Abstract
Dosage compensation in mammals occurs at two levels. In addition to balancing X-chromosome dosage between males and females via X-inactivation, mammals also balance dosage of Xs and autosomes. It has been proposed that X-autosome equalization occurs by upregulation of Xa (active X). To investigate mechanism, we perform allele-specific ChIP-seq for chromatin epitopes and analyze RNA-seq data. The hypertranscribed Xa demonstrates enrichment of active chromatin marks relative to autosomes. We derive predictive models for relationships among POL-II, active mark densities, and gene expression, and suggest that Xa upregulation involves increased transcription initiation and elongation. Enrichment of active marks on Xa does not scale proportionally with transcription output, a disparity explained by nonlinear quantitative dependencies among active histone marks, POL-II occupancy, and transcription. Significantly, the trend of nonlinear upregulation also occurs on autosomes. Thus, Xa upregulation involves combined increases of active histone marks and POL-II occupancy, without invoking X-specific dependencies between chromatin states and transcription.
INTRODUCTION
In many organisms, sex is determined genetically by dimorphic sex chromosomes. In the XY-based system, females are homogametic (XX) and males are heterogametic (XY)1,2 Current evolutionary theories suggest that sex chromosomes evolved from a pair of autosomal homologues, and acquisition of favorable male genes on the Y led to a suppression of recombination, making gradual loss of Y-chromosome material inevitable. Degeneration of the Y would have resulted in a continual series of sudden changes in gene dosage balance not only between male and female Xs, but also between X and autosomes 1. Ohno predicted that two types of dosage compensation schemes must therefore exist 2-4. For mammals, the existence of X-chromosome inactivation (XCI) to silence one of the two X chromosomes in females has been known since 1961 5,6. This mechanism equalizes X-chromosome dosage between the sexes and depends on expression of Xist RNA 7-9 coupled with recruitment of PRC2 complex10-12 . But because XCI creates another level of dosage imbalance, this one between X’s and autosomes of both sexes, a secondary compensatory mechanism must target the active X chromosome (Xa) and double its transcription to restore genome-wide balance.
Several recent studies support the idea of X hyperactivation in mammals. Microarray-based gene expression profiling of mammalian tissues showed that X-linked genes are expressed not at half the average autosomal dose (as would be expected if expression came from a single X) but at nearly the same dose as autosomal genes in both sexes, implying that the Xa is upregulated in both males and females 13,14. These conclusions have been challenged by analysis of RNA-Seq data, which showed that the expression average of X-linked genes was approximately half that of the autosomal average15 A more recent study, however, indicates that this interpretation was confounded by inclusion of silent genes on the X 16.
Here, we take an alternate approach to address whether and how dosage compensation occurs between X and autosomes by investigating chromatin signatures on a genome-wide scale. We carry out allele-specific chromatin immunoprecipitation with deep sequencing (ChIP-seq) for RNA polymerase II (POL-II) and activate chromatin marks and, through a combined analysis with RNA-seq data, we find that Xa upregulation indeed occurs. The data suggest that Xa upregulation occurs at the level of both transcription initiation and elongation and point to nonlinear quantitative dependencies among active histone marks, POL-II occupancies, and transcription output which are not X-specific and are part of a genome-wide mechanism for quantitative control of gene expression.
RESULTS
Confirmation of Xa upregulation
To address how X-linked transcription compares to autosomal transcription in the female soma and whether the differences, if any, could be explained by chromatin mechanisms, we first compared average gene expression of all X-linked and autosomal genes using previously published RNA-seq data from a mouse female fibroblast cell line 17. We calculated gene expression levels as FPKM values (fragments per kilobase per million) for non-overlapping RefSeq mouse genes using TopHat and Cufflinks methods, and found that the total FPKM averages of haploid X and autosomal genes differed only by 22%. This conclusion is consistent with the argument that Xa-hyperactivation does not occur 15.
However, the X-chromosome may harbor more silent genes than autosomes. Reasoning that this difference could confound measurements of average transcriptional output, we categorized genes with respect to their expression status (active vs inactive) and CpG content (high vs low) at the promoters (Supplementary Fig. 1a). A natural FPKM cutoff of ~1.0 for actively expressed genes was suggested by the analyses of dependency between gene expression and POL-II densities across the gene body (Supplementary Fig. 2, see Methods). We observed that almost 57% of X-linked genes were of low CpG content at promoters (LCP) with very little to no transcription (inactive). Indeed, when analysis of the RNA-seq data was performed using only expressed genes, the X-chromosome showed 85% higher mean expression than the average haploid autosome set, with 57% difference in median values (Supplementary Table I). X-hypertranscription was apparent among both active HCP and active LCP genes (Supplementary Fig. 1b). Differences between autosomal and X-linked gene populations were highly significant (Supplementary Table I, Mann-Whitney P-values; Supplementary Fig. 1c). Thus, in agreement with previous reports 13,14,16, the mammalian X is dosage compensated with respect to autosomes.
Allelic ChIP-seq reveals enrichment of active histone marks and POL-II on Xa
Next, we asked whether X:A dosage compensation has a chromatin basis. Over the years, genome-wide profiles of histone marks have been generated by ChIP-on-chip or ChIP-seq. In every case, the analysis was performed either in male cell lines 18,19 or in female lines without the allele-specificity and sequencing depth necessary to distinguish Xa from Xi with sufficient resolution20-22. Here, we performed high-resolution, allele-specific ChIP-seq in a clonal F1 hybrid female mouse fibroblast line that carries one haploid chromosome set of Mus musculus (mus) origin and a second haploid set of Mus castaneous (cast) origins. We examined active chromatin signatures that are associated with the transcription start site (TSS), including phospho-serine-5 RNA polymerase II (POL-II-S5P) and trimethylated histone H3 at lysine 4 (H3K4me3), both of which are associated with transcription initiation; we also examined signatures associated with transcription elongation through the gene body, including phospho-serine-2 POL-II (POL-II-S2P) and trimethylated H3 at lysine 36 (H3K36me3) 23,24. By using paired-end sequencing, we could uniquely align 83-93% of the 17-28 million read-pairs to the genome, yielding 16.5-21.9 million uniquely aligned reads per epitope (Supplementary Table II).
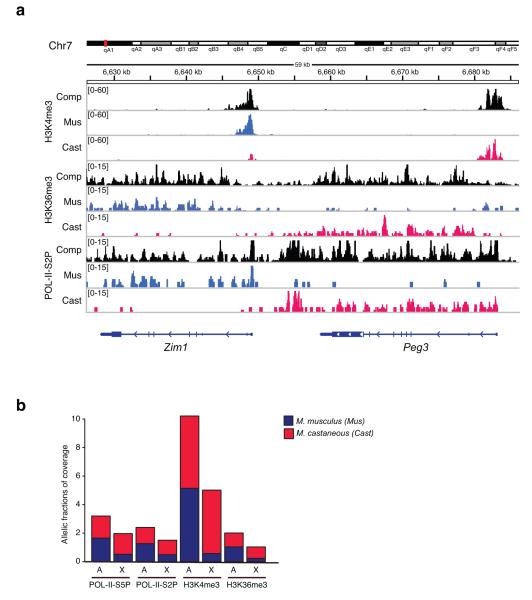
With ~22 million SNP and ~1 million insertion/deletion differences between mus and cast genomes25, ~35% of all read-paired could be assigned to specific alleles. Analysis of monoallelically expressed loci, including the Zim1/Peg3 imprinted domain (Fig. 1a and data not shown), confirmed the allele-specific output of the ChIP-seq. As expected, active marks were found on the paternally expressed Peg3 allele (cast) and the maternally expressed Zim1 allele (mus). Elsewhere in the genome, active marks were evenly distributed between autosomal homologues, as expected (Fig. 1b). By contrast, on the X-chromosome, active marks were predominantly on Xcast (Xa), consistent with occurrence of XCI.
Figure 1. Allele-specific ChIP-seq.
(a) Profiles for POL-II-S5P, H3K4me3, and H3K36me3 are mapped to M. castaneous (cast) or M. musculus (mus) alleles for two imprinted loci, Zim1 52 and Peg3 53 on Chromosome 7. Composite tracks (comp) represent combination of cast, mus and neutral reads. Coverage values are normalized by input and are indicated on the y-axis. (b) X chromosome shows a strong allelic skew in the occupancy of active histone marks and POL-II at the TSS and across the gene body. Barplots show mean composite densities of H3K4me3, H3K36me3, POL-II-S2P and POL-II-S5P on autosomes (A) and X chromosome (X), with proportion of allelic coverage indicated by red (cast), and blue (mus) fractions. Inactive X (Xmus), and active X (Xcast).
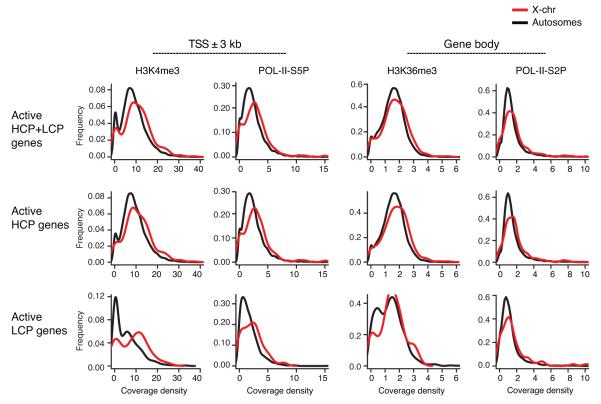
To determine whether Xa-hypertranscription is reflected in enrichment of active chromatin marks, we compared coverages on Xcast (Xa) with those on autosomes of M. castaneous origin. When we calculated medians for coverage densities on active genes, Xa showed 31% more POL-II-S5P and 24% more H3K4me3 coverage around the TSS; Xa also showed 20% more POL-II–S2P and 9% H3K36me3 coverage on the gene bodies (Fig. 2; Supplementary Table I). The enrichments were statistically significant in each category (Supplementary Table I), except for POL-II-S2P and H3K36me3 on LCP active genes, presumably due to smaller sample of LCP active genes on the Xa (n=72). Moreover, distributions of density values for all marks followed a similar shape (Fig. 2), suggesting that enrichment of Xa marks was global and could not be attributed to a distinct subset with exceptionally high coverage.
Figure 2. Distributions of coverage densities for POL-II and active histone modifications on X chromosome and autosomes.
Coverage density values are shown for H3K4me3 and POL-II-S5P at the TSS and for H3K36me3 and POL-II-S2P across the gene bodies as indicated. Distributions are plotted for actively transcribed (HCP+LCP, HCP and LCP) genes. Black line, autosomal genes; red line, X-linked genes.
Nonlinear dependencies between active chromatin marks and transcription output
Interestingly, the enrichment of POL-II and active histone marks on Xa relative to autosomes was not proportional to the nearly 2-fold transcriptional upregulation on Xa (Fig. 2; Supplementary Table I). Indeed, the degree of epitope enrichment was much less than 2-fold. To explain this disparity, we compared POL-II and histone mark densities against transcription output genome-wide. Plotting POL-II-S2P against FPKM (transcription) values revealed a good correlation between gene expression and the elongating form of POL-II, as expected. It also demonstrated a natural separation of active genes from inactive ones (Supplementary Fig. 2: dotted horizontal line, FPKM cutoff of ~1.0) regardless of gene category (HCP, LCP, all genes). Therefore, in subsequent analyses, we used active genes of all three categories to investigate the relationship between chromatin epitope and transcription output.
The relationship between transcription output and chromatin epitopes is shown by scatterplots and point densities for Xa (black line contours) and autosomal (color contours) genes in pairwise comparisons between expression and various chromatin epitopes (Fig. 3 and Supplementary Fig. 3-4). For active HCP M. castaneus alleles, we found pronounced dependencies between gene expression and POL-II-S5P, POL-II-S2P and active histone marks, regardless of whether they were X-linked or autosomal (Fig. 3a-d). Similar strong dependencies were observed between POL-II-S5P and H3K4me3 densities, and between POL-II-S2P and H3K36me3 densities (Fig. 3e-h). Similar trends were also found in the populations of all active (HCP+LCP) and active LCP genes (Supplementary Fig. 3-4). These data showed that Xa and autosomal genes are subject to similar quantitative relationships between active chromatin marks/POL-II and transcription output.
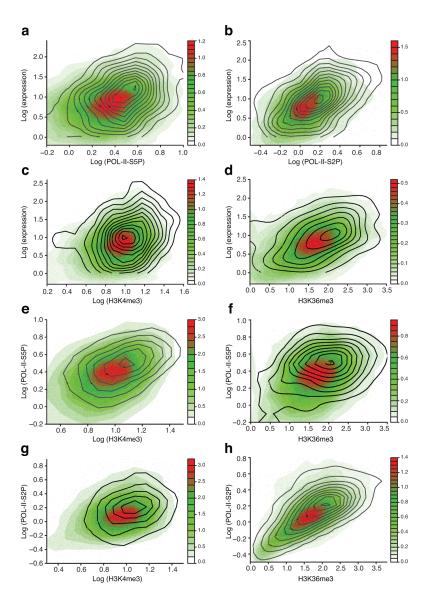
Figure 3. Relationships between levels of gene expression, POL-II, and active histone modifications.
M. castaneous alleles of actively expressed autosomal HCP genes are represented as points, with point density shown by colored contour. Black line contour represents active HCP X-linked M. castaneous alleles (Xa). Expression, POL-II, and histone modification levels are positively correlated, the relationships are non-linear, and X-linked genes follow autosomal trends of dependency, albeit with a shift to higher values. (a,b) POL-II at the TSS (a) and across the gene body (b) vs expression (log-log scale). (c) H3K4me3 at the TSS vs expression (log-log scale). (d) H3K36me3 across the gene body vs expression (linear-log scale). (e) H3K4me3 vs POL-II at the TSS (log-log scale). (f) H3K36me3 across the gene body vs POL-II at the TSS (linear-log scale). (g) H3K4me3 at the TSS vs POL-II across the gene body (log-log scale). (h) H3K36me3 vs POL-II across the gene body (linear-log scale).
These dependencies have several universal features that might provide general insight into the mechanism of Xa upregulation. First, the relationships showed monotonic trends: Increases in POL-II or active histone marks densities were accompanied by corresponding average increases in gene expression. Second, the relationship was nonlinear, as an increase in the input variable (e.g., POL-II-S5P or H3K4me3) resulted in a much higher increase in the readout (e.g., expression). The relationship could be linearized by using log-log scale transformation, which suggested a power-law relationship. This type of dependency is consistent with previously published analysis of correlations between expression and histone mark densities at promoters26. The only exceptions were H3K36me3 comparisons against all other epitopes. For both Xa and autosomal genes, a log scale transformation better linearized the trend for H3K36me3 (Fig. 3f,h and Supplementary Fig. 3-4), implying that relatively smaller changes in H3K36me3 can produce large changes in transcription output. The overall non-linearity suggests potential collective effects in the regulation of POL-II occupancy, changes in chromatin state, and transcriptional activity. Importantly, the non-linear nature of these dependencies allows for signal amplification: For example, a 20-30% increase in POL-II density corresponds to a larger increase in gene expression (Fig. 3a,b).
Third, although Xa and autosomal genes obeyed a common trend of dependency between gene expression and active chromatin/POL-II marks, the Xa scatterplot demonstrated consistent, significant shifts along the autosomal trendline to higher positions on both x- and y-axes (Fig. 3 and Supplementary Fig. 3-4), reflecting the generally higher levels of gene expression, POL-II occupancy, and H3K4me3 and H3K36me3 deposition on Xa relative to autosomes. In comparison to autosomes, Xa genes generally showed better correlations between chromatin epitopes (Fig. 4a), possibly reflecting the generally greater Xa values. Although ranges for these variables on Xa overlapped with autosomes, there was a clear distinction between the two gene populations, consistent with the significant differences in the means/medians of epitope density coverages (Supplementary Table I, Supplementary Fig. 1c and Fig. 1b).
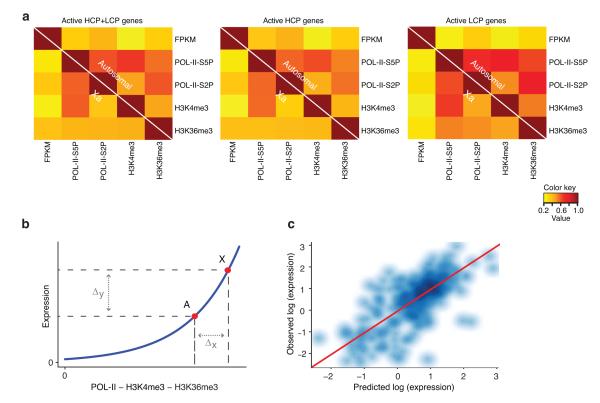
Figure 4. Autosomal relationships between active histone modifications, POL-II, and expression are predictive of X-linked gene expression.
(a) Actively expressed X-linked and autosomal genes show similar patterns of correlation between the levels of active marks and expression. Pearson correlation coefficients between the levels of all marks and expression (FPKM) are shown as heatmaps for actively expressed (HCP+LCP, HCP, and LCP) genes. In each plot, autosomal and X chromosome correlations are shown above and below diagonal, respectively. (b) Active X chromosome loci (X) and the corresponding set of autosomal loci (A) show similar non-linear relationship between active marks and expression (blue curve), which produces a large average expression change in response to smaller changes in the mark occupancy (schematic). (c) Scatterplot of X-linked gene expression values predicted from autosome-based full linear model vs observed X-linked expression (log-log scale). Shades of blue indicate point density. Identity line y=x is shown in red.
Taken together, these data enable us to draw several conclusions regarding Xa upregulation. The similar nonlinear trends of relationships for Xa and autosomal genes suggest that Xa upregulation might be explained by chromatin-based mechanisms that are generally used throughout the genome. On both Xa and autosomes, nonlinearity of the dependencies provide a mechanism for signal amplification. For example, Xa upregulation might be explained simply by increased density of active epigenetic marks associated with increased POL-II occupancy, rather than by Xa-specific rules governing the relationship between histone marks, POL-II, and transcription. Our data also indicate that, unlike in fruitflies where enhanced transcription elongation has been proposed to be the primary mode of Xa upregulation27 the mammalian mechanism displays a strong influence of enhanced transcription initiation, as both POL-II-S5P and H3K4me3 have ~30% increased coverage at the TSS of active Xa genes. At the same time, however, mammalian Xa upregulation is also associated with ~20% increase in POL-II-S2P and ~9% increase in H3K36me3 coverages across gene bodies, implying that elongation is also facilitated on Xa. We therefore propose that Xa upregulation involves both enhanced transcription initiation and elongation that depend on increased trimethylation of H3K4 and H3K36, which amplify gene expression in a nonlinear fashion (Fig. 4b).
Xa hyperactivation is predictable without necessitating Xa-specific principles
We tested this hypothesis by deriving models of gene expression based on autosomal active mark occupancy and applying these models to Xa. The resulting predictions of X-linked gene expression showed accordance with the observed values (Pearson r=0.71)(Fig. 4c). Moreover, autosomal models based on any combination of marks, when applied to an individual autosome, produced the correlation between predicted and observed values similar to that of Xa (Supplementary Table III; See Methods for details). These results suggested that, firstly, quantitative relationships between active histone mark densities, POL-II occupancy, and gene expression had predictive power, consistent with the results of others26. Secondly, trends observed in autosomes were remarkably predictive of X-linked gene expression. Together, these conclusions argue that Xa upregulation is governed by principles that are not unique to X but are applicable throughout the genome.
DISCUSSION
We have tested the hypothesis of Xa upregulation using a novel, unbiased approach and asked whether this arm of dosage compensation is based on chromatin mechanisms similar to those observed on autosomes. By allele-specific ChIP-seq and RNA-seq analysis, we conclude that dosage compensation of Xa does occur, in agreement with previously published studies 13,14,16. The inability of a separate study15 to detect Xa upregulation stems from inclusion of inactive genes. When we considered all genes regardless of expression status, the average difference was masked by unusually high fraction of silent genes (genes with low-CpG content) on the X. Earlier studies 13,14 also included both active and inactive genes; however, microarrays have a lower dynamic range and it is likely that the analysis heavily favored highly transcribed genes.
We then focused on potential mechanisms of Xa upregulation. By analyzing the genome-wide allelic distribution of POL-II and active chromatin marks in the context of gene expression17 we found that Xa was enriched for POL-II-S5P, POL-II-S2P, H3K4me3, and H3K36me3 relative to the haploid autosome set. Scatterplots (Fig. 3 and Supplementary Fig. 3-4) showed a clear rightward and upward shift of Xa contours relative to autosomal contours, along the autosomal trendline to the higher levels of active marks and expression. However, the degree of POL-II enrichment on Xa and the coverage of active histone marks did not scale proportionally with increased transcription output from Xa. This nonlinearity enables signal amplification with small changes in chromatin structure and POL-II density. An unexpected observation was that the nonlinear dependencies applied to autosomes and X alike, as suggested by all pairwise plots (Fig. 3 and Supplementary Fig. 3-4) and by the quality of predicting Xa genes based on the autosome-based models (Fig. 4c and Supplementary Table III). The similar quantitative relationships suggest that chromatin-based processes governing Xa upregulation are used genome-wide, and that dosage compensation of Xa and autosomes do not require X-specific principles to bring about the ~2-fold upregulation. Importantly, while the non-linear power laws apply to both Xa and autosomes, we do not exclude the possibility that Xa-specific factors are required to initiate the chromatin-based enhancement. Such factors could target Xa to a special compartment or lead to association with nuclear pore factors, as has been suggested for Drosophila melanogaster 28.
Our data imply similarities and differences with Xa upregulation in the fruitfly. In D. melanogaster, the X-chromosome dosage between XX and XY individuals is equalized by hypertranscription of the single male X-chromosome29-31. With this mechanism, the fly compensates for differences between X and autosomal gene dosage at the same time it achieves male-to-female X-chromosome balance. This process requires cooperation between MOF histone acetyltransferase32, MSL complexes33-35, and the long noncoding RNA, roX1 and roX2 36,37. Together, they bring about precise chromatin change leading to ~2-fold upregulation of the male X 27,38-45. An alternative mechanism proposes less dependency on MSL-driven hyperactivation and more on the genome’s inherent ability to correct for dosage imbalances – via the so-called “inverse effect” 32,46. This mechanism may also operate in mammalian Xa upregulation.
X hyperactivation in the fruitfly has been proposed to be achieved primarily through enhanced transcription elongation 27,38,43-45. In our system, differences in distributions of POL-II-S5P, POL-II-S2P, H3K4me3, and H3K6me3 argue that Xa upregulation is controlled through transcription initiation, but enhancement of elongation is also a strong possibility. While POL-II-S5P and H3K4me3 coverages increase ~30% at the TSS of Xa genes, POL-II-S2P and H3K36me3 coverages also increase ~20% and ~9%, respectively, across gene bodies. In conclusion, we favor a model in which Xa upregulation is effected by both enhanced transcription initiation and elongation via nonlinear dynamics. Future studies will focus on whether and how Xa-specific factors might be involved in initiating hypertranscription through the chromatin-based mechanisms identified herein.
METHODS
Cell line
To generate EY.T4 clonal hybrid cell line, female mice of Mus musculus (129S1) and male mice of Mus castaneus (CAST/EiJ) origins were crossed, F1 embryos were collected at day 13.5 and mouse embryonic fibroblasts (MEFs) were prepared using female embryos. MEFs were later immortalized by SV-40 T-antigen47 subcloned by limiting dilution, and the chromosome content of each subclone was screened by DNA FISH using chromosome paints and RNA FISH using Xist RNA probe.. In this clone, Xi was of Mus musculus and Xa was of Mus castaneus origins as determined by allele-specific RT-PCR using primers against Xist locus.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as described48 using 2×106 cells and 5-10μg of antibodies per reaction. Antibodies used were as follows: H3K36me3 (ab9050, Abcam), H3K4me3 (ab8580, Abcam), POL-II-S5P (ab5131, Abcam) and POL-II-S2P (ab5095, Abcam) and IgG rabbit serum (I8140, Sigma). ChIP [DNA] was quantitated using Quant-iT Picogreen dsDNA Assay kit (Invitrogen). ChIP products for H3K4me3 and H3K36me3 were verified by PCR using primers against c-fos and c-jun genes.
ChIP library preparation and sequencing
Paired-end Solexa ChIP libraries were prepared as described in Illumina ChIP sequencing manual with minor modifications and using NEB Next DNA sample prep reagent Set 1 (E6000S) (NEB). Input DNA was used as a control and for normalization. Modifications to the Illumina protocol were: 1-30 ng of ChIP products were used as template, 2-paired-end adapters were ligated to end-repaired and A-tailed DNA using T4 DNA ligase (NEB) for 2 hours at 16°C, and 2-Phusion polymerase in GC buffer (Finzyme) was used instead of Phusion polymerase in HF buffer. DNA products were purified with QIAquick spin columns (Qiagen). Concentration, size distribution (400-500bp), and purity of libraries were assessed on DNA1000 Bioanalyzer chip (Agilent). Genome Analyzer II (Illumina) was used to perform 2×36 cycles of paired end sequencing.
Calculation of allele-specific ChIP-seq coverage
Paired-end reads (17-28 million per sample, Supplementary Table II) were aligned to two variant strain genomes (CAST/EiJ and 129S1/SvImJ) of the hybrid EY.T4 cell line. The strain genomes were reconstructed from mm9 reference using catalogued SNPs and indels25. Resulting allelic differences include ~22 million SNPs and ~1 million indels, approximately one modification per 120 bp. Alignment was performed with Novoalign 2.06 (www.novocraft.com) using default parameters with modifications: -i 300 100 –t 180 –R 10 –h 180 180 –v 180., Uniquely aligned pairs with significant score difference between the two strain alignments (>10) were classified as allele-specific and assigned to the higher-scoring allele variant; otherwise they were classified as neutral. Coverage was calculated separately for allelic tracks (cast, mus) and composite track (allelic and neutral combined) based on fragments defined by paired reads (~400 bp on average), discarding duplicate fragments. Positional coverage was normalized input coverage with a pseudocount of 1 and balanced for potential inequality of total input and experiment coverages: nnorm = [(n+1)/(ni+1)] * [Ni/N], where n, ni, are positional coverages and N, Ni are total genome coverages in experiment and input, respectively. For each promoter region and gene body, allelic skew was estimated as the proportion of cumulative coverages at allelic cast/mus tracks, and corresponding fractions of composite coverage were used in further analyses.
Gene set selection
To ensure unique representation of each gene and exclude alternative transcripts, mm9 RefSeq genes were grouped into single-linkage clusters by gene body overlap and the longest gene in each cluster was chosen as a representative. Promoter regions were defined as segments including +/− 3kb from annotated TSS with high- and low-CpG promoters defined by the presence of CpG islands within 1 kb from the promoter. CpG islands were identified by using EMBOSS 6.3.149 on unmasked mm9 genome with default parameters.
Estimation of gene expression levels
Approximately 45 million of 36-bp RNA-seq reads by Yang et al. 17 (NCBI SRA accession SRA010053) were aligned to mm9 genome using TopHat50 with default parameters, except for using arguments --no-novel-juncs and --min-isoform-fraction 0. The resulting alignments were used to calculate FPKM (fragments per kilobase per million) values using Cufflinks51 with default parameters, except for setting --min-isoform-fraction 0. To estimate expression from a single allele, autosomal expression values were divided by 2, whereas X-linked expression values were used in full, given the well-documented inactivation of one X chromosome. As a cutoff for active expression, we chose FPKM of 1.0 corresponding to the boundary between two regimes of FPKM correlation to POL-II density on a gene (Fig. S2): the absence of dependency at low values, consistent with readout fluctuations for silent genes, and positive correlation at higher values, consistent with active transcription. Choosing FPKM cutoff of 0 produced similar values.
Linear models of gene expression
Log-transformed mark densities on autosomes (POL-II on promoters and gene bodies, H3K4me3 on promoters) and untransformed H3K36me3 on gene bodies were used for training a linear regression model that predicts logarithm of gene expression. This model (full model) was applied to the coverage densities on X chromosome, and the resulting expression predictions for X-linked genes were compared to the observed values.
To compare prediction accuracy for X chromosome and for an individual autosome, we trained and tested 19 models in a chromosome-based cross-validation setting. Specifically, a model (partial model) was trained on the autosomal gene set with one autosome removed and applied separately to the removed autosome and X chromosome. Pearson correlation coefficients between predicted and observed logarithms of expression for X chromosome were compared to the distribution of Pearson R for 19 autosomes. This comparison was made for the models based on all possible combinations of marks as predictors.
Logarithmic transformation of mark densities and expression is not defined at zero. To alleviate this problem, we used two approaches. First, we added a pseudocount of 1.0 to each value x so that log (x+1) was calculated. Second, since the fraction of zero values among actively expressed genes was relatively low we tested the removal of genes with zero values from the dataset. Both approaches produced similar results.
Supplementary Material
ACKNOWLEDGEMENTS
We are thankful to members of Lee laboratory for valuable discussions and to B. Chapman, M. Borowsky and T. Ohsumi of the Bioinformatics Core Facility (MGH, Molecular Biology Dept) for their suggestions for ChIP-Seq analysis. This work was supported by the MGH ECOR Medical Discovery Fund (E.Y.), DFG (S.F.P.), and the National Institutes of Health (RO1-GM090278, J.T.L.). J.T.L. is an investigator of the Howard Hughes Medical Institute.
Footnotes
DATABASE ACCESSION NUMBERS XXXX (will be added in proof – awaiting GEO assignment)
AUTHOR CONTRIBUTIONS E.Y. and J.T.L. designed the research; E.Y. and S.F.P. conducted ChIP-seq experiments; R.I.S. performed the bioinformatics analysis; S.F.P. performed allele-specific alignments; E.Y., R.I.S., S.F.P., and J.T.L. analyzed the data; and E.Y., R.I.S., and J.T.L. wrote the paper.
REFERENECES
- 1.Charlesworth B. The evolution of chromosomal sex determination and dosage compensation. Current biology : CB. 1996;6:149–62. doi: 10.1016/s0960-9822(02)00448-7. [DOI] [PubMed] [Google Scholar]
- 2.Ohno S. More about the mammalian X chromosome. Lancet. 1962;2:152–3. doi: 10.1016/s0140-6736(62)90042-9. [DOI] [PubMed] [Google Scholar]
- 3.Ohno S. A phylogenetic view of the X-chromosome in man. Annales de genetique. 1965;8:3–8. [PubMed] [Google Scholar]
- 4.Ohno S. Sex Chromosomes and Sex Linked Genes. Springer Verlag; Berlin: 1967. [Google Scholar]
- 5.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–3. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 6.Lyon MF. Possible mechanisms of X chromosome inactivation. Nat New Biol. 1971;232:229–32. doi: 10.1038/newbio232229a0. [DOI] [PubMed] [Google Scholar]
- 7.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–7. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 8.Brockdorff N, et al. High-density molecular map of the central span of the mouse X chromosome. Genomics. 1991;10:17–22. doi: 10.1016/0888-7543(91)90478-w. [DOI] [PubMed] [Google Scholar]
- 9.Brown M, et al. A recombinant murine retrovirus for simian virus 40 large T cDNA transforms mouse fibroblasts to anchorage-independent growth. Journal of virology. 1986;60:290–3. doi: 10.1128/jvi.60.1.290-293.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva J, et al. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Developmental cell. 2003;4:481–95. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 11.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–5. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 12.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin H, et al. Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS biology. 2007;5:e326. doi: 10.1371/journal.pbio.0050326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen DK, Disteche CM. Dosage compensation of the active X chromosome in mammals. Nature genetics. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- 15.Xiong Y, et al. RNA sequencing shows no dosage compensation of the active X-chromosome. Nature genetics. 2010;42:1043–7. doi: 10.1038/ng.711. [DOI] [PubMed] [Google Scholar]
- 16.Deng X, et al. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nature genetics. 2011 doi: 10.1038/ng.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome research. 2010;20:614–22. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marks H, et al. High-resolution analysis of epigenetic changes associated with X inactivation. Genome research. 2009;19:1361–73. doi: 10.1101/gr.092643.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Neill LP, et al. X-linked genes in female embryonic stem cells carry an epigenetic mark prior to the onset of X inactivation. Human molecular genetics. 2003;12:1783–90. doi: 10.1093/hmg/ddg193. [DOI] [PubMed] [Google Scholar]
- 22.O’Neill LP, Spotswood HT, Fernando M, Turner BM. Differential loss of histone H3 isoforms mono-, di- and tri-methylated at lysine 4 during X-inactivation in female embryonic stem cells. Biological chemistry. 2008;389:365–70. doi: 10.1515/BC.2008.046. [DOI] [PubMed] [Google Scholar]
- 23.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–81. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 24.Schneider R, et al. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nature cell biology. 2004;6:73–7. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 25.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–94. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karlic R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. Histone modification levels are predictive for gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2926–31. doi: 10.1073/pnas.0909344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larschan E, et al. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature. 2011;471:115–8. doi: 10.1038/nature09757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaquerizas JM, et al. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS genetics. 2010;6:e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annual review of genetics. 2008;42:733–72. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- 30.Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annual review of genetics. 2005;39:615–51. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- 31.Cline TW, Meyer BJ. Vive la difference: males vs females in flies vs worms. Annual review of genetics. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
- 32.Bhadra MP, Bhadra U, Kundu J, Birchler JA. Gene expression analysis of the function of the male-specific lethal complex in Drosophila. Genetics. 2005;169:2061–74. doi: 10.1534/genetics.104.036020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer MJ, Richman R, Richter L, Kuroda MI. Sex-specific regulation of the male-specific lethal-1 dosage compensation gene in Drosophila. Genes & development. 1994;8:698–706. doi: 10.1101/gad.8.6.698. [DOI] [PubMed] [Google Scholar]
- 34.Gu W, Szauter P, Lucchesi JC. Targeting of MOF, a putative histone acetyl transferase, to the X chromosome of Drosophila melanogaster. Developmental genetics. 1998;22:56–64. doi: 10.1002/(SICI)1520-6408(1998)22:1<56::AID-DVG6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 35.Lyman LM, Copps K, Rastelli L, Kelley RL, Kuroda MI. Drosophila male-specific lethal-2 protein: structure/function analysis and dependence on MSL-1 for chromosome association. Genetics. 1997;147:1743–53. doi: 10.1093/genetics/147.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meller VH, Wu KH, Roman G, Kuroda MI, Davis RL. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell. 1997;88:445–57. doi: 10.1016/s0092-8674(00)81885-1. [DOI] [PubMed] [Google Scholar]
- 37.Amrein H, Axel R. Genes expressed in neurons of adult male Drosophila. Cell. 1997;88:459–69. doi: 10.1016/s0092-8674(00)81886-3. [DOI] [PubMed] [Google Scholar]
- 38.Gelbart ME, Kuroda MI. Drosophila dosage compensation: a complex voyage to the X chromosome. Development. 2009;136:1399–410. doi: 10.1242/dev.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. The EMBO journal. 1997;16:2054–60. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Molecular cell. 2000;5:367–75. doi: 10.1016/s1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- 41.Smith ER, et al. The drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Molecular and cellular biology. 2000;20:312–8. doi: 10.1128/mcb.20.1.312-318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prestel M, Feller C, Straub T, Mitlohner H, Becker PB. The activation potential of MOF is constrained for dosage compensation. Molecular cell. 2010;38:815–26. doi: 10.1016/j.molcel.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 43.Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes & development. 2006;20:848–57. doi: 10.1101/gad.1400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gilfillan GD, et al. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes & development. 2006;20:858–70. doi: 10.1101/gad.1399406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Legube G, McWeeney SK, Lercher MJ, Akhtar A. X-chromosome-wide profiling of MSL-1 distribution and dosage compensation in Drosophila. Genes & development. 2006;20:871–83. doi: 10.1101/gad.377506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Birchler J, et al. Re-evaluation of the function of the male specific lethal complex in Drosophila. Journal of genetics and genomics = Yi chuan xue bao. 2011;38:327–32. doi: 10.1016/j.jgg.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 47.Brown CJ, et al. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature. 1991;349:82–4. doi: 10.1038/349082a0. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi K, Saitoh S, Yanagida M. Application of the chromatin immunoprecipitation method to identify in vivo protein-DNA associations in fission yeast. Science’s STKE : signal transduction knowledge environment. 2000;2000:pl1. doi: 10.1126/stke.2000.56.pl1. [DOI] [PubMed] [Google Scholar]
- 49.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends in genetics : TIG. 2000;16:276–7. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 50.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim J, Lu X, Stubbs L. Zim1, a maternally expressed mouse Kruppel-type zinc-finger gene located in proximal chromosome 7. Human molecular genetics. 1999;8:847–54. doi: 10.1093/hmg/8.5.847. [DOI] [PubMed] [Google Scholar]
- 53.Kuroiwa Y, et al. Peg3 imprinted gene on proximal chromosome 7 encodes for a zinc finger protein. Nature genetics. 1996;12:186–90. doi: 10.1038/ng0296-186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.