Abstract
Recent studies have begun to reveal that numerous fundamental metabolic pathways in bacteria are regulated by riboswitches residing within certain messenger RNAs. These riboswitches selectively bind metabolites and modulate gene expression in response to changing ligand concentrations. Previously, we provided evidence that the btuB mRNAs of Escherichia coli and Salmonella typhimurium each carry a coenzyme B12-dependent riboswitch that causes repressed translation of the encoded cobalamin-transport protein at elevated coenzyme concentrations. Herein, we use a phylogenetic analysis to define a consensus sequence and secondary structure model for the ligand- binding domain of this riboswitch class. RNA structures that conform to this model are widespread in both Gram-positive and Gram-negative organisms. In addition, we find that the 5′-untranslated region (5′-UTR) of the cobalamin biosynthesis (cob) operon of S.typhimurium carries an RNA motif that matches this consensus sequence. Biochemical and genetic characterization of this motif confirms that the RNA directly binds coenzyme B12, and that it likely serves as a genetic control element for regulating expression of the 25-gene operon for cobalamin production in this pathogen.
INTRODUCTION
Riboswitches serve as ligand-responsive genetic control elements that modulate the expression of certain genes in response to changing concentrations of metabolites. These RNA elements are typically embedded within the 5′-UTR of specific prokaryotic mRNAs, and are composed of two functional and sometimes distinct structural domains (1). One domain serves as a natural aptamer that binds the target metabolite with high selectivity. The other domain is an ‘expression platform’ that harnesses allosteric changes in RNA structure, brought about by aptamer–ligand complex formation, to control expression of the adjacent gene or operon. In prokaryotes, expression platforms have been identified that control transcription termination (2,3) or translation (4).
To date, seven fundamental metabolites are known to be detected by riboswitches, which are responsible for contributing to the genetic regulation of at least 68 genes in Bacillus subtilis (5). These targets include the nucleobases guanine and adenine, the amino acid lysine, and the coenzymes thiamine pyrophosphate, FMN, SAM and B12 (adenosylcobalamin or AdoCbl). Several classes of riboswitches have been shown by database searches to be distributed widely amongst prokaryotic organisms, and some examples have been identified in archeans (4,6–8). Sequence variants of the natural aptamer for thiamine pyrophosphate also have been identified in certain fungal and plant mRNAs, where they bind the coenzyme with affinities that match those of their prokaryotic counterparts (9). These findings indicate that riboswitches are a fundamental and widespread form of genetic control.
Initial confirmation that an mRNA domain directly binds to a metabolite in the absence of proteins was achieved by examining a 202-nucleotide fragment of the 5′-UTR of the btuB gene of Escherichia coli, and similarly by examining the homologous mRNA fragment for Salmonella typhimurium (10). The fragment derived from E.coli binds coenzyme B12 with an apparent dissociation constant of ∼300 nM, while related analogs such as cyanocobalamin and methylcobalamin do not exhibit measurable binding. In the current study, a refined consensus sequence and structural model for the coenzyme B12 class of riboswitches is presented. We establish that the 5′-leader sequence for the cob mRNA conforms to this consensus motif and that it directly binds coenzyme B12 with an apparent KD that matches those of related riboswitches. Furthermore, we identify a variant form of the B12 riboswitch class from B.subtilis that retains ligand-binding and genetic control functions despite the absence of nearly half of the conserved core that is typically indicative of a coenzyme B12 aptamer. The latter observation suggests that this class of riboswitches is composed of modular structural domains and that significant alterations to structure and function can occur through evolution.
MATERIAL AND METHODS
Chemicals and oligonucleotides
Coenzyme B12 was purchased from Sigma and both tritiated coenzyme B12 ([3H]AdoCbl) (11) and PurCbl were gifts from K. L. Brown (Ohio University). DNA oligonucleotides were synthesized by the HHMI Keck Foundation Biotechnology Resource Center at Yale University. DNAs were purified by denaturing (8 M urea) polyacrylamide gel electrophoresis (PAGE) and eluted from the gel by crushing/soaking in 10 mM Tris–HCl (pH 7.5 at 23°C), 200 mM NaCl and 1 mM EDTA. DNAs were recovered from the solution by precipitation with ethanol, resuspended in water and stored at –20°C.
Phylogenetic analyses
The coenzyme B12 riboswitch phylogeny was assembled by searching bacterial genomes for degenerate matches to sequence and structural elements that appeared to be conserved amongst the E.coli and S.typhimurium representatives using the program SequenceSniffer (J.E.B and R.R.B, unpublished algorithm). Most sequences have three or fewer mismatches to a representative motif [GGGAA <<<<< NNNNNNN (50) >>>>> RC << NNNN (2) >> RCNGT (100) CAYY (100) GGGAAG (200) AGYCNGRANAC (1) NGC, where ‘<’ or ‘>’ represent base pairs and numbers in parentheses allow variable insertions with a constrained maximum length].
Analysis of RNA structures by in-line probing
DNA templates for in vitro transcriptions were generated by polymerase chain reaction (PCR) using appropriate DNA primers (see for example the yvrC–lacZ fusion construction below). All RNA constructs were prepared by in vitro transcription using T7 RNA polymerase, and 5′-32P-labeled using methods described previously (2). Approximately 2 nM of end-labeled precursor RNA was subjected to in-line probing by incubating (in the dark to prevent light-mediated degradation of cobalamins) for ∼40 h at 25°C in 50 mM Tris–HCl (pH 8.3 at 25°C), 20 mM MgCl2 and 100 mM KCl in the presence or absence of ligands as indicated. Spontaneous cleavage products were resolved by denaturing 10% PAGE, visualized by PhosphorImager analysis (Molecular Dynamics), and quantitated by ImageQuanNT software. Apparent dissociation constants of RNA–ligand interactions were determined by plotting the normalized fraction cleaved at indicated sites versus the logarithm of ligand concentration in molar units.
Construction of a yvrC–lacZ
A chimeric DNA construct comprising a modified lysC promoter region (A to T and C to A at positions –11 and –31, respectively, relative to the start of transcription) (12) and the first 275 nucleotides of the yvrC transcription template was generated as a HindIII–BamHI fragment upon PCR amplification of B.subtilis chromosomal DNA (strain 1A40; Bacillus Genetic Stock Center, Columbus, OH, USA). The PCR product was cloned into pDG1661 (13) immediately upstream of the lacZ reporter gene. The sequence of the resulting clone was confirmed and was used as a template for subsequent preparation of PCR-derived DNAs for in vitro transcription.
In vivo assay of yvrC–lacZ fusions
Plasmid pDG1661 variants were transformed into B.subtilis strain 1A40 as described previously (5,7,11). Transformants were identified by selecting for chloramphenicol resistance (5 µg/ml) and screening for spectinomycin (100 µg/ml) sensitivity. Transformants were grown with shaking at 37°C in the presence or absence of 100 µM coenzyme B12 in defined media containing 0.5% w/v glucose, 2 g/l (NH4)2SO4, 18.3 g/l K2HPO4·3H2O, 6 g/l KH2PO4, 1 g/l sodium citrate, 0.2 g/l MgSO4·7H2O, 0.5 mM CaCl2, 5 µM MnCl2, 50 µg/ml l-lysine, 50 µg/ml l-methionine, 50 µg/ml l-tryptophan and 5 µg/ml chloramphenicol. Cells were grown to exponential phase (A595 of 0.6) and the level of β-galactosidase was determined as described previously (10).
Scatchard analyses
Scatchard data were generated using Dispo Equilibrium Dialyzers (Harvard Bioscience), wherein chambers a and b are separated by a dialysis membrane with a 5000 molecular weight cut-off. Specifically, 30 µl of equilibration buffer [50 mM Tris–HCl (pH 8.3 at 25°C), 20 mM MgCl2, 100 mM KCl] containing 1 µM 386 cob RNA (G plus 132–386 of the cob 5′-UTR) was added to chamber b. A total of 30 µl equilibration buffer containing [3H]AdoCbl (87.5 mCi/mmol) at concentrations ranging from 1 to 10 µM was placed in chamber a. The reactions were allowed to equilibrate for ∼11 h, at which point aliquots were removed for liquid scintillation counting. Counts per minute (c.p.m.) of chambers a and b were determined by liquid scintillation counting and were utilized for the determination of the concentration of free ([LF]) and bound ([LB]) [3H]AdoCbl using the following relationships:
(c.p.m. b) – (c.p.m. a) = (total c.p.m.)bound
2× (c.p.m. a) = (total c.p.m.)free
The data were assembled into a plot of r {[LB]/(1 µM cob RNA)} versus r/[LF], where the slope is equal to –1/KD and the x intercept reflects the number of ligand binding sites. The data described in the text were derived from three separate experiments after correction for ∼27% tritium that is not in the form of coenzyme B12.
RESULTS AND DISCUSSION
Phylogenetic analysis of coenzyme B12 riboswitches reveals a complex RNA motif
The 5′-UTRs of the btuB genes from E.coli and S.typhimurium each contain a large RNA domain that functions as a metabolite-dependent genetic control element (10). Both RNAs bind coenzyme B12 (5′-deoxy-5′-adenosylcobalamin or AdoCbl) with high affinity and specificity, and thus function as natural aptamers for this important metabolite. These RNAs, along with the 5′-UTR of the cob gene from S.typhimurium, carry a short conserved sequence domain termed the B12 box (12,14), and therefore they likely share structural and functional homology.
Although we proposed a partial secondary structural model for the riboswitch domains present in the btuB mRNAs from E.coli and S.typhimurium (10), the model was not in agreement with that proposed recently for the 5′-UTR of the cob gene (14). To improve upon the structural model, additional examples of the coenzyme B12 class of riboswitches were identified in other organisms. This was achieved through an iterative approach to secondary-structure modeling and comparative sequence analysis using a computer algorithm. The algorithm (SequenceSniffer; J. E. Barrick and R. R. Breaker, unpublished) searches genomic databases for domains that correspond closely to specific sequence and structural constraints, and has been used previously to identify homologs for the TPP (9), guanine (5), SAM (7) and lysine riboswitches (15). More specifically, database searches such as these are effective in revealing similar aptamer domains, as the metabolite-binding portions of riboswitches are the most highly conserved amongst various organisms and amongst different mRNAs within a single organism.
A total of 92 different representatives of the coenzyme B12 class of riboswitches were identified and compared by sequence alignment (Fig. 1; see also the Rfam database for additional representatives) (16). Upon examination of the sequence alignments, a pattern of conserved sequence and secondary-structure elements that are present in all representatives becomes evident (Fig. 2A). The most striking observation is that the B12 box encompasses only a small portion of the conserved sequence and structural domain. The phylogenetic analysis indicates that this complex RNA motif has at least 10 base-paired elements and ∼57 nucleotide positions whose identity is conserved in >90% of the representative sequences.
Figure 1.
Sequence alignment of putative coenzyme B12 riboswitches. Conserved sequence and base pairing elements are highlighted for 43 of 92 representative motifs identified by searching GenBank (see Materials and methods). The domain’s strand orientation (minus strand unless otherwise noted) and nucleotide number are provided (Position) for the 5′ nucleotide of the first interior UG base pair of stem P1 (asterisk). Conserved base-pairing interactions are colored, and are labeled via the secondary structure diagram at the top. Numbers in the alignment represent stretches of nucleotides that are not depicted. The region between P7 and P10 is highly variable, but always contains a putative P8 pairing. Only one example of a riboswitch-like domain is shown for each organism. For organisms with multiple riboswitches, the total number is indicated (#). A full alignment of all sequences is provided as supplementary information and has been submitted to the Rfam database (16). The Sorghum and Leishmania entries (&) are genomic clones from unfinished eukaryotic genomes. As such, these riboswitch sequences might be derived from bacterial contamination. The top four sequences are examined in this study.
Figure 2.
Consensus sequence and secondary-structure model for the metabolite-binding domain of the B12 class of riboswitches. (A) Red nucleotides depict bases whose identities are conserved in at least 90% of the representatives of the phylogeny assembled by database searching (see Fig. 1). (B) Sequence and revised secondary-structure model for the B12 riboswitch from the 5′-UTR of the E.coli btuB gene. In-line structural probing data were taken from a study published previously (10). Although the portion of the RNA corresponding to the most highly conserved domain throughout evolution ends at nucleotide 160, the sequences through nucleotide 202 are required for maximal binding affinity (see Fig. 4). The previously identified B12 box (12,14) resides between nucleotides 140 and 160.
The secondary-structure model proposed previously for the riboswitch from E.coli btuB mRNA (10) corresponds only partially with the new model that is based on phylogenetic data. Specifically, pairing elements P4 (part) and P5 to P8 of the original structural model are retained in the revised model with the modified designations P5 and P8 to P12, respectively (Fig. 2B). In addition, the P9 stem from the original model is retained without designation in the revised model, as it resides outside the domain of highest sequence and structural conservation. In contrast, the revised model includes numerous short base-paired elements that explain the role of nucleotides that were left unfolded in the earlier model, and also that preclude formation of certain other pairing possibilities. Specifically, stems P1 to P3, as predicted in the original model, are replaced and supplemented in the revised model by stems P1 to P4, P6, P7 and P10.
The revised secondary-structure model is similar to a model proposed recently for this same RNA motif (17,18). Most notably, these two models differ with respect to how the B12 box is folded, and on the structural details of the bridge between the left and right halves of the E.coli btuB aptamer. Several lines of evidence support our revised structural model for the B12 aptamer. First, the new secondary-structure model is largely consistent with structural probing data that was presented previously (10). Likewise, mutational analysis supported the formation of a bridging stem element that is retained in the revised model (P8). Thirdly, representatives within the sequence alignment (Fig. 1) exhibit considerable sequence covariation within the stem elements that are common to most B12 riboswitches (Fig. 2B), indicating that these pairing elements are important for riboswitch function. Specifically, the covariation in sequences of proposed stems P3 and P6 (Fig. 1) suggest that these structures are more likely to be formed than the alternative structure proposed recently (17,18).
The revised secondary-structure model is more complex than those of other riboswitches. Interestingly, biochemical evidence (see below) suggests that, at least in some cases, RNA constructs that encompass only the minimal aptamer domain are not sufficient to exhibit maximal binding affinity to coenzyme B12. These latter results indicate that the more highly conserved portion of the riboswitch might not comprise the complete and fully functional aptamer motif. Thus, the likely involvement of more distal RNA elements (such as a possible pseudoknot interaction between the loop of P5 and downstream sequences) adds even greater complexity to the RNA’s structure.
5′-UTRs from the btuB and cob mRNAs of S.typhimurium bind coenzyme B12
Like btuB, genetic expression of the cob mRNA from S.typhimurium is known to be regulated by B12-related compounds (14,19,20). We speculated that the presence of the conserved B12 box also could be an indication that the mRNA that encodes for the majority of the genes required for de novo synthesis of coenzyme B12 might be negatively regulated by the metabolic end product. Consistent with this hypothesis is the observation that our database search, along with another reported recently (17,18), for sequence elements with similarity to the B12 aptamer identified the 5′-UTR of the cob operon from S.typhimurium (Fig. 1).
As expected, the 5′-UTR from S.typhimurium btuB mRNA has considerable sequence and predicted structural similarity to that of the homologous E.coli leader (Fig. 3A). In contrast, the sequence of the corresponding cob 5′-UTR varies significantly from that of btuB. Furthermore, the secondary-structure model proposed recently (14) for the cob RNA differs substantially from our model for the coenzyme B12 aptamer presented in Figure 2. However, we find that the cob 5′-UTR does carry the consensus sequence elements that are indicative of the B12 class of riboswitches, despite the considerable sequence differences between the two RNAs. Furthermore, the cob 5′-UTR can be folded into a secondary structure (Fig. 3B) that carries the major features of the B12 aptamer model (Fig. 2A). The differences in sequence and predicted base-pairing potential reside in portions of the RNA that are not conserved through evolution. Thus, the two leader domains from the btuB and cob mRNAs from S.typhimurium each carry a putative ligand binding aptamer domain for a B12-dependent riboswitch.
Figure 3.
Structure and function of two B12 riboswitch domains from S.typhimurium. Sequence and secondary structure models for the B12 aptamer domains residing in the 5′-UTRs of the btuB mRNA (A) and the cob operon mRNA (B) from S.typhimurium. Constructs are named according to the last nucleotide derived from the respective UTR. Asterisks identify nucleotides that were added to facilitate transcription in vitro. Nucleotides denoted in red correspond to the consensus sequences as depicted in Figure 2A. Corresponding in-line probing analyses for the btuB (C) and cob (D) mRNA leader sequences are presented. T1, –OH and NR identify RNAs that were partially digested with RNase T1 (cleavage after G residues), those incubated at elevated pH, or those that were not reacted, respectively. RNAs in the remaining lanes were incubated for ∼40 h in the absence of ligand (–), or in the presence of 0.1 or 1 mM coenzyme B12. Filled and open arrowheads identify selected RNase T1 digestion products, and sites of coenzyme B12-mediated structure modulation, respectively. The numbered sites of modulation for the btuB sequence were used subsequently for analysis of apparent KD (Fig. 4A).
In-line probing analyses were conducted with fragments of both the btuB and cob mRNA leader sequences from S.typhimurium that correspond to the putative B12 aptamer domains. As indicated previously (10), the btuB mRNA undergoes B12-dependent structure modulation with a pattern that is similar to that observed for the homologous mRNA from E.coli (Fig. 3C). The data also reveal that substantial ligand-dependent structure modulation occurs with the cob mRNA fragment (Fig. 3D). Upon closer inspection, the spontaneous cleavage patterns for both RNAs correspond well with the revised secondary-structure model for the B12 aptamer, wherein the sites of modulation largely correlate with conserved bulges and sites of constant high cleavage correlate with the non-conserved loops of stems. Therefore, we conclude that the 5′-UTR of the cob mRNA directly binds coenzyme B12 without the participation of protein factors, and that the biosynthetic pathway for this metabolite is regulated, at least in part, by a coenzyme B12-responsive riboswitch.
Ligand-binding affinity varies with the length of the 5′-UTR fragment
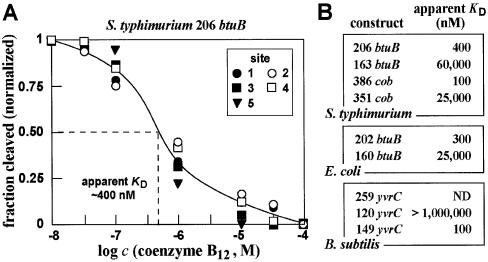
The ligand-dependent changes in spontaneous RNA cleavage can be quantitated and used to derive an apparent KD value for ligand–aptamer complex formation (21). This approach was used previously to obtain an apparent KD value for the interaction of coenzyme B12 with the 202 btuB construct from E.coli of ∼300 nM (10). Similarly, the corresponding 206 btuB construct from S.typhimurium exhibits an apparent KD value of ∼400 nM (Fig. 4A).
Figure 4.
Measurements of binding affinities of various coenzyme B12 aptamers. (A) Representative plot of the ligand concentration-dependent modulation of spontaneous RNA cleavage using the 206 btuB fragment from S.typhimurium. The fraction of RNA cleaved at sites 1–5 (Fig. 3C) was normalized against the fraction cleaved at each site when incubated in the absence of coenzyme B12. Site 5 is not present in the preceding image, but is present in the larger RNA construct used for this KD analysis (data not shown). (B) Comparison of the apparent KD values for various coenzyme B12 aptamer constructs from several bacteria. Values were derived for each construct using the approach described in (A).
The binding affinities for certain riboswitch constructs are known to vary depending upon the length of the construct used. For example, the thiamine pyrophosphate (TPP) aptamers from the thiM and thiC mRNAs of E.coli exhibit the highest affinity for TPP, while larger constructs that carry portions of the expression platform each bind with significantly lower affinity (4). This is likely due to the presence of competing RNA structures that detract from tight ligand binding by reducing preorganization of the aptamer domain. In other words, the allosteric nature of the full riboswitch element (aptamer plus expression platform) might be expected to result in a poorer equilibrium constant for complex formation with its target ligand due to these dynamic but mutually exclusive folds.
We examined several RNA constructs that encompass the minimal consensus sequence and structure for the B12 aptamer to determine whether such molecules might exhibit higher affinity for the coenzyme. Interestingly, the shortened constructs for the two btuB RNAs and for the cob RNA experience a significant (>100-fold) loss of binding affinity when the riboswitch is trimmed to represent only the conserved aptamer domain (Fig. 4B). This suggests that sequence or structural element(s) within the presumed non-conserved domain of the riboswitch aid in proper formation of the more highly conserved aptamer.
In contrast to other instances where extra-consensus sequences detract from binding affinity, the consensus B12 aptamer domain (at least in some cases) makes productive use of flanking sequences to form an aptamer domain with improved affinity. Previously, we had suggested that a pseudoknot might play an important role in the formation of the B12 riboswitch structure of E.coli btuB (10). This speculative proposal was based on the fact that structural probing data were consistent with such an interaction, although mutational analysis was inconclusive. This proposed pseudoknot, which is depicted as a base pairing interaction between the loop of P5 and the loop of a non-conserved hairpin (Fig. 2B), might play a role in supporting the consensus domain in its function as a B12 aptamer. If true, this putative interaction is not obviously present in all other representatives of the phylogeny examined in this study.
Confirmation of coenzyme B12–cob RNA interaction by equilibrium dialysis
Equilibrium dialysis was used to confirm that the cob mRNA sequence directly binds coenzyme B12 in the absence of proteins (data not shown), as had been determined previously (10) for the corresponding btuB mRNA fragment from E.coli. A Scatchard analysis was also carried out for the 386 cob RNA by using equilibrium dialysis with various concentrations of tritiated coenzyme B12 (data not shown). Unfortunately, coenzyme B12 is highly photo-labile, and the data were not of sufficient quality to establish stoichiometry with confidence.
Specifically, we found that 27% of the tritium in a control equilibrium dialysis assay failed to bind the RNA, which is most likely due to breakdown of the compound during assembly of the assay mixtures, despite taking precautions to minimize exposure to light. The uncertainty with respect to the extent of degradation in each independent equilibrium dialysis run likely contributes scatter in the placement of data points on the Scatchard plot. Although the plot should be interpreted with caution, a best-fit line to the data indicates a dissociation constant for the cob aptamer of ∼200 nM, which is similar to that determined by in-line probing (Fig. 4B). The x intercept of this line resides between 0.75 and 1, which is most consistent with a 1:1 relationship between ligand and RNA when in complex.
A variant coenzyme B12 riboswitch occurs in the B.subtilis yvrC mRNA
The phylogenetic survey of domains that resemble B12 riboswitches revealed a single example from B.subtilis (Figs 1 and 5A). This representative resides immediately upstream of an apparent four-gene operon that carries the open reading frames (ORFs) yvrC, yvrB, yvrA and yvqK. Although the functions of these ORFs remain unproven, the yvr genes appear to encode proteins involved in metal import and processing, and thus modulation of their levels of genetic expression in response to coenzyme B12 concentrations seems reasonable since this coenzyme carries a metal ligand.
Figure 5.
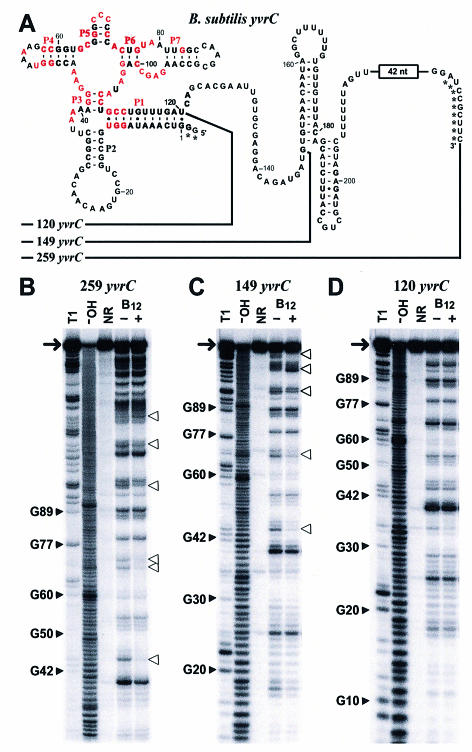
A variant riboswitch aptamer retains selective binding of coenzyme B12. (A) The B12 riboswitch-like domain residing upstream of the yvrC gene of B.subtilis. The 149 yvrC and 120 yvrC constructs carry an additional two nucleotides preceding position 1 to facilitate efficient transcription in vitro. Similarly, the 259 yvrC construct carried an additional sequence (GGAAAAACGGAUACGAAU) residing immediately upstream from nucleotide 1, wherein the first G residue was altered from the natural nucleotide identity. Asterisks identify non-natural nucleotides. (B–D) In-line probing analyses of three RNA fragments of the putative yvrC 5′-UTR of B.subtilis. Details are as described in the legend to Figure 3.
Interestingly, the B.subtilis variant is lacking the P8, P10 and P11 helices, which otherwise would present sequence and structural elements that are conserved in most other representatives (Fig. 2A). The absence of this highly conserved portion of the B12 aptamer makes the yvrC RNA one of the most extreme variants identified in our phylogenetic analysis. Therefore, we conducted several experiments to determine whether the yvrC RNA binds coenzyme B12 and whether the riboswitch-like domain responds genetically to this effector when appended to a reporter gene.
Three RNA constructs were created based on the UTR sequence of B.subtilis yvrC. The two largest constructs, 259 yvrC and 149 yvrC, both exhibit structural modulation upon addition of coenzyme B12, which indicate that they both retain ligand-binding function (Fig. 5B and C) despite the absence of the P8, P10 and P11 helices and the associated conserved nucleotides. However, the 120 yvrC construct, which carries only those nucleotides that correspond closely to the left-most portion of the consensus aptamer motif, fails to bind coenzyme B12 as indicated by the absence of ligand-induced structural modulation (Fig. 5D).
This finding suggests that a structural element that is critical for high-affinity binding resides within nucleotides 121 and 149 of the larger yvrC constructs. In addition, this result suggests that the B12 riboswitch might make use of a two-lobed aptamer, where the left and right halves of the RNA form distinct folds that independently recognize different parts of the ligand. In this model, the novel 29-nucleotide domain that forms the 3′ terminus of 149 yvrC might serve as a functional replacement for the absence of stems P8 to P12 that are found in the E.coli and S.typhimurium btuB RNAs. In a preliminary assessment of this hypothesis, we have examined whether the molecular recognition characteristics of the yvrC and btuB variants might be different. Specifically, the yvrC leader exhibits poorer discrimination against purinyl cobalamin (PurCbl) (10) than does the btuB riboswitch from E.coli (data not shown). Although other explanations for this observation can be envisioned, it is possible that the right half of the aptamer binds the adenosyl moiety of coenzyme B12 and that the 29-nucleotide alternative domain from yvrC recognizes this moiety somewhat differently.
The 5′-UTR of the B.subtilis yvrC mRNA also permits coenzyme B12-dependent modulation of a reporter gene when expressed in vivo. To assess riboswitch function in vivo, a DNA construct carrying the first 275 nucleotides of the yvrC sequence (Fig. 5A) was appended to a β-galactosidase gene in a transcriptional reporter vector. When integrated into B.subtilis, the construct exhibits an ∼3-fold reduction of β-galactosidase activity when coenzyme B12 is added to the culture medium. Specifically, Miller units of 142 and 47 were obtained in the absence and presence of B12 supplementation to the medium, respectively. These findings indicate that the yvrC variant is a functional coenzyme B12-specific riboswitch, despite its weaker correspondence to the sequence and structure of most other representatives in the phylogeny.
CONCLUSIONS
The established diversity of riboswitches in living systems is continuing to expand as biochemical, genetic and bioinformatic approaches are being applied to find and characterize new examples. Phylogenetic analyses reported herein and elsewhere (17,18) indicate that the coenzyme B12 class of riboswitches is widely distributed amongst both Gram- positive and Gram-negative prokaryotes. In addition, our data indicate that RNA domains matching the consensus sequence and structure of this riboswitch class from three different organisms do indeed bind coenzyme B12 and control gene expression in response to high ligand concentrations.
In many instances, genes under the control of a coenzyme B12 riboswitch are involved in the transport of cobalamin compounds or metals, or associated with the biosynthetic pathway for the coenzyme. For example, a coenzyme B12 riboswitch in S.typhimurium modulates expression of the btuB gene encoding a cobalamin transport protein, and a second riboswitch of this class serves as a genetic control element for the 25-gene cob operon. In certain organisms, these riboswitches control the expression of genes that are more distantly related to the coenzyme. Perhaps most striking is the involvement of coenzyme B12 riboswitches in the control of ribonucleotide reductases that do not use this coenzyme. Presumably, when this compound is at a sufficiently high concentration, expression of the coenzyme B12-independent ribonucleotide reductase is reduced to defer this metabolic activity to the coenzyme B12-dependent form of the enzyme.
The potential complexity of riboswitch structures is also apparent by examining the structural model of the coenzyme B12 class of riboswitches. In addition to the numerous short helices and conserved sequence elements, long-range RNA interactions likely augment aptamer function, at least in the case of the btuB variants examined. Given the widespread distribution of riboswitches, RNA appears to have sufficient structural and functional diversity to perform reliably as genetic control elements for genes involved in critical biological processes. The substantial deviation in consensus sequence and structure exemplified by the yvrC RNA and several others indicates that considerable changes in riboswitch structure can occur through evolutionary processes. Although the large-scale alteration of this riboswitch class appears to be somewhat limited, it seems reasonable to speculate that variant riboswitches might exist that respond selectively to closely related metabolites.
Acknowledgments
ACKNOWLEDGEMENTS
We thank K. L. Brown for the gift of tritiated coenzyme B12 and its analogs, and members of the Breaker laboratory for helpful discussions. In addition, we thank Margaret S. Ebert for assistance with preliminary studies of the cob RNA. This work was supported by grants from the National Institutes of Health (GM 559343) and the National Science Foundation (EIA-0129939, EIA-0323510 and EIA-0324045). R.R.B is also grateful for support from the David and Lucile Packard Foundation.
REFERENCES
- 1.Winkler W.C. and Breaker,R.R. (2003) Genetic control by metabolite-binding riboswitches. Chembiochem, 4, 1024–1032. [DOI] [PubMed] [Google Scholar]
- 2.Winkler W.C. Cohen-Chalamish,S. and Breaker,R.R. (2002) An mRNA structure that controls gene expression by binding FMN. Proc. Natl Acad. Sci. USA, 99, 15908–15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mironov A.S., Gusarov,I., Rafikov,R., Lopez,L.E., Shatalin,K., Kreneva,R.A., Perumov,D.A. and Nudler,E. (2002) Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell, 111, 747–756. [DOI] [PubMed] [Google Scholar]
- 4.Winkler W., Nahvi,A. and Breaker,R.R. (2002) Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature, 419, 952–956. [DOI] [PubMed] [Google Scholar]
- 5.Mandal M., Boese,B., Barrick,J.E., Winkler,W.C. and Breaker,R.R. (2003) Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell, 113, 577–586. [DOI] [PubMed] [Google Scholar]
- 6.Rodionov D.A., Vitreschak,A.G., Mironov,A.A. and Gelfand,M.S. (2002) Comparative genomics of thiamin biosynthesis in prokaryotes. J. Biol. Chem., 277, 48949–48959. [DOI] [PubMed] [Google Scholar]
- 7.Winkler W.C., Nahvi,A., Sudarsan,N., Barrick,J.E. and Breaker,R.R. (2003) An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nature Struct. Biol., 9, 701–707. [DOI] [PubMed] [Google Scholar]
- 8.Stormo G.D. and Ji.,Y. (2001) Do mRNAs act as direct sensors of small molecules to control their expression? Proc. Natl Acad. Sci. USA, 98, 9465–9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sudarsan N., Barrick,J.E. and Breaker,R.R. (2003) Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA, 9, 644–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nahvi A., Sudarsan,N., Ebert,M.S., Zou,X., Brown,K.L. and Breaker,R.R. (2002) Genetic control by a metabolite binding mRNA. Chem. Biol., 9, 1043–1049. [DOI] [PubMed] [Google Scholar]
- 11.Brown K.L. and Zou,X. (1999) Thermolysis of coenzymes B12 at physiological temperatures: activation paramenters for cobaltcarbon bond homolysis and a quantitative analysis of the pertubation of the homolysis equilibrium by the ribonucleoside triphosphate reductase from Lactobacillus leichmannii. J. Inorg. Biochem., 77, 185–195. [DOI] [PubMed] [Google Scholar]
- 12.Lundrigan M.D., Koster,W. and Kadner,R.J. (1991) Transcribed sequences of the Escherichia coli btuB gene control its expression and regulation by vitamin B12. Proc. Natl Acad. Sci. USA, 88, 1479–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guérout-Fleury A.M., Frandsen,N. and Stragier,P. (1996) Plasmids for ectopic integration in Bacillus subtilis. Gene, 180, 57–61. [DOI] [PubMed] [Google Scholar]
- 14.Ravnum S. and Andersson,D.I. (2001) An adenosyl-cobalamin (coenzyme-B12)-repressed translational enhancer in the cob mRNA of Salmonella typhimurium. Mol. Microbiol., 39, 1585–1594. [DOI] [PubMed] [Google Scholar]
- 15.Sudarsan N., Wickiser,J.K., Nakamura,S., Ebert,M.S. and Breaker,R.R. (2003) An mRNA structure that controls gene expression by binding lysine. Genes Dev., 17, 2688–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffiths-Jones S., Bateman,A., Marshall,M., Khanna,A. and Eddy,S.R. (2003) Rfam: an RNA family database. Nucleic Acids Res., 31, 439–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodionov D.A., Vitreschak,A.G., Mironov,A.A. and Gelfand,M.S. (2003) Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem., 278, 41148–41159. [DOI] [PubMed] [Google Scholar]
- 18.Vitreschak A.G., Rodionov,D.A., Mironov,A.A. and Gelfand,M.S. (2003) Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA, 9, 1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richter-Dahlfors A.A. and Andersson,D.I. (1992) Cobalamin (vitamin B12) repression of the Cob operon in Salmonella typhimurium requires sequences within the leader and the first translated open reading frame. Mol. Microbiol., 6, 743–749. [DOI] [PubMed] [Google Scholar]
- 20.Richter-Dahlfors A.A., Ravnum,S. and Andersson,D.I. (1994) Vitamin B12 repression of the cob operon in Salmonella typhimurium: translational control of the cbiA gene. Mol. Microbiol., 13, 541–553. [DOI] [PubMed] [Google Scholar]
- 21.Soukup G.A., DeRose,E.C., Koizumi,M. and Breaker,R.R. (2001) Generating new ligand-binding RNAs by affinity maturation and disintegration of allosteric ribozymes. RNA, 7, 524–536. [DOI] [PMC free article] [PubMed] [Google Scholar]