Abstract
A three-dimensional (3D) hierarchical plasmonic nano-architecture has been designed for a sensitive surface-enhanced Raman scattering (SERS) immuno-sensor for protein biomarker detection. The capture antibody molecules are immobilized on a plasmonic gold triangle nano-array pattern. On the other hand, the detection antibody molecules are linked to the gold nano-star@Raman-reporter@silica sandwich nanoparticles. When protein biomarkers are present, the sandwich nanoparticles are captured over the gold triangle nano-array, forming a confined 3D plasmonic field, leading to the enhanced electromagnetic field in intensity and in 3D space. As a result, the Raman reporter molecules are exposed to a high density of “hot spots”, which amplifies the Raman signal remarkably, improving the sensitivity of the SERS immuno-sensor. This SERS immuno-sensor exhibits a wide linear range (0.1 pg/mL to 10 ng/mL), and a low limit of detection (7 fg/mL) toward human immunoglobulin G (IgG) protein in the buffer solution. This biosensor has been successfully used for detection of the vascular endothelial growth factor (VEGF) in the human blood plasma from clinical breast cancer patient samples.
Keywords: Biosensor, Biomarker, Surface-enhanced Raman scattering, Cancer, Surface plasmon, Blood plasma
Protein biomarkers are typically released from cells or organs, which are characteristic of physiologic and pathophysiologic conditions. Prompt monitoring of biomarkers holds great promise for early clinical diagnostics, which facilitates successful treatment of diseases and provides an optimal chance of affecting patient survival. Therefore, immunoassay techniques for biomarker measurement in complex biological samples are of fundamental importance to biomedical research and to diagnosis/prognosis of diseases.1–3 Conventional approaches for biomarker measurement include the enzyme-linked immunosorbent assay (ELISA), radio-immunoassay, western blot and mass spectrometry.4–8 These techniques are, however, complex, laboratory-based, time-consuming, and require experienced personnel to conduct the assay analysis. In addition, the blood serum concentrations of protein biomarkers associated with early-stage cancers and infectious diseases generally range from 10−16 M to 10−12 M.9 However, commercially available immunoassays are typically capable of measuring proteins with a limit of detection (LOD) at the picomolar level,10 which cannot optimally meet the critical need for protein detection. Moreover, these approaches cannot be used as a point-of-care (POC) technique for rapid, high-throughput clinical diagnosis at low cost. Therefore, various biosensors have been developed toward POC testing of biomarkers, including fluorescent, electrochemical and surface plasmon resonance (SPR) devices, etc.11–16 However, the critical need for POC testing remains unmet, in part, because of the challenges in cost and interference from sample matrix background. Therefore, numerous efforts are being made to address these challenges, especially to develop sensors for rapid, accurate detection of biomarkers in real-world biological fluids (i.e., urine, blood, serum and plasma).17–20
Surface-enhanced Raman scattering (SERS) is emerging as a powerful analytical technique for chemical and biological sensing.21–24 Compared with conventional immunoassays based on fluorescence, electrochemistry and ELISA, SERS immunoassays have a lot of advantages.25–27 For example, SERS provides the spectral fingerprint signatures of analytes, which endows SERS sensors with better anti-interference resistance to non-specific molecules in the complex sample matrix as compared to electrochemical and fluorescent sensors. Moreover, it has the multiplexing detection capability with a single laser excitation due to its narrow-band Raman spectral signature and to its wide excitation wavelength. These unique attributes endow the SERS immunoassays ideal for biomarker detection in real-world biological samples. In general, SERS enhancement is attributed to electromagnetic (EM) enhancement and chemical enhancement (CE). The EM enhancement is typically much stronger than the CE enhancement. It is well-known that the EM enhancement is concentrated on the “hot spots”, which are originated from the coupling of the localized surface plasmon resonance (LSPR) fields. Optimization of SERS substrate in size, shape and composition is critical to the improvement in sensitivity and reproducibility of SERS assays.28–31 Our previous studies have shown that the star shape of SERS substrate can concentrate the plasmonic field, and create “hot spots” near the sharp tips due to the lightning-rod effect.25 It is expected that the SERS substrates with a high density of sharp tips will provide high sensitivity for SERS sensing.20,25,32 In addition, several studies have revealed that periodic and aperiodic nanostructures with the enhanced plasmonic field leads to strong SERS enhancement.33–37 However, challenges still remain with the amplification of SERS signals due to the extremely small cross-section of Raman scattering.38 In addition, the application of SERS sensors in clinical samples are still rare.
In the current study, a SERS immuno-sensor is constructed. The capture antibody is first immobilized on the gold triangle nano-array chip. On the other hand, the detection antibody is conjugated with the SERS probe (Au@Raman-reporter@SiO2 sandwich nanoparticle). The antigen (analyte) is sandwiched between the capture antibody (linked to the nano-array chip) and the detection antibody (conjugated to the SERS probe). This SERS sensor is characteristic of a three-dimensional (3D) hierarchical plasmonic nano-architecture, in which the Au nanostar@Raman-reporter@SiO2 sandwich nanoparticles are coupled to a periodic Au triangle nano-array, generating a 3D plasmonic field. Under light excitation, a lot of “hot spots” are created between the triangles in the Au triangle nano-array, and also present between the sharp tips in the Au nano-stars in the SERS probe. When many Au nanostar plasmonic antennas are brought close to the Au triangle nano-array, high-density “hot spots” are generated in a 3D space. The resulting electromagnetic field is enhanced both in the space and in the intensity, which allows the Raman reporter molecules to experience the enhanced electromagnetic field, leading to remarkable amplification of the Raman signal of malachite green isothiocyanate (MGITC), the Raman reporter used. Based on this principle, the developed SERS immuno-sensor can be used for biomarker detection.
For comparative studies, three configurations of plasmonic nano-architectures are employed as the SERS substrates, including (i) the Au nanosphere@MGITC@SiO2 particles coupled on a planar Au film, (ii) the Au nanosphere@MGITC@SiO2 particles coupled on a Au triangle nano-array, and (iii) the Au nanostar@MGITC@SiO2 particles coupled on a Au triangle nano-array. This allows us to investigate the effects of the Au chip (the planar Au film versus the Au triangle nano-array) and the Au core (the Au sphere versus the Au star) on the performance of the SERS immuno-sensor. The three types of sensors are employed to detect the human immunoglobulin G (IgG) in the buffer solution. It is found that the Au nanostar/Au triangle nano-array exhibits the highest sensitivity while the Au nanosphere/Au film is much less efficient. Therefore, the Au nanostar@MGITC@SiO2/Au triangle nano-array system is selected for detection of the vascular endothelial growth factor (VEGF) in human blood plasma of patients. VEGF is selected as the target analyte since it is a well-known protein biomarker for tumor-associated angiogenesis.39–41 VEGF or its receptors are up-regulated in several forms of human cancers. Targeting this protein with administration of a therapeutic antibody is approved by the FDA for treatment of selected malignancies.39–41 In short, this work has demonstrated that the developed SERS immuno-sensor has great promise for detection of biomarkers in clinical blood plasma samples.
RESULTS AND DISCUSSION
Plasmonic nano-structures and their conjugation with antibody
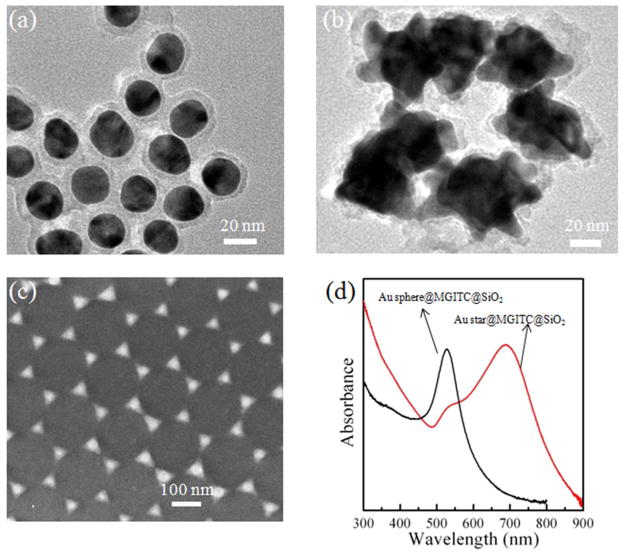
When preparing the Au@MGITC@SiO2 particles,25 the MGITC molecules (Raman reporter) were first adsorbed onto the surface of Au core. A thin silica layer was then coated. As a result, the MGITC molecules were sandwiched between the Au core and the silica shell. The silica shell enables the SERS probe water-soluble and provides a platform for bio-conjugation.25,37 The plasmonic Au core is able to amplify the SERS signal25. The sandwich structure also prevents from leaking of the Raman reporter molecules. In addition, many Raman reporter molecules are concentrated in a single sandwich nanoparticle as the SERS probe. As a result, the SERS signal results from a collection of Raman reporter molecules even for a single antibody-antigen event, which is an effective way to improve the sensitivity. Figure 1(a) and 1(b) show the TEM images of the Au sphere@MGITC@SiO2 and the Au star@MGITC@SiO2 sandwich nanoparticles, respectively. It can be clearly seen that both the Au spheres and stars were coated with a 4–5 nm thick SiO2 layer. Figure 1(d) shows the UV-visible absorption spectra of the Au sphere@MGITC@SiO2 and the Au star@MGITC@SiO2 sandwich nanoparticles. The Au sphere@MGITC@SiO2 and Au star@MGITC@SiO2 nanoparticles had the LSPR absorption bands at 520 nm and 690 nm, respectively. Also, the Au star@MGITC@SiO2 nanoparticles exhibited a strong absorption shoulder at around 530 nm. The sandwich nanoparticles displayed strong SERS peaks of MGITC molecules, as shown in Figure S1 in the Supporting Information. The SERS signal from the Au star@MGITC@SiO2 nanoparticles were much stronger than that from the Au sphere@MGITC@SiO2 nanoparticles, which was due to greater plasmon-induced electromagnetic field enhancement in the Au stars.25 The gap between the adjacent triangle corners in the Au triangle nano-array was about 40 nm (Figure 1(c)). Such a small gap enabled the coupling of LSPR, generating a high density of “hot spots” for the SERS enhancement.
Figure 1.
TEM images of (a) the Au sphere@MGITC@SiO2 sandwich nanoparticles and (b) the Au star@MGITC@SiO2 sandwich nanoparticles, (c) SEM image of the Au triangle nano-array, and (d) UV-visible absorption spectra of the Au sphere@MGITC@SiO2 sandwich nanoparticles and the Au star@MGITC@SiO2 sandwich nanoparticles
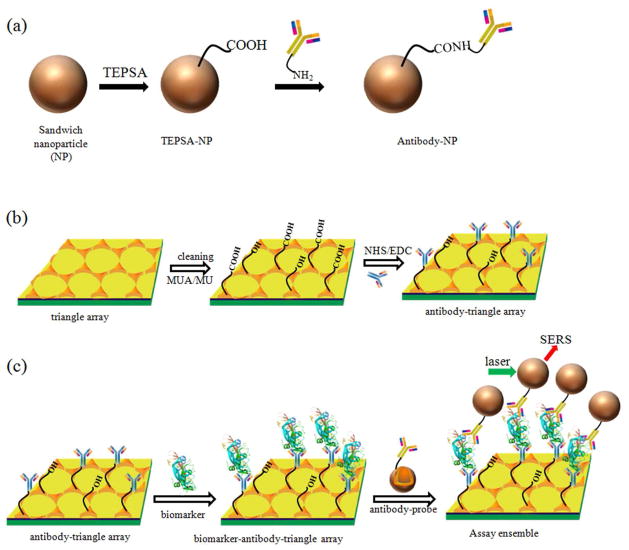
As shown in Figure 2(a) and 2(b), the capture antibody and the detection antibody were conjugated to the nano-array chip and the SERS probe (sandwich nanoparticle) by the carbodiimide chemistry20, respectively. It should be noted that the concentration of antibody solution applied in our protocols was quite high in order to ensure complete coverage of antibody on the substrate surface. Free excessive antibodies were removed by centrifugation and washing with the PBS buffer solution. The successful conjugation was confirmed by the FT-IR and XPS spectra (Figure S2 and S3).
Figure 2.
Schematic illustration of conjugation of (a) the SERS probe (sandwich nanoparticle) to the detection antibody, and (b) the Au triangle nano-array chip to the capture antibody; (c) Schematic illustration of the operating principle of SERS immuno-sensor for biomarker detection. The structure of VEGF biomarker is created by PyMOL with a four-digit code: 1VPF.
Operating principle of SERS immuno-sensor
Figure 2(c) schematically represents the operating principle of SERS immuno-sensor for biomarker detection. The SERS immuno-sensor is designed based on the sandwich-type configuration of antibody/antigen/antibody interaction. The SERS immuno-sensor included two processes performed in a humid chamber. Firstly, the capture antibody-modified Au chip was immersed into a solution containing the analyte. During the incubation, the analyte (biomarker) bound to the capture antibody-modified Au triangle nano-array chip. Excessive analyte was removed by washing with a PBS buffer solution. Next, the biomarker-antibody-Au nano-array chip was incubated in a solution containing the Au star@MGITC@SiO2 nanoparticle conjugated with the detection antibody. Since the antigen (analyte) in the present work had at least two binding sites, it can bind to both the detection antibody and the capture antibody, leading to the formation of the nanoparticle/biomarker/chip sandwich architecture. After washing with a PBS buffer solution, the free sandwich nanoparticles were removed. Finally, the chips were illuminated with the laser; and the SERS signal from the MGITC was recorded.
Comparison of immuno-sensor performance in various sandwich assemblies
In order to optimize the performance of the SERS immuno-sensor, three configurations of plasmonic nano-architectures were employed as the SERS substrates, including (i) the Au nanosphere@MGITC@SiO2 particles coupled on the planar Au film, (ii) the Au nanosphere@MGITC@SiO2 particles coupled on the Au triangle nano-array, and (iii) the Au nanostar@MGITC@SiO2 particles coupled on the Au triangle nano-array. For the sake of optimization, IgG was selected as the protein analyte because the IgG and its antibodies are much less expensive than VEGF and its corresponding antibodies.
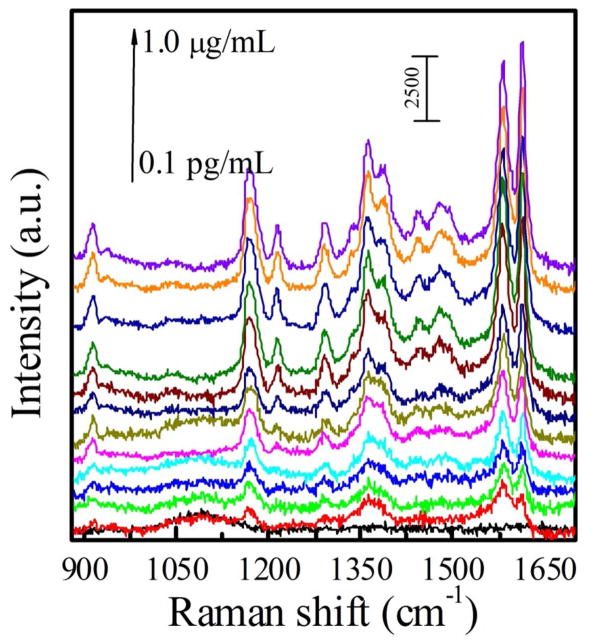
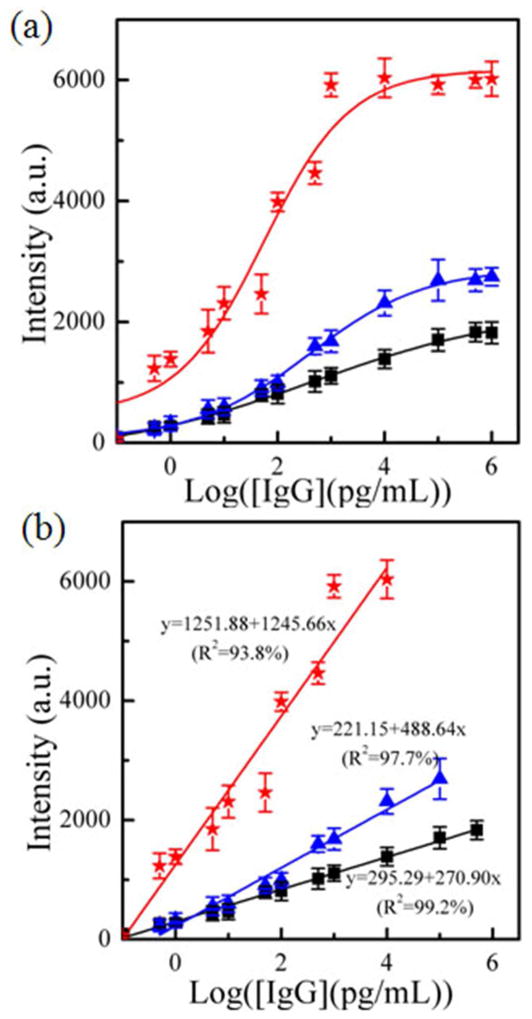
Figure 3 and Figure S4 show the SERS spectra of all three types of sensors that responded to various concentrations of IgG in the PBS buffer solution. The SERS intensity increased with an increase in the IgG concentration. The calibration curves were obtained by plotting the SERS peak intensity at 1578 cm−1 as a function of the IgG concentration (Figure 4). The Au nanostar@MGITC@SiO2 particles on the Au triangle nano-array showed the strongest SERS intensity at the corresponding IgG concentration in comparison with the Au sphere sandwich nanoparticle/Au film and the Au sphere sandwich nanoparticle/Au triangle nano-array. The Au sphere sandwich nanoparticle/Au film displayed the lowest SERS response. Furthermore, the calibration curves showed 5–6 orders of magnitude dynamic linear ranges of the IgG concentration (Table 1). In the linear region, the calibration curves were fitted as y=270.90•x+295.29 (R2=99.2%) for the Au sphere sandwich nanoparticle/Au film, y=488.64•x+221.15 (R2=97.7%) for the Au sphere sandwich nanoparticle/Au triangle nano-array, and y=1245.66•x+1251.88 (R2=93.8%) for the Au star sandwich nanoparticle/Au triangle nano-array, respectively. Here y is the SERS intensity at 1578 cm−1 and x is the logarithmic concentration of IgG (pg/mL). It should be noted that the highest sensitivity was achieved by the Au star sandwich nanoparticle/Au triangle nano-array. In addition, the LOD was obtained on the basis of 3S/N.42,43 The results showed that LOD was 45±3 fg/mL for the Au sphere sandwich nanoparticle/Au film, 25±5 fg/mL for the Au sphere sandwich nanoparticle/Au triangle nano-array, and 7±5 fg/mL for the Au star sandwich nanoparticle/Au triangle nano-array, respectively. In short, the Au star sandwich nanoparticle/Au triangle nano-array system exhibited the best performance in terms of LOD and sensitivity. Furthermore, the performance of the current SERS immuno-sensor was compared with those reported in the literature, including fluorescent, electrochemical, colorimetric and SPR sensors, as listed in Table 2. It can be seen that the present sensor exhibited a lower LOD.
Figure 3.
SERS spectra of the Au star@MGITC@SiO2 sandwich nanoparticle coupled to the Au triangle nano-array chip in various concentrations of IgG in the PBS buffer solution (0.1 pg/mL, 0.5 pg/mL, 1.0 pg/mL, 5.0 pg/mL, 10 pg/mL, 50 pg/mL, 0.1 ng/mL, 0.5 ng/mL, 1.0 ng/mL, 10 ng/mL, 100 ng/mL, 500 ng/mL and 1.0 μg/mL).
Figure 4.
(a) Plots of SERS peak intensity at 1578 cm−1 as a function of the logarithmic concentration of IgG, and (b) the linear range of (a). (■) the Au sphere coupled on the Au film, (
 ) the Au spheres coupled on the Au triangle nano-array, and (
) the Au spheres coupled on the Au triangle nano-array, and (
 ) the Au stars coupled on the Au triangle nano-array.
) the Au stars coupled on the Au triangle nano-array.
Table 1.
Performance of SERS immuno-sensors with different configurations.
| Assay system | Linear range (pg/mL) | Sensitivity (slope of fitted curve) | LOD based on 3S/N (fg/mL) |
|---|---|---|---|
| Au sphere@MGITC@SiO2 on Au film | 0.1–5.0×105 | 270.9 | 45±3 |
| Au sphere@MGITC@SiO2 on Au triangle nano-array | 0.5–1.0×105 | 488.6 | 25±5 |
| Au star@MGITC@SiO2 on Au triangle nano-array | 0.1–1.0×104 | 1245.7 | 7±5 |
Table 2.
Summary of representative sensors for detection of IgG and VEGF.
| Sensing system | Sensing principle | Biomarker | Type of assay | Linear range | LOD | refs |
|---|---|---|---|---|---|---|
| Au array | fluorescence | IgG | direct | 1 μM ~ 1 fM | 0.3 fM | 44 |
| Au/protein | electrochemical | IgG | direct | 10–104 ng/mL | 3 ng/mL | 45 |
| Glass/Au | colorimetry | IgG | sandwich | 1~ 5000 ng/mL | 60 ng/mL | 46 |
| Magnetic NPs | Piezoelectricity | IgG | sandwich | 0.6–34.9 μg/mL | 0.36 μg/mL | 47 |
| Au/aptamer | Voltammetry | VEGF | direct | 50~150 pM | 50 pM | 48 |
| Carbon fiber microelectrode | Voltammetry | VEGF | direct | 10~100 pg/mL | 38 pg/mL | 49 |
| SiO2/protein | Fluorescence | VEGF | direct | 0 ~50 ng/mL | 1.0 ng/mL | 50 |
| Au film | SPR | VEGF | direct | 20 ~ 600 ng/mL | 3 ng/mL | 51 |
3D electromagnetic field enhancement
FDTD simulations were used to explore the origin of the enhanced performance of the Au star/Au triangle array compared to the Au sphere/Au triangle array and the Au sphere/Au film sensor. Although the exact geometry of the Au sphere and star on the top of the triangle array cannot be simulated, the origin of the enhanced SERS can be explored by simulating multiple symmetric positions and examining the average enhancement (Figure S5). For the Au sphere/Au film, the coupled LSPR resulted in the SERS enhancement (|E/E0|4) of ~6,400. When the Au spheres were coupled to the Au triangle nano-array, the SERS enhancement (|E/E0|4) increased slightly to ~8,000. The exact orientation of spheres on the triangle array was unknown, but Figure S5(c) shows that an increased EM field of varying strength existed regardless of position. Although the enhancement magnitude of the LSPR field increased slightly when the Au spheres were coupled to the triangle array instead of the film, the number of possible coupling positions increased greatly.
For the Au sphere/Au film sensor to have a measurable SERS signal, two spheres must attach to the Au film in proximity (Figure S5(b)). The spheres can attach at any position on the film, reducing the probability of coupling except for a high analyte level. In the Au sphere/Au triangle array, the spheres can only attach to the triangle array and not the surrounding area. This ensures a high probability of coupling and measurable SERS signal even at a low analyte concentration. The average SERS signal therefore increases for a given concentration when the Au film is replaced with the Au triangle array even if the LSPR peak is comparable. It should be noted that the Au triangle array alone had similar strength to the coupled Au sphere/Au triangle array (Figure S5(a)). This indicates that the LSPR field was primarily from the Au triangle and not from the Au sphere-Au triangle resonance at 532 nm.
Figure S5(d) shows the electromagnetic field for the Au star/Au triangle array. The SERS enhancement (|E/E0|4) was ~1,000,000, which was significantly higher than either the Au sphere/Au triangle array or the Au sphere/Au film. As shown in our previous work, the 532 nm LSPR mode was originated from coupling of the spikes in the nanostar.25 When the tip of the spike was brought near the triangle, further LSPR coupling occurred between the spikes and the triangle, increasing the SERS signal. The spike-triangle coupling increases the intensity and spatial extension of the local electromagnetic field. The enhanced area of electromagnetic field means that more Raman reporters were subject to the SERS enhancement. Although the exact shape of the star and their positions on the triangle array were unknown, the relatively large size and multiple spikes of the star ensured that strong coupling was present when attached to the triangle nano-array (Figure S5(d)). Therefore, the 3D hierarchical architecture generated a high density of “hot spots”.
The enhanced SERS signal of the Au star/Au triangle array resulted from the increased strength, area and probability of the LSPR coupling. In particular, the improved performance of the Au star/Au array sensor came from the increased probability of coupling between the capture and the signal structures. An effective way to improve the sensor performance is to use a large signal structure (nanostar) with multiple LSPR active features on the surface. This allows for a single capture event to create multiple points of increased SERS signal, which effectively allows one Au nanostar to act as several nanospheres while only requiring one signal analyte to be detected. This process allows the LOD to be further lowered.
Detection of VEGF in clinical blood plasma samples with the SERS immuno-sensor
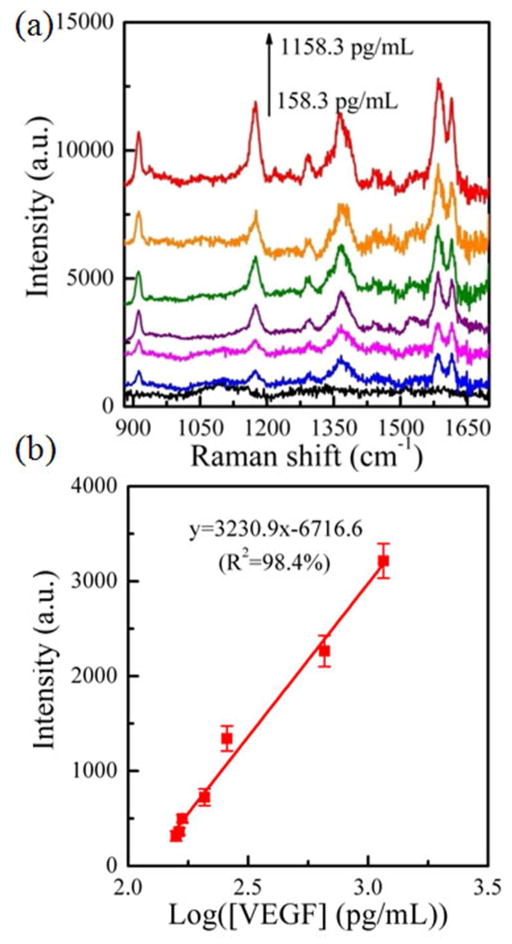
The above-mentioned results showed that Au nanostar coupled to the Au triangle nano-array was the best SERS substrate. Hence the Au star@MGITC@SiO2 sandwich nanoparticle/Au triangle nano-array was selected for detecting VEGF in the human blood plasma. Before testing clinical samples, the SERS immuno-sensor was calibrated with known concentrations of VEGF spiked into blood plasma. Prior to addition of VEGF, the baseline concentration of VEGF in the blood plasma matrices was estimated by a validated commercial ELISA kit. Figure 5 shows the SERS spectra of the Au star sandwich nanoparticle/Au triangle nano-array immuno-sensor system in the presence of various concentrations of VEGF in blood plasma. Obviously, the intensity of SERS peak at 1578 cm−1 gradually increased with an increase in the VEGF concentration. The calibration curve of the SERS peak intensity versus of the logarithmic concentration of VEGF was fitted as y=3230.9•x-6716.6 with the relative coefficient (R2) of 98.4%, where y is the peak intensity at 1578 cm−1, and x is the logarithmic concentration of VEGF. The calibration curve indicated that our SERS assay system can work for VEGF detection in blood plasma.
Figure 5.
(a) SERS spectra of the Au star@MGITC@SiO2 sandwich nanoparticle/Au triangle nano-array immuno-sensor, which responded to various concentrations of VEGF biomarker in blood plasma, and (b) the plot of the intensity of SERS peak at 1578 cm−1 as a function of the logarithmic concentration of VEGF in blood plasma
After calibration, the developed SERS immuno-sensor was used for detection of VEGF in blood plasma from clinical samples of patients with breast cancer. The VEGF concentration in the same clinic samples was also measured using the standard ELISA method. Table 3 compared the mean VEGF concentration values measured by the SERS immuno-sensor and the ELISA for three unknown clinical samples. The results show a high degree of similarity, which suggested that the present SERS sensor was capable of detecting VEGF in clinical samples. Obviously, the SERS immuno-sensor provides significant advantages over the conventional ELISA approach. For example, robust detection in complex matrices, short detection time, fewer washing steps and ease operation are evident with this novel approach.
Table 3.
VEGF concentration in clinical blood plasma samples measured by both the developed SERS immuno-sensor and a standard ELISA method
| Sample No. | [VEGF] via ELISA (pg/mL) | [VEGF] via SERS biosensor (pg/mL) |
|---|---|---|
| #1 | 600.1±12.6 | 585.9±12.5 |
| #2 | 337.7±14.6 | 301±9.6 |
| #3 | 569.6±6.9 | 588.8±15.3 |
CONCLUSIONS
In summary, an ultrasensitive SERS immuno-sensor was developed for protein biomarker detection. The sandwich nanoparticles conjugated with the detection antibody were coupled with Au triangle nano-array functionalized with the capture antibody via the protein biomarker. Coupling of the sandwich nanoparticle-based SERS probes with the Au triangle nano-array created a 3D hierarchical architecture, forming a 3D confined plasmonic field. The resulting 3D plasmonic field was enhanced in intensity and in 3D space, which generated a high density of “hot spots”. Therefore, the Raman signal of the MGITC molecules embedded in the sandwich nanoparticles was greatly amplified. Furthermore, the sandwich nanoparticle-SERS probes containing the Au star core led to much higher sensitivity for the SERS immuno-sensor than the one with the Au sphere as the core; and the Au triangle nano-array endowed much higher sensitivity than the planar Au film. The 3D FDTD simulation confirmed that the enhanced plasmonic field both in intensity and in space contributed to the high sensitivity of the SERS sensor. As a result, the developed SERS immuno-sensor was able to sensitively detect the protein biomarker with a wide dynamic linear range over several orders of magnitude. In addition, the present SERS immuno-sensor was capable of detecting the VEGF biomarker in clinical blood plasma samples. It is believed that the present SERS immuno-sensor can be further developed for point-of-care testing.
METHODS
Chemicals and Materials
Malachite green isothiocyanate (MGITC) was purchased from Molecular Probes, Inc. 3-triethoxylsilylpropyl succinic anhydride (TEPSA) was purchased from Gelest Inc. Goat anti-human IgG polyclonal antibody, IgG from human serum, (3-aminopropyl) trimethoxylsilane (APTMS), 5 M NaCl solution, and sodium silicate stock solution (26.5% SiO2 in 10.6% Na2O), N-hydroxysuccinimide (NHS), 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC), 11-mercaptoundecanoic acid (MUA) and 11-mercapto-1-undecanol (MU) were purchased from Sigma-Aldrich. Na2HPO4 (99.0%) and NaH2PO4 (99.0%) came from Alfa Aesar. Human VEGF monoclonal antibody (capture antibody, cAb) and human VEGF165 biotinylated polyclonal antibody (detection antibody, dAb) were purchased from R&D Systems, Inc. The planar gold film chips were purchased from EMF Corp. (Ithaca, NY). Deionized (D.I.) water was produced by a Milli-Q Millipore system (18.2 MΩ·cm, Millipore Corp.., USA). All solvents were obtained from commercial sources and used without further purification. In addition, citrated blood samples were obtained from patients with breast cancer and control patients without cancer following written informed consent according to the West Virginia University Health Sciences Institutional Review Board guidelines.
Synthesis of sandwich nanoparticles and its conjugation with detection antibody
Au nanosphere and Au nanostar were synthesized according to our previous reports.20,25 Briefly, Au sphere@MGITC@SiO2 and Au star@MGITC@SiO2 sandwich nanoparticles were prepared with the MGITC concentration of ~1.0 ×10−6 M.20,25 The resulting sandwich nanoparticles were dissolved in 200 μL of phosphate buffered saline (PBS) (10 mM Na2HPO4/NaH2PO4, 0.3 M NaCl and pH=7.0) buffer solution for further use. 100 μL of TEPSA was added into 100 μL of PBS solution of the sandwich nanoparticles (Au sphere@MGITC@SiO2 or Au star@MGITC@SiO2) obtained above, and then incubated for 2 h to achieve the carboxyl group-terminated sandwich nanoparticles. After centrifuged and washed with the PBS buffer solution, the carboxyl group-sandwich nanoparticles were incubated for 1 h in a PBS buffer solution containing 50 mM NHS and 200 mM EDC, followed by addition of 100 μL of 1.0 g/L detection antibody (anti-human IgG polyclonal or human VEGF165 biotinylated polyclonal antibody) and incubation overnight. The solution was centrifuged and washed with a PBS buffer solution to remove free excessive antibody, and then the resulting detection antibody-sandwich nanoparticle conjugates were stored in 100 μL of PBS buffer solution for future use.
Gold nano-array fabrication and functionalization of chips with capture antibody
The Au triangle nano-array on a glass slide was fabricated using nanosphere lithography as shown in our previous reports.52,53 A monolayer of hexagonally close-packed polystyrene spheres (200 nm in a diameter) was first self-assembled on a glass slide. A 10 nm thick titanium and a 50 nm thick Au layer were then deposited using e-beam evaporation. Subsequently, the chips were sonicated in ethanol to lift off the polystyrene spheres, leaving an array of Au triangles on the glass slide.
For surface functionalization, the chips (Au planar film and Au triangle nano-array) were firstly cleaned by successive immersion in CH2Cl2, ethanol and D.I. water each for 10 min, and then dried in a vacuum oven at 60 °C for 1 h. The cleaned chips were incubated overnight in an ethanolic solution containing 100 mM MUA and 100 mM MU, and then washed with ethanol to remove free MUA and MU. The resulting MUA/MU modified chips were activated by immersion in a PBS solution containing 50 mM NHS and 200 mM EDC. After washed with a PBS buffer solution, chips were incubated overnight in a PBS buffer solution of 1.0 g/L capture antibody (anti-human IgG polyclonal or human VEGF monoclonal antibody), followed by rigorously washing with PBS buffer solution to remove free capture antibody, and kept in a humid chamber prior to assay.
SERS immuno-sensor
Two types of immunoassay experiments, the IgG immunoassay in a PBS buffer solution and the VEGF immunoassay in blood plasma were conducted.
(a) IgG immunoassay in PBS buffer solution
The SERS immunoassay was carried out in a two-step process. The anti-human IgG-chip was first immersed into a solution of various IgG concentrations (0.1 pg/mL, 0.5 pg/mL, 1.0 pg/mL, 5.0 pg/mL, 10 pg/mL, 50 pg/mL, 0.1 ng/mL, 0.5 ng/mL, 1.0 ng/mL, 10 ng/mL, 100 ng/mL, 500 ng/mL and 1.0 μg/mL), and after incubation for 20 min, the chip was vigorously rinsed with PBS to remove free IgG. Then, the chip was immersed into the solution of anti-human IgG-sandwich nanoparticle conjugates, and incubated for 10 min, followed by rinsing with PBS to remove free sandwich nanoparticles. The resulting sandwich nanoparticle@IgG@chip was subject to the SERS measurement.
(b) VEGF immunoassay in anticoagulant human blood plasma
Firstly, the baseline concentration of VEGF in human blood plasma was determined to be 158.3 pg/mL by a validated commercial ELISA kit (R&D Systems, Minneapolis, MN). ELISA kit for VEGF (R&D system). The human blood plasma was spiked with the VEGF stock solution (1 ng/mL) in human blood plasma to achieve a standard curve, with an upper limit of 1.158 ng/mL. The concentration of VEGF in the plasma was measured with ELISA. Each of the three samples used were completed in triplicate for the ELISA.
For measurement of VEGF with the immuno-sensor, the cAb-chip was incubated for 20 min in human blood plasma of various VEGF concentrations, and then rinsed with PBS buffer solution to remove free VEGF and other molecules. Then, the VEGF-bound cAb-chip was immersed into the solution of dAb-sandwich nanoparticles obtained above, and incubated for 10 min, followed by the same procedure as that for the IgG immunoassay to remove free sandwich nanoparticles. The resulting sandwich nanoparticle@VEGF@chip was subject to the SERS measurement. The resulting curve was used for the calibration curve of VEGF concentration. The measurement for clinical samples followed a similar procedure except for the replacement of VEGF solution.
Instrumentation
The Au triangle array chip was observed under a field-emission scanning electron microscopy (SEM, JEOL JSM-7600F). The structure of nanoparticles was observed with a transmission electron microscopy (TEM, JEOL JEM-2100F) at an acceleration voltage of 200 kV. UV-visible absorption spectra were acquired with a Shimadzu UV-2550 spectrometer. The chemical structure was measured with a Fourier transform infrared (FT-IR) spectrometer (Thermo Nicolet 6700) under the attenuated total reflection (ATR) mode and with a PHI 5000 Versa Probe X-ray photoelectron spectroscopy (XPS) system (Physical Electronics, MN). XPS spectra were calibrated with the reference to the C 1s peak at 284.8 eV. Raman spectra were obtained with a Renishaw invia Raman spectrometer equipped with a 532 nm laser. Three SERS spectra were collected from different sites for each sample and then averaged to represent the SERS results. The maximum laser power on the sample, which was measured by a power meter (Newport, Model-1918-R), was around 0.017 mW; and the accumulation time was 10 s.
Finite difference time domain simulation
Finite difference time domain (FDTD) simulations were performed using the open source MEEP code.54 The dielectric function for Au was a series of four Lorentz sums fitted to the data of Johnson and Christy.55 A background dielectric constant of 1.33 was used to replicate the liquid phase of the sensor. The Au triangle array and nanospheres used for simulations had identical dimensions to those shown in the SEM/TEM image (Figure 1(a) and 1(c)). The absorption spectrum of the Au nanospheres was matched to the experimental UV-visible measurement. The SiO2 shell was found not to have a large impact on the simulation and was left out for computational simplicity. The Au nanostar’s shape was approximated by a sphere of equal size to the core shown in the TEM image (Figure 1(b)), covered by an array of cones. The shape was an idealization of the synthesized nanostar which had a non-symmetric random structure. The simulated and experimental Au nanostar absorption matched and our previous work indicated the electromagnetic field enhancement predicted by this model matches experimental measurements, justifying the approximation used.25 A plane wave, constant wavelength source at 532 nm was utilized. The 3D electromagnetic field was output over several times and normalized against the input source power. The incident wave vector was always perpendicular to the surface. Both the parallel and the perpendicular polarizations were tested. The polarization corresponding to the largest electromagnetic field enhancement was shown in the electromagnetic field visualizations. The visualization was done in the open source MayaVI2 software.
Supplementary Material
Acknowledgments
The resource and facilities used were partially supported by NSF (EPS 1003907), the NASA-WV Space Grant Consortium, the West Virginia University Research Corporation and the West Virginia EPSCoR Office. A portion of this work was performed in the Biospecimen Processing Core Facility, Mary Babb Randolph Cancer Center, West Virginia University, Morgantown, WV, which is supported in part by NIH grant P30 GM103488 and P30 RR 032138. The use of WVU shared facility was acknowledged. S. Cushing is supported by the National Science Foundation Graduate Research Fellowship under Grant No. (1102689). D. Ma is grateful to the financial support from the Natural Sciences and Engineering Research Council of Canada and Fonds de la recherche sur la nature et les technologies. W. Petros is supported by the Mylan Chair of Pharmacology at the WVU Health Sciences Center. L. Gibson is supported, in part, by NIH R01 HL056888, NIH RO1 CA134573, the Alexander B. Osborn Hematopoietic Malignancy and Transplantation Program and the WV Research Trust Fund.
Footnotes
The authors declare no competing financial interest.
Supporting Information. Figures S1–S5 and Table S1. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES AND NOTES
- 1.Arya SK, Bhansali S. Lung Cancer and Its Early Detection Using Biomarker-Based Biosensors. Chem Rev. 2011;111:6783–6809. doi: 10.1021/cr100420s. [DOI] [PubMed] [Google Scholar]
- 2.Ai K, Zhang B, Lu L. Europium-Based Fluorescence Nanoparticle Sensor for Rapid and Ultrasensitive Detection of an Anthrax Biomarker. Angew Chem Int Ed. 2008;47:1– 6. doi: 10.1002/anie.200804231. [DOI] [PubMed] [Google Scholar]
- 3.Trantum JR, Wright DW, Haselton FR. Biomarker-Mediated Disruption of Coffee-Ring Formation as a Low Resource Diagnostic Indicator. Langmuir. 2012;28:2187–2193. doi: 10.1021/la203903a. [DOI] [PubMed] [Google Scholar]
- 4.Kim DM, Noh HB, Park DS, Ryu SH, Koo JS, Shim YB. Immunosensors for Detection of Annexin II and MUC5AC for Early Diagnosis of Lung Cancer. Biosens Bioelectron. 2009;25:456–462. doi: 10.1016/j.bios.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Ambrosi A, Airò F, Merkoçi A. Enhanced Gold Nanoparticle Based ELISA for a Breast Cancer Biomarker. Anal Chem. 2010;82:1151–1156. doi: 10.1021/ac902492c. [DOI] [PubMed] [Google Scholar]
- 6.Wang KY, Chuang SA, Lin PC, Huang LS, Chen SH, Ouarda S, Pan WH, Lee PY, Lin CC, Chen YJ. Multiplexed Immunoassay: Quantitation and Profiling of Serum Biomarkers Using Magnetic Nanoprobes and MALDI-TOF MS. Anal Chem. 2008;80:6159–6167. doi: 10.1021/ac800354u. [DOI] [PubMed] [Google Scholar]
- 7.DeCaprio AP. Biomarkers:3 Coming of Age for Environmental Health and Risk Assessment. Environ Sci Technol. 1997;31:1837–1848. [Google Scholar]
- 8.Van Emon JM, Gerlach CL. A Status Report on Field-portable Immunoassay. Environ Sci Technol. 1995;29:312A–317A. doi: 10.1021/es00007a746. [DOI] [PubMed] [Google Scholar]
- 9.Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, Piech T, Patel PP, Chang L, Rivnak AJ, et al. Single–molecule Enzyme–linked Immunosorbent Assay Detects Serum Proteins at Subfemtomolar Concentrations. Nat Biotechnol. 2010;28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosi NL, Mirkin CA. Nanostructures in Biodiagnostics. Chem Rev. 2005;105:1547–156. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 11.Malhotra R, Patel V, Chikkaveeraiah BV, Munge BS, Cheong SC, Zain RB, Abraham MT, Dey DK, Gutkind JS, Rusling JF. Ultrasensitive Detection of Cancer Biomarkers in the Clinic by Use of a Nanostructured Microfluidic Array. Anal Chem. 2012;84:6249–6255. doi: 10.1021/ac301392g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang F, Haushalter RC, Haushalter RW, Shi Y, Zhang Y, Ding K, Zhao D, Stucky GD. Rare-Earth Upconverting Nanobarcodes for Multiplexed Biological Detection. Small. 2011;7:1972–1976. doi: 10.1002/smll.201100629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ranzoni A, Sabatte G, van IJzendoorn LJ, Prins MWJ. One-Step Homogeneous Magnetic Nanoparticle Immunoassay for Biomarker Detection Directly in Blood Plasma. ACS Nano. 2012;6:3134–3141. doi: 10.1021/nn204913f. [DOI] [PubMed] [Google Scholar]
- 14.Li Z, Wang Y, Wang J, Tang Z, Pounds JG, Lin Y. Rapid and Sensitive Detection of Protein Biomarker Using a Portable Fluorescence Biosensor Based on Quantum Dots and a Lateral Flow Test Strip. Anal Chem. 2010;82:7008–7014. doi: 10.1021/ac101405a. [DOI] [PubMed] [Google Scholar]
- 15.Reddy PJ, Sadhu S, Ray S, Srivastava S. Cancer Biomarker Detection by Surface Plasmon Resonance Biosensors. Clin Lab Med. 2012;32:47–72. doi: 10.1016/j.cll.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Altintas Z, Uludag Y, Gurbuz Y, Tothill IE. Surface Plasmon Resonance Based Immunosensor for the Detection of the Cancer Biomarker Carcinoembryonic Antigen. Talanta. 2011;86:377–383. doi: 10.1016/j.talanta.2011.09.031. [DOI] [PubMed] [Google Scholar]
- 17.Stern E, Vacic A, Rajan NK, Criscione JM, Park J, Ilic BR, Mooney DJ, Reed MA, Fahmy TM. Label-free Biomarker Detection from Whole Blood. Nat Nanotechnol. 2010;5:138–142. doi: 10.1038/nnano.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C, Ma L, Su M. Simultaneous Detection of Multiple Biomarkers with Several Orders of Concentration Difference Using Phase Change Nanoparticles. Anal Chem. 2011;83:2215–2219. doi: 10.1021/ac103102h. [DOI] [PubMed] [Google Scholar]
- 19.Wang C, Ma L, Chen L, Chai KX, Su M. Scanning Calorimetric Detections of Multiple DNA Biomarkers Contained in Complex Fluids Using Phase Change Nanoparticles. Anal Chem. 2010;82:1838–1843. doi: 10.1021/ac902503j. [DOI] [PubMed] [Google Scholar]
- 20.Li M, Zhang J, Suri S, Sooter LJ, Ma D, Wu N. Detection of Adenosine Triphosphate with an Aptamer Biosensor Based on Surface-Enhanced Raman Scattering. Anal Chem. 2012;84:2837–2842. doi: 10.1021/ac203325z. [DOI] [PubMed] [Google Scholar]
- 21.Chon H, Lee S, Son SW, Oh CH, Choo J. Highly Sensitive Immunoassay of Lung Cancer Marker Carcinoembryonic Antigen Using Surface-Enhanced Raman Scattering of Hollow Gold Nanospheres. Anal Chem. 2009;81:3029–3034. doi: 10.1021/ac802722c. [DOI] [PubMed] [Google Scholar]
- 22.Grubisha DS, Lipert RJ, Park HY, Driskell J, Porter MD. Femtomolar Detection of Prostate-Specific Antigen:3 an Immunoassay Based on Surface-Enhanced Raman Scattering and Immunogold Labels. Anal Chem. 2003;75:5936–5943. doi: 10.1021/ac034356f. [DOI] [PubMed] [Google Scholar]
- 23.Han D, Lim SY, Kim BJ, Piao L, Chung TD. Mercury(II) Detection by SERS Based on a Single Gold Microshell. Chem Commun. 2010;46:5587–5589. doi: 10.1039/c0cc00895h. [DOI] [PubMed] [Google Scholar]
- 24.Barhoumi A, Halas NJ. Label-Free Detection of DNA Hybridization Using Surface Enhanced Raman Spectroscopy. J Am Chem Soc. 2010;132:12792–12793. doi: 10.1021/ja105678z. [DOI] [PubMed] [Google Scholar]
- 25.Li M, Cushing SK, Zhang J, Lankford J, Aguilar ZP, Ma D, Wu N. Shape-dependent Surface-enhanced Raman Scattering in Gold–Raman-Probe–Silica Sandwiched Nanoparticles for Biocompatible Applications. Nanotechnology. 2012;23:115501. doi: 10.1088/0957-4484/23/11/115501. [DOI] [PubMed] [Google Scholar]
- 26.He S, Liu KK, Su S, Yan J, Mao X, Wang D, He Y, Li LJ, Song S, Fan C. Graphene-Based High-Efficiency Surface-Enhanced Raman Scattering-Active Platform for Sensitive and Multiplex DNA Detection. Anal Chem. 2012;84:4622–4627. doi: 10.1021/ac300577d. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Harpster MH, Wilson WC, Johnson PA. Surface-Enhanced Raman Scattering Detection of DNAs Derived from Virus Genomes Using Au-Coated Paramagnetic Nanoparticles. Langmuir. 2012;28:4030–4037. doi: 10.1021/la204890t. [DOI] [PubMed] [Google Scholar]
- 28.Li M, Li R, Li CM, Wu NQ. Electrochemical and Optical Biosensors Based On Nanomaterials and Nanostructures: Review. Front Biosci. 2011;S3:1308–1331. doi: 10.2741/228. [DOI] [PubMed] [Google Scholar]
- 29.Zhu Z, Meng H, Liu W, Liu X, Gong J, Qiu X, Jiang L, Wang D, Tang Z. Superstructures and SERS Properties of Gold Nanocrystals with Different Shapes. Angew Chem Int Ed. 2011;50:1593–1596. doi: 10.1002/anie.201005493. [DOI] [PubMed] [Google Scholar]
- 30.Kumari G, Narayana C. New Nano Architecture for SERS Applications. J Phys Chem Lett. 2012;3:1130–1135. doi: 10.1021/jz3001344. [DOI] [PubMed] [Google Scholar]
- 31.Baik SY, Cho YJ, Lim YR, Im HS, Jang DM, Myung Y, Park J, Kang HS. Charge-Selective Surface-Enhanced Raman Scattering Using Silver and Gold Nanoparticles Deposited on Silicon-Carbon Core-Shell Nanowires. ACS Nano. 2012;6:2459–2470. doi: 10.1021/nn204797b. [DOI] [PubMed] [Google Scholar]
- 32.Li M, Gou H, Al-Ogaidi I, Wu N. Nanostructured Sensors for Detection of Heavy Metals: A Review. ACS Sustainable Chem Eng. 2013 doi: 10.1021/sc400019a. [DOI] [Google Scholar]
- 33.Haes AJ, Haynes CL, McFarland AD, Schatz GC, Van Duyne RP, Zou S. Plasmonic Materials for Surface-Enhanced Sensing and Spectroscopy. MRS Bulletin. 2005;30:368–375. [Google Scholar]
- 34.Xu J, Kvasnijka P, Idso M, Jordan RW, Gong H, Homola J, Yu Q. Understanding the Effects of Dielectric Medium, Substrate, and Depth on Electric Fields and SERS of Quasi-3D Plasmonic Nanostructures. Opt Express. 2011;19:20493–20505. doi: 10.1364/OE.19.020493. [DOI] [PubMed] [Google Scholar]
- 35.Caldwell JD, Glembocki O, Bezares FJ, Bassim ND, Rendell RW, Feygelson M, Ukaegbu M, Kasica R, Shirey L, Hosten C. Plasmonic Nanopillar Arrays for Large-Area, High-Enhancement Surface-Enhanced Raman Scattering Sensors. ACS Nano. 2011;5:4046–4055. doi: 10.1021/nn200636t. [DOI] [PubMed] [Google Scholar]
- 36.Zhang XY, Hu A, Zhang T, Lei W, Xue XJ, Zhou Y, Duley WW. Self-Assembly of Large-Scale and Ultrathin Silver Nanoplate Films with Tunable Plasmon Resonance Properties. ACS Nano. 2011;5:9082–9092. doi: 10.1021/nn203336m. [DOI] [PubMed] [Google Scholar]
- 37.Li M, Cushing SK, Liang H, Suri S, Ma D, Wu N. Plasmonic Nanorice Antenna on Triangle Nano-array for Surface-Enhanced Raman Scattering Detection of Hepatitis B Virus DNA. Anal Chem. 2013;85:2072–2078. doi: 10.1021/ac303387a. [DOI] [PubMed] [Google Scholar]
- 38.Christesen SD. Raman Cross-sections of Chemical-agents and Simulants. Appl Spectrosc. 1988;42:318–321. [Google Scholar]
- 39.Tarkowski E, Issa R, Sjögren M, Wallin A, Blennow K, Tarkowski A, Kumar P. Increased Intrathecal Levels of the Angiogenic Factors VEGF and TGF-β in Alzheimer’s Disease and Vascular Dementia. Neurobiol Aging. 2002;23:237–243. doi: 10.1016/s0197-4580(01)00285-8. [DOI] [PubMed] [Google Scholar]
- 40.Ferrara N. Vascular Endothelial Growth Factor: Basic Science and Clinical Progress. Endocr Rev. 2004;25:581–611. doi: 10.1210/er.2003-0027. [DOI] [PubMed] [Google Scholar]
- 41.Storkebaum Erik, Lambrechts Diether, Carmeliet Peter. VEGF: Once Regarded as a Specific Angiogenic Factor, Now Implicated in Neuroprotection. BioEssays. 2004;26:943–954. doi: 10.1002/bies.20092. [DOI] [PubMed] [Google Scholar]
- 42.Gilfrich JV, Birks LS. Estimation of Detection Limits in X-ray Fluorescence Spectrometry. Anal Chem. 1984;56:77–79. [Google Scholar]
- 43.West SD, Turner LG. Determination of Spinosad and Its Metabolites in Meat, Milk, Cream, and Eggs by High-Performance Liquid Chromatography with Ultraviolet Detection. J Agric Food Chem. 1998;46:4620–4627. doi: 10.1021/jf000080q. [DOI] [PubMed] [Google Scholar]
- 44.Zhou L, Ding F, Chen H, Ding W, Zhang W, Chou SY. Enhancement of Immunoassay’s Fluorescence and Detection Sensitivity Using Three-Dimensional Plasmonic Nano-Antenna-Dots Array. Anal Chem. 2012;84:4489–4495. doi: 10.1021/ac3003215. [DOI] [PubMed] [Google Scholar]
- 45.Qiu LP, Wang CC, Hu P, Wu ZS, Shen GL, Yu RQ. A Label-free Electrochemical Immunoassay for IgG Detection Based on the Electron Transfer. Talanta. 2010;83:42–47. doi: 10.1016/j.talanta.2010.08.036. [DOI] [PubMed] [Google Scholar]
- 46.Lei KF, Butt YK. Colorimetric Immunoassay Chip Based On Gold Nanoparticles and Gold Enhancement. Microfluid Nanofluidics. 2010;8:131–137. [Google Scholar]
- 47.Li J, He X, Wu Z, Wang K, Shen G, Yu R. Piezoelectric Immunosensor Based on Magnetic Nanoparticles with Simple Immobilization Procedures. Anal Chim Acta. 2003;481:191–19842. [Google Scholar]
- 48.Zhao S, Yang W, Lai RY. A Folding-Based Electrochemical Aptasensor for Detection of Vascular Endothelial Growth Factor in Human Whole Blood. Biosens Bioelectron. 2011;26:2442–2447. doi: 10.1016/j.bios.2010.10.029. [DOI] [PubMed] [Google Scholar]
- 49.Prabhulkar S, Alwarappan S, Liu G, Li CZ. Amperometric Micro-Immunosensor for the Detection of Tumor Biomarker. Biosens Bioelectron. 2009;24:3524–3530. doi: 10.1016/j.bios.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki Y, Yokoyama K. Construction of a More Sensitive Fluorescence Sensing Material for the Detection of Vascular Endothelial Growth Factor, a Biomarker for Angiogenesis, Prepared by Combining a Fluorescent Peptide and a Nanopillar Substrate. Biosens Bioelectron. 2011;26:3696–3699. doi: 10.1016/j.bios.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 51.Pimková K, Bocková M, Hegnerová K, Suttnar J, nermák J, Homola J, Dyr JE. Surface Plasmon Resonance Biosensor for the Detection of VEGFR-1-a Protein Marker of Myelodysplastic Syndromes. Anal Bioanal Chem. 2012;402:381–387. doi: 10.1007/s00216-011-5395-3. [DOI] [PubMed] [Google Scholar]
- 52.Li H, Low J, Brown KS, Wu NQ. Large-Area Well-Ordered Nanodot Array Pattern Fabricated with Self-Assembled Nanosphere Template. IEEE Sensors J. 2008;8:880–884. [Google Scholar]
- 53.Li H, Wu NQ. A Large-Area Nanoscale Gold Hemisphere Pattern as A Nanoelectrode Array. Nanotechnology. 2008;19:275301. doi: 10.1088/0957-4484/19/27/275301. [DOI] [PubMed] [Google Scholar]
- 54.Oskooi AF, Roundy D, Ibanescu M, Bermel P, Joannopoulos JD, Johnson SG. MEEP: Meep: A Flexible Free-Software Package for Electromagnetic Simulations by the FDTD Method. Comput Phys Commun. 2010;181:687–702. [Google Scholar]
- 55.Johnson PB, Christy RW. Optical Constants of the Noble Metals. Phys Rev B. 1972;6:4370–4379. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.