Abstract
Echinomycin, a member of the quinoxaline family of antibiotics, is known to be a strong inhibitor of RNA synthesis which has been attributed to its ability to bind to double-helical DNA. Here we study the effect of echinomycin upon DNA replication using egg extracts and embryos from Xenopus laevis as well as cultured human cells. Evidence is presented that echinomycin interferes with chromatin decondensation, nuclear assembly and DNA replication. In the absence of transcription and translation, the drug specifically blocks DNA replication in both Xenopus sperm chromatin and HeLa cell nuclei in vitro. By contrast, replication of single-stranded DNA is not inhibited indicating that echinomycin acts by interacting with the DNA and not the replication elongation proteins of chromatin. The addition of the antibiotic to HeLa cells and X.laevis embryos results in anaphase bridges and cell death. Importantly, in X.laevis embryos injected with echinomycin at the two-cell stage the drug specifically inhibits the cell cycle prior to the onset of transcription, suggesting that quinoxaline antibiotics could exert anti- proliferative effects by inhibition of chromosomal DNA replication.
INTRODUCTION
Echinomycin is a quinoxaline antibiotic, previously identified as a potential anti-cancer drug (1). It binds strongly to double-stranded DNA and acts as a molecular staple, sandwiching two base pairs within its U-shaped conformation (2,3). In contrast, echinomycin does not bind to RNA and interacts only very weakly with single-stranded DNA. DNA binding is via the minor groove, causing the helix to lengthen and unwind (4,5). This increases the thermal stability of the double-stranded structure, without causing single-strand or double-strand breaks (6–8).
Work with chromatin has demonstrated that echinomycin interferes with both nucleosome structure (9,10) and transcription (11–15). In contrast, little is known about effects of echinomycin on chromosomal DNA replication or the cell cycle. To understand how echinomycin might interfere with DNA replication in a cellular environment we have taken advantage of cytoplasmic extracts prepared from Xenopus laevis eggs. Such extracts can assemble a variety of DNA templates into nuclear structures and initiate semi-conservative DNA replication in a cell-cycle-regulated manner (16,17). We have also employed cultured cells as well as microinjection of X.laevis embryos to examine the effects of echinomycin during the cell cycle and development.
In this study we demonstrate that echinomycin blocks chromatin decondensation of Xenopus sperm nuclei and inhibits in vitro DNA replication of both Xenopus sperm nuclei and human nuclei by interacting with the DNA and not the replication elongation proteins. When cultured cells are incubated with the drug their progress through the cell cycle is affected, resulting in cell death. Importantly, anaphase bridges accumulate during early development in Xenopus embryos in the presence of the drug, even in developmental stages where RNA is not synthesized. As a result, echinomycin blocks embryos from entering gastrulation. This suggests that inhibition of even a small amount of DNA replication by echinomycin is sufficient to cause anaphase bridges and cell death by mechanisms which appear to be independent of its effects on transcription.
MATERIALS AND METHODS
Echinomycin
Various preparations of echinomycin were used, including samples kindly provided by CIBA-Geigy (Basel) and by Parke-Davis (Ann Arbor, MI, USA). Others were prepared and purified in our own laboratory (18,19). No differences were noted. The antibiotic was dissolved to a concentration of 600 µM in methanol and stored at –20°C. A working stock of 60 µM echinomycin (10% v/v methanol) was made by dilution with water and stored at 4°C. For some experiments a solution at 1 mM concentration was prepared by dissolving the drug in dimethyl sulphoxide (DMSO).
HeLa cell culture
S3 HeLa cells were grown in monolayer culture at 37°C in Gibco Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% Gibco foetal calf serum, 2 mM l-glutamine, 1 U/ml penicillin and 1 µg/ml streptomycin. Cells were synchronized in G1 phase as previously described (20). S3 HeLa cells were plated out at one-tenth confluence in normal medium and left until attached to the dish. The medium was then replaced with medium containing 2.5 mM thymidine and left for 17 h to effect the first block. After this time the cells were washed with phosphate-buffered saline (PBS) and normal medium was added to allow the cells to continue the cell cycle. After 7 h the cells were again exposed to medium containing 2.5 mM thymidine to complete the double block. After a further 17 h the cells were released from the block by removing the medium, washing with PBS and replacing with fresh medium. Following a further release of 6 h and incubation with 25 ng/ml of nocodazole for 6 h, cells were released into fresh medium for 4 h and harvested in G1 phase.
Preparation of HeLa G1 nuclei
HeLa G1 nuclei were prepared essentially according to Krude et al. (20). S3 HeLa cells were washed with ice-cold hypotonic buffer [20 mM HEPES–KOH pH 7.8, 5 mM potassium acetate, 0.5 mM MgCl2, 0.5 mM dithiothreitol (DTT)]. All subsequent steps were carried out at 4°C. The cells were allowed to swell for 5 min in hypotonic buffer and all excess buffer was removed by aspiration. The cells were scraped off the plates into a 15 ml type S Dounce homogenizer (Braun) and cell membranes were broken open using 20–25 strokes of a Teflon pestle (B).
Preparation of low-speed Xenopus egg extract
A low-speed supernatant of Xenopus activated egg extracts (LSS) was prepared essentially according to Blow and Laskey (16). In brief, Xenopus eggs were collected in High Salt Barth (110 mM NaCl, 2 mM KCl, 1 mM MgSO4, 0.5 mM Na2HPO4, 2 mM NaHCO3, 15 mM Tris–HCl pH 7.6), rinsed and then left in distilled water for 10 min at room temperature. Collected eggs were then de-jellied in 2% cysteine–HCl pH 7.8 for 5–10 min, rinsed three times in Barth (88 mM NaCl, 1 mM MgCl2, 15 mM Tris–HCl pH 7.6, 0.5 mM CaCl2) and activated using 0.5 µg/ml calcium ionophore for 5 min at room temperature. Eggs were then rinsed once with Barth without CaCl2 and incubated for 5 min at room temperature. They were finally rinsed three times with ice-cold extraction buffer (50 mM HEPES–KOH pH 7.4, 50 mM KCl, 5 mM MgCl2) supplemented with 2 mM DTT and 10 µg/ml cytochalasin B. All subsequent steps were carried out on ice. Eggs were poured into Beckman SW50 tubes and packed by centrifugation at 1500 r.p.m. for 1 min in a Beckman SW50 rotor. Excess buffer was removed along with degenerated eggs. Eggs were then crushed by spinning at 10 000 r.p.m. for 10 min, after which time the middle cytoplasmic layer was removed with a Pasteur pipette. This cytoplasmic layer was supplemented with 10 µg/ml chymostatin, aprotinin, leupeptin and pepstatin A and recentrifuged at 10 000 r.p.m. for 20 min. The cytoplasmic layer (LSS) was removed through the side of the centrifuge tube with a hypodermic syringe. LSS extracts were supplemented with 2% glycerol and snap-frozen in liquid nitrogen as 20 µl beads.
Replication reactions
Beads of frozen extract were thawed quickly at 23°C and supplemented immediately with an ATP-regenerating system [10 mM creatine phosphate (Boehringer Mannheim), 20 µg/ml creatine phosphokinase (Boehringer Mannheim)] and 250 µg/ml cycloheximide plus either 20 µM biotin-16-dUTP (Boehringer Mannheim) or α[32P]dATP at 2 µCi per 10 µl extract (3000 Ci/mmol) (Dupont NEN). DNA was added to a final concentration of 3–5 ng/µl of demembranated Xenopus sperm chromatin, or 10 ng/µl of activated calf thymus DNA, or 10 ng/µl of single-stranded M13 DNA, or 10 ng/µl of HeLa G1 nuclei.
To measure the incorporation of [α-32P]dATP into macromolecular DNA, duplicate samples of 1 µl were taken from an incubation mix, spotted onto Whatman GF/C glass fibre filters and dried. These were then washed on ice sequentially with 10% trichloroacetic acid (TCA) containing 2% sodium pyrophosphate, 5% TCA and ethanol. The filters were then dried and counted in Optiphase scintillation fluid (Wallace).
MTT viability assay
S3 HeLa cells were grown in flat-bottomed 96-well plates in 200 µl of Gibco DMEM supplemented with 10% Gibco foetal calf serum, 2 mM l-glutamine, 1 U/ml penicillin and 1 µg/ml streptomycin at 37°C. If echinomycin was to be administered, then the medium was replaced with drug-containing medium after the cells had reached the desired density. Cells were plated out in triplicate to provide an average of three wells for each reading. To determine cell survival, a stock solution of MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide, 5 mg/ml] was added to each well being assayed to equal one-tenth the original culture volume. After 4 h the cells were removed from the plate by pipetting the medium up and down and then transferred to separate Eppendorf tubes. The wells were washed out with 50 µl guanidine hydrochloride (6 M solution) which was added to the tubes. After the addition of 300 µl butan-1-ol, each tube was vortex mixed and centrifuged three times. A volume of 200 µl was transferred from the top layer of each tube to single wells in a 96-well plate. The absorbance of the plate was then read at a wavelength of 492 nm.
Injection of Xenopus embryos
Xenopus laevis embryos were de-jellied in cysteine HCl pH 8.0 and developed in 0.1× Modified Barth Saline (MBS) solution: 88 mM NaCl, 1 mM KCl, 2.4 mM NaHCO3, 0.82 mM MgSO4.7H2O, 0.33 mM Ca(NO3)2.4H2O, 0.41 mM CaCl2. 6H2O, 10 mM HEPES–NaOH buffer, 10 mg/l streptomycin sulphate and 10 mg/l benzyl penicillin (21). Developmental stages were determined from the tables produced by Nieuwkoop and Faber (22). Injection into embryos was carried out using a Drummond Nanoject injection apparatus. De-jellied embryos were transferred from 0.1× MBS to 4% Ficoll-400 (Amersham)/1× MBS and left for 10 min before injection. After injection the 4% Ficoll-400/1× MBS was replaced immediately and then at stage 8 the embryos were washed in 2% Ficoll-400/0.2× MBS. Finally the embryos were transferred to 0.1× MBS.
Microscopy
Xenopus egg extract reactions in vitro were diluted in 500 µl of buffer A (15 mM HEPES–KCl pH 7.4, 60 mM KCl, 15 mM NaCl, 10 mM DTT) plus 500 µl of 10% formalin solution or 500 µl of 8% paraformaldehyde solution, and then incubated at room temperature for 10 min. The samples were transferred to scintillation tubes and the nuclei spun on to polylysine-coated coverslips through a 30% sucrose cushion (23). Nuclei were stained with Hoechst 33258 for total DNA content and with fluorescein streptavidin for incorporated biotinylated dUTP.
HeLa cells were grown in 24-well plates on coverslips. After transferring the coverslips to a new 24-well plate containing 1 ml PBS the cells were placed in 500 µl of cold methanol and kept at –20°C for 5 min. The coverslips were then washed twice with PBS followed by 100 µl of propidium iodide solution (5 µg/ml) which was left in contact with the cells for a 30 min incubation at 37°C. After washing three times with PBS the coverslips were mounted on slides.
Xenopus embryos were fixed in MEMFA (0.1 M MOPS pH 7.4, 2 mM EDTA, 1 mM MgSO4, 3.7% formaldehyde) (24) for 2 h at room temperature. They could be stored at –20°C by replacing the MEMFA with methanol. Embryos were embedded in paraffin wax [98% histoplast (Shandon), 2% yellow beeswax] after serial dehydration and then cut into 10 µm sections on a manual microtome. Ribbons of embryo sections were placed on slides which had been treated with Mayer’s albumin, floated on water and dried overnight on a hot plate.
Slides were de-waxed with xylene and then rehydrated through an ethanol series. The slides were covered with 3 mg/ml Hoechst 33258 in PBS Tween (0.1% Tween, 1× PBS) and incubated for 30 min in a dark humidified chamber. After rinsing four times with PBS/Tween the slides were washed once in PBS and mounted in 90% glycerol containing 10% PBS.
RESULTS
Effect of echinomycin upon sperm chromatin decondensation in Xenopus egg extracts
To examine the effect of echinomycin upon DNA replication and the cell cycle we used extracts of Xenopus eggs which replicate DNA in a cell-cycle-dependent manner (16,25). Nuclei formed in the extract are surrounded by intact nuclear envelopes with double membranes and functional nuclear pores (16,17). Such extracts are able to build nuclei and support replication of a variety of DNA templates. Addition of Xenopus sperm chromatin to egg extracts results in a rapid nucleoplasmin-mediated decondensation followed by a slower nuclear-membrane-dependent decondensation (26). Once the DNA has decondensed and an intact nuclear membrane has been built, initiation of DNA replication follows (16,17). DNA replication is semi-conservative, takes place at fixed sites known as replication foci and is strictly regulated to prevent over- or under-replication (23,25).
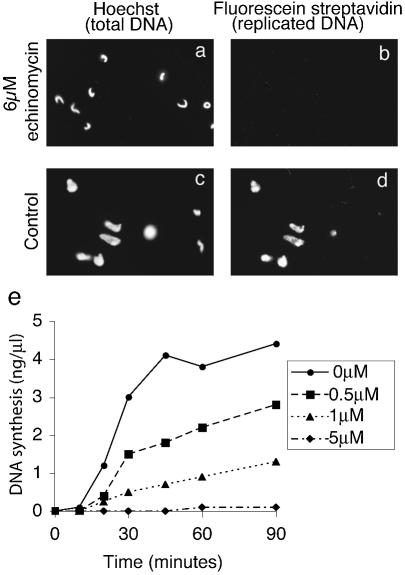
We first asked whether echinomycin interferes with decondensation of Xenopus sperm chromatin following addition to Xenopus egg extracts. At a concentration of 6 µM the drug strongly inhibits nuclear decondensation when compared with control incubations (Fig. 1). In the presence of echinomycin, sperm chromatin is only partially decondensed and nuclear assembly is clearly inhibited as judged by DNA staining (Fig. 1a). Control incubations show that most nuclei have fully decondensed into a rounded nuclear structure (Fig. 1b). Importantly, the partially decondensed chromatin was unable to replicate its DNA as measured by biotin-dUMP incorporation followed by staining with fluorescent labelled streptavidin (Fig. 1c). In contrast, DNA is replicated efficiently in control incubations (Fig. 1d).
Figure 1.
Echinomycin blocks chromosomal decondensation and DNA replication in Xenopus sperm nuclei. Demembranated Xenopus sperm chromatin was incubated in Xenopus egg extract for 45 min with (a and b) 6 µM echinomycin or (c and d) solvent. Total DNA was stained using Hoechst 33258 (a and c). Replicated DNA, having incorporated biotinylated dUMP, was stained with streptavidin fluorescein (b and d). (e) Total DNA synthesis was monitored using incorporation of dAMP from [α-32P]dATP in the presence of different concentrations of echinomycin as shown.
Effect of echinomycin upon Xenopus sperm replication
We next asked if echinomycin can inhibit DNA replication in a concentration-dependent manner. Addition of 5 µM echinomycin to egg extracts resulted in almost complete inhibition of Xenopus sperm DNA replication as measured by the incorporation of radioactivity from [α-32P]dATP (Fig. 1e). In contrast, the addition of solvent as a control showed no such inhibition of DNA synthesis. Low concentrations of drug (0.5 and 1 µM) produced a less significant effect upon DNA replication when compared with the control incubation. Echinomycin therefore only inhibits DNA replication totally at ∼5 µM. This leaky inhibition of nucleotide incorporation at concentrations of ≤1 µM is easily explained on the basis of the known reversibility of echinomycin binding to DNA (27–29).
Echinomycin inhibits synthesis directed by activated calf thymus DNA in Xenopus egg extracts
To determine specifically whether echinomycin can inhibit the elongation phase of DNA synthesis the drug was added to Xenopus egg extracts incubated with activated calf thymus DNA. This substrate provides abundant 3′-hydroxyl strand ends from which DNA can be synthesized. In contrast with Xenopus sperm chromatin, the use of this template removes the need to initiate DNA replication and decondense chromatin. Consequently these experiments provide a means of examining the effect of echinomycin upon the elongation phase of DNA replication.
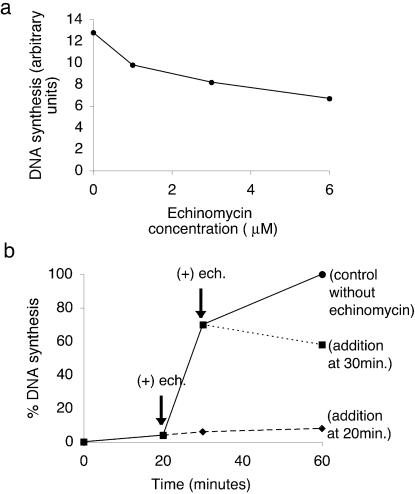
To this end, activated nicked calf thymus DNA was incubated for 75 min in egg extract without drug (control) or with various concentrations of echinomycin ranging from 1 to 6 µM. New DNA synthesis was measured by the incorporation of radioactive nucleotides from [α-32P]dATP. Figure 2a demonstrates that echinomycin does inhibit the incorporation of [32P]dAMP, representing elongation of the calf thymus DNA substrate in a dose-dependent manner, though only about half of the synthesis is inhibited in the experiment illustrated. This is probably because the nucleotide incorporation measured with an activated calf thymus DNA template includes a lot of DNA repair. Repair synthesis is likely to be much less sensitive to echinomycin for reasons explained below where M13 single-stranded DNA replication is considered.
Figure 2.
Echinomycin inhibits synthesis from double-stranded but not single-stranded DNA templates. (a) Nicked calf thymus DNA was incubated in diluted Xenopus egg extract supplemented with [α-32P]dATP for 75 min, together with different concentrations of echinomycin. (b) Demembranated Xenopus sperm chromatin was incubated in Xenopus egg extract supplemented with [α-32P]dATP for 20 and 30 min, after which solvent or 6 µM echinomycin was added and reactions allowed to proceed to 60 min.
Echinomycin inhibits replication elongation of chromosomal DNA in Xenopus egg extracts
The next step was to examine if echinomycin inhibits the elongation step of chromosomal DNA replication. Xenopus sperm chromatin was first incubated in egg extract for 20 min to allow decondensation and nuclear assembly but not initiation of DNA replication. A second preparation of Xenopus sperm chromatin was incubated in egg extracts for 30 min to allow both nuclear assembly and initiation of DNA replication. Echinomycin was then added to each of these incubations at a concentration of 5 µM and DNA replication was monitored by incorporation of [α-32P]dAMP. Figure 2b illustrates that addition of the drug at 20 min severely inhibited DNA replication compared with the control addition. Similar results were also obtained when echinomycin was added at 30 min. At this time point 64% of the input template has replicated, demonstrating that nuclei have already efficiently initiated DNA replication. These results clearly demonstrate that echinomycin blocks the elongation phase of chromosomal DNA replication. Importantly, incubations were performed in the presence of cycloheximide to prevent protein synthesis. Transcription is also absent in eggs used to prepare such extracts (30,31), implying that the effect of echinomycin on DNA replication is independent of transcription as well as translation.
Replication of M13 single-stranded DNA in Xenopus egg extracts is not inhibited by echinomycin
Echinomycin is thought to bind significantly only to double-helical DNA and not to single-stranded DNA (2). If this proposed mechanism of action of echinomycin is correct, then the drug should not be able to bind to M13 single-stranded (ss) DNA and should therefore have no effect upon the replication of this DNA. Incubation of M13 ssDNA (10 µg/ml) with Xenopus egg extracts supplemented with [α-32P]dATP for 60 min resulted in efficient replication, typically up to ∼0.8 µg DNA synthesized per microlitre of extract. However, addition of 5 µM echinomycin to the extract had no effect upon the amount of DNA synthesized (p < 0.05). In contrast, the addition of aphidicolin (25 µg/ml), a potent inhibitor of the replication protein polymerase-α, completely blocked the replication of M13 ssDNA.
These results establish two important points. First, echinomycin is unable to inhibit replication elongation of single-stranded DNA at a concentration which is clearly able to inhibit replication elongation of double-stranded DNA (Fig. 2a and b). Secondly, in contrast with aphidicolin, which inhibits DNA replication by binding directly to polymerase-α (32), echinomycin cannot inhibit DNA replication by binding to replication elongation proteins themselves. If that were the case, then the drug would be expected to block replication irrespective of the DNA template. However, it would clearly be of interest to study the effects of the antibiotic upon the formation of DNA replication initiation complexes.
The failure of echinomycin to inhibit complementary strand synthesis on single-stranded DNA is consistent with negligible or weak binding to the single-stranded DNA template, but it could also reflect the lack of any need for DNA unwinding in such a reaction, as opposed to elongation on double-stranded DNA. Similar considerations would apply to repair synthesis. Mechali and Harland (33) have described in detail the properties of this in vitro system.
Replication of mammalian G1 HeLa nuclei in Xenopus egg extracts
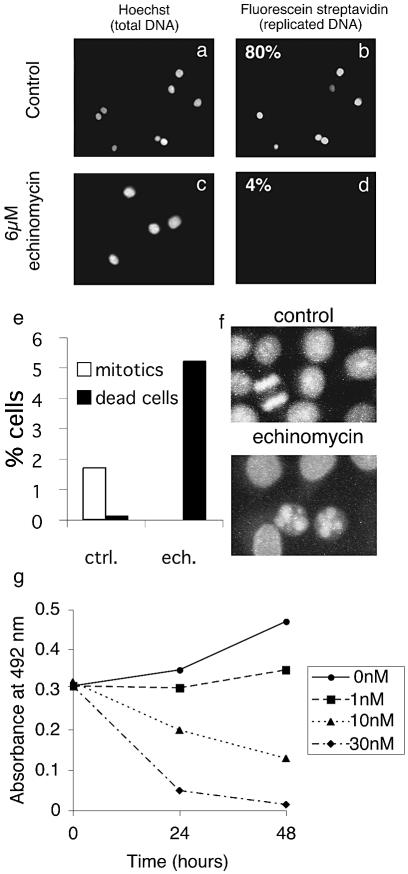
Although echinomycin inhibits DNA replication of Xenopus sperm chromatin, it remains to be established whether the drug can inhibit DNA replication of human chromosomes. To test this we used HeLa G1 nuclei that have assembled an intact nuclear structure but have not initiated DNA replication. Nuclei were incubated in Xenopus egg extract with biotin-dUTP and stained for replication with fluorescent labelled streptavidin. Control incubations demonstrate that these nuclei are able to initiate and elongate DNA replication quite efficiently (Fig. 3b). However, addition of 6 µM echinomycin clearly prevents replication of human chromosomal DNA (Fig. 3d). Elongation of HeLa S-phase nuclei was also found to be inhibited (data not shown).
Figure 3.
Echinomycin inhibits replication of HeLa cell nuclei in Xenopus egg extracts and promotes death of cultured cells. (a–d) G1 HeLa nuclei were incubated for 2 h in Xenopus egg extracts supplemented with biotinylated dUTP. (a and b) Control extracts with solvent added. (c and d) Extracts supplemented with 6 µM echinomycin. Total DNA was stained using Hoechst 33258 (a and c); newly replicated DNA was stained with streptavidin fluorescein (b and d). (e and f) HeLa cells were cultured in the presence of solvent or 50 nM echinomycin for 24 h. Cells were then stained with propidium iodide and the number of mitotic or dead cells counted. (g) HeLa cells were grown at a density of 50 000 cells/well in the presence of solvent or echinomycin for 24 or 48 h and the number of living cells determined using an MTT assay.
Echinomycin causes cell cycle arrest and death of cultured cells
We next wished to address the effect of echinomycin on cultured cells. Addition of echinomycin to proliferating HeLa cells resulted in substantial cell death (Fig. 3e). Following a 24 h incubation with 50 nM echinomycin, the percentage of normal mitotic and interphase cells was sharply reduced while the number of dead cells was increased (Fig. 3e and f, and data not shown). A small fraction of cells also appeared to be undergoing an aberrant mitosis in which chromosomes are unable to segregate correctly (data not shown).
To measure the concentration-dependent effect of echinomycin on cell death we employed an MTT assay which can distinguish between living and dead cells. A concentration of 1 nM echinomycin was found sufficient to inhibit cell growth, while higher concentrations clearly reduced the number of living cells (Fig. 3g). Thus echinomycin is evidently capable of causing cell death at concentrations as low as 10 nM.
Effect of echinomycin on development of Xenopus embryos
A final series of experiments was conducted to investigate whether echinomycin could inhibit the cell cycle in whole X.laevis embryos. Xenopus eggs were fertilized and allowed to develop to a two-cell stage. One femtomole of echinomycin or an equivalent volume of solvent was injected into one or both cells of these embryos which were then monitored for a 3 h period following fertilization. The average volume of one X.laevis embryo, diameter about 1 mm, is 0.5 µl, so injection of 1 fmol of antibiotic into one cell would produce a concentration about 4 nM. Uninjected embryos and control embryos injected with solvent show normal cell division and development up to at least stage 7 (Fig. 4a and c). In contrast, injection of echinomycin results in fewer larger cells (Fig. 4b and c). Animal pole cells from control embryos appear discrete and separate from each other, whereas animal pole cells from echinomycin-injected embryos occasionally appear to be fused together. Importantly, none of the embryos injected with echinomycin went on to develop normally (Fig. 4d). Twenty-one hours following fertilization, 83% of embryos injected with 1.0 fmol of echinomycin had died and the remaining 17% had only developed to stage 9–10. In contrast, 92% of control injected embryos remained viable and developed to stage 11–12. Importantly, all of the embryos injected with 1.0 fmol of echinomycin failed to progress beyond the mid-blastula transition (MBT), at which point development was halted and the embryos eventually died. Embryos injected with 0.5 fmol of echinomycin were able to reach stage 9–10 and then halted development, but were still alive 21 h after fertilization.
Figure 4.
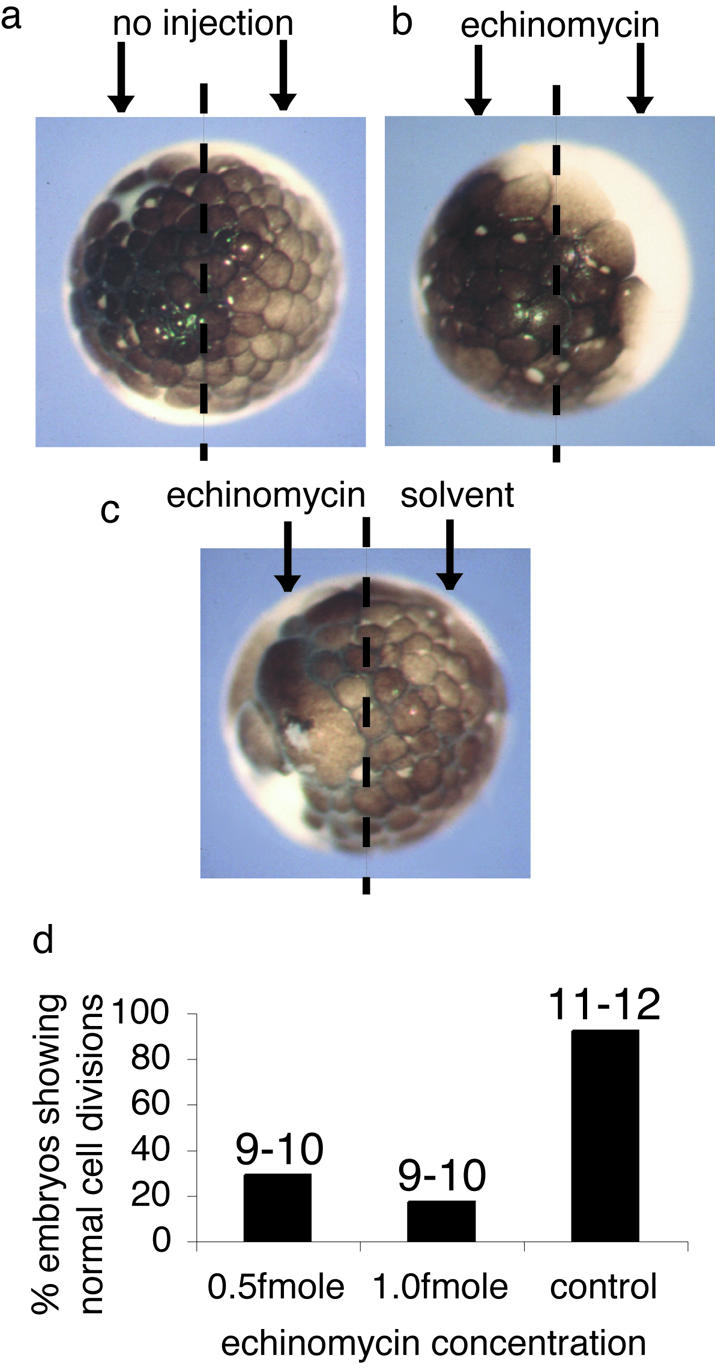
Echinomycin inhibits development of Xenopus embryos. (a) A Xenopus embryo allowed to develop for 3 h following fertilization. (b) A Xenopus embryo allowed to develop to the two-cell stage following fertilization after which 1 fmol of echinomycin was injected into each cell and embryos allowed to develop for 3 h following fertilization. (c) A Xenopus embryo allowed to develop to the two-cell stage after which 1 fmol of echinomycin was injected into one cell and solvent into the other and embryos were allowed to develop for 3 h following fertilization. Images were collected of the animal pole of the embryos (a–c). (d) One hundred embryos were allowed to develop to the two-cell stage after which 0.5 fmol of echinomycin, 1.0 fmol of echinomycin or solvent was injected into both cells and the embryos were monitored throughout development. Numbers above each bar indicate the stage of development reached by the embryos at 21 h when control embryos had progressed to stage 11–12.
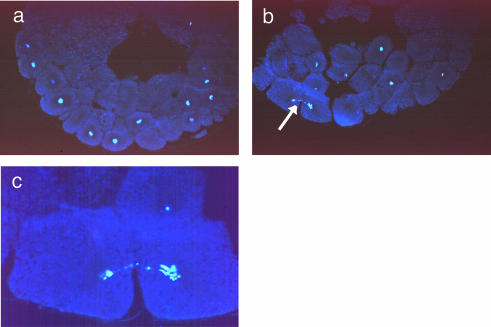
The appearance of the nuclei in embryos injected with echinomycin is quite different from that seen in control injections (Fig. 5). Embryos which were injected with 1 fmol of echinomycin at the two-cell stage and allowed to develop for 3 h following fertilization show nuclei with connecting DNA strands spanning two different cells. Figure 5b illustrates an example typical of many embryos in which this abnormality was photographed. Such connecting DNA strands are characteristic of anaphase bridges. Control embryos show normal mitoses and cell division (Fig. 5a illustrates a normally developing embryo). Magnification of these images clearly reveals the abnormal division of daughter nuclei in echinomycin-injected embryos (Fig. 5c). Anaphase bridges can still be seen to persist at later times of development in drug-injected embryos (data not shown).
Figure 5.
Echinomycin induces anaphase-like bridges and inhibits development beyond the MBT in Xenopus embryos. (a) Section of a Xenopus embryo injected with solvent into both cells at the two-cell stage and allowed to develop for 3.5 h following fertilization, then stained for DNA with Hoechst 33258. (b) Stained section of a Xenopus embryo injected with 1 fmol of echinomycin into both cells at the two-cell stage and allowed to develop for 3.5 h following fertilization. The arrow indicates an anaphase-like bridge. Note the unusually large size of many cells. (c) Magnification of the image shown in (b).
DISCUSSION
In this paper we present evidence for the first time that the antibiotic echinomycin can inhibit chromosomal DNA replication, the cell cycle and embryonic development in vertebrates. The inhibition of DNA replication does not depend upon transcription or translation and does not appear to be the result of direct interaction of echinomycin with replication elongation proteins. Echinomycin is known to act like a molecular staple, binding firmly to double-stranded DNA and impeding unwinding of the double helix. Unwinding of double-stranded DNA is required in both initiation and elongation steps of DNA replication. Our results establish that chromosomal DNA replication is inhibited by echinomycin using a biological system that allows us to distinguish between the effects of echinomycin on transcription and its effect on DNA replication when bound to DNA.
Echinomycin-induced cell cycle defects
Echinomycin has previously been shown to bind tightly to double-stranded but not single-stranded DNA (2). Our initial finding that the antibiotic is a potent inhibitor of chromosomal decondensation demonstrates that it can block an early process essential to all eukaryotic cells emerging from mitosis and meiosis. In Xenopus egg extracts, chromosomal DNA remains condensed and unable to form replication-competent nuclei in the presence of echinomycin (Fig. 1). This result is readily explained by the tight binding of echinomycin to DNA. Previous studies have shown that the drug causes DNA to rotate by half a turn on the surface of nucleosome core particles in vitro (9). Further studies have also demonstrated that echinomycin will prevent nucleosome formation and can cause DNA to dissociate from nucleosomes (10). This known interference of echinomycin with chromatin structure can satisfactorily explain the impaired chromatin decondensation observed in Xenopus egg extracts.
HeLa cells and Xenopus embryos exposed to echinomycin, however, do not show a clear defect in chromosome decondensation upon exit from mitosis. Instead, a more visible defect of anaphase-like bridges was observed (Fig. 5). This dramatic effect of low concentrations of echinomycin on proliferating cells is also explicable on the basis that the drug is able to prevent unwinding of double-helical DNA (34). By interfering with DNA unwinding, echinomycin would inhibit the completion of DNA replication, as seen in Xenopus egg extracts. Partially replicated chromosomes would not be able to separate properly during mitosis. Normally, checkpoints prevent the formation of anaphase bridges by delaying chromosome separation in the presence of unreplicated DNA. These checkpoints are notably absent in tumorigenic cells and during the early cell divisions of embryonic development (i.e. the systems used in most of the experiments described here) (35). Accordingly, this highly visible effect of echinomycin can be observed in our experiments and may explain why echinomycin has a more potent effect upon rapidly replicating tumorigenic cells (36). The increased death of HeLa cells grown in the presence of echinomycin further supports the ability of echinomycin to interfere with the cell cycle in vertebrates (Fig. 3). This too may be mediated via non-repairable cell cycle defects caused by incomplete DNA replication.
Echinomycin has also been shown to interfere with transcription in microbial cells and cell-free systems (11–15). To distinguish transcription from DNA replication, we have used extracts from Xenopus eggs which do not engage in transcription prior to the MBT (30,31). Cycloheximide was also added to inhibit translation. Our results demonstrate that the effect of echinomycin upon DNA replication does not depend upon transcription or new protein synthesis. Although transcription and translation are abundant in experiments using proliferating cells, the appearance of anaphase-like bridges suggests that one of the effects of echinomycin in dividing cells is to inhibit chromosomal DNA replication. This is further supported by the reduced effect of echinomycin upon slowly replicating cells (36).
Inhibition of embryonic development
Microinjection of 1.0 fmol of echinomycin into two-cell stage Xenopus embryos resulted in cells cycling more slowly and ultimately cell death. The experiments using Xenopus egg extracts further support the conclusion that this cell cycle arrest is due to inhibition of DNA replication rather than transcription or translation. Furthermore, this interpretation would explain the observation of anaphase-like bridges in Xenopus embryos exposed to echinomycin prior to the onset of transcription.
During normal embryonic development the MBT is characterized in part by initiation of transcription in the embryo. The failure of echinomycin-injected embryos to develop beyond the MBT and form a yolk plug could be a consequence of both the inhibition of RNA transcription after MBT and accumulation of errors within nuclei caused by the presence of echinomycin during DNA replication. Check points sensing defects in DNA have recently been shown to become active at the MBT, preventing embryos developing beyond this point (35). Similar results have also been reported in embryos injected with aphidicolin, an inhibitor of DNA replication, where embryos also displayed a lack of cell invagination (37).
The potent effect of low concentrations of echinomycin (4–50 nmol) in HeLa and Xenopus cells compared with the micromolar concentrations used in vitro may reflect the inability of cells to tolerate small amounts of unreplicated DNA. It is not unreasonable to suggest that cells or embryos cannot live with 98% DNA replication, and the segregation effects we have observed are consistent with this explanation. Nevertheless we cannot absolutely rule out the possibility of other, perhaps more direct, effects on the cell cycle, nor indeed other actions independent of DNA binding. However, in contrast with the very sensitive responses of intact cells and embryos, the in vitro Xenopus replication assays are not able to detect very low levels of interference with DNA replication.
Therapeutic use of echinomycin
Echinomycin has been tested in several phase II clinical trials to treat various cancers. The ability of the antibiotic as well as some derivatives of echinomycin to show potent anti-tumour activity (36,38,39) makes our effort to understand its mechanism of action all the more important. Here we have presented evidence that the effect of echinomycin on vertebrate cells occurs at least in part via inhibition of DNA replication. This results in abnormal mitoses and cell death. Combined with our demonstration that echinomycin inhibits chromatin decondensation as well as the previously reported inhibition of transcription, the results provide a clearer understanding of why echinomycin acts as a potent antitumour drug. Further studies of the effect of echinomycin and its derivatives on multicellular organisms may help to improve its utility as an agent for specifically killing rapidly dividing cells.
Acknowledgments
ACKNOWLEDGEMENTS
We are very grateful to Professor R. A. Laskey and Sir John Gurdon for constant advice and criticism as well as for allowing unhindered access to the facilities of their laboratories plus expert guidance in performing the experiments. We thank Joe Makkerh, Tony Mills, Andrew Mitchell, Julie Morgan and Hannah Wilkinson for invaluable technical assistance and Guillermo de la Cueva-Mendez for critical reading of the manuscript. This work was supported by grants from the Cancer Research Campaign, the Wellcome Trust and the European Union (Framework 5).
REFERENCES
- 1.Katagiri K., Yoshida,T. and Sato,K. (1975) Quinoxaline antibiotics. In J.W.Corcoran and F.E.Hahn (eds), Antibiotics: Mechanism of Action of Antimicrobial and Antitumour Agents. Springer-Verlag, Heidelberg, Germany, Vol. 3, pp. 234–251. [Google Scholar]
- 2.Waring M.J. and Wakelin,L.P.G. (1974) Echinomycin: a bifunctional intercalating antibiotic. Nature, 252, 653–657. [DOI] [PubMed] [Google Scholar]
- 3.Waring M.J. (1979) In F.E.Hahn (ed.) Antibiotics: Mechanism of Action of Antieukaryotic and Antiviral Compounds. Springer-Verlag, Heidelberg, Vol. 5, Part 2, pp. 173–194. [Google Scholar]
- 4.Wang A.H., Ughetto,G., Quigley,G.J. and Rich,A. (1986) Interactions of quinoxaline antibiotic and DNA: the molecular structure of a triostin A-d(GCGTACGC) complex. J. Biomol. Struct. Dynam., 4, 319–342. [DOI] [PubMed] [Google Scholar]
- 5.Quigley G.J., Ughetto,G., van der Marel,G.A., van Bloom,J.H., Wang,A.H. and Rich,A. (1986) Non-Watson–Crick G.C. and A.T. base pairs in a DNA–antibiotic complex. Science, 232, 1255–1258. [DOI] [PubMed] [Google Scholar]
- 6.Wakelin L.P.G. and Waring,M.J. (1976) The binding of echinomycin to deoxyribonucleic acid. Biochem. J., 157, 721–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang C.-H., Prestayko,A.W. and Crooke,S.T. (1982) Bifunctional intercalation of antitumour antibiotics BBM-928A and echinomycin with deoxyribonucleic acid. Effects of intercalation on deoxyribonucleic acid degradative activity of bleomycin and phleomycin. Biochemistry, 21, 3704–3710. [DOI] [PubMed] [Google Scholar]
- 8.Leng F., Chaires,J.B. and Waring,M.J. (2003) Energetics of echinomycin binding to DNA. Nucleic Acids Res., 31, 6191–6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Low C.M.L., Drew,H.R. and Waring,M.J. (1986) Echinomycin and distamycin induce rotation of nucleosome core DNA. Nucleic Acids Res., 14, 6785–6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leslie K.D. and Fox,K.R. (2002) Interaction of Hoechst 33258 and echinomycin with nucleosomal DNA fragments containing isolated ligand binding sites. Biochemistry, 41, 3484–3497. [DOI] [PubMed] [Google Scholar]
- 11.Ward D.C., Reich,E. and Goldberg,I.H. (1965) Base specificity in the interaction of polynucleotides with antibiotic drugs. Science, 149, 1259–1263. [DOI] [PubMed] [Google Scholar]
- 12.Sato K., Shiratori,O. and Katagiri,K. (1967) The mode of action of quinoxaline antibiotics. Interaction of quinomycin A with DNA. J. Antibiot. (Tokyo), 20, 270–276. [PubMed] [Google Scholar]
- 13.Gause G.G. Jr, Loshkareva,N.P. and Zbarsky,I.B. (1968) Effect of olivomycin and echinomycin on initiation and growth of RNA chains catalyzed by RNA polymerase. Biochim. Biophys. Acta, 166, 752–754. [DOI] [PubMed] [Google Scholar]
- 14.Fok J. and Waring,M.J. (1972) Breakdown of pulse-labelled RNA in Bacillus megaterium revealed by exposure to the antibiotics mithramycin, chromomycin and nogalamycin. Mol. Pharmacol., 8, 65–74. [PubMed] [Google Scholar]
- 15.Waring M.J. and Makoff,A. (1974) Breakdown of pulse-labelled ribonucleic acid and polysomes in Bacillus megaterium: actions of streptolydigin, echinomycin and triostins. Mol. Pharmacol., 10, 214–224. [PubMed] [Google Scholar]
- 16.Blow J.J. and Laskey,R.A. (1986) Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell, 47, 577–587. [DOI] [PubMed] [Google Scholar]
- 17.Newport J. (1987). Nuclear reconstruction in vitro: stages of assembly around protein-free DNA. Cell, 48, 205–217. [DOI] [PubMed] [Google Scholar]
- 18.Gauvreau D. and Waring,M.J. (1982) Quantitative determination of echinomycin by disc agar diffusion assay. Eur. J. Appl. Microbiol. Biotechnol., 15, 104–110. [Google Scholar]
- 19.Gauvreau D. and Waring,M.J. (1984) Directed biosynthesis of novel derivatives of echinomycin by Streptomyces echinatus. Part I. Effect of exogenous analogues of quinoxaline-2-carboxylic acid on the fermentation. Can. J. Microbiol., 30, 439–450. [DOI] [PubMed] [Google Scholar]
- 20.Krude T., Jackman,M., Pines,J. and Laskey,R.A. (1997) Cyclin/Cdk-dependent initiation of DNA replication in a human cell-free system. Cell, 88, 109–119. [DOI] [PubMed] [Google Scholar]
- 21.Gurdon J.B. (1977) Methods for nuclear transplantation in amphibia. Methods Cell Biol., 16, 125–139. [DOI] [PubMed] [Google Scholar]
- 22.Nieuwkoop P.D. and Faber,J. (1967) A Normal Table of Xenopus laevis (Daudin). North-Holland, Amsterdam. [Google Scholar]
- 23.Mills A.D., Blow,J.J., White,J.G., Amos,W.B., Wilcock,D. and Laskey,R.A. (1989) Replication occurs at discrete foci spaced throughout nuclei replicating in vitro. J. Cell Sci., 94, 471–477. [DOI] [PubMed] [Google Scholar]
- 24.Hemmati-Brivanlou A. and Harland,R.M. (1989) Expression of an engrailed-related protein is induced in the anterior neural ectoderm of early Xenopus embryos. Development, 106, 611–617. [DOI] [PubMed] [Google Scholar]
- 25.Leno G.H., Downes,C.S. and Laskey,R.A. (1992) The nuclear membrane prevents replication of human G2 nuclei but not G1 nuclei in Xenopus egg extract. Cell, 69, 151–158. [DOI] [PubMed] [Google Scholar]
- 26.Philpott A., Leno,G.H. and Laskey,R.A. (1991) Sperm decondensation in Xenopus egg cytoplasm is mediated by nucleoplasmin. Cell, 65, 569–578. [DOI] [PubMed] [Google Scholar]
- 27.Fox K.R. and Waring,M.J. (1981) Kinetics of dissociation of quinoxaline antibiotics from DNA. Biochim. Biophys. Acta, 654, 279–286. [DOI] [PubMed] [Google Scholar]
- 28.Fox K.R., Wakelin,L.P.G. and Waring,M.J. (1981) Kinetics of the interaction between echinomycin and DNA. Biochemistry, 20, 5768–5779. [DOI] [PubMed] [Google Scholar]
- 29.Fletcher M.C. and Fox,K.R. (1996) Visualising the dissociation of sequence selective ligands from individual binding sites on DNA. FEBS Lett., 380, 118–122. [DOI] [PubMed] [Google Scholar]
- 30.Gurdon J.B. (1974) The Control of Gene Expression in Animal Development. Oxford University Press, Oxford. [Google Scholar]
- 31.Newport J. and Kirschner,M. (1982) A major developmental transition in early Xenopus embryos. I: Characterization and timing of cellular changes at the midblastula stage. Cell, 30, 675–686. [DOI] [PubMed] [Google Scholar]
- 32.Ikegami S., Taguchi,T. and Ohashi,M. (1978) Aphidicolin prevents mitotic cell division by interfering with the activity of DNA polymerase-α. Nature, 275, 458–459. [DOI] [PubMed] [Google Scholar]
- 33.Mechali M. and Harland,R.M. (1982) DNA synthesis in a cell-free system from Xenopus eggs: priming and elongation on single-stranded DNA in vitro.Cell, 30, 93–101. [DOI] [PubMed] [Google Scholar]
- 34.Bachur N.R., Johnson,R., Yu,F., Hickey,R., Applegren,N. and Malkas,L. (1993) Antihelicase action of DNA-binding anticancer agents: relationship to guanosine–cytidine intercalator binding. Mol. Pharmacol., 44, 1064–1069. [PubMed] [Google Scholar]
- 35.McGarry T.J. (2002) Geminin deficiency causes a Chk1-dependent G2 arrest in Xenopus. Mol. Biol. Cell, 13, 3662–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dolma S., Lessnick,S.L., Hahn,W.C. and Stockwell,B.R. (2003). Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell, 3, 285–296. [DOI] [PubMed] [Google Scholar]
- 37.Newport J. and Dasso,M. (1989) On the coupling between DNA replication and mitosis. J. Cell Sci. Suppl., 12, 149–160. [DOI] [PubMed] [Google Scholar]
- 38.Lathan B. and Von Hoff,D.D. (1984) Cytotoxic activity of echinomycin in a human tumor cloning system. Cancer Drug Deliv., 1, 191–198. [DOI] [PubMed] [Google Scholar]
- 39.Park Y.S., Kim,Y.H., Kim,S.K. and Choi,S.J. (1988) A new antitumor agent: methyl sulfonium perchlorate of echinomycin. Bioorg. Med. Chem. Lett., 8, 731–734. [DOI] [PubMed] [Google Scholar]