Abstract
A computer program, ARAGORN, identifies tRNA and tmRNA genes. The program employs heuristic algorithms to predict tRNA secondary structure, based on homology with recognized tRNA consensus sequences and ability to form a base-paired cloverleaf. tmRNA genes are identified using a modified version of the BRUCE program. ARAGORN achieves a detection sensitivity of 99% from a set of 1290 eubacterial, eukaryotic and archaeal tRNA genes and detects all complete tmRNA sequences in the tmRNA database, improving on the performance of the BRUCE program. Recently discovered tmRNA genes in the chloroplasts of two species from the ‘green’ algae lineage are detected. The output of the program reports the proposed tRNA secondary structure and, for tmRNA genes, the secondary structure of the tRNA domain, the tmRNA gene sequence, the tag peptide and a list of organisms with matching tmRNA peptide tags.
INTRODUCTION
tRNA and tmRNA genes
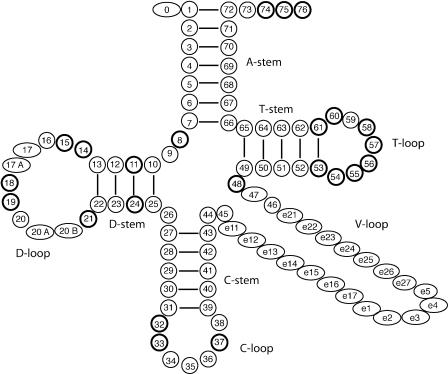
The secondary structure of tRNA molecules typically takes the form of a cloverleaf (Fig. 1) comprising (clockwise from the top) an amino-acyl acceptor stem (A-stem), TψC-stem (T-stem), TψC loop (T-loop), variable loop (V-loop), anticodon stem (C-stem), anticodon loop (C-loop), one spacer base, dihydrouridine stem (D-stem), dihydrouridine loop (D-loop) and two spacer bases. A standard numbering system has been adopted for the base positions (Fig. 1), derived from the structure of a yeast tRNAPhe (1). Within this canonical structure, there exist several consensus sequences where the bases are invariant or semi-invariant across most tRNA genes. These are believed to play an important role in the formation of the L-shaped tertiary structure of the tRNA molecule (1).
Figure 1.
Canonical tRNA cloverleaf secondary structure and numbering system. Diagram redrawn from Sprinzl et al. (12) with permission. Stem and loop descriptions have been added and the C-stem uppermost bond line has been removed to give a 5 bp C-stem, as in figure 4 of Eddy and Durbin (7).
In eukaryotes, two such sequences, GTGGCNNAGT- - -GGT-AGNGC (with ‘-’ hereafter denoting a gap, which can be filled with any base, or none at all; N, any nucleotide) starting at position 7 on the A-stem, and GGTTCGANTCC starting at position 52 on the T-stem, correspond to the A and B box intragenic transcription promoter signals for RNA polymerase III (2). These consensus sequences are also highly conserved in prokaryotes, reflecting their importance to the structure and function of most tRNAs. Another consensus sequence, YTNNNR [Y: C or T (pyrimidines); R: A or G (purines)], starts at position 32 in the C-loop, and contains the triplet anticodon (represented by NNN in the above sequence). The T at position 33 is highly conserved (3).
tmRNAs are named for their dual tRNA-like and mRNA-like function (4). The canonical tmRNA secondary structure consists of a tRNA domain at the 5′ and 3′ ends surrounding an internal region consisting of stem–loops and pseudo-knots. The tRNA domain contains alanyl-tRNA synthetase recognition signals, T-loop, shortened V-loop and extended C-stem, but the D-stem and D-loop present in a typical tRNA are replaced by a single non-base-pairing loop. Recently, tmRNAs have been identified that are encoded in two parts (5). In some of these permuted genes, the T-loop consensus motif subset GTTC has diverged toward GGGC. Previously, tmRNA genes have not been found in chloroplasts from the ‘green’ lineage of algae and higher plants. However, two tmRNA genes in the chloroplasts of Mesostigma viride and Nephroselmis olivacea, primitive members of this lineage, have been discovered by Williams and Gueneau de Novoa (6).
tRNA search algorithms
One of the most sensitive and selective tRNA detection algorithms is the tRNA-CM covariance model (7), which is a probabilistic representation of a typical tRNA secondary structural profile and primary sequence consensus. tRNA-CM was proven to be both very sensitive and selective, and can detect tRNA genes with insertions or deletions. However, tRNA-CM is extremely computationally intensive, because the computation time is proportional to the third power of total tRNA sequence length (7).
There have been several implementations of heuristic search algorithms. tRNAscan (3) searches for occurrences of part of the B box promoter signal in the T-loop, and then attempts to construct a canonical tRNA cloverleaf structure around each occurrence. The Eufindtrna algorithm (8), designed mainly for eukaryotic tRNA genes, does not attempt to construct a secondary structure, but searches for the RNA polymerase III A and B box promoter signals and a string of four or more T bases downstream of the B box, which forms part of the ρ-independent transcription termination signal. The absence of structure prediction implies that EufindtRNA might be more sensitive to detecting pseudogenes with mutations in the A-, D-, C- or T-stems than other heuristic methods.
The current benchmark search algorithm, designed specifically for the human genome project, is tRNAscan-SE (9). tRNAscan-SE combines the speed of heuristic algorithms with the sensitivity and selectivity of covariance models. It uses modified versions of tRNAscan and EufindtRNA to perform the search and to supply candidate tRNA sequences to a covariance model for analysis. The covariance model must only analyse a small fraction of the total sequence, greatly improving search speed. Both tRNAscan and EufindtRNA are implemented with relaxed threshold parameters to increase sensitivity, and the transcription termination signal search is removed from EufindtRNA to improve prediction of prokaryotic tRNA genes.
In a recent report from Tsui and co-workers (10), a novel procedure for tRNA detection is presented, based on calculation of the tRNA cloverleaf folding free energy change. Their approach appears promising, but it is our impression that development into a ready-to-use tRNA detector is incomplete and not yet ready for general implementation.
Aim
The purpose of this study is to develop a heuristic algorithm to search in silico for tRNA genes and tmRNA genes concurrently. Furthermore, it should be comparable in sensitivity, selectivity and speed with the current benchmark algorithm for tRNA detection, tRNAscan-SE (9). The program should be user friendly with a limited number of (user) parameter settings, produce results that are easy to interpret, and a website should be available for the user to perform on-line analysis. The ARAGORN program successfully fulfills all of these requirements, with the exception of selectivity where tRNAscan-SE performs better. Unlike tRNAscan-SE, the taxonomic lineage of the input sequence does not need to be specified to achieve maximum search sensitivity. However, it should be noted that, by default, ARAGORN does not search for tRNA genes with C-loop introns, but can be configured to detect tRNA genes with C-loop introns from one to 3000 bases long. In this case, the position of the intron in the C-loop is predicted. In contrast, tRNAscan-SE has a default intron plus V-loop length of 116 bases, and there is no upper bound on intron length other than poor search speed. All introns are assumed to be situated at the same position within the C-loop. When it comes to speed, ARAGORN configured with an upper intron limit of 100 bases is more than five times faster than tRNAscan-SE in the G + C content range of 40–60%.
MATERIALS AND METHODS
Search algorithm
The ARAGORN heuristic detection algorithm searches for partially mismatched, non-gapped, occurrences of the sequence GTTC, which is a subset of the B box consensus sequence. Around each hit, the algorithm attempts to construct a T-loop from five to nine bases long and a T-stem from 4 to 5 bp long. To detect tRNA genes, the sequence is searched from 28 to 85 bases upstream of this T-stem for the sequence motif TRGYNAA, a subset of the A box consensus sequence which allows for a D-stem from 3 to 4 bp long. Around the motif, a D-loop from five to 11 bases long and containing the sequence A- - - - -GG-R is constructed. A 7–9 bp long A-stem is constructed using the sequence from two to three bases upstream of the D-stem and immediately downstream of the T-stem. The length of the V-loop is allowed to vary from three to 25 bases, upstream of the T-stem. Finally, the 3′ end of the C-stem is constructed by searching between the D- and T-stems for a sequence that is complementary to the 5′ end of the C-stem, immediately downstream of the D-stem and spacer base. Limited use of tertiary structure contact between the T-loop, V-loop and D-loop is made. If position 55 in the T-loop is a non-consensus G, then a non-consensus TT at positions 18 and 19 in the D-loop is given an improved score.
The tmRNA search algorithm is based on the BRUCE program (11). Two additional criteria are used to determine the suitability of a candidate tmRNA sequence: the ability of the downstream end of the tag peptide sequence to fold into a hairpin structure and the presence of a hairpin structure upstream of the tag peptide, which may be part of a pseudo-knot.
Testing the algorithm
In the tRNAscan-SE paper from 1997, Lowe and Eddy tested their new algorithm using 589 cytoplasmic tRNA sequences from the 1995 release of the Sprinzl database of verified tRNA sequences (12), and reported improved performance compared with tRNAscan and EufindtRNA. The performance of the tRNA-CM covariance model was slightly better, but tRNAscan-SE was much faster. The ARAGORN algorithm described here is tested against tRNAscan-SE (version 1.23) using a wider set of 1290 cytoplasmic tRNA sequences from the current Sprinzl database (at http://www.uni-bayreuth.de/departments/biochemie/trna/), which was latest updated in January 1999. Similarly to Lowe and Eddy, we divide this set into three different sets of tRNA genes, from Archaea (161 sequences), Bacteria (686 sequences) and Eukaryota (443 sequences), respectively. It should be noted that neither algorithm makes use of the flanking regions of the tRNA genes such as promoters. For archaeal and bacterial sequences, tRNAscan-SE is invoked using the –A and –B switches, respectively, to load the specific covariance model for each lineage. The default model is used for eukaryotic sequences.
Tsui and co-workers (10) performed limited tests of their folding energy algorithm against tRNAscan-SE using a small range of sequenced bacterial, archeal and eukaryotic genomes. We compare ARAGORN with tRNAscan-SE on three sequenced genomes, the archaeal genome of Methanococcus jannaschii, the bacterial genome of Escherichia coli O157:H7 and the eukaryotic genome of Saccharomyces cerevisiae.
To investigate speed and selectivity (represented as the number of false positives identified), seven sequences with G + C contents of 20, 30, 40, 50, 60, 70 and 80% were randomly generated with neutral overall A + T and G + C skew. The length of each sequence was 10 Gigabases (Gb).
The ARAGORN tmRNA search algorithm is tested on 221 complete tmRNA sequences from the tmRNA website at http://www.indiana.edu/~tmrna/ (13), the 57 bacterial genomes used in the evaluation of the BRUCE software (11) and seven randomly generated 100 Mb sequences with G + C contents as above.
RESULTS
tRNA genes
A subset of the latest release (January 1999) of the Sprinzl compilation of tRNA gene sequences (12) is used to test the sensitivity of the tRNA search algorithm. tRNA genes are separated into different test sets according to lineage, and sensitivity is calculated as the fraction of tRNA genes detected (Table 1). For detection of archaeal and eubacterial tRNA genes, the results indicate that ARAGORN achieves a slightly better sensitivity than tRNAscan-SE (100 versus 99.4%, and 99.7 versus 99.4%, respectively). For eukaryotic tRNA genes, the results indicate a slightly lower sensitivity than tRNAscan-SE (98.2 versus 98.6%). The combined sensitivity is slightly above that achieved with tRNAscan-SE (99.2 versus 99.1%). For all three lineages, the results are so close as to be considered comparable. ARAGORN detects all seven of the tRNAs from the 1995 release of the Sprinzl database that the original implementation of tRNAscan-SE missed (9). ARAGORN and tRNAscan-SE are also tested on three sequenced genomes (Table 2). Here, ARAGORN is configured with a maximum intron size of 100 nucleotides, which roughly corresponds to the tRNAscan-SE default setting. The results are consistent with the Sprinzl results. For the eubacterial genome E.coli O157:H7, ARAGORN detects one more tRNA than tRNAscan-SE, indicating a slightly greater sensitivity. This extra tRNA has an identical sequence to the tRNA detected by the Tsui folding energy algorithm that tRNAscan-SE also missed (10). For the archaeal genome M.jannaschii, ARAGORN and tRNAscan-SE both detect the same number of tRNAs, and for the eukaryotic genome S.cerevisiae, ARAGORN detects one less tRNA than tRNAscan-SE, indicating a slightly lower sensitivity. The missed tRNA gene has anticodon triplet GTC and lies at position 519 095–519 165 on chromosome 14. In these genome comparisons, ARAGORN is between eight and 22 times faster than tRNAscan-SE depending on the covariance model used for tRNAscan-SE. The genome of Epifagus virginiana chloroplast contains a tRNA gene lying across the origin of counting, and so will appear as split between the beginning and ending of the genome sequence. ARAGORN successfully detects this tRNA gene.
Table 1. Sprinzl database tRNA detection rates for ARAGORN and tRNAscan-SEa.
| Test set | No. of tRNAs | No. of tRNAs detected | Detection rate (%) | ||
|---|---|---|---|---|---|
| ARAGORN | tRNAscan-SE | ARAGORN | tRNAscan-SE | ||
| Archaea | 161 | 161 | 160 | 100 | 99.4 |
| Bacteria | 686 | 684 | 682 | 99.7 | 99.4 |
| Eukaryota | 443 | 435 | 437 | 98.2 | 98.6 |
| Combined | 1290 | 1280 | 1279 | 99.2 | 99.1 |
atRNAscan-SE version 1.23.
Table 2. Whole genome tRNA detection rates for ARAGORN and tRNAscan-SEa.
| Lineage | Genome | No. of tRNAs detected | Search time (s)b | ||
|---|---|---|---|---|---|
| ARAGORNc | tRNAscan-SEd | ARAGORNc | tRNAscan-SEd | ||
| Archaea | M.jannaschii | 37 | 37 | 1.4 | With –A 24 |
| Bacteria | E.coli O157:H7 | 104 | 103 | 5.2 | With –B 112 |
| Eukaryota | S.cerevisiae | 274 | 275 | 11 | Default 114 |
atRNAscan-SE version 1.23.
bTested on an AMD Athlon, 1.6 GHz, 1024 Mb RAM with Linux.
cARAGORN run with a maximum intron size of 100 nucleotides and the –t switch (tRNA detection only). The intron size roughly corresponds to the default used by tRNAscan-SE.
dThe –A and –B switches invoke the specific covariance model for each lineage. This increases the search time. Run with –G (general model), the search time decreases to 15 s for M.jannaschii and 43 s for E.coli. However, in E.coli, the number of tRNAs detected decreases by 1. The default model in tRNAscan-SE is the eukaryotic model.
To test selectivity, ARAGORN (configured with a maximum intron size of 100 nt) is applied to seven randomly generated sequences, each 10 Gb long, with G + C contents of 0.2, 0.3, 0.4, 0.5, 0.6, 0.7 and 0.8, respectively, and neutral overall A + T and G + C skew (Table 3). The false-positive rate is calculated as the number of false positives divided by the total length of searched sequence in Gb. For sequences with a G + C content of 0.2–0.5, between 0.6 and 3.5 false positives per Gb are reported. However, for G + C contents of 0.6 or above, the false-positive rate increases. At 0.6, the rate is 14 per Gb, at 0.7, 78 per Gb, and at 0.8, 397 false positives per Gb. However, it should be noted that larger eukaryotic genomes most often are found in the G + C content range of 40–60%. In this range, ARAGORN has a much better selectivity than either the original tRNAscan (330 per Gb) or Eufindtrna (230 per Gb), as reported by Lowe and Eddy (9). The same authors reported a false-positive rate of less than 0.00007 per Mb (corresponding to 0.07 per Gb) in the original release of tRNAscan-SE (9). In the current tests, tRNAscan-SE achieves the best selectivity, predicting only 2 false positives at the 20% G + C content level (data not shown). The ARAGORN results indicate that the number of false positives rises with rising G + C content, which is not unexpected considering the high G + C content in tRNAs. The search speed also decreases as G + C content increases. Nevertheless, in the G + C content range of 40–60%, ARAGORN is more than five times as fast as tRNAscan-SE (data not shown).
Table 3. ARAGORN tRNA detection selectivity for random sequences.
| G + C content (%) | Length (Gb) | No. of false positivesa | Selectivity (per Gb) | Search speed (Mb/s)a,b |
|---|---|---|---|---|
| 20 | 10 | 6 | 0.6 | 0.98 |
| 30 | 10 | 7 | 0.7 | 1.11 |
| 40 | 10 | 17 | 1.7 | 1.13 |
| 50 | 10 | 35 | 3.5 | 1.07 |
| 60 | 10 | 143 | 14.3 | 0.91 |
| 70 | 10 | 778 | 77.8 | 0.73 |
| 80 | 10 | 3969 | 396.9 | 0.51 |
aARAGORN run with a maximum intron size of 100 nucleotides and the –t switch (tRNA detection only). The intron size roughly corresponds to the default used by tRNAscan-SE.
bTested on an AMD Athlon, 1.6 GHz, 1024 Mb RAM with Linux.
tmRNA genes
ARAGORN detects all 229 complete tmRNA sequences, including the Dehalococcoides ethenogenes tmRNA, which BRUCE misses (11). ARAGORN was also rerun on the 57 bacterial genomes that were sequenced at the time of publication of the BRUCE software. All tmRNAs were found without any reports of false positives. As with BRUCE, it must be cautioned that the tests cannot be definitive until there exists independent verification of all tmRNA genes. ARAGORN predicts the same peptide tags as presented in the tmRNA database, with two exceptions. Similarly to BRUCE, a longer peptide tag of AKTAPEAELALAA is predicted for the tmRNA gene from Aquifex aeolicus. A shorter tag of ANDSNFAAVAKAA is predicted for the tmRNA gene from Francisella tularensis. ARAGORN detects the recently discovered tmRNA genes in the chloroplasts of the two ‘green’ algae species, M.viride and N.olivacea. The predicted peptide tags are ANNILPFNRKTAVAV for M.viride and TTYHSCLEGHLS for N.olivacea.
Similarly to above, to test selectivity, ARAGORN was applied to seven randomly generated 100 Mb sequences with G + C contents of 0.2, 0.3, 0.4, 0.5, 0.6, 0.7 and 0.8, respectively, and neutral overall A + T and G + C skew (Table 4). For sequences with a G + C content of 0.2, 0.3, 0.4 and 0.6, no false positives are detected. At a G + C content of 0.5, the false-positive rate is 0.01 per Mb, at 0.7 the rate is 0.03 per Mb, and at a G + C content of 0.8, the rate increases to 0.07 false positives per Mb. These results indicate that the number of false positives rises with rising G + C content, which is also not unexpected considering the high G + C content in the tRNA domain of tmRNAs. The search speed also decreases as G + C content increases.
Table 4. ARAGORN tmRNA detection selectivity for random sequences.
| G + C content (%) | Length (Mb) | No. of false positivesa | Selectivity (per Mb) | Search speed (Mb/s)a,b |
|---|---|---|---|---|
| 20 | 100 | 0 | < 0.01 | 0.90 |
| 30 | 100 | 0 | < 0.01 | 0.85 |
| 40 | 100 | 0 | < 0.01 | 0.73 |
| 50 | 100 | 1 | 0.01 | 0.59 |
| 60 | 100 | 0 | < 0.01 | 0.42 |
| 70 | 100 | 3 | 0.03 | 0.24 |
| 80 | 100 | 7 | 0.07 | 0.11 |
aARAGORN run with the –m switch (tmRNA detection only).
bTested on an AMD Athlon, 1.6 GHz, 1024 Mb RAM with Linux.
Availability, options and output
ARAGORN is written in C. The source code can be downloaded from the website at http://bioinfo.thep.lu.se. The website also contains a user interface to the program allowing the user to upload a sequence and run the program on a server.
ARAGORN accepts as input a file with one or more nucleotide sequences in FASTA format. By default, ARAGORN assumes that each sequence has a circular topology (search wraps around ends), that both strands should be searched, that the progress of the search is not reported, both tRNA and tmRNA genes are detected, and tRNA genes containing C-loop introns are not detected. These settings can be changed individually to linear topology (no wrapping), search of the sense strand only, report of search progress, detection of only tRNA genes, detection of only tmRNA genes, or detection of tRNA genes with C-loop introns from one to 3000 bases long. For each candidate tRNA, secondary structure, anticodon position and amino acid isoacceptor species are predicted (Fig. 2). If the tRNA contains a C-loop intron, the predicted intron position, length and sequence are reported. The isoacceptor species is based on the universal genetic code. tmRNA output is identical to the BRUCE program. An abbreviated output format is also available. In this case, for each sequence in the input file, only the sequence name and tab delimited information about each gene detected in the sequence are given. It should be noted that when searching through files consisting of one or more short sequences containing single tmRNA genes, ARAGORN will report two tmRNA genes for each sequence; one non-permuted and one permuted, unless linear topology is specified.
Figure 2.
Example output of the computer program ARAGORN from a search of eubacterial tRNA genes in the Sprinzl database (12).
DISCUSSION
The results for the Sprinzl tRNA gene database indicate that ARAGORN is an effective tRNA search program, with sensitivity comparable with or better than other current heuristic tRNA search algorithms, especially with eubacterial and archeal genomes, and a sensitivity comparable with tRNAscan-SE. ARAGORN could be regarded as the next stage in the development of purely heuristic tRNA search algorithms. The results for the randomly generated sequences indicate that ARAGORN is strongly selective, but selectivity will be expected to degrade for genomes with an extraordinarily high G + C content, leading to detection of more false positives. Previously published tests for the BRUCE program indicate that ARAGORN is also an effective tmRNA search program (11). Our results show that the sensitivity of the tmRNA detection algorithm is further improved.
We have here developed a computer program for concurrent detection of tRNA and tmRNA genes, which has previously not been available in other algorithms. We see several advantages of releasing a new algorithm. (i) ARAGORN is a general tRNA prediction algorithm, and does not require the user to know whether the search sequence is bacterial, eukaryotic or archaeal to achieve maximum search sensitivity if an appropriate maximum C-loop intron size is set. Increasing the maximum intron size will reduce search speed but increase the likelihood of detecting any tRNA genes with long C-loop introns. (ii) The output of the de facto standard algorithm for tRNA prediction in long sequences, tRNAscan-SE, may now be compared with the output of an independently developed algorithm of similar sensitivity. In many cases (e.g. genome sequencing projects), tRNAscan-SE, thanks to its outstanding performance, may be the only tRNA prediction tool used. However, there is a danger in using one algorithm exclusively. If any part of the algorithm is suboptimal in a particular circumstance or for a particular set of tRNAs, this could go unnoticed without independent verification of the prediction. Here, ARAGORN and tRNAscan-SE may complement each other by either giving a higher probability of a tRNA identification when reporting the same result, or suggesting a deeper investigation when the results disagree. (iii) ARAGORN is much faster than tRNAscan-SE. (iv) ARAGORN is the first program that predicts both tRNAs and tmRNAs concurrently. Up till now, no tmRNAs have been annotated in most bacterial genome sequencing projects, and have a tendency to go unnoticed in sequence analysis. Using ARAGORN will help rectify omission of these genes. (v) Matching the tmRNA peptide tag to known tmRNA tags can potentially assist the identification of unkown sequences and the investigation of evolutionary descent. (vi) ARAGORN reports the tRNA secondary structure in an intuitive way, as a cloverleaf diagram. tRNAscan-SE also reports secondary structure; however, the linear representation of the secondary structure is not as easy to interpret. (vii) ARAGORN is available on the Internet (http://bioinfo.thep.lu.se). This website allows input sequences of up to 15 Mb. The main tRNAscan-SE websites (e.g. http://www.genetics.wustl.edu/eddy/tRNAscan-SE/) have a sequence size limitation of 100 kb, which is well below the size of a prokaryotic genome.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Siv Andersson for valuable comments and David Ardell for sharing his knowledge of tRNAs. Thanks also to Tolkien.
REFERENCES
- 1.Kim S.H., Suddath,F.L., Quigley,G.J., McPherson,A., Sussman,J.L., Wang,A.H.J., Seeman,N.C. and Rich,A. (1974) Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science, 185, 435–440. [DOI] [PubMed] [Google Scholar]
- 2.Sharp S.J., Schaak,J., Cooley,L., Burke,D.J. and Söll,D. (1985) Structure and transcription of eukaryotic tRNA genes. CRC Crit. Rev. Biochem., 19, 107–144. [DOI] [PubMed] [Google Scholar]
- 3.Fichant G.A. and Burks,C. (1991) Identifying potential tRNA genes in genomic DNA sequences. J. Mol. Biol., 220, 659–671. [DOI] [PubMed] [Google Scholar]
- 4.Muto A., Ushida,C. and Himeno,H. (1998) A bacterial RNA that functions as both a tRNA and an mRNA. Trends Biochem. Sci., 23, 25–29. [DOI] [PubMed] [Google Scholar]
- 5.Keiler K.C., Shapiro,L. and Williams,K.P. (2000) tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: a two-piece tmRNA functions in Caulobacter.Proc. Natl Acad. Sci. USA, 97, 7778–7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gueneau de Novoa P. and Williams,K.P. (2004) The tmRNA Website: reductive evolution of tmRNA in plastids and other endosymbionts. Nucleic Acids Res., 32, D104–D108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eddy S.R. and Durbin.R. (1994) RNA sequence analysis using covariance models. Nucleic Acids Res., 22, 2079–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pavesi A., Conterio,F., Bolchi,A., Dieci,G. and Ottonello,S. (1994) Identification of new eukaryotic tRNA genes in genomic DNA databases by a multistep weight matrix analysis of transcription control regions. Nucleic Acids Res., 22, 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowe T.M. and Eddy,S.R. (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res., 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsui V., Macke,T. and Case,D.A. (2003) A novel method for finding tRNA genes. RNA, 9, 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laslett D., Canback,B. and Andersson,S. (2002) BRUCE: a program for the detection of transfer-messenger RNA genes in nucleotide sequences. Nucleic Acids Res., 30, 3449–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sprinzl M., Horn,C., Brown,M., Ioudovitch,A. and Steinberg,S. (1998) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res., 26, 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams K.P. (2002) The tmRNA Website: invasion by an intron. Nucleic Acids Res., 30, 179–182. [DOI] [PMC free article] [PubMed] [Google Scholar]