described[12, 27]. Bioactive substrates were prepared in PPB to a final concentration of 60 mg/liter Micrococcus lysodeikticus (Sigma M3770) and the assays to determine the enzymatic bioactivity were done at pH 7.4 for a time period of 10 min at 450 nm. Prior to performing the assay, samples were incubated with the bioactive substrate at room temperature for 1 min before the decreases in absorbance at 450 nm (ΔA450) were recorded.
II.e.1. Solution-State Bioactivity of HEWL
The solution-state activity assays were determined by estimating the activity rate of HEWL when injected immediately into the custom designed cuvette containing bare substrates without any adsorbed protein. For the bioactive substrate concentration and the experimental conditions that were used in our current study, the activity rate was found to be linear for each of the surfaces over the protein mass range of 0.1 μg – 30 μg, suggesting that the protein’s active site was not saturated by the bioactive substrate at these concentrations (see section S.5 of the supporting information).
The specific activities of the proteins in solution were subsequently calculated by normalizing ΔA450 to the amount of protein in solution, with the values found to be constant over the working mass range used in these studies.
II.e.2. Adsorbed-State Bioactivity of HEWL
Due to concerns that the bioactivity assay may have some unappreciated influence on the layers of adsorbed protein, bioactivity assays were only performed at the end of the experiment for the HEWL layers after the 15 h equilibration period under pure buffer conditions. The bioactivities of the adsorbed proteins were then assayed after the completion of the CD analyses. The amount of adsorbed protein was quantified by the layer’s absorbance at 205 nm (A205), both before and after the bioactivity assays were performed to ensure that the bioactivity assays did not cause a measureable amount of the protein to be desorbed from the adsorbent surface. The specific activities of the adsorbed proteins were then calculated by normalizing the ΔA450 absorbance values by the total amount of protein adsorbed on the surface (Qads* area of adsorbent surface). The relative bioactivities (%) of the adsorbed proteins were then determined by normalizing the measured adsorbed-state specific activity to the protein’s solution-state specific activity.
II.f. Statistical Analysis
The results from this study are presented as the mean values ± 95% confidence intervals (C.I.). The statistical significance of differences between mean values for different samples/conditions was evaluated using either the Student’s t test or a nonparametric sign test [28, 29], with values of p < 0.05 being considered as statistically significant.
III.RESULTS AND DISCUSSION
III.a. Surface Characterization
Table 1 presents the results analyzed by the characterization techniques applied to the surfaces used in this study. All of the measured values reported in Table 1 fall within the expected range [20-24].
Table 1.
Surface characterization: Atomic composition, static contact angle, film thickness, and surface roughness analyses for each surface. (Mean ± 95% C.I., N = 3.)
| Surface Moiety |
C (%) | S (%) | N (%) | O (%) | Roughness (nm) |
Contact Angle (°) |
Thickness (nm) |
|---|---|---|---|---|---|---|---|
| GLASS ** | 25.0 (2.0) | * | < 1.0 | 49.0 (2.0) | < 10.0 | 23 (4) | NA |
| PMMA | 76.0 (1.0) | * | * | 24.0 (1.0) | < 1.5 | 63 (3) | 90 (10) |
| HDPE | 96.0 (3.0) | * | * | 3.0 (3.0) | < 8.0 | 97 (5) | 100 (10) |
indicates a negligible value;
Fused glass slide also contains Zn (<1%), Al (<1%) and Si (22.0±1.0%). The presence of extra carbon composition is believed to be originating from surface contamination due to the exposure of samples to air after the cleaning procedure. These are the typical adventitious and unavoidable hydrocarbon impurities that adsorb spontaneously from ambient air onto the GLASS surfaces [24].; NA refers to the thickness of the GLASS slides described in II.a.1.
III.b. Areal Density of Adsorbed Protein and Protein-Protein Effects
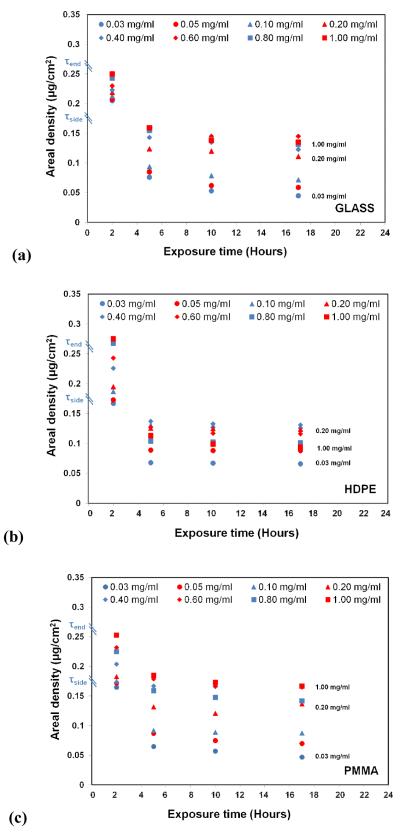
Figure 1 presents a plot of the areal density of the adsorbed HEWL on each of our three surfaces for each solution concentration. As these plots clearly show, 2 h of exposure to the protein solutions resulted in very similar areal densities for each surface, which fall within the areal densities corresponding to the theoretical limits for a saturated surface for a monolayer of HEWL organized in a close-packed side-on orientation (0.17 μg/cm2) and close-packed end-on orientation (0.26 μg/cm2) [13]. It is also important to note that the surface densities at this time-point generally increased with increasing solution concentration for each surface between these two theoretical values, with protein-protein interaction effects considered to increase in magnitude with increased surface density. The fact that the distribution of initial areal densities of adsorbed HEWL was quite similar for each of our three materials also indicates that, on average, the protein-protein interaction effects were initially quite similar for each type of surface.
Figure 1.
Effect of varying exposure time on the surface density of HEWL adsorbed on (a) glass, (b) HDPE, and (c) PMMA surfaces. (Exposure time point (e.g., n hours; n≥2) represents 2 h exposure under the designated protein solution concentration followed by (n-2) hours of equilibration in PPB) (N=3; averaged 95% C.I. = ±0.032 μg/cm2 for areal density measurements). τside (0.17 μg/cm2 and τend (0.26 μg/cm2) refers to the theoretical full surface coverage of HEWL for adsorption in ‘side-on’ and ‘end-on’ orientations, respectively [13]. The decrease in areal density on each of the surface from 2-5 h, 5-10 h, and 10-17 h are statistically significant at p < 0.05 per the non-parametric sign test [28, 29] (see Table S.2. of the supporting information for the raw data in Fig 1).
As indicated by the 5 h time point in Figure 1, when the layers of adsorbed lysozyme were allowed to relax for three hours under pure buffer solution conditions, the areal densities of each layer spontaneously decreased to values at or below that for a close-packed side-on orientation. Following these shifts, the areal densities were still found to be widely distributed, thus continuing to provide a range of protein-protein interactions for each surface type. The areal densities then appeared to stabilize with relatively little further change with continued exposure time. However, comparison of the mean values of the concentrations on each of the surfaces between the 5-10 h and 10-17 h time points using a nonparametric sign test [28, 29] reveals that the consistent slight decreases in areal density over time do actually represent a statistically significant difference (p < 0.05), thus showing that the lysozyme continued to desorb from the surface at a very slow rate. Most importantly, the results for each time point during this equilibration phase of the experiments continued to provide a broad range of areal densities, which we assume to proportionally correspond to a broad range of protein-protein interaction effects within the adsorbed protein layers.
III.c Adsorption-Induced Changes in Protein Secondary Structure
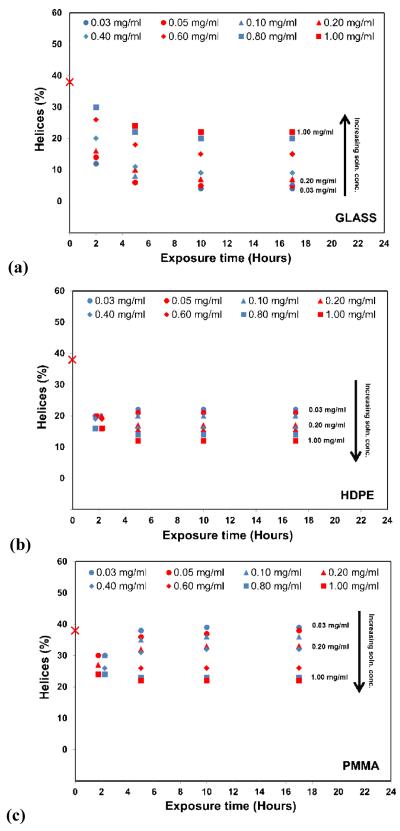
The influence of adsorption conditions on the secondary structure of the adsorbed HEWL is presented in Figure 2. The percent helical structure shown in Figure 2 corresponding to the 0 h exposure times represents the native helical content of the protein in solution (38 ± 2% helix), with the subsequent time points representing the average helical structure of the protein layers in the adsorbed state. The 2 h time point represents the structure of the saturated layers of the adsorbed protein after 2 h exposure to their respective protein solution concentrations followed by rinsing in pure buffer to remove loosely bound proteins, while the time points after 2 h represent the time given the protein to equilibrate following adsorption while immersed in pure buffer solution (e.g., 5 h time point represents 2 h exposure under the designated protein solution concentration followed by 3 hours of equilibration in PPB).
Figure 2.
Helical content of adsorbed HEWL on (a) glass, (b) HDPE, and (c) PMMA surface as a function of exposure time. Symbols represent the different protein solution concentrations that were used to adsorb the HEWL to each surface. Zero time point represents the native helical structure of HEWL in solution, which was 38% (± 2%), consistent with the reported secondary structure [30]. (N = 3, averaged 95% C.I. values = ±4 % helicity for each data point). Data points for the 2 h time point for (b) and (c) have been shifted slightly for visualization due to overlap. The black arrows indicate the direction of increasing solution concentration from which the protein was adsorbed. (see Table S.4. of the supporting information for the raw data in Fig. 2.)
As shown in Figure 2, adsorption of HEWL to each surface following 2 h exposure to the protein solution resulted in a significant reduction of helical secondary structure for each surface and for each solution concentration, which reflects the combined influences of protein-surface interaction, protein-protein interaction, and internal protein stability effects upon HEWL. Comparisons of the adsorption response at 2 h between these three surfaces show some interesting differences. In particular, the solution concentration from which HEWL was adsorbed had a very strong influence on the adsorbed structure on glass, with greater helicity being retained for adsorption from increased solution concentration. In distinct contrast to this, while a significant loss in helical structure upon adsorption also occurred on the HDPE and PMMA surfaces, the range of the influence of solution concentration was much reduced. In addition, the increase in solution concentration had very different effects on the protein’s structure on these surfaces compared to the glass surface. On PMMA, there was actually significantly reduction in the helical structure for the HEWL layers adsorbed from solutions of higher protein concentration, while there was no significant difference in the drop in the percent helical structure between layers adsorbed from different solution concentrations for the HDPE surface.
The data shown in Figure 2 for the exposure times of 5, 10, and 17 h represent the structural response of HEWL during the 15 hours of equilibration in the pure buffer following the 2 h adsorption period. As shown in Figure 2, the structure of the HEWL on each of these surfaces underwent significant further changes between the 2 h and 5 h overall exposure times, but then generally tended to stabilized at the 5 h time point. Subsequent changes in the percent helicity between the 5 h, 10 h, and 17 h time points was not significantly different (p > 0.05) for any of our surfaces except for the transition from the 5 h to the 10 h time exposures times for HEWL on glass, which showed slight but still statistically significant further decreases in helicity (p < 0.01; nonparametric sign test [28, 29]).
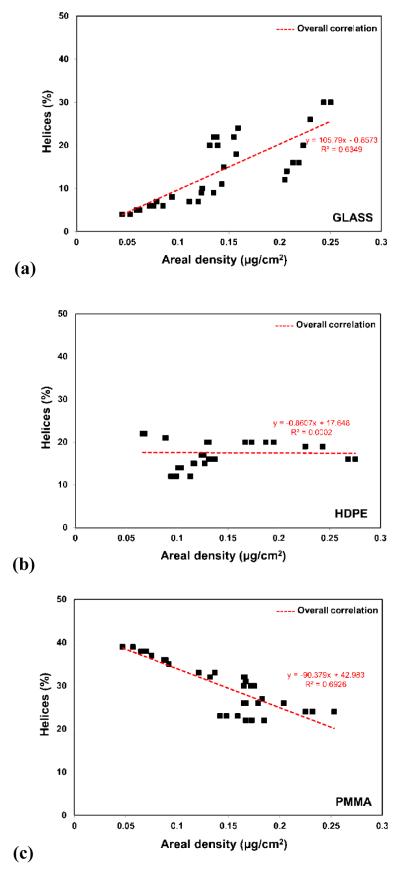
As with the 2 h results, comparisons of the structural behavior of HEWL between our three surfaces for the 5, 10, and 17 h time points suggest that HEWL behaved distinctly differently on each different type of surface, with the influence of the protein solution concentration from which the HEWL was adsorbed having the opposite effect on PMMA as it did on glass, with relatively little effect on HDPE. In order explore the influence of protein-protein interaction effects more directly based on our assumption that the degree of protein-protein interaction effects within the adsorbed HEWL layers is directly proportional to the areal density of the layer, the values of the percent helical structure for the data shown in Figure 2 were replotted against the areal densities for the HEWL layers from Figure 1, with these plots presented in Figure 3.
Figure 3.
HEWL % helicity (y-axis) vs. areal densities (x-axis) of the adsorbed HEWL layers on (a) glass, (b) HDPE, and (c) PMMA. (N=3; averaged 95% C.I. values of ±4 % for helicity and ±0.032 μg/cm2 for areal density.) (Table S.5. in supporting information provides the raw data in Fig. 3.)
The data presented in Figure 3 provide a much clearer picture of the influence of protein-protein interaction effects (as represented by the areal density) on the stability of the adsorbed HEWL (as represented by the percent helicity) for each of our three surfaces. On the glass surface, increased areal density of the adsorbed protein layer is clearly shown to stabilize the protein against protein-surface interaction-induced unfolding with only a 20% decrease in the native-state % helicity (38% to 30%) for the highest areal density (0.25 μg/cm2, reflecting a close-packed end-on structure; see Figure 2), while at the lowest areal density (0.045 mg/cm2, reflecting an areal density 4x lower than that for a close-packed side-on structure) caused the helicity to decrease all the way down to only 4% (i.e., 90% loss in helicity). As shown in Figure 3.b, changes in areal density of HEWL on HDPE had minimal influence on its helical structure with the percent helicity ranging from 12 to 22% with an average of about 18% helicity (53% loss in % helicity). As the most interesting (and unexpected) result, protein-protein interaction effects are shown to have the opposite effect on HEWL on the PMMA surface compared with glass. On the PMMA surface, when protein-protein interaction effects were decreased by the displacement of proteins from the surface from their initial saturated condition, HEWL actually refolded to regain the percentage of helicity lost following the initial adsorption process to attain a percent helicity equal to its native-state solution structure.
Based on these combined results, we hypothesize the following general molecular-level relationships between protein-surface, internal protein stability, and protein-protein interaction effects in order to explain the observed behavior presented in Figure 3. On surfaces with a large density of hydrogen bondable and ionic groups, such as glass, protein-surface interaction effects will occur in the form of competition of the surface hydrogen bondable groups with the hydrogen bonds that stabilize the secondary helical structure of the protein [31], thus tending to destabilize the helical structures of the protein on the surface. protein-protein interaction effects, in turn, tend to restrict the conformational freedom of neighboring proteins thus providing a stabilizing force that helps to inhibit the unfolding process induced by protein-surface interaction effects. Therefore under low areal density conditions when protein-protein interaction effects are minimized, protein-surface interaction effects tend to overcome internal protein stability effects, leading to substantial unfolding of the protein on this type of surface. Similarly, under high areal density conditions, protein-protein interaction effects couple with the internal protein stability effects to help stabilize the protein, thus limiting the degree of unfolding that occurs.
In contrast to surfaces with a high density of hydrogen-bondable and ionic groups on the surface, highly hydrophobic surfaces, such as HDPE, which do not have hydrogen bonding capability, have the potential to strongly interact with the hydrophobic amino acid residues that typically stabilize a protein’s tertiary structure [31]. Because of the substantial thermodynamic driving force behind hydrophobic interactions in aqueous solution, this type of protein-surface interaction effect dominates over both internal protein stability and protein-protein interaction effects for HEWL, with protein-protein interaction effects then having relatively little influence on the degree of protein unfolding that occurs upon adsorption.
As a third category of surface chemistry, surfaces with moderate density of hydrogen-bondable and/or charged groups have only moderate capability to form hydrogen bonds with the protein, while also tending to be only moderately hydrophobic. These types of surfaces can thus be expected to exhibit relatively weak protein-surface interaction effects, with only moderate tendency to disrupt both the hydrogen bonds that stabilize the helical secondary structures of the adsorbed protein and moderate tendency to compete with the hydrophobic interactions that tend to stabilize the protein’s tertiary structure. We propose that the presence of high protein-protein interaction effects on a protein adsorbed on this type of surface will apply additional in-plane compressive forces on the protein that will tend to destabilize its structure, thus causing protein-protein interaction effects to contribute to the unfolding of the protein. However, if the strong protein-protein interaction effects contributing to this destabilization are then removed, a protein with high internal protein stability (such as HEWL) will tend to overcome the weak protein-surface interaction effects working to unfold the protein on the surface, resulting in a tendency for the protein to actually refold back towards its native structure. Although speculative at this time, this hypothesized interplay between internal protein stability, protein-surface interaction, and protein-protein interaction effects is consistent with the results obtained from this present study.
III.d Adsorption-Induced Changes in Protein Bioactivity
The key element in most applications where protein-surface interactions are important is the bioactive state of the adsorbed protein. Protein bioactivity is primarily determined by the structure and accessibility of a protein’s bioactive site, both of which are influenced by the combination of protein-surface interaction, protein-protein interaction, and internal protein stability effects in an adsorbed protein layer. In this section, results for the influence of these interactions on the conformation and bioactivity of adsorbed HEWL on our three material surfaces are presented.
III.d.1 Relationship between the Conformation and Bioactivity of Adsorbed HEWL under Varying Protein-Protein Interaction and Protein-Surface Interaction Conditions
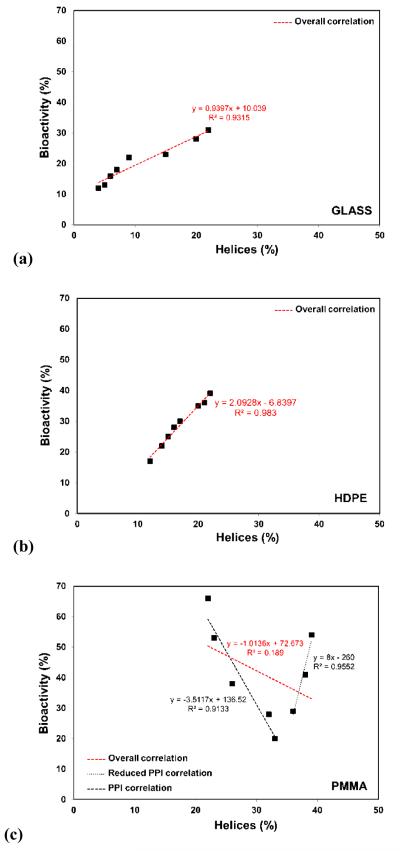
Figure 4 presents the bioactivities of the adsorbed HEWL expressed as a percentage of its solution bioactivity vs. its percent helicity for each of our three surfaces for the layers of adsorbed HEWL at the 17 h exposure time (i.e., 2 h immersion in protein solution followed by 15 h equilibration in pure buffer solution).
Figure 4.
Bioactivity vs. percent helicity for adsorbed HEWL on (a) glass, (b) HDPE, and (c) PMMA for 17 h exposure time period. Three separate correlation lines given in (c) represent the correlation between bioactivity and percent helical structure of lysozyme for percent helicity < 35%, > 35%, and overall. (N=3; averaged 95% C.I. values of ±4% for helicity and ±9% for bioactivity.)
As shown in Figures 4.a (glass) and 4.b (HDPE), HEWL loses more than 60% of its bioactivity following adsorption, with an apparent direct linear correlation between the retained relative bioactivity of adsorbed HEWL and its percent helicity. These results suggest that on both of these surfaces, HEWL bioactivity is primarily being influenced by the structure of the bioactive site, with loss in helical structure from the native state structure, which is 38% helix, reflecting the degree of conformational distortion of the bioactive site. Comparing these results to the results shown in Figure 3 (% helicity vs. areal density), suggests that on glass, protein-protein interaction effects primarily influence HEWL bioactivity by acting to inhibit protein-surface interaction-induced unfolding, which in turn helps preserve the structure of the bioactive site. Alternatively, on HDPE, for which the adsorption response was found to be dominated by protein-surface interaction effects, the loss in percent helicity of HEWL results in loss of bioactivity, presumably through concomitant structural distortions in the structure of the bioactive site, but with little influence from protein-protein interaction effects.
The bioactive response of HEWL vs. percent helicity when adsorbed on PMMA was again quite surprising and distinctly different from the behavior of HEWL on either glass or HDPE. As indicated from the results presented in Figure 3, when protein-protein interaction effects were minimized, HEWL was able to regain a percent helicity that was not significantly different than that of its native structure (38% helicity). As shown in Figure 4.c, the adsorbed HEWL retained up to 55% of its solution-state bioactivity under these conditions, but with its percent bioactivity rapidly decreasing to as low as only 20% for relatively small decreases in % helicity from that point. A strong correlation is indicated between bioactivity and helicity as the % helicity decreased, but only down to a value of about 35%. The apparent correlation then reversed, with bioactivity rapidly increasing as helicity further decreased down to a value as low as about 20%. Thus, as indicated in Figure 4c, while a low correlation is found between the overall results for bioactivity and % helicity, the data suggests two subdomains within the overall data set that exhibit strong correlation, but in the opposite directions to one another. Surprisingly, HEWL exhibited the highest percent bioactivity at its lowest percent helicity, which, as shown in Figure 3, corresponds to conditions of the strongest protein-protein interaction effects. These results suggest that the loss in bioactivity of HEWL on PMMA is not just a result of adsorption-induced structural changes in its bioactive site. Other factors are evidently playing a substantial role, such as possibly the influence of protein-protein interaction effects on the protein’s orientation on the surface and the subsequent accessibility of the bioactive site for the substrate used in the bioactive assay. It is also, of course, possible that the combination of protein-surface interaction, protein-protein interaction, and internal protein stability effects on this surface result in structural distortions of the bioactive site in HEWL that are not directly reflected by the overall helicity of the protein on this surface. At this time, we can only speculate on explanations for this behavior and further studies are required to provide additional understanding of these intriguing results.
IV. CONCLUSION
A new experimental approach has been developed and applied to study the combined influence of protein-surface interactions, protein-protein interactions, and internal protein stability on the conformational behavior and bioactivity of adsorbed protein. In this paper, we present the first application of the developed methods to characterize the adsorption response of HEWL on glass, HDPE and PMMA surfaces, with these surfaces selected to provide three characteristically different molecular mechanisms for their interactions with the protein.
The results from the structural studies indicate that protein-protein interaction effects tend to (1) stabilize the structure of HEWL on a silica glass surface, which can be expected to exhibit strong hydrogen bond and electrostatic interactions with proteins; (2) have little influence on the structure of HEWL on strongly hydrophobic surfaces, such as HDPE; and (3) actually destabilize the structure of HEWL on a PMMA surface, which has only moderate hydrogen bonding and hydrophobic character. Furthermore, the results of the bioactivity studies indicate that protein-protein interaction effects play a less direct role on the bioactivity of adsorbed HEWL through their influence on the adsorbed protein’s structure, with bioactivity reducing in direct proportion to the degree of adsorption-induced disruption of the protein’s structure, as indicated for the glass and HDPE surfaces. However, protein-protein interaction effects may also influence adsorbed-state bioactivity by affecting the accessibility of the protein’s bioactive site, either by directly blocking access or by influencing the orientation of the protein such that access to the bioactive site is blocked by the surface itself. We speculate that these later effects may be responsible for the lack of a clear overall correlation between bioactivity and adsorbed structure of HEWL on PMMA.
Supplementary Material
Highlights.
Influence of surface chemistry and protein-protein effects on protein adsorption
Protein-protein effects are strongly influenced by surface chemistry
Protein-protein effects strongly influence adsorbed protein structure
Protein-protein effects strongly influence adsorbed protein bioactivity
Acknowledgements
This project received support from the Defense Threat Reduction Agency-Joint Science and Technology Office for Chemical and Biological Defense (Grant no. HDTRA1-10-1-0028). The facilities used were also supported by NIH Grants 5P20RR021949-04 and 8P20GM103444-04. We also would like to thank Ms. Megan Grobman, Dr. Lara Gamble, and Dr. David Castner of NESAC/BIO at the University of Washington for assistance with surface characterization with XPS under the funding support by NIH NIBIB (grant # EB002027).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Information Available This information is available free of charge via the Internet at http://pubs.acs.org. The supporting information contains (i) adsorption procedure, (ii) effect of adsorption time and equilibration time on the surface density of adsorbed HEWL, (iii) calibration and instrument performance, (iv) effect of exposure time on the structure of adsorbed HEWL, (v) estimating solution state bioactivity of the HEWL, and (vi) raw data of adsorbed HEWL responses under varying protein-protein interaction conditions
REFERENCES
- [1].Latour RA. Molecular simulation of protein-surface interactions: Benefits, problems, solutions, and future directions. Biointerphases. 2008;3:FC2–FC12. doi: 10.1116/1.2965132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rabe M, Verdes D, Seeger S. Understanding protein adsorption phenomena at solid surfaces. Advances in Colloid and Interface Science. 2011;162:87–106. doi: 10.1016/j.cis.2010.12.007. [DOI] [PubMed] [Google Scholar]
- [3].Vogler EA. Protein adsorption in three dimensions. Biomaterials. 2012;33:1201–1237. doi: 10.1016/j.biomaterials.2011.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zoungrana T, Findenegg GH, Norde W. Structure, stability, and activity of adsorbed enzymes. J. Colloid Interf. Sci. 1997;190:437–448. doi: 10.1006/jcis.1997.4895. [DOI] [PubMed] [Google Scholar]
- [5].Norde W, Giacomelli CE. Conformational changes in proteins at interfaces: From solution to the interface. Macromolecular Symposia. 1999;145:125–136. [Google Scholar]
- [6].Norde W, Lyklema J. Interfacial behaviour of proteins, with special reference to immunoglobulins. A physicochemical study. Adv. Colloid Interfac. 2012;179:5–13. doi: 10.1016/j.cis.2012.06.011. [DOI] [PubMed] [Google Scholar]
- [7].Hlady V, Buijs J. Protein adsorption on solid surfaces. Curr. Opin. Biotech. 1996:72–77. doi: 10.1016/s0958-1669(96)80098-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Seigel RR, Harder P, Dahint R, Grunze M. On-line detection of nonspecific protein adsorption at artificial surfaces. Anal. Chem. 1997;69:3321–3328. doi: 10.1021/ac970047b. [DOI] [PubMed] [Google Scholar]
- [9].Latour RA. Biomaterials: Protein-Surface Interactions. In: Wnek GE, Bowlin GL, editors. The Encyclopedia of Biomaterials and Bioengineering. Vol. 1, Informa Healthcare. New York, NY: 2008. pp. 270–284. [Google Scholar]
- [10].Szott LM, Horbett TA. Protein interactions with surfaces: cellular responses, complement activiation, and newer methods. Curr. Opin. Chem. Biol. 2011;15:677–682. doi: 10.1016/j.cbpa.2011.04.021. [DOI] [PubMed] [Google Scholar]
- [11].Horbett TA, Brash JL, Norde W. Proteins at Interfaces III State of the Art, ACS Symposium Series. American Chemical Society; Washington DC: 2012. [Google Scholar]
- [12].Sivaraman B, Fears KP, Latour RA. Investigation of the Effects of Surface Chemistry and Solution Concentration on the Conformation of Adsorbed Proteins Using an Improved Circular Dichroism Method. Langmuir. 2009;25:3050–3056. doi: 10.1021/la8036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wertz CF, Santore MM. Adsorption and reorientation kinetics of lysozyme on hydrophobic surfaces. Langmuir. 2002;18:1190–1199. [Google Scholar]
- [14].Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- [15].van der Veen M, Stuart MC, Norde W. Spreading of proteins and its effect on adsorption and desorption kinetics. Colloids and Surfaces B-Biointerfaces. 2007;54:136–142. doi: 10.1016/j.colsurfb.2006.08.017. [DOI] [PubMed] [Google Scholar]
- [16].Sreerama N, Woody RW. Computation and analysis of protein circular dichroism spectra. Numerical Computer Methods, Pt D. 2004;383:318–351. doi: 10.1016/S0076-6879(04)83013-1. [DOI] [PubMed] [Google Scholar]
- [17].Greenfield NJ. Using circular dichroism spectra to estimate protein secondary structure. Nature Protocols. 2006;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Norde W. Driving forces for protein adsorption at solid surfaces. Biopolymers at Interfaces. 1998;75:27–54. [Google Scholar]
- [19].Fears KP, Latour RA. Assessing the Influence of Adsorbed-State Conformation on the Bioactivity of Adsorbed Enzyme Layers. Langmuir. 2009;25:13926–13933. doi: 10.1021/la900799m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Banik I, Kim KS, Yun YI, Kim DH, Ryu CM, Park CS, Sur GS, Park CE. A closer look into the behavior of oxygen plasma-treated high-density polyethylene. Polymer. 2003;44 [Google Scholar]
- [21].Oiseth SK, Krozer A, Lausmaa J, Kasemo B. Ultraviolet light treatment of thin high-density polyethylene films monitored with a quartz crystal microbalance. Journal of Applied Polymer Science. 2004;92 [Google Scholar]
- [22].Ton-That C, Shard AG, Daley R, Bradley RH. Effects of annealing on the surface composition and morphology of PS/PMMA blend. Macromolecules. 2000;33 [Google Scholar]
- [23].De Geyter N, Morent R, Van Vlierberghe S, Dubruel P, Leys C, Gengembre L, Schacht E, Payen E. Deposition of polymethyl methacrylate on polypropylene substrates using an atmospheric pressure dielectric barrier discharge. Progress in Organic Coatings. 2009;64 [Google Scholar]
- [24].Serra J, González P, Liste S, Serra C, Chiussi S, León B, Pérez-Amor M, Ylänen HO, Hupa M. FTIR and XPS studies of bioactive silica based glasses. Journal of Non-Crystalline Solids. 2003;332:20–27. [Google Scholar]
- [25].Sharma S, Berne BJ, Kumar SK. Thermal and Structural Stability of Adsorbed Proteins. Biophysical Journal. 2010;99:1157–1165. doi: 10.1016/j.bpj.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. MEASUREMENT OF PROTEIN USING BICINCHONINIC ACID. Analytical Biochemistry. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- [27].Fears KP, Sivaraman B, Powell GL, Wu Y, Latour RA. Probing the Conformation and Orientation of Adsorbed Enzymes Using Side-Chain Modification. Langmuir. 2009;25:9319–9327. doi: 10.1021/la901885d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Snedecor GW, Cochran WG. Statistical Methods. Iowa State University Press; Ames, Iowa: 1989. Shortcut and nonparametric methods. Chapter 8. [Google Scholar]
- [29].Whitley E, Ball J. Statistics review 6: Nonparametric methods. Critical Care. 2002;6:509–513. doi: 10.1186/cc1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Refaee M, Tezuka T, Akasaka K, Williamson MP. Pressure-dependent Changes in the Solution Structure of Hen Egg-white Lysozyme. Journal of Molecular Biology. 2003;327:857–865. doi: 10.1016/s0022-2836(03)00209-2. [DOI] [PubMed] [Google Scholar]
- [31].Branden C, Tooze J. Introduction to protein structure. Garland Pub.; 1991. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.