Abstract
A number of antibodies have been developed that induce lethal iron deprivation (LID) by targeting the transferrin receptor 1 (TfR1/CD71) and either neutralizing transferrin (Tf) binding, blocking internalization of the receptor and/or inducing its degradation. We have developed recombinant antibodies targeting human TfR1 (ch128.1 and ch128.1Av), which induce receptor degradation and are cytotoxic to certain malignant B-cells. We now show that internalization of TfR1 bound to these antibodies can lead to its sequestration and degradation, as well as reduced Tf uptake, and the induction of a transcriptional response consistent with iron deprivation, which is mediated in part by downstream targets of p53. Cells resistant to these antibodies do not sequester and degrade TfR1 after internalization of the antibody/receptor complex, and accordingly maintain their ability to internalize Tf. These findings are expected to facilitate the rational design and clinical use of therapeutic agents targeting iron import via TfR1 in hematopoietic malignancies.
Keywords: Transferrin, antibody, iron deprivation
Introduction
Import of iron into cells is primarily accomplished through the uptake of its carrier protein, transferrin (Tf), into acidified recycling vesicles via the transferrin receptor 1 (TfR1/CD71) [1,2]. The subsequent transport of iron from acidified vesicles into the cytoplasm is then accomplished by the divalent metal transporter 1 (DMT1), while the Tf–TfR complex is returned to the cell surface [1-4]. In general, TfR1 is required by proliferating cells and is expressed at high levels in certain malignancies, in some of which it is associated with disease grade and progression [2,5]. Hematopoietic malignancies express abnormally high levels of TfR1 to meet their high demand for iron and maintain their rapid rate of proliferation [2,5]. Accordingly, TfR1 is an attractive target for anti-cancer agents, particularly those that aim to induce lethal iron deprivation in malignant hematopoietic cells.
A number of therapeutic agents designed to target TfR have been shown to induce lethal iron deprivation (LID) and/or deliver toxic compounds, pro-drugs or nucleic acids into malignant cells [3,6-13]. Many of these agents are actively being developed for the treatment of various malignancies. A variety of antibodies with specificity for human TfR1, capable of directly inhibiting cell proliferation and/or inducing apoptosis in a number of malignant hematopoietic cells, have been developed since the early 1980s [2,6,9-12,14]. Anti bodies specific for TfR1 offer a potentially safer and more effective alternative to the use of pleiotropic iron chelators for the targeted disruption of cellular iron import in malignant cells. The use of therapeutic antibodies targeting human TfR1 in hematopoietic malignancies for the induction of LID has advanced greatly in recent years. A number of these antibodies remain under investigation and are promising candidates for the treatment of hematopoietic malignancies alone or in combination with other therapies [2,6,10]. Despite the development of promising therapies targeting iron homeostasis, TfR1, and other hallmarks of cancer in malignant cells, better treatments are urgently needed for incurable hematopoietic malignancies, such as multiple myeloma and aggressive lymphomas [15], and could be offered through the design and development of novel targeted therapies.
We developed a mouse/human chimeric antibody with human immunoglobulin 3 (IgG3) constant regions and the variable regions of a mouse monoclonal antibody specific for human TfR1 (128.1) [6], previously known as anti-hTfR IgG3 (ch128.1) [14]. We also developed a fusion protein composed of ch128.1 genetically fused to chicken avidin at its carboxy terminus, previously known as anti-hTfR IgG3-Av (ch128.1Av) [9,14]. This antibody–avidin fusion protein exists as a dimer in solution due to the non-covalent interaction between its avidin moieties, can be used as a delivery vehicle for biotinylated compounds into cells expressing TfR1, and is intrinsically toxic (anti-proliferative/pro-apoptotic) to a variety of malignant B cells, including multiple myeloma and lymphoma cells [9,14,16-18]. The binding of Tf or the hemochromatosis protein (HFE) to TfR1 is not inhibited by ch128.1 or ch128.1Av, and these antibodies did not cross-react with TfR2 [14,17], a second version of the receptor expressed primarily in hepatocytes and the gut endothelium [1,2]. ch128.1Av and, to a lesser extent, ch128.1 induce degradation of TfR1 in sensitive cells, and their cytotoxic effect is blocked in the presence of iron salts. However, the mechanism by which these antibodies block iron uptake and induce LID in malignant hematopoietic cells requires further study, particularly in light of their differential toxicity to a variety of malignant B-cells expressing high levels of TfR1.
The main goals of the present work are to further investigate the mechanisms of LID induced in malignant hematopoietic cells by antibodies that bind TfR1 but do not inhibit its binding to Tf, and to understand the differences between cells that are sensitive and resistant to treatment with these antibodies. We show that although the internalization of these antibodies is canonical (both clathrin and dynamin dependent), ch128.1Av and, to a lesser extent, ch128.1 inhibit the function of TfR1 by inducing its sequestration, which we confirm is associated with a subsequent degradation of the receptor in sensitive cells. This reduction in the levels of TfR1 on the cell surface results in decreased Tf uptake, and the induction of LID in malignant hematopoietic B-cells, which is mediated in part by downstream targets of p53. We also describe cellular phenotypes associated with resistance to treatment with these antibodies. These studies are expected to aid in the future design of therapeutic agents aimed at exploiting iron homeostasis for the treatment of incurable B-cell malignancies through the targeting of TfR1.
Materials and methods
Cells and reagents
The Epstein–Barr virus (EBV) positive human B-lymphoblastoid cell lines IM-9 and ARH-77, and the plasmacytoid cell line U266, isolated from a patient with multiple myeloma, were purchased from the American Type Culture Collection (ATCC, Manassas, VA). Cells derived from a patient with Burkitt lymphoma and positive for EBV (Akata +) were a kind gift from Dr. Richard Ambinder (Johns Hopkins School of Medicine), while the isogenic B-lymphoblastoid cell lines TK6 (p53 wild type) and NH32 (p53 mutant at codon 237 of exon 7) [19], differing in their expression of the p53 protein, were kind gifts of Drs. Daniel H. Appella and Ana I. Robles (National Institutes of Health). All cell lines were maintained in standard tissue culture conditions, RPMI 1640 supplemented with 10% fetal bovine serum and penicillin/streptomycin, as previously described [16,18,20]. Tf-Alexa® (excitation 488 nm), CMPTX® (excitation 596 nm) and Zenon® (excitation 596 nm) were obtained from Life Technologies (Carlsbad, CA), calcein and the iron chelator desferrioxamine (DFO) from Sigma-Aldrich Corp. (St. Louis, MO) and the dynamin inhibitor Dynasore® from Tocris Biosciences (Ellsville, MO). The cell-permeable iron chelator salicylaldehyde isonicotinoyl hydrazone (SIH) was a kind gift of Dr. Prem Ponka (McGill University) [21], and the antibodies ch128.1 and ch128.1Av were produced and purified as previously described [9,14].
Cell proliferation and viability assays
IM-9 and U266 cells were incubated with 10 nM ch128.1Av, 10 nM ch128.1, 50 μM DFO or buffer in tissue culture conditions for 24 h or 48 h in 24- or 96-well plates. In proliferation assays based on [3H]-thymidine incorporation, 16 h prior to the end of each time point, an aliquot of cells treated with each agent was removed from 24-well plates and seeded in a new 96-well plate to evaluate the inhibition of proliferation by the [3H]-thymidine incorporation assay, as previously described [16]. Cell viability WST-1 assays (Hoffmann-La Roche Ltd, Basel, Switzerland) were conducted according to the manufacturer’s instructions in 96-well plates, at least in triplicate, and their colorimetric readings obtained using a DTX880 Multimode Detector (Beckman Coulter Inc., Brea, CA). Experiments were repeated at least twice. Data from treatment conditions were normalized to values obtained from control cells.
Confocal microscopy of Tf uptake in vitro
Cells labeled with the live cell tracker CMPTX®, according to the manufacturer’s instructions, were treated with Tf-Alexa® at a concentration of 50 μg/mL in serum-free medium for 30 min at 37 °C. Alternatively, unlabeled cells were incubated with 10 nM of ch128.1 or ch128.1Av bound to Zenon® as indicated by the manufacturer for 1 h or 6 h. All cells were then washed and incubated with fluorescent Tf as described above. Cells were washed, fixed with 2% paraformaldehyde, mounted on slides in 100% glycerol and imaged using a confocal microscope (Carl Zeiss, Inc., Oberkochen, Germany) equipped with an oil-immersion NA 1.4 60× objective and saved after a 2 × digital enhancement, or using an open perfusion chamber (volume 200 μL) on a Nikon Diaphot microscope coupled to a custom confocal imaging system, as previously described [22]. Briefly, excitation from a 475 nm diode laser was delivered via scanning mirrors to the specimen through a 60 × lens. Fluorescence emission was gathered through a dichroic mirror and 535 bandpass filter to a photomultiplier tube (Hamamatsu, Bridgewater, NJ), and images were acquired at four frames per minute by an image acquisition board (Bitflow Raven) controlled by Video Savant software with a maximum image resolution of 980 × 730 pixels. Experiments were repeated at least twice.
Inhibition of Tf and/or antibody uptake
Cells were incubated for 30 min in the presence of hypertonic sucrose (0.4 M) at 4 °C, or with increasing concentrations (0-400 μM) of the dynamin inhibitor Dynasore® [23] to prevent TfR internalization. Cells were then incubated with Tf-Alexa® or Zenon®-bound antibodies at the concentrations indicated above for 30 min at 37 °C. Experiments were repeated at least twice. Confocal microscopy images of these cells were obtained as described in the section above on Tf uptake in vitro.
Inhibition of Tf uptake in vivo
Cells (5 × 106) were labeled with CMPTX® according to the manufacturer’s instructions and incubated with 50 μg of ch128.1 or ch128.1Av for 30 min at 37 °C and then injected into the peritoneal cavity of C.B-17 SCID (severe combined immune deficiency)-Beige mice for 2 h or 48 h. This was performed for three animals per condition for IM-9 and U266 cells during the 48 h incubation experiments, and one animal per condition for all other cells and treatments. This xenograft model was used given the human nature of the cells and the fact that the technique has been successfully employed in a number of tumor targeting studies with other antibodies or chemotherapeutic agents [24,25]. Immediately after euthanasia, cells were recovered from the peritoneal cavity of the mice by subsequent washes with 5—10 mL of phosphate buffered saline (PBS). Recovered cells were then incubated with Tf-Alexa® at a concentration of 50 μg/mL in serum-free medium for 30 min at 37 °C. Cells were simultaneously kept in vitro at 37 °C for 2 h or 48 h, then washed with PBS and incubated with Tf-Alexa® alongside the cells recovered from mouse xenografts. Confocal microscopy images of these cells were then obtained for the 2 h incubations as described in the section on Tf uptake in vitro; fluorescence intensity of internalized Tf in all conditions was also analyzed by flow cytometry as described below. Experimental protocols were approved by the University of California, Los Angeles (UCLA) Institutional Animal Care and Use Committee (IACUC), and all local and national guidelines on the care of animals were strictly adhered to. C.B-17 SCID-Beige mice were bred and maintained in the Defined-Flora Association for Assessment and Accreditation of Laboratory Animal Care-Accredited Animal Facility of the Department of Radiation Oncology at UCLA.
Flow cytometry of Tf uptake and cell surface binding
Cells treated with antibodies or buffer (PBS) control for the time shown were washed and pulse-labeled with Tf-Alexa® as specified above, then washed, fixed and run on a FACScan cytometer (Becton Dickinson, Franklin Lakes, NJ). In testing competition of Tf with ch128.1 or ch128.1Av, cells were incubated for 30 min at 4 °C with 100 nM of the antibodies followed by 1 h incubation with Tf-Alexa® also at 4 °C. Fluorescence associated with cell surface binding of Tf-Alexa® was then determined. Flow cytometry analysis of cell viability and apoptosis is discussed in detail in the Supplementary Methods. Analyses of the distributions of fluorescence intensities collected (channel FL1) of approximately 10,000 cells per condition were conducted using the WinMDI application (version 2.8).
Labile iron pool detection
Cells treated with 10 nM ch128.1Av or 50 μM DFO for 24 h or 48 h were incubated with 100 nM calcein, washed and exposed to an excess (200 μM) SIH [21], as previously described [26], and this was repeated at least twice. After 20 min, calcein fluorescence was detected using a DTX880 Multimode Detector (Beckman Coulter) with exitation 480 nm, emission 530 nm filters.
Immunoblot of whole-cell TfR1 or p53 levels
Whole cell lysates were collected in cell lysis buffer (100 mM Tris-HCl pH 6.8, 4% sodium dodecyl sulfate [SDS], 10% glycerol) containing complete protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN), and quantified using the bicinchoninic acid (BCA) protein assay (Thermo Fischer Scientific, Walnut, CA) as instructed by the manufacturer. Proteins (25 μg total protein per well) were separated by SDS-polyacrylamide gel electrophoresis (PAGE) in a 4–12% Bis-Tris NuPage gel (Life Technologies). Proteins were then transferred to Whatman Protran nitrocellulose membranes (Thermo Fisher Scientific) and probed with antibodies specific for either human TfR1 (clone H68.4 from Sigma) or human p53 (produced by Millipore Corp., Billerica, MA); anti-human glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Cell Signaling Technology, Boston, MA) was used as a loading control according to the manufacturer’s instructions, repeated at least twice, showing similar results. Digital chemiluminescent images were obtained using the FluorChem Megapixel High Performance Fluorescence, Chemiluminescence and Visible Imaging System (Cell Biosciences, Santa Clara, CA). Levels of TfR for three independent experiments were analyzed and quantified by densitometry analysis.
Microarray hybridization and data quality control
Samples of total mRNA from control or 10 nM ch128.1Av treated IM-9 and U266 cells collected prior to treatment as well as after 1 h, 3 h, 9 h and 24 h of incubation were quantified, their integrity evaluated using a Bioanalyzer system (Life Technologies), and hybridized onto HumanRef-8 v2 Expression BeadChips (Illumina, Inc., San Diego, CA). Global gene expression profiles for these samples were collected using the BeadArray software package (Illumina). Quality control, preprocessing and data normalization of whole genome expression profiles were conducted using the R language for statistical computing (http://www.r-project.org) and the Lumi [27] package supplied by the Bioconductor Project (http://www.bioconductor.org) as well as the MATLAB Bioinformatics Toolbox (The MathWorks, Inc., Natick, MA). This is described in more detail in the Supplementary Methods, in which we also describe the methods for microarray analysis, data filtering and in silico transcription factor analysis. Array raw data and associated information are currently available from the public Gene Expression Omnibus (GEO) database under data series GSE14754.
Quantitative polymerase chain reaction
Genes whose expression changed the most after treatment, compared to time-matched controls, in array data from both sensitive and resistant cells were chosen for validation using quantitative polymerase chain reaction (PCR). Samples of 5 μg of the mRNA used for microarray studies at 1 h and 24 h were reverse transcribed using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Carlsbad, CA) according to the manufacturer’s instructions. Quantitative real-time PCR reactions were performed using an ABI TaqMan 7900 instrument (Applied Biosystems) with TaqMan® probe sets for β-glucuronidase (GUSB; housekeeping control), TP53, TFRC, CDKN1A, FDXR, GADD45A, TP53I3, KLF6 and IL6R in triplicate following the manufacturer’s instructions, repeated three times.
Transient inhibition of p53 expression using siRNA
IM-9 cells (106) were electroporated with a single pulse of 260 V and a capacitance of 1050 μF with 50 nM siRNA using the Gene Pulser XcellTM (Bio-Rad, Hercules, CA). The p53 siRNA/siAbTM Assay kit (Millipore) was used for these studies according to the manufacturer’s instructions, repeated at least twice.
Computational and statistical analysis
Fluorescence intensity measurements from confocal microscopy images of cells were conducted using the National Insitutes of Health (NIH) ImageJ software (rsbweb.nih.gov/ij/) and analysis code implementing functions from the image processing toolbox of the MATLAB suite, as discussed in more detail in the Supplementary Methods. Graphs were generated using Microsoft Excel software (Redmond, WA) in which statistical analyses including two-tailed Student’s t-tests were also conducted.
Results
TfR1 targeting and antibody induced cytotoxicity
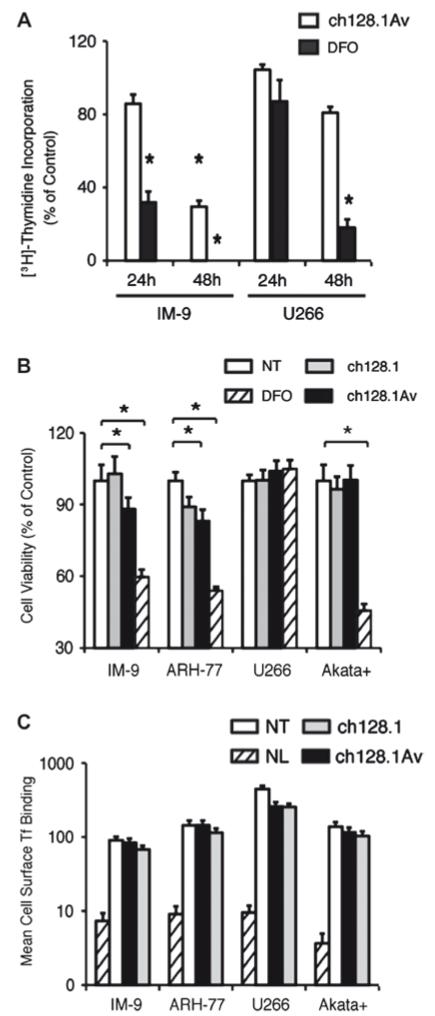
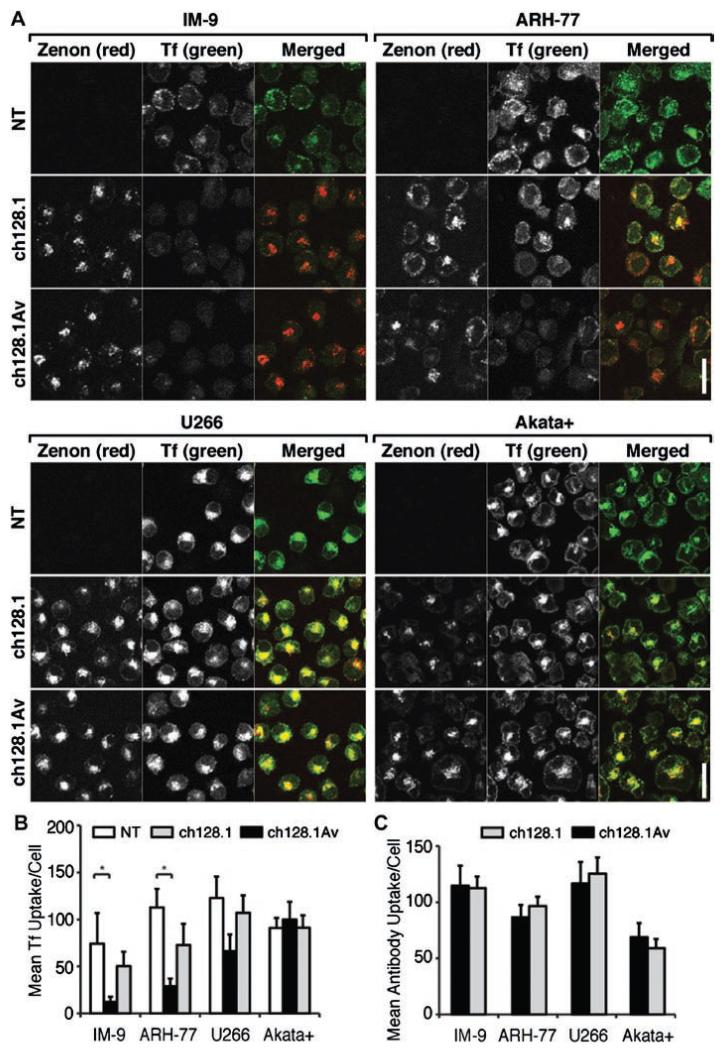
We have previously shown that ch128.1Av and to a lesser extent ch128.1 are cytotoxic to a variety of malignant hematopoietic cells [14,18], indicated by their reduced viability, lack of proliferation, and induced apoptosis in response to 72-96 h treatment with this fusion protein. Of all cells tested, the most sensitive to treatment with ch128.1Av are IM-9 cells, closely followed by ARH-77. The most resistant cells to ch128.1Av under similar conditions have been U266 cells. Treatment of sensitive cells with similar concentrations of ch128.1Av for a period of 48 h induces a significant anti-proliferative/pro-apoptotic effect [Fig. 1(A) and Supplementary Fig. 1]. However, since the present study focuses on early events in the induction of LID, the majority of our experiments were conducted during the first 24 h of treatment, during which the loss of cell viability in sensitive cells induced by the fusion protein is limited, despite their sensitivity at these early times to the iron chelator DFO [Fig. 1(B)]. Consistent with previous observations [14,17], exposure of cells to a high concentration (100 nM) of ch128.1Av did not prevent the binding of Tf to TfR1 expressed on their surface [Fig. 1(C)], confirming that ch128.1Av inhibits TfR1 function downstream of receptor binding at the cell surface. This is consistent with our observation that sensitive cells (IM-9 and ARH-77) internalized less fluorescently labeled Tf after treatment with ch128.1Av [Figs. 2(A) and 2(B)]. This effect was observed in sensitive cells as early as 1 h after treatment [Supplementary Figs. 2(A) and 2(B)] and was time dependent, becoming more evident after longer (6 h) exposure to ch128.1Av. A similar, although not significant, effect was observed after treatment in sensitive cells treated with ch128.1 [Fig. 2(B)]. In contrast, under similar conditions, resistant cells internalized fluorescent Tf at levels comparable to those of untreated cells [Figs. 2(A) and 2(B)]. The amount of fluorescently labeled ch128.1Av or ch128.1 internalized by cells was not generally indicative of their sensitivity or resistance to treatment [Fig. 2(C) and Supplementary Fig. 2(C)]. However, the intracellular distribution of the internalized antibodies in sensitive cells suggested their sequestration within as of yet unidentified subcellular compartments [Fig. 2(A) and Supplementary Fig. 2(A)]. These results indicate that ch128.1Av acts by reducing the intake of Tf in a time-dependent manner, without preventing its binding to TfR1.
Figure 1.
Effect of antibodies targeting human TfR1 on malignant B cells. (A) The rate of proliferation of cells treated with 10 nM ch128.1Av or 50 μM DFO for 24 or 48 h was evaluated by [3H]-thymidine incorporation, shown as a percentage of the incorporation observed in cells treated with buffer alone. Data shown are the average of quadruplicates and are representative of three independent experiments (*p < 0.05). (B) The percentage of viability of a panel of cells treated 24 h with buffer as a control (no treatment: NT), 50 μM DFO, 10 nM ch128.1 or 10 nM ch128.1Av is shown, with error bars indicating standard deviations (*p < 0.05). (C) Binding of fluorescent Tf to TfR on the cell surface for 1 h at 4 °C of the same panel of cells after pre-incubation for 30 min at 4 °C with 100 nM ch128.1, 100 nM ch128.1Av or buffer as a control (NT), determined by flow cytometry is shown. Background fluorescence in the absence of Tf (NL) is shown as a hatched bar. Error bars indicate standard deviations of fluorescence intensity distributions from flow cytometry measurements.
Figure 2.
Effect of antibodies on Tf uptake and internalization of TfR1. (A) Confocal images of cells incubated with fluorescent Zenon® labeled 10 nM ch128.1, 10 nM ch128.1Av or buffer as a control for 6 h followed by fluorescent Tf for 30 min at 37 °C. The white bars indicate 20 μm. (B) Quantification of the average internalization of green fluorescent Tf per cell in each condition and (C) of the average uptake of red fluorescent Zenon® labeled ch128.1 or ch128.1Av per cell from confocal images at 6 h is shown. Error bars indicate standard deviations (*p < 0.05).
Inhibition of TfR1 function in xenograft models of hematopoietic malignancies
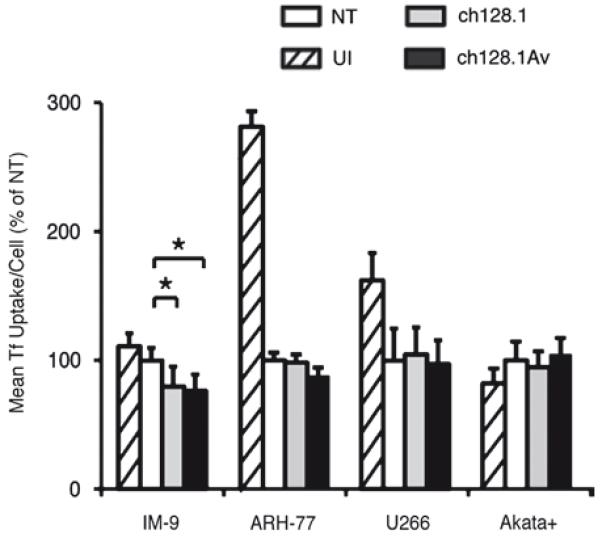
Given the potent inhibition of TfR1 function and induction of LID by ch128.1Av in sensitive but not resistant cells in vitro, we explored the consistency of this effect in vivo. We measured the uptake of fluorescent Tf by cells recovered 2 h or 48 h after injection into the peritoneal cavity of SCID-Beige mice in the presence of 50 μg ch128.1Av, 50 μg ch128.1, or buffer alone. Tf uptake was used as a measure of TfR1 function in recovered cells, analogous to our in vitro measurements in these cells after treatment with ch128.1Av. Both sensitive and resistant cells extracted from the peritoneal cavity of mice after engraftment retained the ability to internalize fluorescently labeled Tf, as expected, albeit at sometimes lower levels than cells maintained in vitro for the equivalent period of time (Fig. 3). Consistent with our in vitro results, IM-9 cells recovered from the peritoneal cavities of mice treated with ch128.1Av or ch128.1 showed decreased internalization of Tf at 2 h (Supplementary Fig. 3) or 48 h post-injection (Fig. 3). A similar effect was observed in ARH-77 cells, but was not significant. In agreement with our in vitro observations, resistant cells (U266 and Akata+) retained the ability to internalize Tf after treatment with ch128.1Av or ch128.1 (Fig. 3 and Supplementary Fig. 3).
Figure 3.
Inhibition of TfR1 function in vivo in a panel of malignant hematopoietic cells. Cells labeled with CMPTX® red, injected in the peritoneal cavity of SCID-Beige mice in the presence of buffer as a control (NT), 50 μg ch128.1 or 50 μg ch128.1Av for 48 h were recovered, and exposed to a pulse of green fluorescent Tf for 30 min at 37 °C, with buffer treated un-injected (UI) cells shown as controls. The quantification of fluorescent Tf uptake analyzed by flow cytometry from both un-injected and those recovered 48 h post-injection is shown. Error bars indicate standard deviations (*p < 0.05).
Effect on TfR1 trafficking and induction of iron deprivation
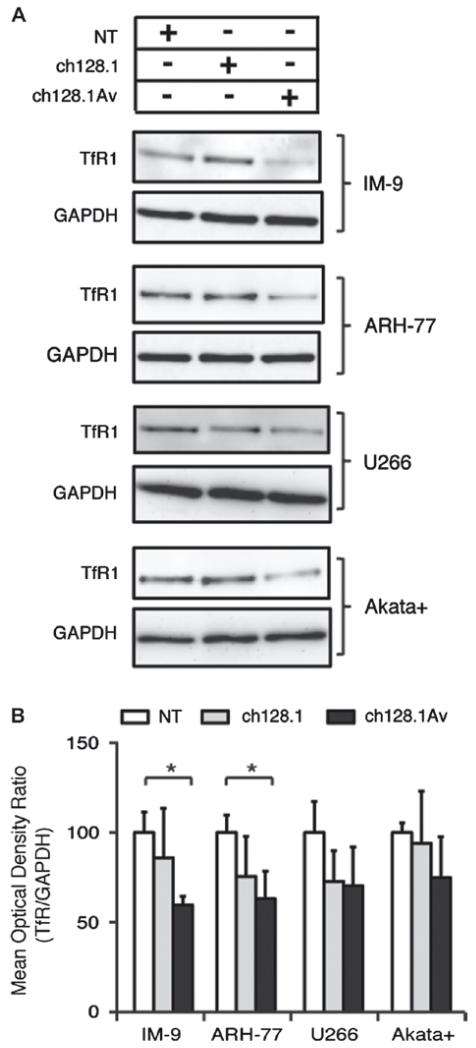
We next sought to determine whether, upon binding TfR1, internalization of ch128.1Av (tetravalent) or ch128.1 (bivalent) is dependent on the canonical pathway of clathrin/dynamin mediated TfR1 internalization. We found that the internalization of Tf and ch128.1Av bound to TfR1 was both clathrin (Supplementary Fig. 4) and dynamin (Supplementary Fig. 5) dependent, and was not significantly different between sensitive and resistant cells, suggesting the pathway of receptor internalization is unchanged by treatment with ch128.1Av. In light of previous results highlighting the induced degradation of TfR1 by ch128.1Av in highly sensitive cells [14], we further investigated events downstream of receptor internalization that might be affected by treatment with ch128.1Av in both sensitive and resistant cells, such as receptor recycling and degradation. We observed a significant reduction of whole-cell TfR1 levels, as compared to those of GAPDH, in sensitive cells (IM-9 and ARH-77) after 12 h treatment with 10 nM ch128.1Av (Fig. 4), and reduced levels of cell surface TfR1 after 2–6 h treatment of IM-9 cells under similar conditions, determined by cell surface binding of fluorescent Tf [Supplementary Fig. 6(A)]. The same effect was not consistently observed and was not significant in the case of ch128.1 under these conditions (Fig. 4). In contrast, resistant cells such as U266 and Akata+ showed a non-significant decrease in their levels of TfR1 protein after similar treatment with ch128.1Av (Fig. 4), and no decrease in the amount of cell surface TfR after similar treatment for 2–6 h [Supplementary Fig. 6(B)]. Since sensitive cells treated with ch128.1Av showed a reduction in cell surface TfR1 available for Tf binding as well as degradation of TfR1, we are unable to distinguish which of these two processes is primarily responsible for the reduction in Tf uptake. However, it is most likely that both play a significant role. Consistent with the disparity observed in the inhibition of TfR1 function by ch128.1Av in sensitive and resistant cells, we observed a significant reduction in the labile iron pool of IM-9 cells after treatment with ch128.1Av for 24 h and 48 h, but not in U266 cells under similar conditions [Supplementary Figs. 6(C) and 6(D)]. The labile iron levels of IM-9 and to a lesser extent U266 cells were both reduced after treatment with DFO under similar conditions [Supplementary Figs. 6(C) and 6(D)]. These results suggest that the end-point of TfR1 targeting by ch128.1Av in sensitive cells is iron deprivation induced by a severe reduction in Tf-bound iron uptake. However, unlike DFO, ch128.1Av does not reduce the labile iron stores of resistant cells, as it is unable to efficiently sequester and degrade TfR1 in these cells.
Figure 4.
Antibody-induced TfR1 degradation in vitro. (A) Whole-cell levels of TfR1 in a panel of malignant B cells after 12 h treatment with buffer as a control (NT), ch128.1 or ch128.1Av. (B) Quantification of whole-cell levels of TfR1 after 12 h treatment with buffer as a control (NT), ch128.1 or ch128.1Av. The ratio of TfR1, detected from a compilation of 3 independent experiments with respect to the level of GAPDH is shown, with all treatments compared to buffer treated cells. Error bars indicate standard deviations (*p < 0.05).
Transcriptional response to iron deprivation induced in sensitive cells
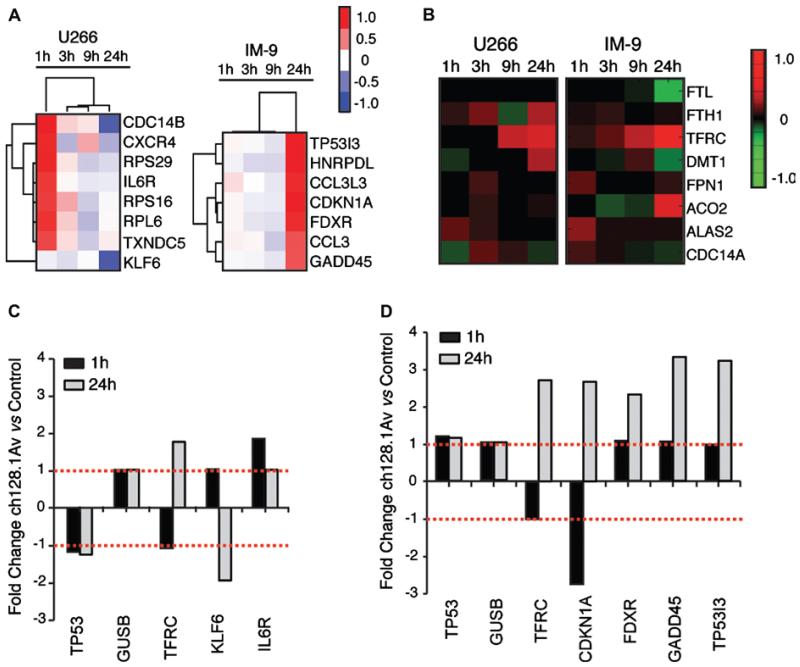
To better understand the events downstream of LID induced by targeting of TfR1 in the most sensitive and most resistant cells, we investigated the transcriptional events associated with antibody treatment in vitro. In a time-course comparative study, we observed the changes in gene expression in IM-9 and U266 cells at 1 h, 3 h, 9 h and 24 h after treatment with ch128.1Av. Whole-genome expression profiling of IM-9 cells treated with ch128.1Av showed time-dependent changes in gene expression that were maximal at the 24 h time point [Figs. 5(A) and 5(D)]. In contrast, no similar trend was detected in U266 cells, whose changes in gene expression were maximal at 1 h after treatment with ch128.1Av [Figs. 5(A) and 5(C)]. Genes whose expression increased in IM-9 cells 24 h after treatment included CDKN1A (encoding p21), GADD45 (encoding the growth arrest and DNA damage inducible protein 45A), TP53I3 (encoding the p53 inducible protein 3) and FDXR (encoding the mitochondrial flavoprotein ferredoxin reductase) [Fig. 5(A)]. We hypothesized that transcriptional changes induced in IM-9 cells at 24 h of treatment with ch128.1Av were a result of iron deprivation. This was confirmed by the observation that a number of genes whose expression changed after treatment (Supplementary Table I) have been previously described to be affected in response to iron deprivation using iron chelating agents such as DFO, or in response to 48 h treatment with a lower dose (2.5 nM) of ch128.1Av [28-31]. The expression of genes whose mRNA transcripts contain an iron regulatory element (IRE) motif, including TFRC (TfR1), DMT1, ACO2 (mitochondrial aconitase), and FTL (ferritin light chain), changed significantly in IM-9 cells treated with ch128.1Av [Fig. 5(B)]. Expression of these genes in U266 cells either did not change or changed inversely after treatment with ch128.1A [Figs. 5(A) and 5(B)], with the exception of the TfR1 transcript, whose expression increased after treatment in both IM-9 and U266 cells. These results thus support the hypothesis that the transcriptional response triggered by ch128.1Av is correlated to the induction of lethal iron deprivation in sensitive cells.
Figure 5.
Transcriptional changes in IM-9 and U266 cells treated with ch128.1Av. (A) Expression profiles of U266 (left) and IM-9 (right) cells treated with 10 nM ch128.1Av sampled at 1 h, 3 h, 9 h and 24 h compared to their time-matched controls are shown. Hierarchical clustering of genes and treatment times is based on their expression changes, as determined by an un-centered correlation algorithm. Genes shown are those with the greatest time-dependent variance in ch128.1Av treated IM-9 and U266 cells compared to their time-matched controls (p < 0.05). The color bar shows fold changes from low (blue) to high (red) expression with respect to controls on a log base 2 scale; gene names and descriptions are listed in Supplementary Table I. (B) Heat map of differentially expressed IRE-containing genes in U266 (left) and IM-9 (right) cells at 1 h, 3 h, 9 h and 24 h compared to time-matched controls. The accompanying color bar indicates fold changes from low (green) to high (red) expression with respect to controls on a log base 2 scale. (C, D) Quantitative RT-PCR validation of select differentially expressed genes from panel A in U266 (C) and IM-9 (D) cells treated with 10 nM ch128.1Av compared to time-matched controls at 1 h and 24 h; fold changes are shown on a log base 2 scale.
Cytotoxicty resulting from antibody-induced iron deprivation is p53-associated
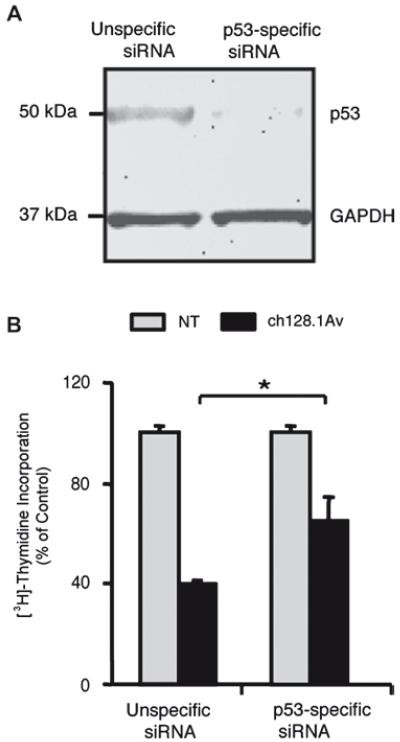
A pattern of changes in gene expression similar to that observed in IM-9 cells (p53 wild type) after treatment with ch128.1Av has also been previously associated with the activity of the tumor suppressor protein p53 [29]. Genes that changed most notably in IM-9 cells in response to treatment with ch128.1Av, and are known downstream targets of p53, include CDKN1A, TP53I3, GADD45 and FDXR. An in silico analysis of transcription factor binding sites of the promoter regions of genes whose transcript levels changed significantly in IM-9 cells after treatment was also conducted, and indicated a significant enrichment of genes in this list with p53 binding sites in their promoter regions (Supplementary Fig. 7). As expected, no similar enrichment of genes with p53 binding sites in their promoter regions was observed in U266 cells (data not shown). However, no changes in the expression of p53 transcripts were observed in IM-9 or U266 cells in response to ch128.1Av [Figs. 5(C) and 5(D)]. To determine the contribution of p53 to the cytotoxic effect of ch128.1Av, we decreased the level of p53 protein expressed by IM-9 cells, via p53-specific siRNA [Fig. 6(A)]. This decrease in p53 expression was consistent with a decrease of approximately 20% in the inhibition of proliferation induced by ch128.1Av after 24 h of treatment [Fig. 6(B)], confirming that the toxic effect of iron deprivation induced by ch128.1Av in sensitive cells is mediated at least in part by the presence of functional p53. A pair of isogenic malignant lymphoblastoid cell lines, which differ only in their expression of p53 (TK6 expressing wild type p53 and NH32 a p53 homozygous mutant at codon 237 of exon 7), also showed differential sensitivity to treatment with ch128.1Av. TK6 cells showed greater cytotoxicity at 24 h and 48 h treatment with ch128.1Av than did NH32 cells [Supplementary Figs. 8(A) and 8(B)], as well as a greater induction of apoptosis [Supplementary Fig. 8(C)]. These results suggest that toxicity induced by ch128.1Av is in part p53-dependent, and that functional p53 is required but not sufficient for the cytotoxicity triggered by antibody-induced iron deprivation.
Figure 6.
The role of p53 in antibody-induced LID. (A) An immunoblot of p53 expression in IM-9 cells 24 h after electroporation with a non-specific siRNA or p53-targeted siRNA; GAPDH is shown as a loading control. (B) [3H]-thymidine incorporation of cells with and without siRNA-mediated reduction of p53 expression treated with 10 nM ch128.1Av for 48 h or 24 h after electroporation. Data is shown as a percent of control treated cells, with error bars indicating standard deviations of triplicate samples (*p < 0.05). Immunoblot and proliferation assay data were obtained from the same experiment.
Discussion
A number of therapeutic antibodies targeting TfR have been designed to exploit the reliance of hematopoietic malignancies on iron [2,6,9-12,14]. In the present study we further characterize the molecular events induced by targeting of TfR1 by the non-neutralizing antibodies ch128.1 and ch128.1Av that contribute to cell death as a result of LID. We describe the cellular and molecular mechanisms that underlie this antibody-induced LID in a panel of malignant hematopoietic cells, and identify mechanisms of resistance to ch128.1Av and potentially other antibodies targeting TfR1.
Sensitivity of cells to antibodies that target TfR1 can be generally attributed to one or more of the following: antibody neutralization of ligand binding, blocking of receptor internalization, receptor sequestration/degradation and/or the activation of pro-apoptotic/anti-proliferative events [6,10,12,14,32]. Resistance, on the other hand, could be attributed in part to the expression of TfR1 variants, low levels of cell surface expression of TfR1, the availability of alternative routes of iron import including TfR2, or the aberrant expression of pro-proliferative/anti-apoptotic molecules that may resist the induction of a cytotoxic response after iron deprivation is induced [6,10,14,33]. Our present studies indicate that despite the interaction of resistant malignant cells with ch128.1Av, these cells maintain a high level of Tf uptake and do not experience a decrease in their intracellular iron stores. Thus, the possibility that competition might exist between ch128.1Av and Tf only under certain conditions or with variants of the receptor expressed exclusively by sensitive cells is unlikely. In addition, no expression of TfR2 or other iron-import proteins was observed in sensitive or resistant cells (data not shown). Our results suggest that alternative routes of iron import are not required for resistance in certain cells, since TfR1 remains functional in resistant cells after treatment with either ch128.1Av or ch128.1. Altogether, our results suggest that resistance to non-neutralizing antibodies specific for TfR1 stems from the inability of certain cells to sequester and/or degrade the receptor despite its interaction with the antibodies. It is possible that these mechanisms of sensitivity and resistance may also apply to certain neutralizing antibodies. However, these effects are more difficult to study in the case of neutralizing antibodies given that their cytotoxic activity relies at least in part on their ability to inhibit the binding of Tf to TfR1 [10,34]. In these cases, one would have to take into account the changing availability of Tf under physiologically relevant situations, and the varying density of receptors on target cells.
The mechanisms that facilitate the cytotoxicity of antibodies in vitro, such as the sequestration and degradation of TfR1 induced by ch128.1Av, may not be as effective in the complex in vivo environment of growing tumor cells [35]. Likewise, the resistance mechanisms employed by certain cells may not function in vivo as they would in vitro, and in some cases, resistant cells may be sensitized by additional or missing factors in vivo. Varying sensitivities to the inhibition of TfR1 function by ch128.1Av have been observed in our panel of cell lines in isolates from peritoneal xenografts. Importantly, these results show a striking similarity to the observed effects of ch128.1Av on TfR1 function in vitro. Although these experiments may not be an accurate indicator of antibody efficacy in vivo, they detail the cellular response to TfR1 targeting by antibodies in vivo and the inhibition of receptor function up to 2 days after the inoculation of tumor xenografts in a way that is similar to the mechanisms we observed in vitro. Further studies are needed to determine whether the inhibition of TfR1 function observed in response to treatment with ch128.1Av, and to a lesser extent with ch128.1, in sensitive cells in vivo will translate into the induction of lethal iron deprivation and inhibition of tumor growth.
Our data suggest that the outcome of TfR1 targeting by non-neutralizing antibodies is the induction of LID. The response of sensitive cells to targeting of TfR1 with antibodies such as ch128.1Av is similar to that observed with DFO and other iron chelators, an observation that is supported by changes in the expression of mRNA transcripts in sensitive and resistant cells after treatment with ch128.1Av. However, the effect of ch128.1Av on transcriptional regulation of iron-related genes was not as rapid or robust as the previously reported changes in mRNA transcript expression observed in different cells in response to iron chelators such as DFO [29], at times as early as 30 min after addition of the iron chelating agent and up to six-fold higher than control measurements after 20 h [36]. However, despite the expected slower and less pronounced transcriptional response we have observed after treatment of sensitive cells (IM-9) with ch128.1Av compared to that previously reported for pleiotropic chelators, greater selectivity in cell targeting is expected from the use of therapeutic antibodies to achieve LID [29,30]. Additionally, transcriptional changes induced in sensitive cells after 24 h treatment with ch128.1Av suggest a role for p53 in the cytotoxic response to antibody-induced iron deprivation. However, the lack of change in p53 transcripts suggests that only a conversion of latent to active p53 could be responsible for the effect observed on its downstream targets, instead of de novo synthesis of the protein. The involvement of p53 or p53-related proteins in the response to iron deprivation in cells treated with a variety of iron chelating agents has been recently documented [30]. However, a direct correlation has yet to be established between TfR1 targeting using antibodies and the activation of p53 target genes. The link between antibody-induced LID and the increased expression of a significant amount of p53 target genes could exist through the activity of p53 itself and/or other p53-associated proteins and their targets, such as p53R2 [30], given that ch128.1Av remains cytotoxic to IM-9 cells whose expression of p53 protein has been reduced using siRNA. In a pair of isogenic cell lines, one expressing wild type p53 (TK6) and the other an inactive mutant of p53 (NH32), ch128.1Av showed greater selective cytotoxicity against cells expressing the wild type p53. However, the fact that in cells expressing both artificially low levels of p53 (after siRNA treatment) and an inactivated mutant of p53, a cytotoxic effect is still observed after treatment with ch128.1Av, suggests that this treatment may also be effective against malignancies with defects in p53.
Our data on the inhibition of TfR1 function by ch128.1Av, and to a lesser extent ch128.1, in a subset of malignant hematopoietic cells support a model of action in which these antibodies induce LID beginning with the sequestration and/or degradation of TfR1, followed by the induction of cytotoxic iron deprivation that is mediated in part by downstream targets of p53. We have found potential avenues of resistance to this approach to the targeting of TfR1, including the differential regulation of receptor trafficking, allowing a subset of treated cells to retain normal receptor function and Tf uptake after treatment, as well as a lack of key molecules that play a role in the cytotoxic response to iron deprivation, such as functional p53. These resistance mechanisms are not mutually exclusive, but both can potentially be overcome in future applications through the rational design of therapies consisting of antibodies targeting TfR1, alone or in combination with other therapeutic agents. Further studies are still required in a wider variety of cell lines both in vitro and in vivo to determine the universality of our findings, as well as the limitations of the present mechanism of TfR1 targeting for the induction of LID. A wide variety of incurable hematopoietic malignancies, including multiple myeloma and aggressive lymphomas, still require adequate therapies and could potentially benefit from the further development of these treatments.
Supplementary Material
Acknowledgements
We thank Dr. Matthew Schibler (UCLA Advanced Light Microscopy Facility at the California NanoSystems Institute, CNSI), Rafaela Quintero (UCLA) and Giovanni Coppola (UCLA) for their technical assistance.
This work was supported in part by NIH/NCI grants R01CA107023, K01CA138559, R01CA57152, U54CA143931, R25GM0638, the training grant T32CA009120, NIH Fogarty AITRP-AIDS Malignancies Program D43TW000013S1, the UC MEXUS-CONACYT Fellowship Program, the Howard Hughes Medical Institute Gilliam Fellowship, the Whitcome Fellowship of the Molecular Biology Institute at UCLA and the Fletcher Jones Foundation. D. Casero was partially supported by a post-doctoral grant from the Spanish Foundation of Science and Technology (FECYT) and CNSI-UCLA. The UCLA JCCC Flow Cytometry Core Facility is supported by the NIH Awards CA16042 and AI28697, the Jonsson Cancer Center, the UCLA AIDS Institute and the UCLA School of Medicine.
Footnotes
Potential conflict of interest: Disclosure forms provided by the authors are available with the full text of this article at www.informahealthcare.com/lal.
Supplementary material available online
Details of analyses and figures and tables giving further results
References
- [1].Ponka P, Lok CN. The transferrin receptor: role in health and disease. Int J Biochem Cell Biol. 1999;31:1111–1137. doi: 10.1016/s1357-2725(99)00070-9. [DOI] [PubMed] [Google Scholar]
- [2].Daniels TR, Delgado T, Rodriguez JA, et al. The transferrin receptor part I: biology and targeting with cytotoxic antibodies for the treatment of cancer. Clin Immunol. 2006;121:144–158. doi: 10.1016/j.clim.2006.06.010. [DOI] [PubMed] [Google Scholar]
- [3].Ciechanover A, Schwartz AL, Lodish HF. Sorting and recycling of cell surface receptors and endocytosed ligands: the asialo glycoprotein and transferrin receptors. J Cell Biochem. 1983;23:107–130. doi: 10.1002/jcb.240230111. [DOI] [PubMed] [Google Scholar]
- [4].Larrick JW, Enns C, Raubitschek A, et al. Receptor-mediated endocytosis of human transferrin and its cell surface receptor. J Cell Physiol. 1985;124:283–287. doi: 10.1002/jcp.1041240217. [DOI] [PubMed] [Google Scholar]
- [5].Das Gupta A, Shah VI. Correlation of transferrin receptor expression with histologic grade and immunophenotype in chronic lymphocytic leukemia and non-Hodgkin’s lymphoma. Hematol Pathol. 1990;4:37–41. [PubMed] [Google Scholar]
- [6].White S, Taetle R, Seligman PA, et al. Combinations of anti-transferrin receptor monoclonal antibodies inhibit human tumor cell growth in vitro and in vivo: evidence for synergistic antiproliferative effects. Cancer Res. 1990;50:6295–6301. [PubMed] [Google Scholar]
- [7].Kasibhatla S, Jessen KA, Maliartchouk S, et al. A role for transferrin receptor in triggering apoptosis when targeted with gambogic acid. Proc Natl Acad Sci USA. 2005;102:12095–12100. doi: 10.1073/pnas.0406731102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Horonchik L, Wessling-Resnick M. The small-molecule iron transport inhibitor ferristatin/NSC306711 promotes degradation of the transferrin receptor. Chem Biol. 2008;15:647–653. doi: 10.1016/j.chembiol.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ng PP, Dela Cruz JS, Sorour DN, et al. An anti-transferrin receptor-avidin fusion protein exhibits both strong proapoptotic activity and the ability to deliver various molecules into cancer cells. Proc Natl Acad Sci USA. 2002;99:10706–10711. doi: 10.1073/pnas.162362999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Callens C, Moura IC, Lepelletier Y, et al. Recent advances in adult T-cell leukemia therapy: focus on a new anti-transferrin receptor monoclonal antibody. Leukemia. 2008;22:42–48. doi: 10.1038/sj.leu.2404958. [DOI] [PubMed] [Google Scholar]
- [11].Daniels TR, Delgado T, Helguera G, et al. The transferrin receptor part II: targeted delivery of therapeutic agents into cancer cells. Clin Immunol. 2006;121:159–176. doi: 10.1016/j.clim.2006.06.006. [DOI] [PubMed] [Google Scholar]
- [12].Crepin R, Goenaga AL, Jullienne B, et al. Development of human single-chain antibodies to the transferrin receptor that effectively antagonize the growth of leukemias and lymphomas. Cancer Res. 2010;70:5497–5506. doi: 10.1158/0008-5472.CAN-10-0938. [DOI] [PubMed] [Google Scholar]
- [13].Peer D, Karp JM, Hong S, et al. Nanocarriers as an emerging platform for cancer therapy. Nat Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- [14].Ng PP, Helguera G, Daniels TR, et al. Molecular events contributing to cell death in malignant human hematopoietic cells elicited by an IgG3-avidin fusion protein targeting the transferrin receptor. Blood. 2006;108:2745–2754. doi: 10.1182/blood-2006-04-020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Laubach JP, Richardson PG, Anderson KC. The evolution and impact of therapy in multiple myeloma. Med Oncol. 2010;27(Suppl. 1):S1–S6. doi: 10.1007/s12032-010-9442-2. [DOI] [PubMed] [Google Scholar]
- [16].Daniels TR, Ng PP, Delgado T, et al. Conjugation of an anti transferrin receptor IgG3-avidin fusion protein with biotinylated saporin results in significant enhancement of its cytotoxicity against malignant hematopoietic cells. Mol Cancer Ther. 2007;6:2995–3008. doi: 10.1158/1535-7163.MCT-07-0330. [DOI] [PubMed] [Google Scholar]
- [17].Rodriguez JA, Helguera G, Daniels TR, et al. Binding specificity and internalization properties of an antibody-avidin fusion protein targeting the human transferrin receptor. J Control Release. 2007;124:35–42. doi: 10.1016/j.jconrel.2007.08.020. [DOI] [PubMed] [Google Scholar]
- [18].Ortiz-Sanchez E, Daniels TR, Helguera G, et al. Enhanced cytotoxicity of an anti-transferrin receptor IgG3-avidin fusion protein in combination with gambogic acid against human malignant hematopoietic cells: functional relevance of iron, the receptor, and reactive oxygen species. Leukemia. 2009;23:59–70. doi: 10.1038/leu.2008.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Xia F, Wang X, Wang YH, et al. Altered p53 status correlates with differences in sensitivity to radiation-induced mutation and apoptosis in two closely related human lymphoblast lines. Cancer Res. 1995;55:12–15. [PubMed] [Google Scholar]
- [20].Lu TP, Lai LC, Lin BI, et al. Distinct signaling pathways after higher or lower doses of radiation in three closely related human lymphoblast cell lines. Int J Radiat Oncol Biol Phys. 2010;76:212–219. doi: 10.1016/j.ijrobp.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Simunek T, Boer C, Bouwman RA, et al. SIH—a novel lipophilic iron chelator—protects H9c2 cardiomyoblasts from oxidative stress-induced mitochondrial injury and cell death. J Mol Cell Cardiol. 2005;39:345–354. doi: 10.1016/j.yjmcc.2005.05.008. [DOI] [PubMed] [Google Scholar]
- [22].Beltran-Parrazal L, Lopez-Valdes HE, Brennan KC, et al. Mitochondrial transport in processes of cortical neurons is independent of intracellular calcium. Am J Physiol Cell Physiol. 2006;291:C1193–1197. doi: 10.1152/ajpcell.00230.2006. [DOI] [PubMed] [Google Scholar]
- [23].Kirchhausen T, Macia E, Pelish HE. Use of dynasore, the small molecule inhibitor of dynamin, in the regulation of endocytosis. Methods Enzymol. 2008;438:77–93. doi: 10.1016/S0076-6879(07)38006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Feo-Zuppardi FJ, Taylor CW, Iwato K, et al. Long-term engraftment of fresh human myeloma cells in SCID mice. Blood. 1992;80:2843–2850. [PubMed] [Google Scholar]
- [25].Markman M. Intraperitoneal antineoplastic drug delivery: rationale and results. Lancet Oncol. 2003;4:277–283. doi: 10.1016/s1470-2045(03)01074-x. [DOI] [PubMed] [Google Scholar]
- [26].Breuer W, Epsztejn S, Cabantchik ZI. Dynamics of the cytosolic chelatable iron pool of K562 cells. FEBS Lett. 1996;382:304–308. doi: 10.1016/0014-5793(96)00190-1. [DOI] [PubMed] [Google Scholar]
- [27].Lin SM, Du P, Huber W, et al. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36:e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Daniels TR, Neacato II, Rodriguez JA, et al. Disruption of HOX activity leads to cell death that can be enhanced by the interference of iron uptake in malignant B cells. Leukemia. 2010;24:1555–1565. doi: 10.1038/leu.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Saletta F, Rahmanto YS, Noulsri E, et al. Iron chelator-mediated alterations in gene expression: identification of novel iron-regulated molecules that are molecular targets of hypoxia-inducible factor-1 alpha and p53. Mol Pharmacol. 2010;77:443–458. doi: 10.1124/mol.109.061028. [DOI] [PubMed] [Google Scholar]
- [30].Richardson DR, Kalinowski DS, Lau S, et al. Cancer cell iron metabolism and the development of potent iron chelators as anti-tumour agents. Biochim Biophys Acta. 2009;1790:702–717. doi: 10.1016/j.bbagen.2008.04.003. [DOI] [PubMed] [Google Scholar]
- [31].Gao J, Richardson DR. The potential of iron chelators of the pyridoxal isonicotinoyl hydrazone class as effective antiproliferative agents, IV: the mechanisms involved in inhibiting cell-cycle progression. Blood. 2001;98:842–850. doi: 10.1182/blood.v98.3.842. [DOI] [PubMed] [Google Scholar]
- [32].Suzuki E, Daniels TR, Helguera G, et al. Inhibition of NF-kappaB and Akt pathways by an antibody-avidin fusion protein sensitizes malignant B-cells to cisplatin-induced apoptosis. Int J Oncol. 2010;36:1299–1307. doi: 10.3892/ijo_00000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Taetle R, Castagnola J, Mendelsohn J. Mechanisms of growth inhibition by anti-transferrin receptor monoclonal antibodies. Cancer Res. 1986;46:1759–1763. [PubMed] [Google Scholar]
- [34].Moura IC, Lepelletier Y, Arnulf B, et al. A neutralizing monoclonal antibody (mAb A24) directed against the transferrin receptor induces apoptosis of tumor T lymphocytes from ATL patients. Blood. 2004;103:1838–1845. doi: 10.1182/blood-2003-07-2440. [DOI] [PubMed] [Google Scholar]
- [35].Tacchini L, Bianchi L, Bernelli-Zazzera A, et al. Transferrin receptor induction by hypoxia. HIF-1-mediated transcriptional activation and cell-specific post-transcriptional regulation. J Biol Chem. 1999;274:24142–24146. doi: 10.1074/jbc.274.34.24142. [DOI] [PubMed] [Google Scholar]
- [36].Piccinelli P, Samuelsson T. Evolution of the iron-responsive element. RNA. 2007;13:952–966. doi: 10.1261/rna.464807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Templeton DM, Liu Y. Genetic regulation of cell function in response to iron overload or chelation. Biochim Biophys Acta. 2003;1619:113–124. doi: 10.1016/s0304-4165(02)00497-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.