Abstract
Previous studies in our lab have identified Pre-B-cell colony enhancing factor as a novel biomarker in acute lung injury. This study continues to elucidate the underlying molecular mechanism of Pre-B-cell colony enhancing factor (PBEF) in the pathogenesis of acute lung injury in pulmonary cell culture models. Our results revealed that IL-1β induced PBEF expression in pulmonary vascular endothelial cells at the transcriptional level and a -1535 T-variant in the human PBEF gene promoter significantly attenuated its binding to an IL-1β induced unknown transcription factor. This may underlie the reduced expression of PBEF and thus the less susceptibility to acute lung injury in those -1535T carriers. Furthermore, overexpression of PBEF significantly augmented IL-8 secretion and mRNA expression by more than 6 fold and 2 fold in A549 cells and HPAEC, respectively. It also significantly augmented IL-1β mediated cell permeability by 44% in A549 cells and 65% in endothelial cells. The knockdown of PBEF expression significantly inhibited IL-1β-stimulated IL-8 secretion and mRNA level by 60% and 70%, respectively; and the knockdown of PBEF expression also significantly attenuated IL-1β-induced cell permeability by 29% in epithelial cells and 24% in endothelial cells. PBEF expression also affected the expression of two other inflammatory cytokines (IL-16 and CCR3 genes). These results suggest that PBEF is critically involved in pulmonary vascular and epithelial inflammation and permeability, which are hallmark features in the pathogenesis of acute lung injury. This study lend further support that PBEF is a potential new target in acute lung injury.
Keywords: Pulmonary inflammation, permeability, PBEF, Acute lung injury, IL-1β, IL-8, Epithelial cells, Endothelial cells, Inflammatory cytokines
INTRODUCTION
Acute lung injury (ALI) and its more severe form, acute respiratory distress syndrome (ARDS), are characterized by alveolar inflammation, injury and increased pulmonary capillary permeability. The clinical consequence is an impaired gas exchange and ultimate respiratory failure (Leaver and Evans, 2007; Levitt and Matthay, 2006). Despite considerable progress in basic and clinical researches, the mortality and morbidity of ALI/ARDS remain high since the etiology and molecular pathogenesis are still incompletely understood.
In our previous study on animal models of ALI, we identified pre-B-cell colony enhancing factor (PBEF) as a significantly upregulated gene in ALI (Ye et al., 2005). We discovered single nucleotide polymorphisms in the human PBEF gene promoter. Moreover we found that carriers of the haplotype GC from SNPs T-1001G and C-1543T had a 7.7-fold higher risk of ALI. The T variant from the SNP C-1535T resulted in a significant decrease in the transcription rate (1.8-fold) by the reporter gene assay (Ye et al., 2005). Our findings were confirmed and extended in a separate and larger population (>1000 patients) by Bajwa et al. (2006). We further found that a reduction in PBEF protein expression by siRNA significantly attenuated pulmonary artery endothelial cell barrier dysfunction induced by the potent edemagenic agent, thrombin, reflected by reductions in transendothelial electric resistance (Ye et al., 2005). Taken together, these results strongly indicate PBEF as a potential novel candidate gene and biomarker in ALI.
The objective of this study was to elucidate the molecular mechanisms underlying PBEF in the pathogenesis of acute lung injury using human pulmonary cells as in vitro cell models. Since increased vascular and epithelial cell inflammation and permeability processes are important features of ALI, we assessed the effect of PBEF knockdown with PBEF siRNA and PBEF overexpression on basal and IL-1β-mediated human pulmonary epithelial cell (A549) and human pulmonary artery endothelial cell (HPAEC) IL-8 production and permeability by in vitro cell permeability assay. We also examined the role of PBEF expression on the expression of other inflammatory cytokines such as IL-16 and CCR3.
MATERIALS AND METHODS
Materials
A rabbit anti-human IL-8 polyclonal antibody (Cat. No. sc-7922) and a mouse anti-human β-actin monoclonal antibody (Cat. No. A1978) were obtained from Sigma-Aldrich (St. Louis, MO, USA). A rabbit anti-human PBEF polyclonal antibody was from the Bethyl Laboratories, Inc. (Cat. No. A300-372A, Montgomery, TX, USA). A recombinant human IL-1β (Cat. No. 201-LB) was from R&D Systems Inc. (Minneapolis, MN, USA). Superscript III Reverse Transcriptase (Cat. No. 18080044) and Platinum Taq DNA polymerase (Cat. No. 10966018) was from Invitrogen (Carlsbad, CA, USA). Tricine was purchased from Sigma-Aldrich (Cat. No. T0377, St. Louis, MO, USA). Sources of other key reagents are specified in relevant texts.
Cell culture
Human A549 cell, a lung carcinomatous type II alveolar epithelial cell line, was obtained from ATCC (Cat. No. CCL-185™, Manassas, VA, USA) and maintained in DMEM supplemented with 10% fetal bovine serum, 2 mM glutamine, penicillin/streptomycin. Primary human pulmonary artery endothelial cells (HPAEC, Cat. No. CC-2530) were obtained from Cambrex Bio Science Inc. (Walkersville, MD, USA) and maintained in EGM™-2 Endothelial Cell Medium-2 (Cat. No. CC-4176). All cells were cultured at 37°C in a humidified atmosphere of 5% CO2, 95% air. Cells from each primary flask were detached with 0.25% trypsin, resuspended in fresh culture media, and seeded into 6-well plates for Western blotting or RT-PCR analysis or seeded into the culture inserts for in vitro cell permeability assays.
Inhibition of newly synthesized PBEF mRNA
HPAEC were grown to confluence and incubated in the presence or absence of IL-1β (10 ng/ml) plus or minus 5 μg/ml actinomycin D, a transcription inhibitor for 4 h. After treatment, total cellular RNA was analyzed by a semi-quantitative RT-PCR for the expression of PBEF mRNA. Briefly, total RNA (0.5 μg) from each sample was used as a starting material in the reverse transcriptase reaction. The primers for human PBEF cDNA and human ribosomal protein S18 (RPS18) gene, used as a house-keeping gene control, were designed based on published Genbank mRNA sequences: NM_005746.2 and K03432, respectively. The primer sequences for the human PBEF were as follows: PBEF-5: 5′-AAGCTTTTTAGGGCCCTTTG-3′, PBEF-3: 5′-AGGCCATGTTTTATTTGCTGA-3′. The PCR product size was 319 bp. The primer sequences for the human 18S rRNA were: M18R5: ATGGCCGTTCTTAGTTGGTG, M18R3: GGGACTTAATCAACGCAAGC, PCR product size: 154bp. The primers were synthesized by Integrated DNA Technologies (Coralville, IA). After the reverse transcriptase reaction, one-tenth of the RT product was used in a typical PCR run of 25 cycles. The PCR products were separated on agarose gel electrophoresis and visualized by ethidium bromide staining. The band image was acquired using the Alpha Imager and analyzed by the AlphaEase™ Stand Alone Software (Alpha Innotech Corp., San leandro, CA). Statistical comparative analysis of integrated density value (mean ± SEM) between control and ALI groups was performed using the unpaired t test.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed essentially as described by Wang et al. (1998) except the probe was labeled with Biotin instead of a radioisotope and most reagents were provided by Pierce (Rockford, IL). Two 30-mer probes: -1535T, 5′-tacattggtgagtgcTtggtgataccttcc-3′ and -1535C, 5′-tacattggtgagtgcCtggtgataccttcc-3′, were synthesized and 5′ biotinylated by Integrated DNA Technologies (Coralville, IA). They differ in one nucleotide as indicated by the capital font at -1535 position of the PBEF gene promoter. Nuclear extracts from HPAEC with or without IL1-β(10ng/ml) treatment for 4 h were prepared using NE-PER® Nuclear and Cytoplasmic Extraction Reagents (Pierce, Rockford, IL). Halt™ Protease Inhibitor Cocktail (Pierce, Rockford, IL) was added into the nuclear extract to inhibit potential protein degradation by endogenous proteases. Frozen aliquots of the nuclear proteins were stored at −70° C for future use. The protein concentration of nuclear extracts was determined by the methods of Bradford (Bradford, 1976). DNA-binding reactions and chemiluminescent nucleic acid detection were carried out according to the supplier’s instruction in LightShift® Chemiluminescent EMSA Kit (Pierce, Rockford, IL). The images were acquired and recorded using The FluroChem® Imaging System (Alpha Innotech Corporation, San Leandro, CA).
Transfection of PBEF siRNA into human A549 cells
PBEF stealth siRNA was designed based on the human PBEF cDNA reference sequence (NM_005746.1) using the BLOCK-iT™ RNAi Designer (Invitrogen, Carlsbad, CA, USA). Using GFP-labeled non-specific siRNA, we first optimized the conditions for human A549 cells transfection and achieved >90% transfection efficiency using the Lipofectamine 2000 reagent (Cat. No. 11668-019, Invitrogen, Carlsbad, CA, USA). To transfect PBEF stealth siRNA into human A549 cells, cells were seeded for 24 h in the regular growth medium (without antibiotics) so that they would be 80–90% confluent at the time of transfection. For each transfection in 24-well plates, 50 pmol PBEF stealth siRNA was diluted in 50 μl Opti-MEM I without serum and gently mixed with 1 μl Lipofectamine 2000 diluted in the 50 μl Opti-MEM I (Cat. No. 31985-062, Invitrogen, Carlsbad, CA, USA). After incubated for 15 min at room temperature, PBEF stealth siRNA & Lipofectamine 2000 complexes were added to each well. Cell culture plates were gently mixed by rocking back and forth. The amount of PBEF stealth siRNA and Lipofectamine 2000 were adjusted according to the different sizes of cell culture plates. Transfected cells were further incubated at 37°C for 48 hours until the treatment with IL-1β was performed. Transfection of PBEF siRNA into HPAEC was carried out as described before by Ye et al. (2005).
Preparation and expression of the PBEF-overexpressing construct pCAGGS-mPBEF
A mouse PBEF (mPBEF) coding cDNA sequence was amplified from a mouse lung total RNA (Cat. No. 736511, Stratagene, La Jolla, CA, USA) by RT-PCR using the following primer pair: ttagaattcgccaccatgaatgctgcggcagaagc;tagaattcctaATGGTGATGGTGATGATGcaaatgaggtgccacgtcctgct. The bold letter indicates the optimized Kozac sequence. The capitalized letter part is His tag sequence. The underlined sequences are EcoRI adaptors. The amplified mouse PBEF cDNA was digested with EcoRI, subcloned into the unique EcoRI site of pCAGGS vector, i.e., pCAGGS-mPBEF, and then sequence-verified. In this construct, mouse PBEF expression was driven by a chicken beta-actin/rabbit beta-globin hybrid promoter (AG) with an enhancer from the human cytomegalovirus immediate early promoter (CMV-IE). Overexpression of PBEF in A549 cells was carried out by a transient transfection of pCAGGS-mPBEF into A549 cells. Briefly, one day before transfection, A549 cells were plated in 6-well plate at 5 × 105 cells/well in 2 ml of growth medium without antibiotics. On the day of transfection, cells were at 95% confluence. For each well, 4 μg plasmid DNA was transfected using Lipofectamine 2000 (Cat. No. 11668-019, Invitrogen, Carlsbad, CA, USA) according to the supplier’s instructions. Cell medium and cell lysate proteins were harvested at different time points of interest after the transfection, respectively.
Isolation of RNA and RT-PCR analysis
Total RNA from in vitro cultured cells was isolated using the TRIZOL solution (Cat. No. 15596-018, Invitrogen, Carlsbad, CA, USA) according to the supplier’s instruction. RT-PCR was performed using the following procedures. 1 μg total RNA was reverse transcribed using an Invitrogen reverse transcriptase (Superscript III, Cat. No. 18080-044) with random hexamers at 50 °C for 1 h followed by 70 °C for 15 min and 4 °C for 5 min in a 20 μl reaction volume. Each PCR reaction from the cDNA template (2 μl RT product) was performed using gene specific primers (Table 1) with 94 °C for 3 min, then 32 cycles at 94 °C for 1 min, 55 °C for 1 min and 72 °C for 1 min, followed by 72 °C for 7 min for the final extension. β-actin was used as a house-keeping gene control. PCR products were separated on a 1.5 % agarose gel and stained by Ethidium Bromide (0.5 μg/ml). The band image was acquired using an Alpha Imager and analyzed by the AlphaEase™ Stand Alone Software (Alpha Innotech Corp., San leandro, CA, USA).
Table 1.
Primers and products sizes
| Product | 5′ Primers | 3′ Primers | Size (bp) | Accession No. |
|---|---|---|---|---|
| PBEF | AAGCTTTTTAGGGCCCTTTG | AGGCCATGTTTTATTTGCTGACAAA | 319 | NM_005746 |
| IL-8 | ATGACTTCCAAGCTGGCCGT | CCTCTTCAAAAACTTCTCCACACC | 297 | NM_000584 |
| β-actin | CAAACATGATCTGGGTCATCTTCTC | GCTCGTCGTCGACAACGGCTC | 487 | NM_001101 |
| IL-16 | TAGTGCCAAGGTACAAACAGGTG | GGGTCTCAAACTCAGATGCCTAT | 280 | NM_172217 |
| CCR3 | AGCCCCTAAAGCAGCACTAA | TGATAGCTTAGGCGTCACCA | 228 | NM_001837 |
Western blotting
Western blotting analysis was performed as described before (McGlothlin et al., 2005; Ye et al., 2005) with a minor modification. Briefly, after washing with PBS, cells were lysed with 500 μl of cell lysis buffer containing 10 mM Tris (pH 7.4), 1% Triton X-100, 0.5% Nonidet P-40, 150 mM NaCl, 1 mM EDTA, 0.2 mM EGTA, 0.2 mM vanadate, 0.2 mM PMSF, and 0.5% protease inhibitor cocktail. Total cell lysates were cleared by centrifugation, boiled with the same amount of 4 × SDS sample buffer for 5 min. Total proteins of cell lysates were quantified using the BCA Protein Assay Kit (Pierce Biotechnology, Inc., Rockford, IL, USA). An equal amount of total protein from each sample was then subjected to 16.5% Tris/tricine polyacrylamide gel electrophoresis. The separated proteins were transferred to PVDF membranes by electrotransfer. The blots were subsequently blocked with 5% bovine serum albumin in phosphate-buffered saline containing 0.1% Tween 20 (TBS-T) at room temperature for 1 h and then incubated at 4°C overnight with a primary antibody of interest. After washing three times for 10 min each with TBS-T, the membrane was incubated with a horseradish-peroxidase-linked secondary antibody of interest at room temperature for 1 h. The blots were then visualized with the ECL Western blot detection system (Cat. No. RPN2106, Amersham Bosciences, Buckinghamshire, UK). The same membrane was re-probed with an anti-human β-actin antibody. β-actin was used as an internal control. The band image was acquired using an Alpha Imager and analyzed by the AlphaEase™ Stand Alone Software (Alpha Innotech Corp., San Leandro, CA, USA). Band density on Western blot images was used as a measure of assayed protein level.
In Vitro Cell Permeability Assay
The in Vitro Cell Permeability Assay was carried out according to the manufacturer’s instructions (Cat. No. ECM640, Millipore, Billerica, MA, USA). Briefly, cells were seeded to the culture inserts of permeability chamber (1.0 × 106 cells/ml), which were coated by collagen. Then, cells were incubated at 37°C, 5% CO2, until a monolayer formed. After IL-1β (Cat. No. 636-R1, R&D systems, Minneapolis, MN, USA) was added, cells were incubated for another 18 hours in a 37°C, 5% CO2 tissue culture incubator. Monolayers cultured with either basal medium or growth medium, and blank inserts were included as controls. Finally, 150 μl of FITC-Dextran was added to each insert for 5 min at room temperature, and then 100 μl of the solution in the bottom chamber was transferred to a 96-well plate. The plate was read in a TriStar Multimode Reader (LB 941, Berthold Technologies GnbH & Co. KG, Bad Wildbad, Germany) with a 485 nm excitation and 535 nm emission wavelength. The data were expressed in relative fluorescent units.
Statistical analysis
Statistical analyses were performed using the SigmaStat (ver 3.5, Systat Software, Inc., San Jose, CA, USA). Results are expressed as means ± SD of 4 samples from at least two independent experiments. Stimulated samples were compared with controls by unpaired Student’s t test. P < 0.05 was considered statistically significant.
RESULTS
Dose-response and time-course of IL-1β induced PBEF protein expression in A549 cells
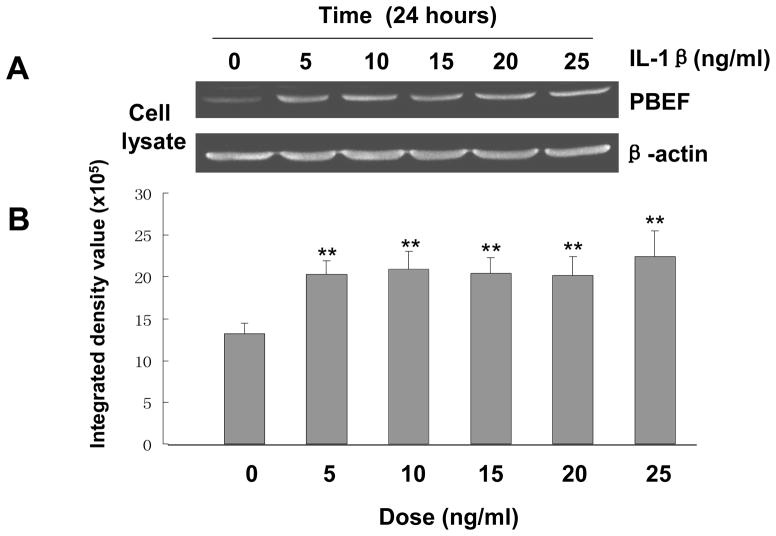
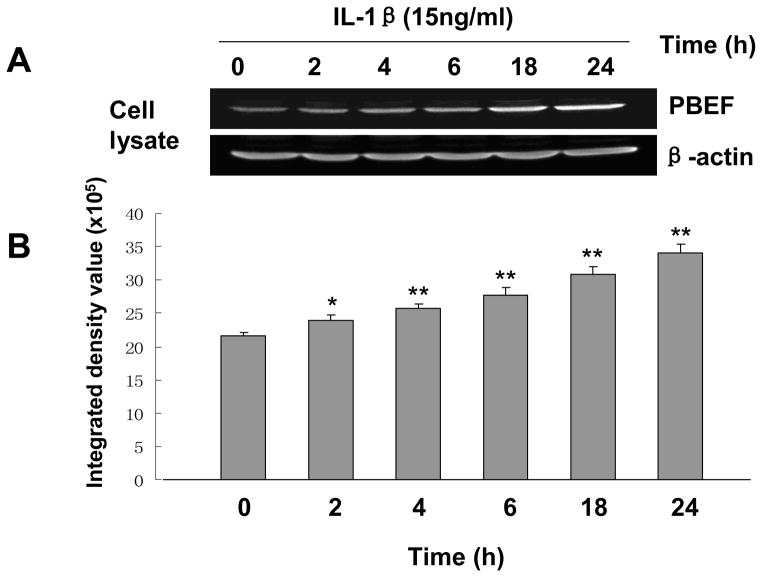
Since there was no previous publication on whether IL-1β can induce PBEF expression in lung epithelial cells, we first performed a pilot experiment showing the induction of PBEF expression by IL-1β in A549 cells (data not shown) before we determined the dose response and time course to optimize the experimental conditions of IL-1β induced PBEF expression in A549 cells. Results in Figure 1 show that with different dose treatments of IL-1β (5 to 25 ng/ml), cell lysate PBEF expression was significantly increased compared to control group at each tested dosage. In the time course experiment (Figure 2), PBEF protein is up to higher level at 24 h after IL-1β treatment [3.4 × 106± 1.4 × 105vs. 2.17 × 106 ± 0.3 × 105 (control), n = 3, **p < 0.01 for 24 h]. These data indicate that IL-1β significantly increased PBEF protein expression in a dose-dependent & time-dependent manner in A549 cells under our experimental conditions.
Fig. 1.
Dose-response of IL-1β induced PBEF protein expression in A549 cells. A. A representative western blotting image of PBEF and β-actin protein detections. After starving for serum overnight, A549 cells were stimulated with different doses of IL-1β, as indicated, for 24 hours. Equal amount of total cell lysate protein from each sample was separated by 16.5% SDS-PAGE and immunodetected by western blotting using anti-human PBEF or β-actin antibodies. Protein band images on the western blotting were acquired using the Alpha Imager (Alpha Innotech Corp., San Leandro, CA). The displayed image is one of three replicate samples and is typical of those obtained. B. Quantitation of cell lysate PBEF protein level by densitometry analysis. The protein band intensity on the western blotting was analyzed by the AlphaEase Stand Alone software (Alpha Innotech Corp., San Leandro, CA). The integrated density value of each band was used as a measure of the protein level. Results from each group are presented as mean ± SD of 3 samples. Statistical comparative analyses of PBEF levels between the control (IL-1β, 0 ng/ml) and different treatment groups (IL-1β, 5–25 ng/ml) were performed using the unpaired t test. **, p<0.01.
Fig. 2.
Time-course of IL-1β induced PBEF protein expression in A549 cells. A. A representative western blotting image of PBEF and β-actin protein detections. After starving for serum overnight, A549 cells were stimulated with IL-1β (15 ng/ml) at different time points (h) as indicated. Equal amount of total cell lysate protein from each sample was separated by 16.5% SDS-PAGE and immunodetected by the western blotting using anti-human PBEF or β-actin antibodies. β-actin was used as a house-keeping gene and loading control. Protein band images on the western blotting were acquired using the Alpha Imager (Alpha Innotech Corp., San Leandro, CA). The displayed image is one of three replicate samples and is typical of those obtained. B. Quantitation of cell lysate PBEF protein level by densitometry analysis. Cell lysate PBEF protein levels were quantified by densitometry analyses in the same way as described above in Figure 1. *, p<0.05; **, p<0.01.
IL-1β induction of PBEF expression at mRNA level in A549 cells
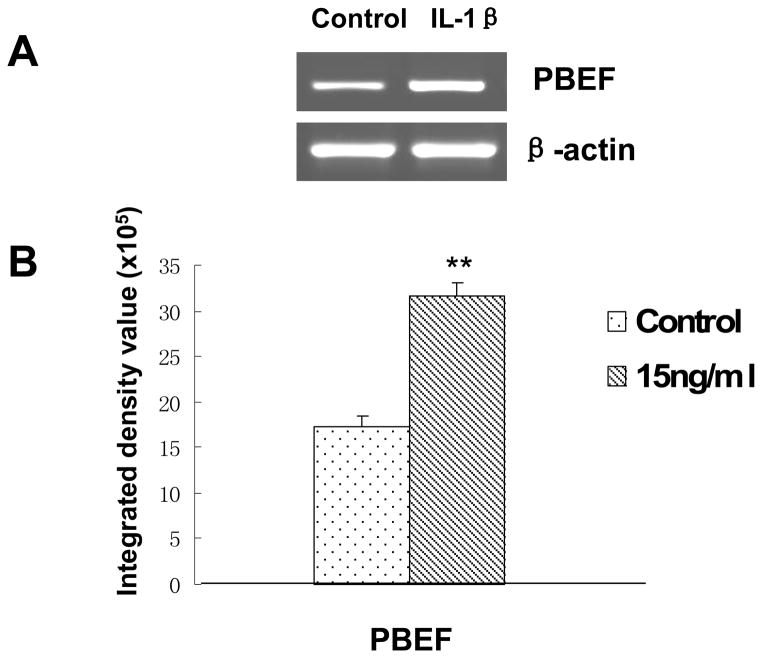
To investigate whether IL-1β also augments PBEF expression at the mRNA level in A549 cells, we performed a semi-quantitative RT-PCR analysis of PBEF mRNA level in IL-1β induced A549 cells. As presented in Figure 3, PBEF mRNA level in IL-1β treated A549 cells were significantly increased compared to the control groups[3.17 × 106 ± 1.44 × 105 vs. 1.73 × 106 ±1.20 × 105 (control), n = 3, **P < 0.01].
Fig. 3.
Effects of IL-1β treatment on the mRNA expression of PBEF in A549 cells. A. A representative gel image of PBEF and β-actin mRNA detections. After starving for serum overnight, A549 cells were stimulated with IL-1β (15 ng/ml) for 4 h. Total cell RNA was reverse-transcribed, amplified by PCR using the gene specific primers (Table 1), separated by 1.5% agarose electrophoresis and visualized with ethidium bromide. The gel image was captured using the Alpha Imager (Alpha Innotech Corp., San Leandro, CA). β-actin was used as a house-keeping gene control. The displayed image is one of three replicate samples and is typical of those obtained. B. Quantitation of PBEF mRNA levels by densitometry analysis. The mRNA band intensity on the gel image was analyzed by the AlphaEase Stand Alone software (Alpha Innotech Corp., San Leandro, CA). The integrative density value of each band was used as a measure of the mRNA level. Results from each group are presented as mean ± SD of 3 samples. Statistical comparative analyses of PBEF mRNA levels between the control and IL-1β treated group were performed using the unpaired t-test.;**, p<0.01.
IL1-β induces the transcription of the PBEF gene
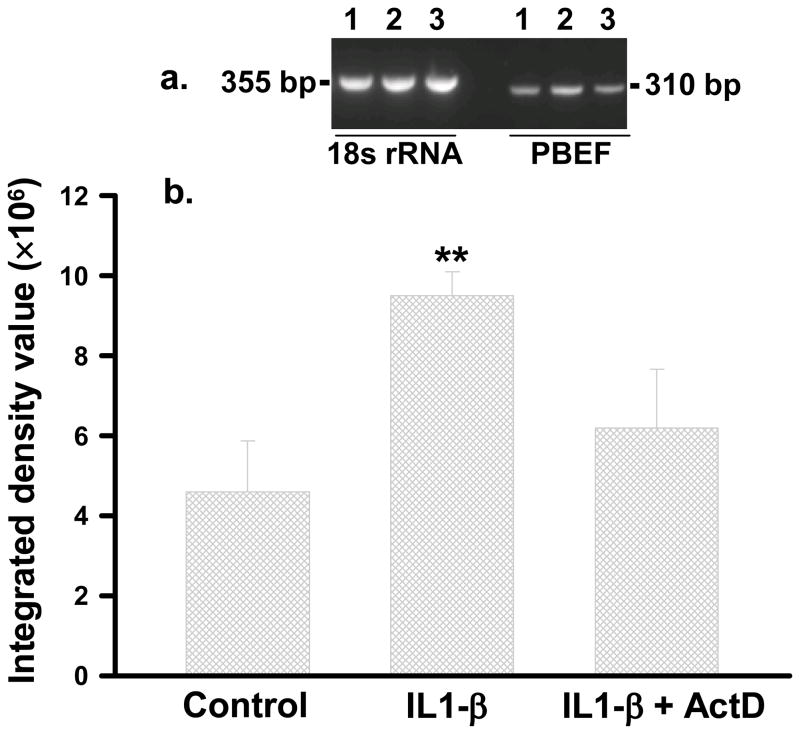
IL1-β similarly increased PBEF mRNA level in HPAEC. We further determined whether this effect was due to the increased transcription or mRNA stability of PBEF expression. HPAEC were treated for 4 h with IL1-β (10ng/ml) in the presence or absence of actinomycin D (5μg/ml), a transcription inhibitor. Cellular mRNA levels were assayed by a semi-quantitative RT-PCR. A representative gel pattern of HPAEC PBEF mRNA by a semi-quantitative RT-PCR is displayed in Figure 4 (panel a). Integrated density values of each group are presented in bar graphs in Figure 4 (panel b, Control: 4.6 × 106 ± 12.7 × 105; IL1-β: 9.5 × 106 ± 6.0 × 105; IL1-β + act D: 6.2 × 106 ± 14.6 × 105). IL1-β alone increased PBEF mRNA level by 2.07 fold (n=4, p<0.01) vs. control. Simultaneous treatment of HPAEC with IL1-β plus act D prevented the IL1-β induction of PBEF mRNA, supporting that IL1-β induces the transcription of PBEF gene.
Fig. 4.
RT-PCR semi-quantification of PBEF mRNA in HPAEC treated without or with either IL1-β or IL1-β + Act D. HPAEC were treated without or with either IL1-β (10 ng/ml) or IL1-β + Act D (5μg/ml). Total RNA was reverse-transcribed, amplified by PCR using gene specific primers, separated on 2% agarose electrophoresis, analyzed by an Alpha Innotech densitometer. a, A representative image. 1-control; 2-IL1-β; 3- IL1-β +Act D; b, Densitometry analyses. **, p<0.01.
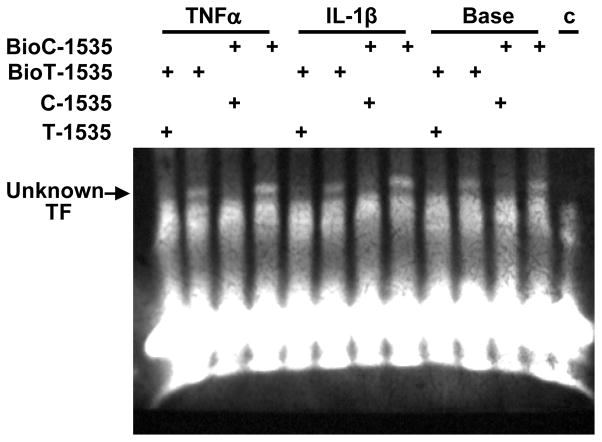
EMSA analysis of the human PBEF promoter -1535Tand -1535C 30-mer oligonucleotides
We reported previously that the T variant in the human PBEF promoter SNP C-1535T resulted in a significant decrease of the luciferase reporter gene expression (3). We examined whether the T-variant altered the binding affinity to any transcription factor, which may underlie its effect on the decrease in the transcription of PBEF gene expression. As presented in Figure 5, EMSA analysis revealed that -1535 T-variant in the human PBEF gene promoter has less binding to an unknown transcription factor than the common -1535 C-allele, this difference became more pronounced after the IL1-β or TNFα treatment. The bindings were specific since their unlabeled “cold” oligos could totally compete out the binding.
Fig. 5.
Electrophoretic Mobility Shift Assay on -1535C and -1535 T Binding to an unknown Transcription Factor (TF). HPAEC were treated without (Base) or with either IL-1β (10ng/ml) or TNFα (10 ng/ml) for 4 h. 10 μg nuclear extract protein in each sample was incubated with either biotin labeled c-1535 (Bio C-1535) or BioT-1535 oligo in the absence or presence of their unlabeled 200 fold excess counterparts. The bindings were visualized by a chemiluminecence. C, no oligo control.
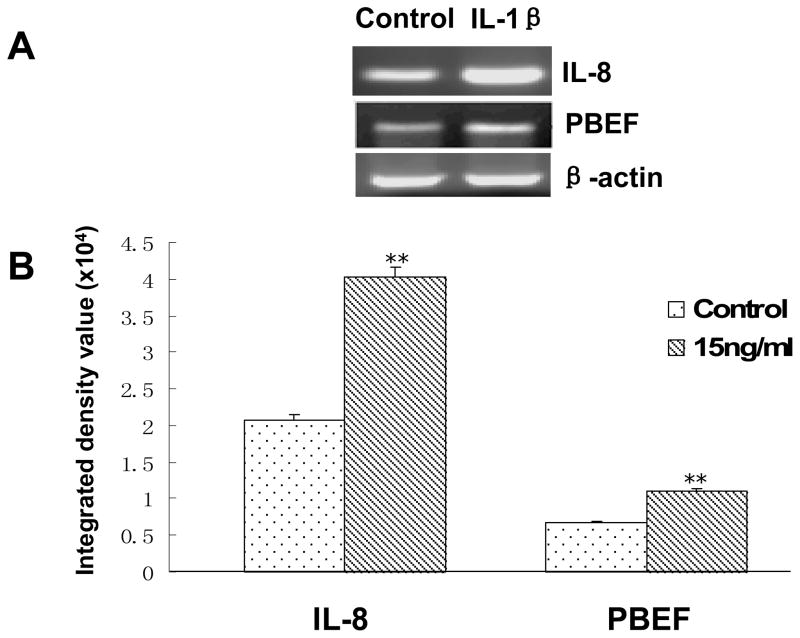
IL-1β induction of IL-8 and PBEF at the mRNA level in HPAEC
IL-8 is a known IL1-β inducible biomarker in acute lung injury. PBEF may directly or indirectly affect IL-1 β induced IL-8 expression during the inflammatory process. To this end, we examined whether IL-1β could also affect IL-8 and PBEF expression at the mRNA levels in HPAEC, we quantified IL-8 and PBEF mRNA levels in IL-1β treated primary HPAEC. As shown in Figure 6, IL-8 and PBEF mRNA levels in IL-1β treated primary HPAEC significantly increased compared to the control group [4.03 × 104 ± 1.4 × 103 vs. 2.07 × 104 ± 0.7 × 103 (control), n = 4, **P < 0.01; 1.10 × 104 ± 0.4 × 103vs. 0.67 × 104 ± 0.2 × 103 (control), n = 4, **P < 0.01; respectively]. These results indicate that IL-1β also induces IL-8 and PBEF expression at the mRNA level in HPAEC.
Fig. 6.
Effects of the IL-1β stimulation on IL-8 & PBEF mRNA expressions in HPAEC. A. A representative gel image of IL-8, PBEF and β-actin mRNA detections. After starving for serum overnight, HPAEC were stimulated with IL-1β(15 ng/ml) for 4 h. Total cell RNA was reverse-transcribed, amplified by PCR using the gene specific primers (Table 1), separated by 1.5% agarose electrophoresis and visualized with ethidium bromide. The gel image was captured using the Alpha Imager (Alpha Innotech Corp., San Leandro, CA). β-actin was used as a house-keeping gene control. The displayed image is one of four replicate samples and is typical of those obtained. B. Quantitation of IL-8 and PBEF mRNA levels by the densitometry analysis. The mRNA band intensity on the gel image was analyzed by the AlphaEase Stand Alone software (Alpha Innotech Corp., San Leandro, CA). The integrative density value of each band was used as a measure of the mRNA level. Results from each group are presented as mean ± SD of 4 samples from two separate experiments. Statistical comparative analyses of IL-8 and PBEF mRNA levels between the control and IL-1β treated group were performed using the unpaired t test. **, p<0.01.
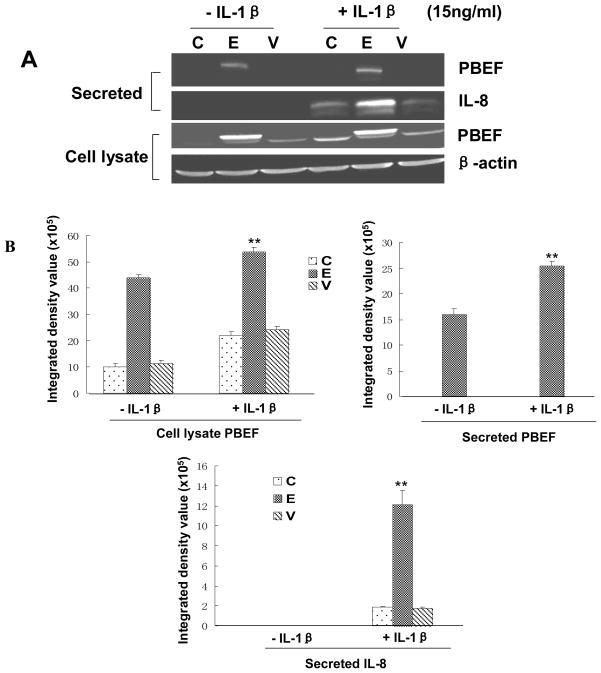
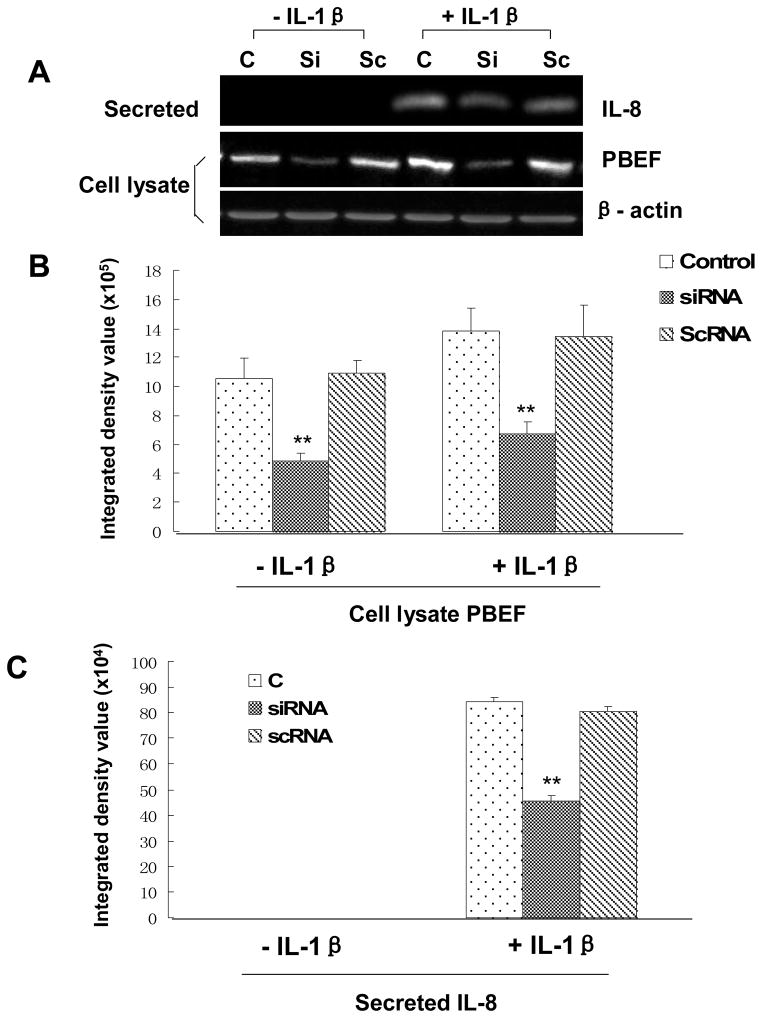
PBEF overexpression augmented IL-8 secretion from A549 cells and HPAEC
We first tested whether the pCAGGS-mPBEF construct could indeed increase PBEF expression in A549 cells. As presented in Fig. 7, the overexpression of PBEF in A549 cells transiently transfected with pCAGGS-mPBEF was achieved under basal conditions (- IL-1β), as evidenced in the medium [15.98 × 105 ± 1.1 × 104 vs. 0.0 ± 0.0 (vector & control), n = 3, **P < 0.01]. In IL-1β-stimulated conditions, PBEF level in the medium was further increased compared to the basal conditions [25.43 × 105 ±0.9 × 104 vs. 15.98× 105 ± 1.1 × 104, n = 3, **P < 0.01]. Similar observations were obtained with PBEF protein expression in the cell lysate of A549 cells transiently transfected with pCAGGS-mPBEF while the pCAGGS vector control group did not show any change in PBEF protein expression level. These results clearly show that pCAGGS-mPBEF vector increased PBEF expression in both basal and IL-1β activated A549 cells.
Fig. 7.
Effects of PBEF overexpression on IL-8 secretion in A549 cells. A. A representative western blotting image of IL-8, PBEF and β-actin protein detections. After starving for serum overnight, A549 cells were transfected with the vehicle control (C), pCAGGS-mPBEF vector (E), or pCAGGS vector (V) for 48 hours before subjected to the treatment without or with IL-1β (15 ng/ml) for 24 hours. Secreted IL-8 and PBEF, cell lysate PBEF and β-actin protein were immunodetected as described in Figure 1. B. Quantitation of cell lysate and secreted PBEF as well as secreted IL-8 levels protein levels by the densitometry analysis. Cell lysate & secreted PBEF and IL-8 protein levels in both control and PBEF overexpressing groups were quantified by densitometry analyses in the same way as described above in Figure 1.**, p<0.01.
With a significant increase of PBEF expression in A549 cells by pCAGGS-mPBEF vector, we next examined the effect of PBEF overexpression on the IL-1β-induced increases in IL-8 secretion from A549 cells. Figure 7 demonstrated that PBEF overexpression significantly increased IL-8 secretion from A549 cells compared to the control group [12.09 × 105 ± 1.43 × 105 vs. 1.85 × 105 ± 1.0 × 104 (control), n = 3, **P < 0.01]. A similar phenomenon was also observed in HPAEC as presented in Fig. 8. PBEF overexpression significantly increased IL-8 secretion from HPAEC compared to the control group [25.18 × 105 ±1.03 × 105 vs. 10.59 × 105 ± 0.31 × 105 (control), n = 3, **P < 0.01]. These results indicate that overexpression of PBEF could augment IL-8 secretion in both basal and IL-1β-stimulated A549 cells and HPAEC.
Fig. 8.
Effects of PBEF overexpression on IL-8 secretion in HPAEC. A. A representative western blotting image of IL-8, PBEF and β-actin protein detections. HPAEC were treated n the same way as those in A549 cells as described above in Figure 5. Secreted IL-8, cell lysate PBEF and β-actin protein were immunodetected in the same way as described above in Figure 1. B. Quantitation of cell lysate PBEF protein levels by densitometry analysis. C. Quantitation of secreted IL-8 levels by densitometry analysis. Secreted IL-8 and cell lysate PBEF protein levels were quantified by densitometry analyses in the same way as those in A549 cells as described above in Figure 5. **, p<0.01.
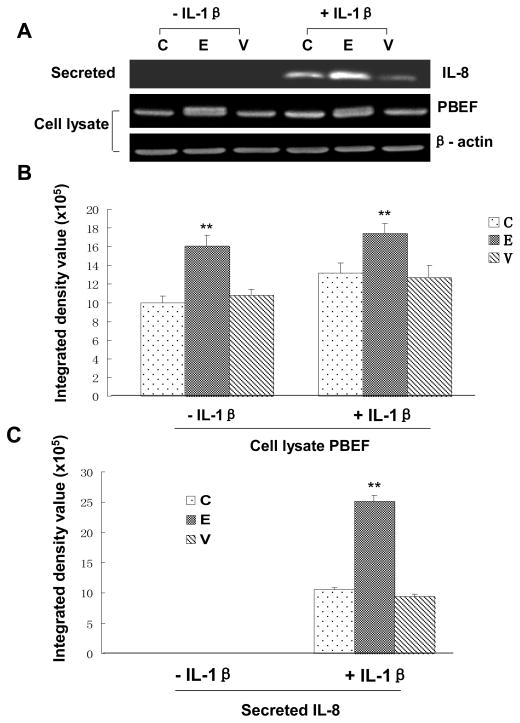
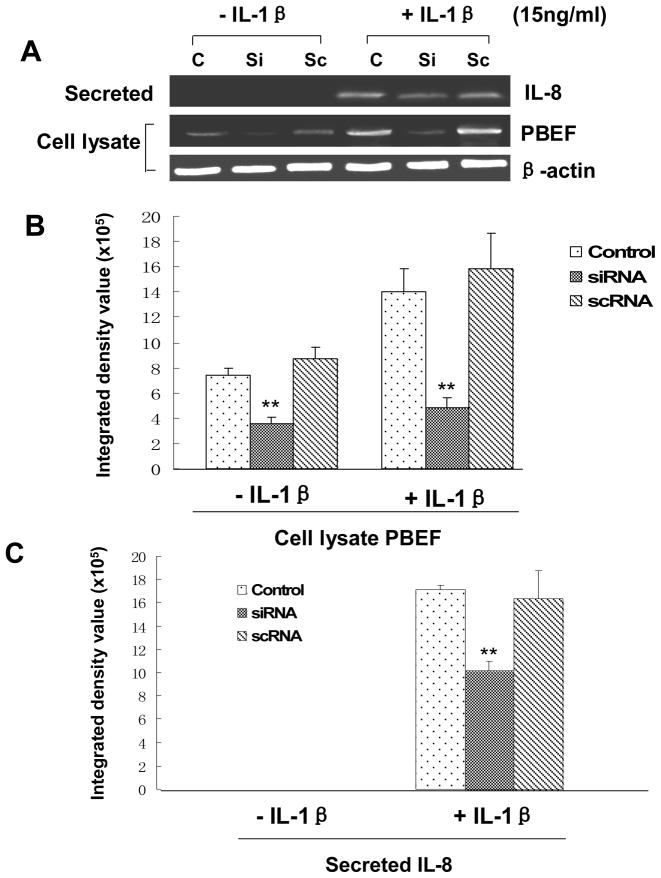
PBEF silencing attenuated IL-8-induced increases in IL-8 secretion from A549 cells and HPAEC
Since PBEF overexpression could augment IL-1β induced IL-8 secretion from A549 cells, we wanted to know whether PBEF silencing would decrease IL-8 secretion from A549 cells. Firstly, we performed a pilot experiment and found that PBEF stealth siRNA indeed knocked down PBEF expression in A549 cells. As presented in Fig. 9, under basal conditions (- IL-1β), PBEF siRNA significantly knocked down PBEF protein expression in A549 cells [3.58 × 105 ± 0.46 × 105 (siRNA) vs. 7.40 × 105 ± 0.54 × 105 (control), n = 3, **P < 0.01]. Scrambled RNA had no significant effect on the PBEF protein expression [8.68 × 105 ±0.93 × 105 (scRNA) vs. 7.40 × 105 ± 0.54 × 105 (control), n = 3, NS]. PBEF siRNA had no effect on the protein expression level of β-actin, a house-keeping gene serving as a control. In the treatment group (+IL-1β), PBEF siRNA significantly blunted the IL-1β effect [4.77 × 105 ± 0.90 × 105 (siRNA) vs. 14.05 × 105 ± 1.77 × 105 (control), n = 3, **P < 0.01] while PBEF protein level in the scRNA group had no change relative to the control group[15.83 × 105 ± 2.82 × 105 (scRNA) vs. 14.05 × 105 ± 1.77 × 105 (control), n = 3, NS]. This result indicates that PBEF siRNA can knock down PBEF expression in both basal and IL-1β activated A549 cells.
Fig. 9.
Effects of PBEF knock down on IL-8 secretion in A549 cells. A. A representative western blotting image of IL-8, PBEF and β-actin protein detections. After starving for serum overnight, A549 cells were transfected with the vehicle control (C), scrambled siRNA (Sc), or PBEF stealth siRNA (Si) for 48 h before subjected to treatment without or with IL-1β (15 ng/ml) for 24 hours. Secreted IL-8, cell lysate PBEF and β-actin protein were immunodetected as described in Figure 1 B. Quantitation of cell lysate PBEF protein levels by densitometry analysis. C. Quantitation of secreted IL-8 levels by densitometry analysis. Cell lysate PBEF and secreted IL-8 protein levels in various treatments of A549 cells were quantified by densitometry analyses in the same way as described above in Figure 1.**, p<0.01.
With an efficient knock down of PBEF in A549 cells by PBEF siRNA, we next examined the effect of PBEF knockdown on the IL-1β-induced increase of IL-8 secretion from A549 cells. Fig. 9 demonstrated that PBEF silencing significantly decreased IL-8 secretion from A549 cells in IL-1β-stimulated conditions compared to the control group [10.18 × 105 ± 0.82 × 105 (siRNA ) vs. 17.14 × 105 ± 0.35 × 105 (control), n = 3,**P < 0.01], while secreted IL-8 level in the scRNA group has no change relative to the control group. [16.38 × 105 ± 0.24 × 105 (scRNA ) vs. 17.14 × 105 ± 0.35 × 105 (control) n = 3, NS]. A similar phenomenon was also observed in HPAEC as presented in Fig. 10. PBEF knock down significantly decreased IL-8 secretion from HPAEC compared to the control group [15.23 × 105 ±1.47 × 105vs. 26.23 × 105 ± 0.23 × 105 (control), n = 3, **P < 0.01]. These results indicate that PBEF is involved in IL-8 expression and secretion under both basal and IL-1β induced conditions in A549 cells and HPAEC.
Fig. 10.
Effects of PBEF knock down on IL-8 secretion in HPAEC. A. A representative western blotting image of IL-8, PBEF and β-actin protein detections. HPAEC were treated in the same way as those in A549 cells as described above in Figure 7. Secreted IL-8, cell lysate PBEF and β-actin protein were immunodetected in the same way as described above in Figure 1. B. Quantitation of cell lysate PBEF protein levels by densitometry analysis. C. Quantitation of secreted IL-8 levels by densitometry analysis. Secreted IL-8 and cell lysate PBEF protein levels were quantified by densitometry analyses in the same way as those in A549 cells as described above in Figure 7. **, p<0.01.
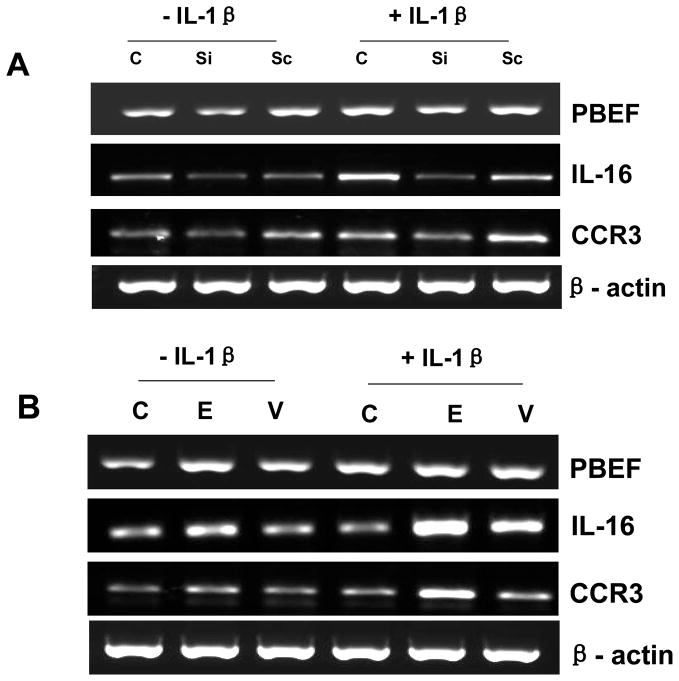
Effect of PBEF expression on IL-16 and CCR3 gene expression
To further investigate the relationship between PBEF and other inflammatory cytokines and the function of PBEF in the inflammation pathway, we examined the effect of PBEF expression on IL-16 and CCR3 gene expression. We selected IL-16 and CCR3 because based on our unpublished observation, knockdown of PBEF expression significantly inhibited the expression of genes involved in inflammation including IL-16 and CCR3 (Ye et al., unpublished observation). Results in Fig. 11 showed that knockdown of PBEF in A549 cells could attenuate IL-1β-stimulated increase of IL-16 and CCR3 gene expression, while the overexpression of PBEF in A549 cells could promote IL-1β-stimulated increase of IL-16 and CCR3 gene expression at the mRNA level. Taken together, these data implicate PBEF might be an important signal transducer or initiator in the inflammation pathway.
Fig. 11.
RT-PCR analyses of PBEF, IL-16, CCR3 and β-actin mRNA levels in A549 cells. A549 cells were grown and transfected with the vehicle control (C), PBEF stealth siRNA (Si), scrambled siRNA (Sc), pCAGGS-mPBEF vector (E), or pCAGGS vector (V) for 48 h before subjected to the treatment without or with IL-1β(15 ng/ml) for 4 hours. Total cell RNA was reverse-transcribed, amplified by PCR using the gene specific primers (Table 1), separated by 1.5 % agarose electrophoresis and visualized with ethidium bromide. A. A representative RT-PCR gel image of PBEF, IL-16, CCR3 and β-actin mRNA detections in PBEF-knockdown A549 cells. B. A representative RT-PCR gel image of PBEF, IL-16, CCR3 and β-actin mRNA detections in PBEF-overexpression A549 cells.
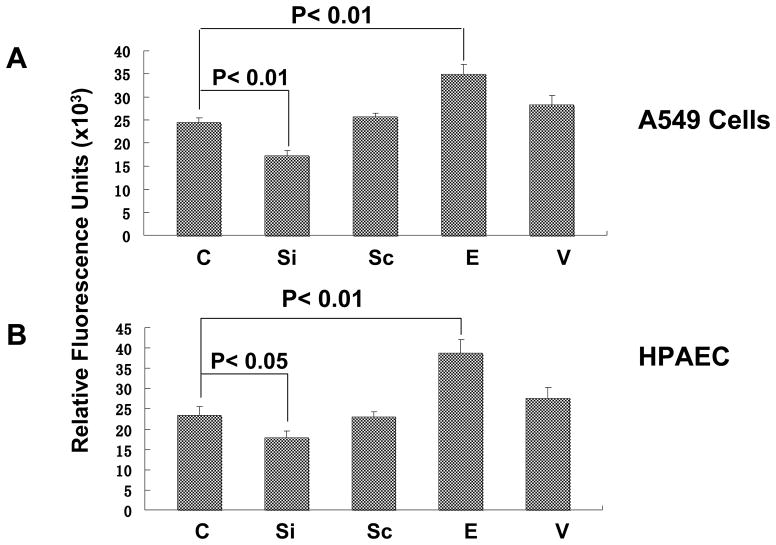
PBEF expression affected cell permeability in A549 cells and HPAEC
To assess whether PBEF expression affected epithelial cell and HPAEC permeability, we performed in vitro monolayer cell permeability assay in A549 cells and HPAEC transfected with PBEF stealth siRNA, pCAGGS-mPBEF vector and other controls with IL-1β treatment. As presented in Fig. 12, PBEF siRNA significantly decreased IL-1β induced A549 cell permeability level in the PBEF siRNA treated group compared to the IL-1β alone treatment (17333.33 ±1030.24 vs. 24333.33 ±820.56 relative fluorescence units, n = 4, **P < 0.01). Similar observations were obtained in HPAEC, whose cell permeability level in the PBEF siRNA group significantly decreased compared to the IL-1β alone treatment (17833.33 ± 1120.2 vs. 23505 ± 1690.5 relative fluorescence units, n = 4, *P < 0.05). Scrambled RNA had no effect on the IL-1β induced cell permeability in both A549 cells and HPAEC. These results indicate PBEF siRNA significantly attenuated the IL-1β induced barrier-disruption and higher permeability in epithelial cells and endothelial cells. Figure 12 also demonstrated that PBEF overexpression significantly promoted IL-1β induced A549 cell permeability level in the pCAGGS-mPBEF transfected group compared to the IL-1β alone treatment (34950.09 ± 2093.69 vs. 24333.33 ±820.56 relative fluorescence units, n = 4, **P < 0.01). Similar observations were obtained in HPAEC, whose cell permeability level in the PBEF overexpression group significantly increased compared to the IL-1β alone treatment (38833.33 ± 2644.86 vs. 23505 ± 1690.5 relative fluorescence units, n = 4, **P < 0.01). These results indicate that overexpression of PBEF in A549 cells and HPAEC augmented IL-1β induced lung epithelial cell barrier dysfunction and thus increased cell permeability in vitro.
Fig. 12.
Effects of PBEF expression on in vitro cell permeability of A549 cells and HPAEC. A549 cells or HPAEC were transfected with the vehicle control (C), PBEF stealth siRNA (Si), scrambled siRNA (Sc), pCAGGS-mPBEF vector (E), or pCAGGS vector (V) for 48 h and then seeded onto the culture inserts of permeability assay chambers. The cells were then subjected to the treatment with 15 ng/ml IL-1β for 18 hours. Then, FITC-Dextran was added to each insert for 5 min at room temperature. FITC-Dextran leaking out into the bottom chamber was assayed using a fluorometer. Relative fluorescence units in each group were used as an indicator of cell permeability. A. Effects of PBEF expression on IL-1β-mediated increase of in vitro cell permeability of A549. B. Effects of PBEF expression on IL-1β-mediated increase of in vitro cell permeability of HPAEC. *P < 0.05, **P < 0.01.
DISCUSSION
The data from this study indicate that IL-1β significantly induced the PBEF expression at a transcriptional level in HPAEC. PBEF overexpression augmented IL-8 secretion from A549 cells and HPAEC. The knockdown of PBEF expression by PBEF siRNA significantly blunted IL-1β induced IL-8 secretion and its production in A549 cells and HPAEC. These results support that PBEF is critically involved in pulmonary inflammation.
ALI/ARDS is characterized by an acute inflammatory process in the airspaces and lung parenchyma. Evidence from several clinical studies indicates that a complex network of inflammatory cytokines and chemokines play a major role in mediating, amplifying, and perpetuating the lung injury process (Goodman et al., 2003). The pro-inflammatory cytokines IL-1β, TNFα and IL-8 have been identified in bronchoalveolar lavage fluids (BALF) from ARDS patients. Because IL-1β can stimulate the production of a variety of chemotactic cytokines such as interleukin-8 (IL-8), epithelial cell neutrophil activator, monocyte chemotactic peptide, and macrophage inflammatory peptide-1α, IL-1β has earned a position of prominence at the head of the inflammatory cytokine cascade (Goodman et al., 2003). In clinical fluids there are strong relationships between IL-1β and clinical lung injury severity and outcome in patients with ARDS. These clinical studies indicate that IL-1β probably has an important role in the early pathogenesis of ALI and ARDS (Park et al., 2001). Among the IL-1β induced pro-inflammatory cytokines, IL-8 is regarded as one of the most important mediators in the pathogenesis of ARDS. In broncho-alveolar fluid, IL-8 levels significantly increased in patients with ARDS and correlated with the development of ARDS in at-risk patients (Adams et al., 2001; Hudson, 1982). IL-8 has been identified as one of biomarkers of ALI/ARDS mortality (Ware, 2005). In fact, IL-8 was first purified and molecularly cloned as a neutrophil chemotactic factor from lipopolysaccharide-stimulated human mononuclear cell supernatants (Matsushima et al., 1988). Since then, studies of models of acute inflammation have established IL-8 as a key mediator in neutrophil mediated acute inflammation (Mukaida, 2003). In acid aspiration- and endotoxemia-induced ARDS in rabbits, IL-8 is produced in the lungs (Folkesson et al., 1995; Yokoi et al., 1997). In both models, the abrogation of IL-8 activity reduces neutrophil infiltration as well as tissue damage. It was assumed that locally produced IL-8 can suppress neutrophil apoptosis (Dunican et al., 2000), induce neutrophil migration into the lungs and damage to lung tissues, including alveolar epithelial barrier function (Modelska et al., 1999). The data in this study indicate that IL-1β significantly induced the PBEF expression at both mRNA and protein levels in A549 cells and HPAEC (Figures 1–3, 6). Our results also revealed that a transcriptional regulation mechanism is at least in part responsible for the IL-1β induced upregulation of the PBEF gene expression (Figure 4). Furthermore, we found that a -1535 T-variant in the human PBEF gene promoter had significantly attenuated its binding to an IL-1β induced unknown transcription factor compared to its common -1535 C-allele (Fig. 5). Previously, we had shown that this -1535 T-variant was associated with a lower susceptibility to ALI than that of -1535C-allele and the T variant from the SNP C-1543T resulted in a significant decrease in the transcription rate (1.8-fold; p<0.01) by the reporter gene assay (Ye et al., 2005). The altered transcriptional factor binding may underlie the reduced expression of PBEF and thus less susceptibility to acute lung injury in those -1535T carriers. Our findings in this study suggest that PBEF may be a target of IL-1β involved in the inflammatory process during the pathogenesis of ALI. The knockdown of PBEF expression by PBEF siRNA significantly blunted IL-1β-stimulated IL-8 secretion and its production in A549 cells and HPAEC (Figures 6–10), and PBEF overexpression augmented IL-8 secretion from A549 cells (Fig. 7). Our results (Fig. 11) also showed that the knockdown of PBEF in A549 cells attenuated IL-1β-stimulated increase of IL-16 and CCR3 gene expression, while the overexpression of PBEF in A549 cells promoted IL-1β-stimulated increase of IL-16 and CCR3 gene expression at mRNA level. IL-16 and CCR3 are among multiple cytokines which play a critical role in orchestrating and perpetuating inflammation in asthma and chronic obstructive pulmonary disease (COPD) although their roles in acute lung injury were not well characterized yet. IL-16 is a T-cell chemoattractant produced by leukocytes. Wang et al reported that IL-16 was a critical factor in the development of inflammation-mediated renal injury and may be a therapeutic target for prevention of ischemia-reperfusion injury of the kidney (Wang et al., 2008). A plethora of studies have also implicated IL-16 in exacerbation of infectious, immune-mediated, and autoimmune inflammatory disorders, including atopic dermatitis, irritable bowel syndrome, systemic lupus erythematosus, neurodegenerative disorders, and viral infections (Glass et al., 2006). CCR3 is the principal mediator of eosinophil chemotaxis and is expressed on a variety of inflammatory cells. These cells include basophils, mast cells and T-helper-2 lymphocytes, and resident tissue cells such as airway epithelium. Animal studies suggest that CCR3 is a prominent mediator of allergic responses and that antagonizing the receptor will lead to a reduction in airway inflammation (De Lucca, 2006). IL-16 and CCR3 may be among the repertoire of bioactive molecules regulated by the PBEF gene and implicated in the pathogenesis of acute lung injury or other inflammatory diseases. These data implicate PBEF might be an important “master” signal transducer or initiator in the inflammation pathway to regulate the synthesis of IL-8 or other inflammatory cytokines and PBEF could play a critical role as an inflammatory cytokine during the pathogenesis of ALI.
Both endothelial and epithelial injuries were observed by earlier seminal ultrastructural studies of the lung in patients dying with ALI secondary to sepsis (Bachofen and Weibel, 1977; 1982). The vascular endothelial cell monolayer serves as a dynamic, semiselective barrier that regulates transport of fluid and macromolecules between blood and the interstitium. A variety of physical, inflammatory, and bioactive stimuli alter the endothelial barrier, leading to formation of paracellular gaps thereby increasing vessel permeability and compromising organ function (Csortos et al., 2007; Dudek and Garcia, 2001; Mehta and Malik, 2006). Increased vascular permeability contributes to the profound pathophysiological derangements in ALI (De Lucca, 2006). Epithelial injury in the lungs is one of the hallmarks of ALI in humans (Liener et al., 2002). During the process of acute inflammation, endothelial permeability was increased rapidly. If alveolar epithelium is also damaged, the change in both endothelial and epithelial permeability could lead to major alveolar flooding with high-molecular-weight proteins, with prolonged changes in gas exchange and a much higher likelihood of disordered repair (Parsons et al., 2005). Normally, the lung alveolar epithelium is an extremely tight barrier that restricts the movement of proteins and liquid from the interstitium into the alveolar spaces. In ALI, impaired alveolar epithelial function in the lungs is a marker of poor outcome of ALI (Thomas et al., 2005). A number of inflammatory cytokines including IL-1β and IL-8 can induce or aggravate the inflammation of endothelial and epithelial cells, leading to their barrier dysfunctions (Leone et al., 2007). Results in Figure 12 of this study have demonstrated that overexpression of PBEF augmented both basal and IL-1β induced increase in the cell permeability of both A549 cells and HPAEC while the knockdown of PBEF expression by PBEF siRNA significantly attenuated IL-1β-induced cell permeability of A549 cells and HPAEC, suggesting that PBEF may have an important role in both endothelial and epithelial cell barrier regulation. A549 cells are a Type II tumor epithelial cell line. While the type II epithelial cell only covers 7% of the alveolar surface area it constitutes 67% of the epithelial cell number within the alveoli (Crapo et al., 1982) pointing to its biochemical importance. The permeability increase of A549 cells in the IL-1β-stimulated condition involved both paracellular permeability, with gap formation visualized by actin cytoskeleton staining, and basement membrane permeability (Lacherade et al., 2001). This IL-1β mediated effect may be partly due to its induction of IL-8 production. IL-8 was demonstrated to induce the actin fiber formation and intercellular gap formation of endothelial cells (Schraufstatter et al., 2001). IL-8 could have the same effect on epithelial barrier function. Our previous study found that reductions in PBEF protein expression significantly attenuated endothelial barrier dysfunction induced by the potent edemagenic agent, thrombin, reflected by reductions in transendothelial electric resistance (Ye et al., 2005). Furthermore, we found that reductions in PBEF protein expression blunted thrombin-mediated increases in Ca2+ entry, polymerized actin formation, and myosin light chain phosphorylation, events critical to the thrombin-mediated permeability response (Ye et al., 2005). It remains to be elucidated whether similar mechanisms operate to account for the attenuating effects of PBEF knockdown on IL-1β induced epithelial and endothelial cell barrier dysfunction. It should be mentioned that a similar observations as found in A549 cells were also obtained in a primary human lung small airway epithelial cells (Cat. No. CC-2547, Lonza, Walkersville, MD, USA) (data not shown).
In summary, our results demonstrated that a transcriptional regulation mechanism is at least in part responsible for the IL-1β induced upregulation of the PBEF gene expression and an altered transcriptional factor binding may underlie the reduced expression of PBEF and thus less susceptibility to acute lung injury in those -1535T carriers. We found that the overexpression of PBEF significantly augmented IL-8 secretion and mRNA expression by more than 6 fold and 2 fold, respectively, in A549 and HPAEC. It also significantly augmented IL-1β mediated cell permeability by 44% in A549 cells and 65% in endothelial cells. The knockdown of PBEF expression significantly inhibited IL-1β-stimulated IL-8 secretion and mRNA level by 60% and 70%, respectively; and the knockdown of PBEF expression also significantly attenuated IL-1β-induced cell permeability by 29% in epithelial cells and 24% in endothelial cells. Furthermore, we found that PBEF expression could also affect the expression of IL-16 and CCR3 genes, two inflammatory cytokines. These results suggest that PBEF may play a critical role as an inflammatory cytokine in the development of pulmonary inflammation and dysregulation of pulmonary vascular endothelial and alveolar epithelial cell barriers, which may be an important mechanism underlying PBEF in the pathogenesis of ALI. These results lend further support that PBEF may represent a new diagnostic and therapeutic target in ALI.
Acknowledgments
This work was in part supported by National Heart, Lung, and Blood Institute Grants HL 080042 (Ye, SQ), 2P01HL058064-070001 (Garcia, JGN) and the University of Missouri-Columbia Start-up fund.
References
- Adams JM, Hauser CJ, Livingston DH, Lavery RF, Fekete Z, Deitch EA. Early trauma polymorphonuclear neutrophil responses to chemokines are associated with development of sepsis, pneumonia, and organ failure. J Trauma. 2001;51:452–456. doi: 10.1097/00005373-200109000-00005. [DOI] [PubMed] [Google Scholar]
- Bachofen M, Weibel ER. Alterations of the gas exchange apparatus in adult respiratory insufficiency associated with septicemia. Am Rev Respir Dis. 1977;116:589–615. doi: 10.1164/arrd.1977.116.4.589. [DOI] [PubMed] [Google Scholar]
- Bachofen M, Weibel ER. Structural alterations of lung parenchyma in the adult respiratory distress syndrome. Clin Chest Med. 1982;3:35–56. [PubMed] [Google Scholar]
- Bajwa EK, Yu C, Gong MN, Thompson BT, Christiani DC. PBEF gene polymorphisms influence the risk of developing ARDS. Am J Respir Crit Care Med. 2006;3:A272. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(7):248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Crapo JD, Barry BE, Gehr P, Bachofen M, Weibel ER. Cell number and cell characteristics of the normal human lung. Am Rev Respir Dis. 1982;126:332–337. doi: 10.1164/arrd.1982.126.2.332. [DOI] [PubMed] [Google Scholar]
- Csortos C, Kolosova I, Verin AD. Regulation of vascular endothelial cell barrier function and cytoskeleton structure by protein phosphatases of the PPP family. Am J Physiol Lung Cell Mol Physiol. 2007;293(4):L843–854. doi: 10.1152/ajplung.00120.2007. [DOI] [PubMed] [Google Scholar]
- De Lucca GV. Recent developments in CCR3 antagonists. Curr Opin Drug Discov Devel. 2006;9(4):516–524. [PubMed] [Google Scholar]
- Dudek SM, Garcia JG. Cytoskeletal regulation of pulmonary vascular permeability. J Appl Physiol. 2001;91(4):1487–1500. doi: 10.1152/jappl.2001.91.4.1487. [DOI] [PubMed] [Google Scholar]
- Dunican AL, Leuenroth SJ, Grutkoski P, Ayala A, Simms HH. TNFalpha-induced suppression of PMN apoptosis is mediated through interleukin-8 production. Shock. 2000;14(3):284–288. doi: 10.1097/00024382-200014030-00007. [DOI] [PubMed] [Google Scholar]
- Folkesson HG, Matthay MA, Hebert CA, Broaddus VC. Acid aspiration-induced lung injury in rabbits is mediated by interleukin-8-dependent mechanisms. J Clin Invest. 1995;96:107–116. doi: 10.1172/JCI118009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass WG, Sarisky RT, Vecchio AM. Not-so-sweet sixteen: the role of IL-16 in infectious and immune-mediated inflammatory diseases. J Interferon Cytokine Res. 2006;26(8):511–520. doi: 10.1089/jir.2006.26.511. [DOI] [PubMed] [Google Scholar]
- Goodman RB, Pugin J, Lee JS, Matthay MA. Cytokine-mediated inflammation in acute lung injury. Cytokine Growth Factor Rev. 2003;14(6):523–535. doi: 10.1016/s1359-6101(03)00059-5. [DOI] [PubMed] [Google Scholar]
- Hudson LD. Causes of the adult respiratory distress syndrome--clinical recognition. Clin Chest Med. 1982;3:195–212. [PubMed] [Google Scholar]
- Lacherade JC, Van LA, Planus E, Escudier E, D’Ortho MP, Lafuma C, Harf A, Delclaux C. Evaluation of basement membrane degradation during TNF-alpha-induced increase in epithelial permeability. Am J Physiol Lung Cell Mol Physiol. 2001;281(1):134–143. doi: 10.1152/ajplung.2001.281.1.L134. [DOI] [PubMed] [Google Scholar]
- Leaver SK, Evans TW. Acute respiratory distress syndrome. BMJ. 2007;335(7616):389–394. doi: 10.1136/bmj.39293.624699.AD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone AK, Chun JA, Koehler CL, Caranto J, King JM. Effect of proinflammatory cytokines, tumor necrosis factor-alpha and interferon-gamma on epithelial barrier function and matrix metalloproteinase-9 in Madin Darby canine kidney cells. Cell Physiol Biochem. 2007;19(1–4):99–112. doi: 10.1159/000099198. [DOI] [PubMed] [Google Scholar]
- Levitt JE, Matthay MA. Treatment of acute lung injury: historical perspective and potential future therapies. Semin Respir Crit Care Med. 2006;27(4):426–437. doi: 10.1055/s-2006-948296. [DOI] [PubMed] [Google Scholar]
- Liener UC, Brückner UB, Knöferl MW, Steinbach G, Kinzl L, Gebhard F. Chemokine activation within 24 hours after blunt accident trauma. Shock. 2002;17:169–172. doi: 10.1097/00024382-200203000-00002. [DOI] [PubMed] [Google Scholar]
- Matsushima K, Morishita K, Yoshimura T, Lavu S, Kobayashi Y, Lew W, Appella E, Kung HF, Leonard EJ, Oppenheim JJ. Molecular cloning of a human monocyte-derived neutrophil chemotactic factor (MDNCF) and the induction of MDNCF mRNA by interleukin 1 and tumor necrosis factor. J Exp Med. 1988;167(6):1883–1893. doi: 10.1084/jem.167.6.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlothlin JR, Gao L, Lavoie T, Simon BA, Easley RB, Ma SF, Rumala BB, Garcia JG, Ye SQ. Molecular cloning and characterization of canine pre-B-cell colony-enhancing factor. Biochem Genet. 2005;43(3–4):127–41. doi: 10.1007/s10528-005-1505-2. [DOI] [PubMed] [Google Scholar]
- Mehta D, Malik AB. Signaling mechanisms regulating endothelial permeability. Physiol Rev. 2006;86:279–367. doi: 10.1152/physrev.00012.2005. [DOI] [PubMed] [Google Scholar]
- Modelska K, Pittet JF, Folkesson HG, Courtney V, Matthay MA. Acid-induced lung injury. Protective effect of anti-interleukin-8 pretreatment on alveolar epithelial barrier function in rabbits. Am J Respir Crit Care Med. 1999;160:1450–1456. doi: 10.1164/ajrccm.160.5.9901096. [DOI] [PubMed] [Google Scholar]
- Mukaida N. Pathophysiological roles of interleukin-8/CXCL8 in pulmonary diseases. Am J Physiol Lung Cell Mol Physiol. 2003;284(4):L566–577. doi: 10.1152/ajplung.00233.2002. [DOI] [PubMed] [Google Scholar]
- Park WY, Goodman RB, Steinberg KP, Ruzinski JT, Radella F, 2nd, Park DR, Pugin J, Skerrett SJ, Hudson LD, Martin TR. Cytokine balance in the lungs of patients with acute respiratory distress syndrome. Am J Respir Crit Care Med. 2001;164:1896–1903. doi: 10.1164/ajrccm.164.10.2104013. [DOI] [PubMed] [Google Scholar]
- Parsons PE, Eisner MD, Thompson BT, Matthay MA, Ancukiewicz M, Bernard GR. Lower tidal volume ventilation and plasma cytokine markers of inflammation in patients with acute lung injury. Crit Care Med. 2005;33:1–6. doi: 10.1097/01.ccm.0000149854.61192.dc. [DOI] [PubMed] [Google Scholar]
- Schraufstatter IU, Chung J, Burger M. IL-8 activates endothelial cell CXCR1 and CXCR2 through Rho and Rac signaling pathways. Am J Physiol Lung Cell Mol Physiol. 2001;280(6):L1094–1103. doi: 10.1152/ajplung.2001.280.6.L1094. [DOI] [PubMed] [Google Scholar]
- Thomas R, Martin NH, Morio N, Gustavo MB. Apoptosis and epithelial injury in the lungs. Proc Am Thorac Soc. 2005;2:214–220. doi: 10.1513/pats.200504-031AC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Diao H, Guan Q, Cruikshank WW, Delovitch TL, Jevnikar AM, Du C. Decreased renal ischemia-reperfusion injury by IL-16 inactivation. Kidney Int. 2008;73(3):318–326. doi: 10.1038/sj.ki.5002692. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xiao L, Thiagalingam A, Nelkin BD, Casero RA., Jr The identification of a cis-element and a trans-acting factor involved in the response to polyamines and polyamine analogues in the regulation of the human spermidine/spermine N1-acetyltransferase gene transcription. J Biol Chem. 1998;273(51):34623–30. doi: 10.1074/jbc.273.51.34623. [DOI] [PubMed] [Google Scholar]
- Ware LB. Prognostic determinants of acute respiratory distress syndrome in adults: impact on clinical trial design. Crit Care Med. 2005;33(3 Suppl):217–222. doi: 10.1097/01.ccm.0000155788.39101.7e. [DOI] [PubMed] [Google Scholar]
- Ye SQ, Simon B, Maloney JP, Zambelli-Weiner A, Gao L, Grant A, Easley RB, McVerry B, Tuder RM, Standiford T, Brower R, Barnes K, Garcia JGN. Pre-B-cell colony-enhancing factor as a potential novel biomarker in acute lung injury. Am J Respir Crit Care Med. 2005;171 (2):361–370. doi: 10.1164/rccm.200404-563OC. [DOI] [PubMed] [Google Scholar]
- Ye SQ, Zhang LQ, Adyshev D, Usatyuk PV, Garcia A, Lavoie TL, Verin AD, Natarajan V, Garcia JGN. Pre-B-cell-colony-enhancing factor is critically involved in thrombin-induced lung endothelial cell barrier dysregulation. Microvasc Res. 2005;70 (3):142–51. doi: 10.1016/j.mvr.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Yokoi K, Mukaida N, Harada A, Watanabe Y, Matsushima K. Prevention of endotoxemia-induced acute respiratory distress syndrome-like lung injury in rabbits by a monoclonal antibody to IL-8. Lab Invest. 1997;76:375–384. [PubMed] [Google Scholar]