Abstract
Pluripotent stem cells are attractive candidates as a cell source for regenerative medicine and tissue engineering therapies. Current methods of differentiation result in low yields and impure populations of target phenotypes, with attempts for improved efficiency often comparing protocols that vary multiple parameters. This basic science study focused on a single variable to understand the effects of two- versus three- dimensional culture on directed differentiation. We compared mouse embryonic stem cells (ESCs) differentiated on collagen type I-coated surfaces (SLIDEs), embedded in collagen type I gels (GELs), and in suspension as embryoid bodies (EBs). For a systematic analysis in these studies, key parameters were kept identical to allow for direct comparison across culture configurations. We determined that all three configurations supported differentiation of ESCs and that the kinetics of differentiation differed greatly for cells cultured in 2D versus 3D. SLIDE cultures induced overall differentiation more quickly than 3D configurations, with earlier expression of cytoskeletal and extracellular matrix proteins. For 3D culture as GELs or EBs, cells clustered similarly, formed complex structures, and promoted differentiation towards cardiovascular phenotypes. GEL culture, however, also allowed for contraction of the collagen matrix. For differentiation towards fibroblasts and smooth muscle cells which actively remodel their environment, GEL culture may be particularly beneficial. Overall, this study determined the effects of dimensionality on differentiation and helps in the rational design of protocols to generate phenotypes needed for tissue engineering and regenerative medicine.
Keywords: collagen, differentiation, embryoid body, embryonic stem cell, regenerative medicine, three-dimensional culture, tissue engineering
INTRODUCTION
Despite numerous advances in tissue engineering and regenerative medicine, cell sourcing remains a significant hurdle for the translation of therapies from the bench to clinic [Gimble et al., 2007; Peister et al., 2011]. Regenerative medicine and tissue engineering approaches, including the creation of tissues for transplantation and in vitro disease models, benefit from continuously available, functional, and pure cellular phenotypes. Pluripotent stem cells are considered good candidates for regenerative medicine applications due to their ability to self-renew and potential to become any cell in the adult body. For example, embryonic stem cells can spontaneously differentiate in vitro into the three germ lineages (ectoderm, mesoderm, or endoderm), from which will arise all somatic cell types [Martin, 1981; Thomson et al., 1998; Itskovitz-Eldor et al., 2000].
Embryonic stem cells have been differentiated to cells of all three germ lineages in both two and three dimensional physical microenvironments. The common configurations of cells on a monolayer (refs for 2D differentiation), embedded in protein gels [Bosnakovski et al., 2006; Gerecht et al., 2007] and in suspension [Itskovitz-Eldor et al., 2000; Dang et al., 2002] have both advantages and disadvantages. Stem cells cultured on adherent surfaces can be presented with bound proteins [Nishikawa et al., 1998; Schenke-Layland et al., 2007] and well-controlled exogenous physical cues, such as cyclic tension [Saha et al., 2006; Doyle et al., 2009] and shear stress [Ahsan and Nerem, 2010; Nikmanesh et al., 2012; Wolfe et al., 2012]. Yet, culture in this 2D configuration restricts cell growth to a single geometric plane. Suspension culture, which allows for the formation of cell clusters during spontaneous differentiation (or embryoid bodies: EBs), can mimic cellular interactions reminiscent of in vivo development processes [Boheler et al., 2002] but only allows external stimuli in the form of soluble factors or hydrodynamic forces [Fuchs et al., 2012]. Encapsulation of cells within scaffolds or hydrogels, however, enables both the presentation of proteins [Gerecht et al., 2007; Oh et al., 2012; Trappmann et al., 2012] and the application of mechanical cues [Powers et al., 2002; Cullen et al., 2007], while maintaining the cells in the rounded configuration often prevailing in vivo (Reviewed in [Devolder and Kong, 2012]). Although these different 2D and 3D modalities have been used in conjunction with other exogenous cues to promote directed differentiation, there has not yet been a systematic analysis to determine the fundamental effects of dimensionality on differentiation.
This study characterizes the overall differentiation of pluripotent stem cells cultured in both two- and three- dimensions. In particular, mouse embryonic stem cells (ESCs) were differentiated in 2D on collagen type I-coated slides and compared to both the 3D analog of ESCs embedded within collagen type I hydrogels and standard EB differentiation. Differentiation kinetics for the three culture configurations were evaluated by gene expression of germ lineage markers and cytoskeletal proteins, as well as higher throughput screens for general differentiation patterns. This type of systematic study of culture dimensionality enables more informed choices when targeting specific phenotypes in vitro for tissue engineering and regenerative medicine applications.
MATERIALS AND METHODS
Expansion of Mouse Embryonic Stem Cells
Mouse D3 embryonic stem cells (ESCs) and embryonic fibroblasts (MEFs) were purchased from ATCC and cultured as described previously [Ahsan and Nerem, 2010; Wolfe et al., 2012]. Briefly, ESCs were initially expanded on mitotically arrested MEFs and stored in liquid nitrogen. Prior to experiments, ESCs were thawed and cultured on gelatin-coated tissue culture plastic. Culture medium consisted of Dulbecco’s Modification of Eagles Medium (DMEM) supplemented with 15% ES-qualified fetal bovine serum (Invitrogen), 2 mM L-glutamine, 0.1 mM non-essential amino acids, 1000 U/ml leukemia inhibitory factor (ESGRO® from EMD Millipore), and 0.1 mM penicillin/streptomycin (Thermo Scientific, Inc.).
Two and Three Dimensional Differentiation Systems
Cells were differentiated in three physical configurations: adherent, gel, and suspension culture (Figure 1A). Differentiation medium for all systems consisted of culture medium except without leukemia inhibitory factor. For adherent culture, considered a two-dimensional (2D) culture system (Figure 1A, TOP), glass slides were coated with 3.5 μg/cm2 collagen type I (MP Biomedicals©) for at least one hour and were then seeded with 1 x104 ESCs/cm2 (SLIDE). SLIDE samples were maintained at 37°C/5% CO2 for 2, 4, or 6 days. To present cells with collagen type I but in a three dimensional (3D) configuration, 0.75 x106 ESCs were embedded in collagen type I (2 mg/mL) that was base-neutralized to generate 0.75 mL gels (Figure 1A; MIDDLE). GEL samples were maintained in free-floating conditions for 4, 8, or 12 days with medium (25 mL/construct) changed every other day. Using a standard differentiation protocol, ESCs were cultured as embryoid bodies (Figure 1A; BOTTOM) by initially plating 1 x106 cells in a 100 mm diameter non-tissue culture treated dish with 10 mLs of medium (EB). For EB culture, dishes and medium were changed daily after day 2 and maintained for up to 12 days. Phase images were taken during culture (SLIDE samples) or after histological processing (EB and GEL samples). Macroscopic pictures of GEL samples were quantified using ImageJ software.
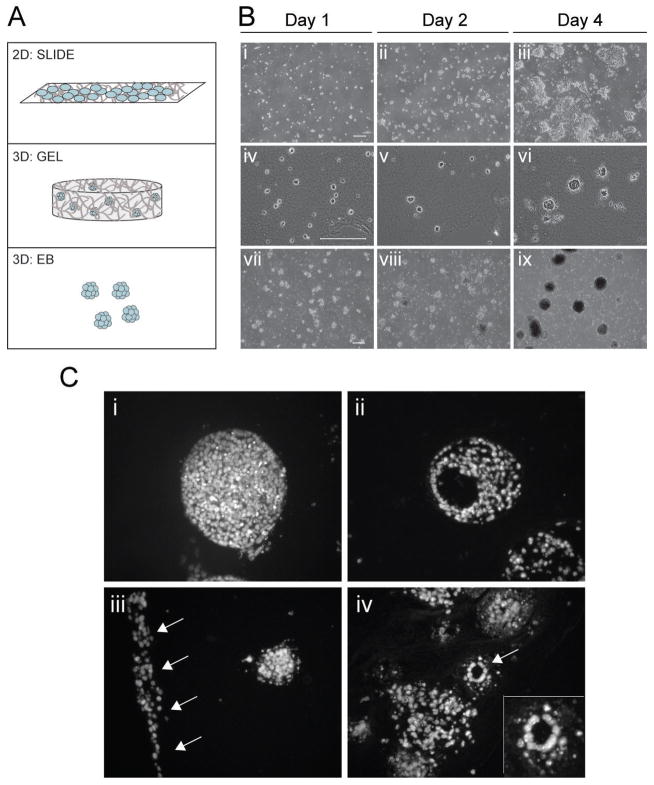
Figure 1. Initial cell distribution and structures supported by the 2D and 3D models.
(A) ESCs were seeded on glass slides coated with collagen type I in the 2D model (SLIDE). In the 3D models, ESCs were either seeded in collagen type I constructs (GEL) or grown in suspension (EB) for 4 days. (B) Images of ESCs cultured for 1, 2, and 4 days as SLIDEs (i-iii), GELs (iv-vi), and EBs (vii-ix). (C) In GEL samples at days 8 and 12, cell bodies of defined boundaries (i) and those with cavities (ii) were found. Elongated bodies of cells (iii) and lumen-like structures (arrow in iv; insert shows higher magnification) were also observed. SLIDE and EB phase images were taken of live cells, while GEL images were histological sections stained with Hoechst to indicate nuclei. Scale bars represent 200 μm.
Gene Expression Analysis
At the end of culture, samples were analyzed for mRNA expression, utilizing either standard real-time PCR or PCR arrays. RNA was isolated using the Qiagen RNeasy Kit (for SLIDE and EB samples) or the RNeasy Lipit Tissue Kit (for GEL samples) and then quantified using a Nanodrop® spectrophotometer for each sample. Standard analysis of mRNA levels for each sample was done on cDNA converted from 1 μg RNA (Invitrogen Superscript® III First-strand synthesis) and analyzed using SYBR® Green (Applied Biosystems) on a StepOnePlus™ PCR System. Primers were custom designed (Primer Express® Software v3.0) for octamer-binding protein 4 (OCT4), alpha-fetoprotein (AFP), Brachyury (BRACHY-T), nestin (NES), actin alpha 1 (ACTA1), actin alpha 2 (ACTA2), tubulin (TUBA1B), keratin 8 (KRT8), vimentin (VIM), lamin (LMNA), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Forward and reverse primers are listed in Supplemental Table 1. Gene expression levels were quantitated using standard curves and are reported normalized to GAPDH expression.
The Mouse Embryonic Stem Cell RT2 Profiler™ PCR Array (SA Biosciences, of Qiagen) was used to profile the expression of genes involved in the maintenance of pluripotency and the general differentiation of embryonic stem cells. Eight different groups were analyzed: undifferentiated ESCs, SLIDEs at Day 4; GELs at Day 4, 8, and 12; and EBs at Day 4, 8, and 12. For each sample, the 84 analyzed genes were normalized to a set of housekeeping genes (listed in Supplemental Table 2). Analysis across groups (n=3 independent samples per group) was performed using Matlab (Mathworks) and Ingenuity® Pathway Analysis (Redwood City, CA) software. A binary tree consisting of nested subsets to show relative similarities across experimental samples was created using an algorithm of Euclidean distance and average linkage [Herrero et al., 2001]. Further analysis was performed clustering on experimental group and/or gene to produce colored heatmaps indicating relative expression levels. The mean expression levels of genes are indicated in black, with relative upregulation and downregulation shown in red and green, respectively, and color saturation (dark hues) used for values three or more standard deviations away from the mean.
Statistical Analysis
Results are presented as mean ± standard error of the mean. Experimental samples were analyzed via student’s t-test or analysis of variance (ANOVA) using Matlab.
RESULTS
Cell Growth in Differentiation Systems
All three differentiation systems (SLIDE, GEL, and EB culture; Figure 1A) successfully supported cell growth over a period of days. Initial cell density was sufficiently low to ensure that conditions started as single or few cells for all models, as illustrated by images at Day 1 (Figure 1Bi, iv, vii). With subsequent culture over four days, cell clusters on SLIDEs, in GELs, and as EBs all continued to increase in size and number indicating support for cell proliferation (Figure 1B). Clusters in 2D culture generated multiple cell layers but preferentially grew along the plane of the slide. SLIDE samples were limited to 6 days of differentiation because cell confluency ultimately became such that the initially dominant trait of cell-matrix binding was overcome by cell-cell interaction. Clusters in either GEL or EB samples grew radially, as would be expected from 3D culture. Higher magnification images of fluorescently labeled nuclei in histology cross-sections provided a more detailed view of the range of morphological arrangements of the clusters and cell-cell interactions in GEL samples (Figure 1C). Cell organization in GEL samples was similar to that seen in standard EB culture, commonly including bodies with defined boundaries (Figure 1Ci) that occasionally contained cavities (Figure 1Cii). Unique to cells grown in GELs, however, were elongated bodies along the edges of the gel construct (Figure 1Ciii, arrows). A few instances also included lumen-like structures where cells were cuboidal in shape arranged in a circular arrangement (Figure 1Civ, arrow & insert).
Pluripotency and Germ Lineage Differentiation
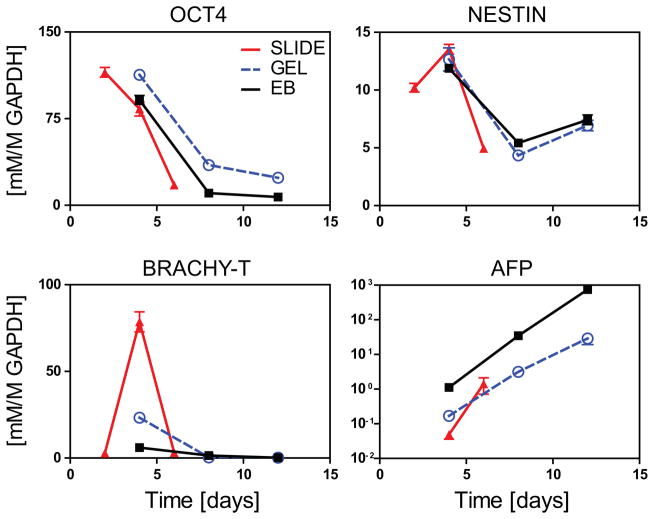
Representative markers of pluripotency and germ lineage specification were assessed using real time rtPCR (Figure 2). Here the expression patterns of OCT4 (pluripotency), NESTIN (ectoderm), and BRACHY-T (mesoderm) were largely similar across culture conditions. For SLIDE, GEL, and EB culture, OCT4 expression markedly decreased after 4 days indicating loss of pluripotency. All three groups also showed dynamic levels of NESTIN and BRACHY-T, for which expression was highest at day 4. Expression of AFP, a marker of endoderm, instead steadily increased with time for all groups, but with levels at all timepoints 1–2 orders of magnitude higher in EB samples. Taken together, this data indicated that ESCs differentiate in SLIDE, GEL, and EB cultures. To better characterize the breadth of differentiation, a higher throughput approach was subsequently used.
Figure 2. Gene expression of pluripotent and lineage markers for SLIDE, GEL, and EB samples.
Samples were evaluated for pluripotency (OCT4) and specification towards the three germ lineages (ectoderm – NESTIN; mesoderm – BRACH-T; and endoderm – AFP). Real time rtPCR values for genes of interest were normalized to the housekeeping gene GAPDH. SLIDE samples (RED, triangle) were assessed at days 2, 4, and 6, while GEL (BLUE, circle) and EB samples (BLACK, square) were evaluated at days 4, 6, and 8. Data presented are mean±SEM for n=3.
ESC Differentiation Patterns
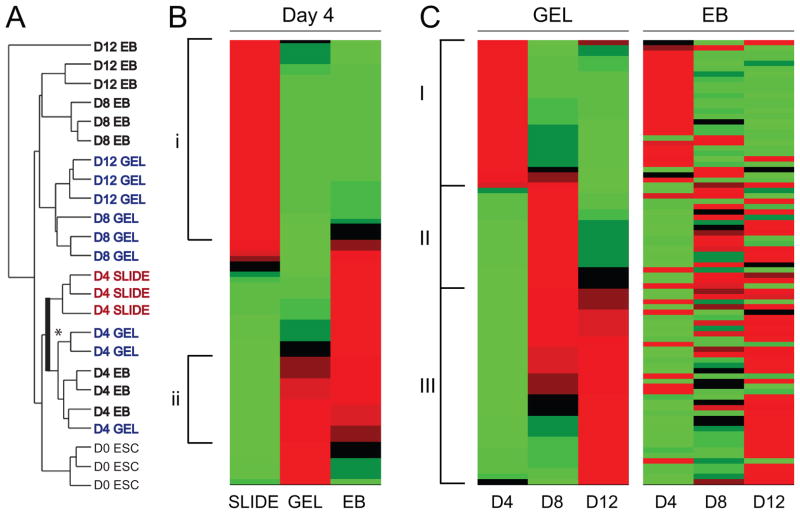
To obtain an overall sense for the patterns of differentiation of ESCs grown in SLIDE, GEL, and EB culture systems, the Mouse Embryonic Stem Cell PCR array (SA Biosciences of Qiagen) was used to determine gene expression of eighty four genes of pluripotency and differentiation (listed in Supplemental Table 2). Hierarchical cluster analysis showed that samples from the same group generally clustered together (Figure 3A), indicating that the chosen PCR array was suitable for interpreting effects of culture system and duration. The hierarchical tree indicated that effects due to culture duration dominated more during early differentiation, while samples from the same culture paradigm clustered together at later timepoints. Specifically, at day 4 it was seen that for cells grown on SLIDEs, differentiation was greater (further from ESCs) than for cells grown in GELs or as EBs. Closer inspection of the branching points then showed that cells cultured in 3D (either in GELs or as EBs) clustered on a separate branch than those cultured on 2D SLIDEs (indicated in bold and with an asterisk in Fig 3A). At later time points, sample associations in 3D configurations were stronger based on culture system (GEL vs EB) rather than culture duration (Day 8 vs Day 12). Overall, this indicates that culture configuration and duration strongly define the general differentiation pattern of cells and that configuration effects become increasing important for extended culture durations.
Figure 3. Hierarchical clustering and heat maps based on gene expression from a PCR array.
Expression of 84 genes were grouped using a hierarchical algorithm to clusters values according to similarity. (A) A hierarchical tree indicating relative similarity between D0 ESCs, SLIDEs at Day 4, GELs at Day 4, 8, and 12, and EBs at Day 4, 8, and 12. (B) A heat map displaying gene–clustered data for SLIDE, GEL and EB samples after 4 days of culture. Distinct groupings of genes: (i) genes classified as those upregulated in SLIDE samples only and (ii) genes similarly regulated in GEL and EB samples. (C) Heat maps indicating differentiation over time for GEL and EB samples. Distinct groupings of genes for GEL samples: (I) genes most highly expressed at day 4 with subsequent loss in expression with time, (II) genes transiently expressed with highest levels at day 8, and (III) genes for which expression increases with time from day 4 to 12. The EB heat map is shown with the gene order of the GEL heat map. The colors indicate level of gene expression: BLACK is the mean across samples, RED indicates expression higher than the mean, and GREEN indicates expression below the mean. For each group n=3 independent samples.
Heat maps of the averages of normalized gene expression levels (to values for undifferentiated ESCs) of SLIDE, GEL, and EB samples were created to spatially represent general differentiation patterns. Evaluation of all three differentiation systems at Day 4 showed a distinct difference in patterning between 2D and 3D culture (Figure 3B). At this time point, 35 of 84 genes were upregulated only in SLIDE samples (Figure 3B-i), while a separate set of genes (Figure 3B-ii) were similarly highly expressed in both GEL and EB samples. These results expound on the tree analysis showing the relative differentiation of SLIDE, GEL and EB samples by revealing that the differentiation pattern, as reflected by individual genes, are more closely related between GEL and EB samples than to SLIDE samples.
Heat maps for the independent 3D culture systems displayed similar patterns of differentiation kinetics. When GEL samples at days 4, 8, and 12 were analyzed, groups aligned temporally and, as one would expect, genes fell in one of three categories: highly expressed genes at day 4 that were then downregulated with increased culture time (Figure 3C-I); low expressing genes at day 4 that were only transiently upregulated (Figure 3C-II); and genes that were monotonically upregulated with culture time over 12 days (Figure 3C-III). When the gene order was fixed to maintain those same categories and used to analyze EB samples, groups again aligned temporally. Furthermore, the overall differentiation pattern was similar between GEL and EB samples though certain individual genes transitioned differently. Taken together, clustering analysis indicated that all systems supported differentiation but that differentiation patterns were distinct for 2D versus 3D cultures.
Functional Gene Groupings
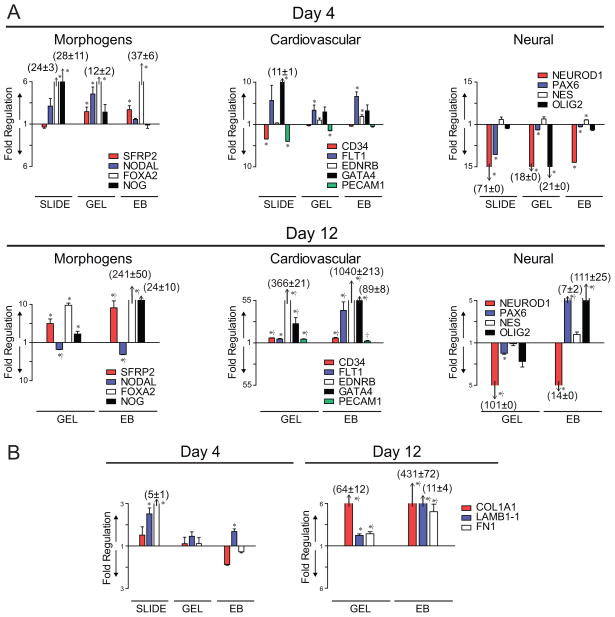
Specific groupings of genes were identified using the PCR array data and IPA software. Functional categories that were further analyzed included Developmental Morphogens, Cardiovascular Differentiation, Neural Differentiation, and Matrix Proteins (Figure 4). Fold regulation is represented for early (Day 4) and later (Day 12) differentiation compared to undifferentiated ESCs, such that bar length above the x axis indicates level of higher expression and the bar length below the x-axis indicates level of lower expression. (Acronyms for genes are listed in Supplemental Table 2.)
Figure 4. Relative gene expression of functional groups at Day 4 and 12.
Genes of the PCR Array results were grouped based on function: (A) Developmental signals, Cardiovascular differentiation, and Neural differentiation, as well as (B) Extracellular Matrix Proteins. Relative gene expression compared to Day 0 ESCs are shown for Day 4 and 12, for which the length of the bar corresponds to the magnitude of expression. For the genes of interest, bars above the y=1 axis represent an upregulation and below represent a downregulation. Asterisks (*) indicate a significant difference (p ≤ 0.05) compared to the corresponding Day 0 ESC expression level. Crosses (†) on the Day 12 plots indicate a significant difference (p ≤ 0.05) between the Day 4 and Day 12 expression levels for that gene. Data presented are mean±SEM for n=3.
Development relies on creating morphogenic gradients, in part by expression of SFRP2, NODAL, FOXA2, and NOG (Figure 4A). SFRP2, soluble frizzled-related protein 2 which modulates Wnt signaling, was upregulated after 4 days of culture in both 3D culture conditions (GEL and EB samples) with respect to undifferentiated ESCs, with significant greater increases in expression observed after 12 days in EB culture. NODAL, a member of the TGF-β superfamily essential for mesodermal formation and axial patterning in the developing embryo [Nostro et al., 2008; Willems and Leyns, 2008], was instead only transiently upregulated in GEL samples at day 4, with a significant downregulation observed at day 12 in both GEL and EB samples. FOXA2, a transcription factor required for notochord formation and endodermal differentiation [Ang and Rossant, 1994; Weinstein et al., 1994; Yamanaka et al., 2007], was upregulated in all groups at day 4 and increased another 7-fold in EB cultures by day 12. NOG, an antagonist of the TGF-β superfamily, instead was only significantly elevated in 2D slide culture at day 4, but was significantly upregulated in both 3D culture conditions at day 12. The three separate culture conditions distinctly regulated expression of morphogens, but with more similar trends in overall expression between the 3D culture conditions. The ability to continue culture of the 3D systems for extended durations is likely valuable in developing and sustaining morphogen spatial gradients during differentiation in vitro. The choice of the specific 3D culture system would then be application specific as the GEL cultures maintained similar gene expression levels for up to 12 days and EB cultures allowed for a more dynamic expression profile with time.
Expression of cardiovascular markers was evaluated to determine differentiation towards a mesodermal phenotype (Figure 4A). At day 4 in both 2D and 3D culture systems, CD34 and PECAM1, markers strongly associated with the hematopoietic and endothelial phenotypes respectively, were similarly regulated by the different culture conditions. At day 4, expression was only significantly affected by culture on SLIDEs. With added culture time, however, both markers were subsequently significantly upregulated in both GELs and EBs. Flt1 was found to be upregulated only in GEL and EB culture and was elevated at both day 4 and 12. ENDRB, an endothelin receptor, was modestly upregulated in EB samples at day 4, but was markedly upregulated by ~350- and ~1000-fold in GEL and EB samples at day 12. GATA4, a transcription factor implicated in myocardial differentiation, was expressed an order of magnitude higher on slide culture at day 4 compared to undifferentiated ESCs, though was expressed at even higher levels in GEL and EB samples but not until day 12. Overall, it was found that 3D culture is supportive of cardiovascular differentiation, particularly with extended culture durations. EB culture, however, seemingly promotes the greatest level of cardiovascular differentiation.
Neural differentiation was assessed by looking at NEUROD1, PAX6, NES, and OLIG2 (Figure 4A). In general, neural differentiation was not significant at day 4 under any culture condition as most markers were expressed at levels either similar to, or even markedly lower than, undifferentiated ESCs. NEUROD1 and NES levels remained very low even at day 12, but PAX6 and OLIG2 were markedly upregulated in EB samples. Thus, neural differentiation of ESCs is not readily supported on glass slides or in GELs, but instead requires sustained culture in EBs.
Collagen type I (COL1A1), Laminin (LAMB1-1), and Fibronectin (FN1) expression was evaluated as an indication of the overall expression of matrix proteins (Figure 4B). ESCs on collagen type I-coated glass slides significantly expressed higher levels of both LAMB1-1 and FN1, without any change in COL1A1 expression, compared to undifferentiated ESCs. While at day 4 expression levels were not changed in the 3D culture conditions, by day 12 all three genes were significantly upregulated in both GEL and EB samples. Thus, all three culture models support expression of matrix proteins though the kinetics of expression are delayed in 3D compared to 2D configurations.
Matrix Remodeling
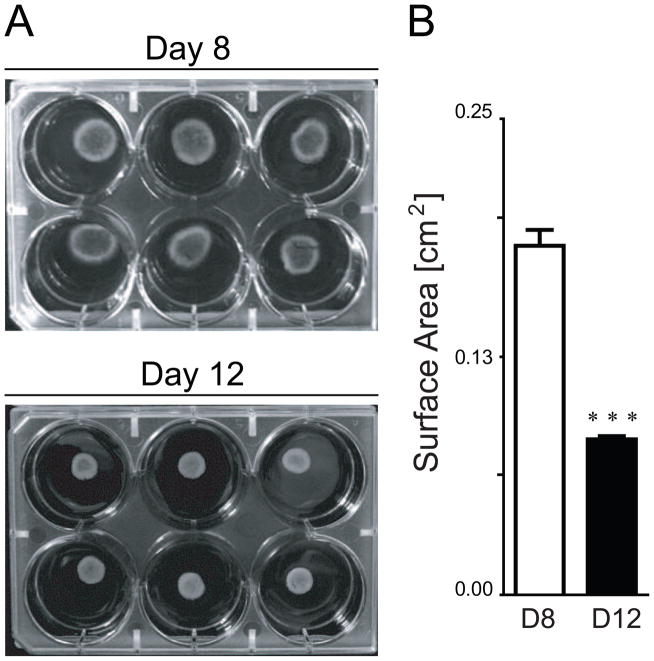
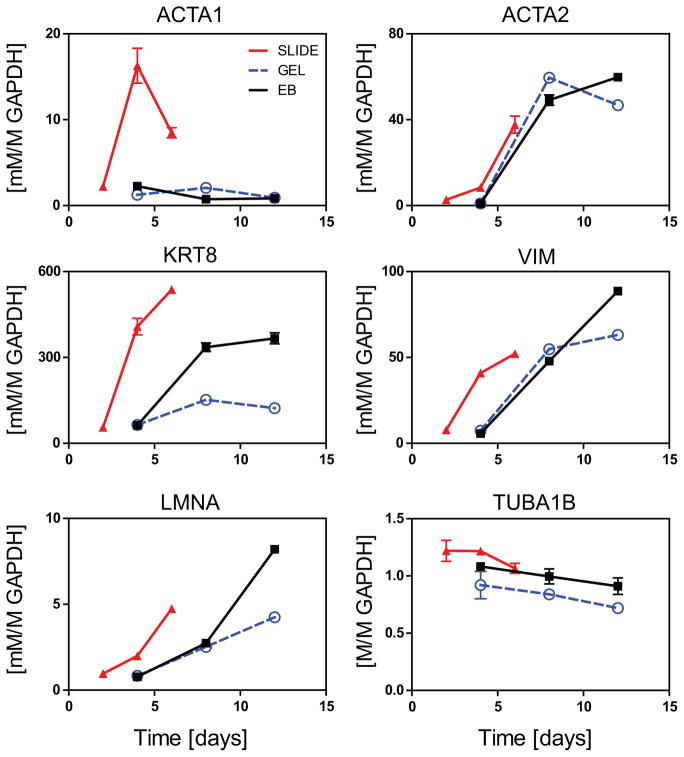
Collagen gels serve as a scaffold that allows for cell-based remodeling during culture. ESCs cultured in gels were observed to significantly compact the volume after approximately a week. Macroscopic images were taken and surface area was quantified using image analysis (Figure 5). A significant (p<0.001) decrease (50%) in surface area was observed between days 8 and 12. Since cytoskeletal proteins are often implicated in cell-generated traction forces, we used standard PCR to quantify gene expression of microfilaments, intermediate filaments, and microtubules over time for samples from all three culture models (Figure 6). Cytoskeletal protein expression in all models was extremely low at early differentiation time points, with the exception of TUBA1B. Expression of microfilaments ACTA1 and ACTA2 were differentially regulated, where ACT1A was only upregulated in SLIDE samples and ACTA2 was upregulated with time in both 2D and 3D culture systems. All intermediate filaments (KRT8, VIM, and LMNA) were expressed earlier for cells on SLIDEs than in 3D culture. For both GEL and EB samples, expression of the assessed intermediate filaments were markedly higher at days 8 and 12 compared to day 4, though EB culture always had the promoted higher expression levels at day 12. Conversely, microtubule expression, as indicated by TUBA1B, was not affected by either culture condition or time. Taken together, these results indicate that 2D SLIDE culture promotes accelerated expression of cytoskeletal proteins compared to 3D culture and that increases in expression for GEL samples is concomitant with observed changes in hydrogel compaction.
Figure 5. Collagen gel compaction from Day 8 to Day 12 of culture.
(A) Image of GELs cultured for 8 and 12 days show a decrease in surface area. (B) Quantification of the images reveal a significant difference in surface area across days (***, p<0.001). Data presented are mean±SEM for n=6.
Figure 6. Gene expression of cytoskeletal genes for SLIDE, GEL, and EB samples.
SLIDE samples (RED, triangle) were assessed at days 2, 4, and 6, while GEL (BLUE, circle) and EB samples (BLACK, square) were evaluated at days 4, 6, and 8. Gene expression are shown for microfilaments (ACTA1 and ACTA2), intermediate filaments (KRT8, VIM, and LMNA), and microtubules (TUBA1B) all normalized to the housekeeping gene GAPDH. Data presented are mean±SEM for n=3.
DISCUSSION
In these studies, we compared mouse embryonic stem cells (ESCs) differentiated on collagen type I-coated surfaces (SLIDEs), embedded in collagen type I gels (GELs), and in suspension as embryoid bodies (EBs) to determine the effects of culture dimension on differentiation. The extracellular protein collagen type I was selected due to its capability to induce embryonic stem cells to mesodermal phenotypes and its potential to mitigate tumorigenicity of cells delivered in vivo [Krawetz et al., 2012; Liu et al., 2012]. Many parameters (e.g. initial single cell seeding, days of expansion, and media formulation) were kept identical to allow for direct comparison across the three culture configurations. Based on this systematic approach, we were able to determine that all three configurations supported differentiation of ESCs and that the kinetics of differentiation differed greatly for cells cultured in two versus three dimensions. Two dimensional adherent cultures induced overall differentiation more quickly than 3D configurations; in particular, expression of cytoskeletal and extracellular matrix proteins emerged sooner on SLIDES. For 3D culture as GELs or EBs, the differentiation patterns and cell cluster morphology were similar. There were some differences, however in the rate and level of cardiovascular differentiation and expression of developmental morphogens. Additionally, the ability of differentiated ESCs to markedly contract the collagen hydrogels is likely indicative of differences in the ability to generate traction forces between GEL and EB cultures.
EB and GEL culture largely supported morphogen expression and cardiovascular differentiation. Both 3D culture systems are based on cell expansion in clusters, where organization is not homogenous within the cluster. This allows for distinct spatial regions for simultaneous but heterogenous differentiation, which is also a key aspect in gastrulation when distinct germ layers form during early development [Chenoweth et al., 2010]. Furthermore, GELs may allow for more complex structures since the encapsulation step does not preferentially select for cells that are able to bind to matrix or other cells, as in the case of 2D SLIDE and 3D EB culture, respectively. In both 3D models, however, the observed expression of developmental markers (e.g. SFRP2, NODAL, and NOG) is consistent with the timing of germ lineage specification both in our studies and those by others for embryoid bodies [Kurosawa et al., 2003; Wang et al., 2007]. Extension of culture in GELs also allowed for asymmetric organization into elongated bodies and lumen-like structures. Such self-assembly may be useful in the formation of complex functional tissues or vascular networks.
For cells in cluster configurations, multi-directional cell-cell connections are present. As these types of connections are important regulators of vascular and cardiac cell homeostasis [Bershadsky, 2004; Chen et al., 2004], this may contribute to the support of cardiovascular differentiation in 3D. In addition, cluster configurations have implications in the number of cell-cell connections and cadherin-mediated binding, both of which have been implicated in stem cell fate decisions [Rodriguez et al., 2004; Xu et al., 2010]. While overconfluent adherent cultures can also be dominated by cell-cell interactions, the switch from cell-matrix to cell-cell dominated interactions may complicate data interpretation and even alter differentiation kinetics. Thus, for cells cultured in 3D as either GELs or EBs, differentiation may be mediated by similar mechanisms.
The differentiation patterns between GELs and EBs were found to be similar without the addition of exogenous factors. The use of additional soluble cues, including cytokines, are readily presented to cells either as EBs or in GELs through changes in medium composition or presentation of degradable microspheres [Carpenedo et al., 2009]. In hydrogels, however, there is also the potential to present bound proteins and create gradients to promote differentiation to challenging phenotypes. While our differentiation modalities, which were unbiased towards any particular phenotype, did not induce meaningful neural differentiation, researchers have used tethering of biomolecules to direct neural cell migration in a polymeric scaffold [Elisseeff et al., 2000; Lutolf and Hubbell, 2005; Curley and Moore, 2011]. In addition to spatial gradients, hydrogels also allow for the use of other physical factors that are known to influence differentiation, such as substrate stiffness [Engler et al., 2006] [Engler et al., 2006; Sun et al., 2012] and applied physical forces [Yamamoto et al., 2005; Sumanasinghe et al., 2006; McMahon et al., 2008; Ahsan and Nerem, 2010]. Cell-generated contraction forces also add a dynamic mechanical environmental factor in collagen gels. These differentiating pluripotent cells were able to remodel the volume and density of the gels, as has been observed with more mature phenotypes such as fibroblasts [Hinz et al., 2001; Grinnell, 2003], smooth muscle cells [Kanda and Matsuda, 1994; Lee et al., 1995], and adult stem cells [Awad et al., 2000]. Such a contractile permissive environment may in fact facilitate differentiation towards mechanically active cell phenotypes. Combining the above aspects, hydrogels readily allow for the control of numerous elements of the microenvironment, which can then provide higher efficiency in differentiation towards target phenotypes.
SLIDES, of the three groups tested, had the most accelerated loss of pluripotency and overall differentiation. Adherent slide culture, as a means of promoting cell-matrix interactions, is often used to study mechanotransduction. In this modality, applied fluid shear stress [Shyy and Chien, 2002; Li et al., 2005] or tensile strain [Lee et al., 1999] impose well-defined mechanical forces to terminally differentiated cell populations. Recent studies in stem cell differentiation have used similar systems [Yamamoto et al., 2005; Sumanasinghe et al., 2006; Riha et al., 2007; Kearney et al., 2010], as well as ones that modified the underlying substrate [Engler et al., 2006]. All of these systems alter the force balance across the membrane (via transmembrane proteins such as integrins) to modulate cytoskeletal tension, known to play a role in mechanotransduction to the nucleus [Wang et al., 2009; Mendez and Janmey, 2012]. Especially noteworthy in this system is that changes in expression of cytoskeletal elements were observed to be highly dynamic, indicating the potential to be mechanoresponsive. Thus, use of adherent cultures systems with pluripotent stem cells may be a valuable model to study mechanotransduction in pluripotent stem cells during early stages of differentiation.
Cells respond to the initially presented microenvironment by secretion of molecules and synthesis of proteins that then help to remodel the cellular environment. Cells in adherent cultures, with collagen type I presented on only a single surface, were found here to have high expression levels of cytoskeletal elements and extracellular matrix proteins. These categories of proteins not only change the surrounding protein composition but also change the mechanisms by which the cell can interact with the microenvironment. When cells were instead surrounded with collagen type I as embedded in gels, the gene expression for synthesis of matrix proteins was dampened, perhaps due to a decrease in the need to establish a protein-based microenvironment. In addition, it was observed that as culture time progressed to 12 days the differences between EB and GEL culture became greater. This may be a reflection of an increasingly different microenvironment between the two groups since the cells embedded in collagen were actively contracting the gel and dramatically remodeling their microenvironment. Thus, the selection of culture paradigm may need to consider the process of active remodeling, which may impact both autocrine signals in cell differentiation and matrix properties for tissue engineering.
Current methods of differentiation result in low yields and impure populations of target phenotypes and attempts for improved efficiency have been largely through comparison of discrete protocols. This basic science study focused on a single variable to understand the effects of dimensionality on directed differentiation. Various markers of differentiation were evaluated and reported for both 2D and 3D culture conditions. Due to differences in stem cell phenotype and differentiation mechanisms, these studies need to be replicated in human embryonic and induced pluripotent stem cells for translation to medical therapies. Our results, however, will help in the rational design of protocols to generate a number of specific phenotypes, which ultimately may require complex paradigms that include the sequential use of multiple distinct culture configurations and include growth factors, heterotypic co-culture, and physical force.
Supplementary Material
Acknowledgments
This work was initiated at the Georgia Institute of Technology and completed at Tulane University. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number P20GM103629. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
LIST OF ABBREVIATIONS
- ESC
Mouse Embryonic stem cell
- EB
Embryoid Body
- 2D
Two dimensional
- 3D
Three dimensional
- OCT4
Octamer-binding protein 4
- AFP
Alpha-fetoprotein
- BRACHY-T
Brachyury
- NES
Nestin
- ACTA1
Actin alpha 1
- ACTA2
Actin alpha 2
- TUBA1B
Tubulin
- KRT8
Keratin 8
- VIM
Vimentin
- LMNA
Lamin
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- ANOVA
Analysis of variance
- rtPCR
Reverse transcriptase polymerase chain reaction
- mRNA
Messenger RNA
- SFRP2
Secreted frizzled-related protein 2
- FOXA2
Forkhead box A2
- NOG
Noggin
- CD34
CD34 antigen
- PECAM1
Platelet/endothelial cell adhesion molecule 1
- GATA4
GATA binding protein 4
- ENDRB
Endothelin receptor type B
- NEUROD1
Neurogenic differentiation 1
- PAX6
Paired box gene 6
- OLIG2
Oligodendrocyte transcription factor 2
- COL1A1
Collagen type I
- LAMB1-1
Laminin
- FN1
Fibronectin
References
- Ahsan T, Nerem RM. Fluid shear stress promotes an endothelial-like phenotype during the early differentiation of embryonic stem cells. Tissue Eng Part A. 2010;16(11):3547–3553. doi: 10.1089/ten.tea.2010.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang SL, Rossant J. HNF-3 beta is essential for node and notochord formation in mouse development. Cell. 1994;78(4):561–574. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- Awad HA, Butler DL, Harris MT, Ibrahim RE, Wu Y, Young RG, Kadiyala S, Boivin GP. In vitro characterization of mesenchymal stem cell-seeded collagen scaffolds for tendon repair: effects of initial seeding density on contraction kinetics. J Biomed Mater Res. 2000;51(2):233–240. doi: 10.1002/(sici)1097-4636(200008)51:2<233::aid-jbm12>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Bershadsky A. Magic touch: how does cell-cell adhesion trigger actin assembly? Trends Cell Biol. 2004;14(11):589–593. doi: 10.1016/j.tcb.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Boheler KR, Czyz J, Tweedie D, Yang HT, Anisimov SV, Wobus AM. Differentiation of pluripotent embryonic stem cells into cardiomyocytes. Circ Res. 2002;91(3):189–201. doi: 10.1161/01.res.0000027865.61704.32. [DOI] [PubMed] [Google Scholar]
- Bosnakovski D, Mizuno M, Kim G, Takagi S, Okumura M, Fujinaga T. Chondrogenic differentiation of bovine bone marrow mesenchymal stem cells (MSCs) in different hydrogels: influence of collagen type II extracellular matrix on MSC chondrogenesis. Biotechnol Bioeng. 2006;93(6):1152–1163. doi: 10.1002/bit.20828. [DOI] [PubMed] [Google Scholar]
- Carpenedo RL, Bratt-Leal AM, Marklein RA, Seaman SA, Bowen NJ, McDonald JF, McDevitt TC. Homogeneous and organized differentiation within embryoid bodies induced by microsphere-mediated delivery of small molecules. Biomaterials. 2009;30(13):2507–2515. doi: 10.1016/j.biomaterials.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CS, Tan J, Tien J. Mechanotransduction at cell-matrix and cell-cell contacts. Annu Rev Biomed Eng. 2004;6:275–302. doi: 10.1146/annurev.bioeng.6.040803.140040. [DOI] [PubMed] [Google Scholar]
- Chenoweth JG, McKay RD, Tesar PJ. Epiblast stem cells contribute new insight into pluripotency and gastrulation. Dev Growth Differ. 2010;52(3):293–301. doi: 10.1111/j.1440-169X.2010.01171.x. [DOI] [PubMed] [Google Scholar]
- Cullen DK, Lessing MC, LaPlaca MC. Collagen-dependent neurite outgrowth and response to dynamic deformation in three-dimensional neuronal cultures. Ann Biomed Eng. 2007;35(5):835–846. doi: 10.1007/s10439-007-9292-z. [DOI] [PubMed] [Google Scholar]
- Curley JL, Moore MJ. Facile micropatterning of dual hydrogel systems for 3D models of neurite outgrowth. J Biomed Mater Res A. 2011;99(4):532–543. doi: 10.1002/jbm.a.33195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang SM, Kyba M, Perlingeiro R, Daley GQ, Zandstra PW. Efficiency of embryoid body formation and hematopoietic development from embryonic stem cells in different culture systems. Biotechnol Bioeng. 2002;78(4):442–453. doi: 10.1002/bit.10220. [DOI] [PubMed] [Google Scholar]
- Devolder R, Kong HJ. Hydrogels for in vivo-like three-dimensional cellular studies. Wiley Interdiscip Rev Syst Biol Med. 2012;4(4):351–365. doi: 10.1002/wsbm.1174. [DOI] [PubMed] [Google Scholar]
- Doyle AM, Nerem RM, Ahsan T. Human mesenchymal stem cells form multicellular structures in response to applied cyclic strain. Ann Biomed Eng. 2009;37(4):783–793. doi: 10.1007/s10439-009-9644-y. [DOI] [PubMed] [Google Scholar]
- Elisseeff J, McIntosh W, Anseth K, Riley S, Ragan P, Langer R. Photoencapsulation of chondrocytes in poly(ethylene oxide)-based semi-interpenetrating networks. J Biomed Mater Res. 2000;51(2):164–171. doi: 10.1002/(sici)1097-4636(200008)51:2<164::aid-jbm4>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Fuchs C, Scheinast M, Pasteiner W, Lagger S, Hofner M, Hoellrigl A, Schultheis M, Weitzer G. Self-organization phenomena in embryonic stem cell-derived embryoid bodies: axis formation and breaking of symmetry during cardiomyogenesis. Cells Tissues Organs. 2012;195(5):377–391. doi: 10.1159/000328712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerecht S, Burdick JA, Ferreira LS, Townsend SA, Langer R, Vunjak-Novakovic G. Hyaluronic acid hydrogel for controlled self-renewal and differentiation of human embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104(27):11298–11303. doi: 10.1073/pnas.0703723104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimble JM, Katz AJ, Bunnell BA. Adipose-derived stem cells for regenerative medicine. Circ Res. 2007;100(9):1249–1260. doi: 10.1161/01.RES.0000265074.83288.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinnell F. Fibroblast biology in three-dimensional collagen matrices. Trends Cell Biol. 2003;13(5):264–269. doi: 10.1016/s0962-8924(03)00057-6. [DOI] [PubMed] [Google Scholar]
- Herrero J, Valencia A, Dopazo J. A hierarchical unsupervised growing neural network for clustering gene expression patterns. Bioinformatics. 2001;17(2):126–136. doi: 10.1093/bioinformatics/17.2.126. [DOI] [PubMed] [Google Scholar]
- Hinz B, Celetta G, Tomasek JJ, Gabbiani G, Chaponnier C. Alpha-smooth muscle actin expression upregulates fibroblast contractile activity. Mol Biol Cell. 2001;12(9):2730–2741. doi: 10.1091/mbc.12.9.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itskovitz-Eldor J, Schuldiner M, Karsenti D, Eden A, Yanuka O, Amit M, Soreq H, Benvenisty N. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol Med. 2000;6(2):88–95. [PMC free article] [PubMed] [Google Scholar]
- Kanda K, Matsuda T. Mechanical stress-induced orientation and ultrastructural change of smooth muscle cells cultured in three-dimensional collagen lattices. Cell Transplant. 1994;3(6):481–492. doi: 10.1177/096368979400300605. [DOI] [PubMed] [Google Scholar]
- Kearney EM, Farrell E, Prendergast PJ, Campbell VA. Tensile strain as a regulator of mesenchymal stem cell osteogenesis. Ann Biomed Eng. 2010;38(5):1767–1779. doi: 10.1007/s10439-010-9979-4. [DOI] [PubMed] [Google Scholar]
- Krawetz RJ, Taiani JT, Wu YE, Liu S, Meng G, Matyas JR, Rancourt DE. Collagen I scaffolds cross-linked with beta-glycerol phosphate induce osteogenic differentiation of embryonic stem cells in vitro and regulate their tumorigenic potential in vivo. Tissue Eng Part A. 2012;18(9–10):1014–1024. doi: 10.1089/ten.TEA.2011.0174. [DOI] [PubMed] [Google Scholar]
- Kurosawa H, Imamura T, Koike M, Sasaki K, Amano Y. A simple method for forming embryoid body from mouse embryonic stem cells. J Biosci Bioeng. 2003;96(4):409–411. doi: 10.1016/S1389-1723(03)90148-4. [DOI] [PubMed] [Google Scholar]
- Lee AA, Delhaas T, McCulloch AD, Villarreal FJ. Differential responses of adult cardiac fibroblasts to in vitro biaxial strain patterns. J Mol Cell Cardiol. 1999;31(10):1833–1843. doi: 10.1006/jmcc.1999.1017. [DOI] [PubMed] [Google Scholar]
- Lee RT, Berditchevski F, Cheng GC, Hemler ME. Integrin-mediated collagen matrix reorganization by cultured human vascular smooth muscle cells. Circ Res. 1995;76(2):209–214. doi: 10.1161/01.res.76.2.209. [DOI] [PubMed] [Google Scholar]
- Li YS, Haga JH, Chien S. Molecular basis of the effects of shear stress on vascular endothelial cells. J Biomech. 2005;38(10):1949–1971. doi: 10.1016/j.jbiomech.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Liu Y, Goldberg AJ, Dennis JE, Gronowicz GA, Kuhn LT. One-step derivation of mesenchymal stem cell (MSC)-like cells from human pluripotent stem cells on a fibrillar collagen coating. PLoS One. 2012;7(3):e33225. doi: 10.1371/journal.pone.0033225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23(1):47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78(12):7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon LA, Reid AJ, Campbell VA, Prendergast PJ. Regulatory effects of mechanical strain on the chondrogenic differentiation of MSCs in a collagen-GAG scaffold: experimental and computational analysis. Ann Biomed Eng. 2008;36(2):185–194. doi: 10.1007/s10439-007-9416-5. [DOI] [PubMed] [Google Scholar]
- Mendez MG, Janmey PA. Transcription factor regulation by mechanical stress. Int J Biochem Cell Biol. 2012;44(5):728–732. doi: 10.1016/j.biocel.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikmanesh M, Shi ZD, Tarbell JM. Heparan sulfate proteoglycan mediates shear stress-induced endothelial gene expression in mouse embryonic stem cell-derived endothelial cells. Biotechnol Bioeng. 2012;109(2):583–594. doi: 10.1002/bit.23302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa SI, Nishikawa S, Hirashima M, Matsuyoshi N, Kodama H. Progressive lineage analysis by cell sorting and culture identifies FLK1+VE-cadherin+ cells at a diverging point of endothelial and hemopoietic lineages. Development. 1998;125(9):1747–1757. doi: 10.1242/dev.125.9.1747. [DOI] [PubMed] [Google Scholar]
- Nostro MC, Cheng X, Keller GM, Gadue P. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell. 2008;2(1):60–71. doi: 10.1016/j.stem.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SA, Lee HY, Lee JH, Kim TH, Jang JH, Kim HW, Wall I. Collagen three-dimensional hydrogel matrix carrying basic fibroblast growth factor for the cultivation of mesenchymal stem cells and osteogenic differentiation. Tissue Eng Part A. 2012;18(9–10):1087–1100. doi: 10.1089/ten.TEA.2011.0360. [DOI] [PubMed] [Google Scholar]
- Peister A, Woodruff MA, Prince JJ, Gray DP, Hutmacher DW, Guldberg RE. Cell sourcing for bone tissue engineering: amniotic fluid stem cells have a delayed, robust differentiation compared to mesenchymal stem cells. Stem Cell Res. 2011;7(1):17–27. doi: 10.1016/j.scr.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers MJ, Domansky K, Kaazempur-Mofrad MR, Kalezi A, Capitano A, Upadhyaya A, Kurzawski P, Wack KE, Stolz DB, Kamm R, Griffith LG. A microfabricated array bioreactor for perfused 3D liver culture. Biotechnol Bioeng. 2002;78(3):257–269. doi: 10.1002/bit.10143. [DOI] [PubMed] [Google Scholar]
- Riha GM, Wang X, Wang H, Chai H, Mu H, Lin PH, Lumsden AB, Yao Q, Chen C. Cyclic strain induces vascular smooth muscle cell differentiation from murine embryonic mesenchymal progenitor cells. Surgery. 2007;141(3):394–402. doi: 10.1016/j.surg.2006.07.043. [DOI] [PubMed] [Google Scholar]
- Rodriguez JP, Gonzalez M, Rios S, Cambiazo V. Cytoskeletal organization of human mesenchymal stem cells (MSC) changes during their osteogenic differentiation. J Cell Biochem. 2004;93(4):721–731. doi: 10.1002/jcb.20234. [DOI] [PubMed] [Google Scholar]
- Saha S, Ji L, de Pablo JJ, Palecek SP. Inhibition of human embryonic stem cell differentiation by mechanical strain. J Cell Physiol. 2006;206(1):126–137. doi: 10.1002/jcp.20441. [DOI] [PubMed] [Google Scholar]
- Schenke-Layland K, Angelis E, Rhodes KE, Heydarkhan-Hagvall S, Mikkola HK, Maclellan WR. Collagen IV induces trophoectoderm differentiation of mouse embryonic stem cells. Stem Cells. 2007;25(6):1529–1538. doi: 10.1634/stemcells.2006-0729. [DOI] [PubMed] [Google Scholar]
- Shyy JY, Chien S. Role of integrins in endothelial mechanosensing of shear stress. Circ Res. 2002;91(9):769–775. doi: 10.1161/01.res.0000038487.19924.18. [DOI] [PubMed] [Google Scholar]
- Sumanasinghe RD, Bernacki SH, Loboa EG. Osteogenic differentiation of human mesenchymal stem cells in collagen matrices: effect of uniaxial cyclic tensile strain on bone morphogenetic protein (BMP–2) mRNA expression. Tissue Eng. 2006;12(12):3459–3465. doi: 10.1089/ten.2006.12.3459. [DOI] [PubMed] [Google Scholar]
- Sun Y, Villa-Diaz LG, Lam RH, Chen W, Krebsbach PH, Fu J. Mechanics regulates fate decisions of human embryonic stem cells. PLoS One. 2012;7(5):e37178. doi: 10.1371/journal.pone.0037178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Trappmann B, Gautrot JE, Connelly JT, Strange DG, Li Y, Oyen ML, Cohen Stuart MA, Boehm H, Li B, Vogel V, Spatz JP, Watt FM, Huck WT. Extracellular-matrix tethering regulates stem-cell fate. Nat Mater. 2012;11(8):742. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- Wang H, Gilner JB, Bautch VL, Wang DZ, Wainwright BJ, Kirby SL, Patterson C. Wnt2 coordinates the commitment of mesoderm to hematopoietic, endothelial, and cardiac lineages in embryoid bodies. J Biol Chem. 2007;282(1):782–791. doi: 10.1074/jbc.M606610200. [DOI] [PubMed] [Google Scholar]
- Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol. 2009;10(1):75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- Weinstein DC, Ruiz i Altaba A, Chen WS, Hoodless P, Prezioso VR, Jessell TM, Darnell JE., Jr The winged-helix transcription factor HNF–3 beta is required for notochord development in the mouse embryo. Cell. 1994;78(4):575–588. doi: 10.1016/0092-8674(94)90523-1. [DOI] [PubMed] [Google Scholar]
- Willems E, Leyns L. Patterning of mouse embryonic stem cell-derived pan-mesoderm by Activin A/Nodal and Bmp4 signaling requires Fibroblast Growth Factor activity. Differentiation. 2008;76(7):745–759. doi: 10.1111/j.1432-0436.2007.00257.x. [DOI] [PubMed] [Google Scholar]
- Wolfe RP, Leleux J, Nerem RM, Ahsan T. Effects of shear stress on germ lineage specification of embryonic stem cells. Integr Biol (Camb) 2012;4(10):1263–1273. doi: 10.1039/c2ib20040f. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhu X, Hahm HS, Wei W, Hao E, Hayek A, Ding S. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc Natl Acad Sci U S A. 2010;107(18):8129–8134. doi: 10.1073/pnas.1002024107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Sokabe T, Watabe T, Miyazono K, Yamashita JK, Obi S, Ohura N, Matsushita A, Kamiya A, Ando J. Fluid shear stress induces differentiation of Flk–1-positive embryonic stem cells into vascular endothelial cells in vitro. Am J Physiol Heart Circ Physiol. 2005;288(4):H1915–1924. doi: 10.1152/ajpheart.00956.2004. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y, Tamplin OJ, Beckers A, Gossler A, Rossant J. Live imaging and genetic analysis of mouse notochord formation reveals regional morphogenetic mechanisms. Dev Cell. 2007;13(6):884–896. doi: 10.1016/j.devcel.2007.10.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.