Abstract
During meiosis II in the yeast Saccharomyces cerevisiae, the cytoplasmic face of the spindle pole body changes from a site of microtubule initiation to a site of de novo membrane formation. These membranes are required to package the haploid meiotic products into spores. This functional change in the spindle pole body involves the expansion and modification of its cytoplasmic face, termed the outer plaque. We report here that SPO21 is required for this modification. The Spo21 protein localizes to the spindle pole in meiotic cells. In the absence of SPO21 the structure of the outer plaque is abnormal, and prospore membranes do not form. Further, decreased dosage of SPO21 leaves only two of the four spindle pole bodies competent to generate membranes. Mutation of CNM67, encoding a known component of the mitotic outer plaque, also results in a meiotic outer plaque defect but does not block membrane formation, suggesting that Spo21p may play a direct role in initiating membrane formation.
INTRODUCTION
During the course of cellular differentiation, cells of many different types undergo rearrangements of the secretory pathway to generate new membrane compartments (Jones and Fawcett, 1966; Burgoyne and Morgan, 1993; Arvan and Castle, 1998; Spritz, 1999). A particularly striking example of this phenomenon is the process of spore formation in the budding yeast, Saccharomyces cerevisiae, which requires the de novo formation of a new membrane compartment, the prospore membrane (Moens, 1971; Moens and Rapport, 1971; Byers, 1981).
Spore formation occurs in a series of steps, first described in cytological studies in the electron microscope (EM; Moens, 1971; Moens and Rapport, 1971). The first cytologically evident step in spore formation is modification of the spindle pole bodies (SPBs) during the second meiotic division. The SPB is the yeast equivalent of the centrosome and is embedded in the nuclear envelope; thus it has separate nuclear and cytoplasmic faces. At meiosis II, the cytoplasmic face of each SPB, termed the outer plaque, expands laterally forming a convex, laminar structure. This modified outer plaque then serves as the initiation site for the formation of the prospore membrane. The prospore membrane forms as a flattened sac, which extends during anaphase II to engulf the adjacent nuclear lobe. After nuclear division, the ends of the prospore membrane fuse, enclosing the daughter nucleus inside two continuous membranes. Mature spores then form by the deposition of spore wall material in the lumen between the newly formed double membrane (Lynn and Magee, 1970).
Growth of the prospore membrane occurs by the fusion of secretory vesicles within the cytoplasm (Neiman, 1998). However, the mechanism by which vesicles are redirected from the plasma membrane to these intracellular sites remains obscure. The outer plaque of the SPB serves as both the initiation site for the prospore membrane and as the site of attachment of the prospore membrane to the nucleus. The outer plaque is, therefore, likely to play an important role in the initial delivery and fusion of vesicles to the prospore membrane. In fact, studies of strains forming dyads, 2-spored asci, directly implicate the SPB in regulating prospore membrane formation (Davidow et al., 1980).
Under some conditions, haploid dyads are formed in which the haploid spores are exclusively nonsisters (Davidow et al., 1980; Okamoto and Iino, 1981), that is, the cell packages 1 daughter nucleus from each of the meiosis II spindles into spores, forming nonsister dyads (NSDs). The basis for NSD formation has been demonstrated to be the modification of only 2 of the 4 spindle pole bodies, 1 on each spindle, which leads to the formation of only 2 prospore membranes (Davidow et al., 1980; Okamoto and Iino, 1982). Thus, there appears to be a direct correlation between SPB modification and prospore membrane formation.
During vegetative growth the outer plaque of the SPB serves as a nucleation site for cytoplasmic microtubules. The known components of the outer plaque in mitotic cells include the γ-tubulin complex, Tub4p, Spc97p, and Spc98p, which seeds microtubule growth (Sobel and Snyder, 1995; Geissler et al., 1996; Marschall et al., 1996; Spang et al., 1996; Knop et al., 1997). The γ-tubulin complex binding protein Spc72p (Knop and Schiebel, 1998) has also been identified as a component of the outer plaque in vegetative cells, as have two additional proteins, Nud1p, and Cnm67p (Wigge et al., 1998). Of these, CNM67 is induced during sporulation, and cnm67 mutants are sporulation defective (Brachat et al., 1998; Chu et al., 1998). However, no role for any of these genes in prospore membrane formation has been reported.
In this article we describe the characterization of SPO21, a gene required for meiosis-specific modification of the SPB outer plaque. In the absence of SPO21, outer plaque structure is abnormal and formation of prospore membranes is blocked. The Spo21 protein localizes specifically to meiotic spindle poles, suggesting a direct role for Spo21p in the formation of prospore membranes.
MATERIALS AND METHODS
Yeast Strains and Media
Standard yeast genetic methods and media were used (Rose et al., 1990). Strain AN120 (MATa/MATα ura3/ura3 leu2/leu2 trp1-hisG/trp1-hisG his3/his3 arg4-NspI/ARG4 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 rme1::LEU2/RME1) has been described (Neiman et al., 2000). Strains AN161 (cnm67Δ::his5+/cnm67Δ::his5+), AN180 (spo21Δ::his5+/ spo21Δ::his5+), and AN230 (spo21::GFP/spo21::GFP) were constructed as follows: in each case targeted integration was used to introduce the deletion or fusion into AN120 by transformation. The resulting heterozygous diploids, AN160, AN178, and AN229, were then dissected and haploid segregants were mated to create the strains AN161, AN180, and AN230, respectively. AN231 (spo21Δ/ spo21::GFP) was made by mating appropriate segregants from dissections of AN178 and AN229. To construct strain AN254 (MATa/MATα ura3/ura3 leu2/leu2 trp1-hisG/trp1-hisG his3/his3 lys2/lys2 hoΔ::LYS2/hoΔ::LYS2 cnm67Δ::his5+/cnm67Δ::his5+ spo21Δ::TRP1/ spo21Δ::TRP1) strains AN160–9A (MATa ura3 leu2 trp1-hisG his3 lys2 hoΔ::LYS2 cnm67Δ::his5+) and AN1064 (MATα ura3 leu2 trp1-hisG his3 lys2 hoΔ::LYS2 spo21Δ::TRP1) were crossed, and appropriate haploid segregants were mated.
The primers and templates used to generate the PCR products for transformation were as follows: for cnm67Δ, primers ANO157 (5′TCT ACA CAT ACA AAA AAT AAT CAC TAG TAA ATA GTG ACA GGT CTT TTG TAC ATC CCC GGG CTG CAG GAA TTC) and ANO158 (5′CTT AAT TTT TAG TTA CAA TTA CAA CAA TTT ATC TAT TGA CTC CGT TAA TAA AAA GTC GAC GGT TAC GAT AAG) were used to amplify the S. pombe his5+ gene from pME3 (Wach et al., 1997); for spo21Δ, primers ANO172 (5′CTA AAG GCA TAT TAA AGA TCT ATT AAA GAT CTA TTA AAG CTT TCT GCT ACC AGT CCC GGG CTG CAG GAA TTC) and ANO173 (5′TAT ATG TAT ATA TAG AAT ATT AAG GAT TAT AAA AGA ATG TGT AGC TGT TGA GGT GTC GAC GGT ATC GAT AAG) were used to amplify the S. pombe his5+ gene from pME3; for spo21::GFP, the primers ANO193 (5′CTC CAG CAC TCC GTA TAA ACA AAG CCA AAG ACA AGT TCC GCA CTC CAT CAA GCG GAT CCC CGG GTT AAT TAA) and ANO194 (5′TAT GTA TAT ATA GAA TAT TAA GGA TTA TAA AAG AAT TGT TAG CTG TTG AGG TGA ATT CGA GCT CGT TTA AAC) were used to amplify a cassette containing a carboxy-terminal GFP tag and the S. pombe his5+ from plasmid pFA6a-GFP(S65T)-HIS3 MX6 (Longtine et al., 1998).
For time course studies, cultures were sporulated as described (Neiman, 1998). To induce dyad formation by interrupted sporulation (Srivastava et al., 1983) cells were removed from sporulation medium after 3 h, pelleted, washed once in H2O, and resuspended in H2O, and the incubation was resumed.
Plasmids
The plasmid pRS316-SPO21::GFP2 was constructed as follows: SPO21 was amplified from genomic DNA by PCR with the use of the oligonucleotides MNO106 (5′GCG GCG GCG GCC GCT GGT TGG TTG GAA GCA TAG GTA C) and MNO107 (5′GCG GCG CTC GAG TTA ATG GTT TCT TCG GCA ACC CTG). The amplified product, which carries the SPO21 coding region as well as 300 bp of upstream and downstream sequence, was digested with NotI and XhoI and cloned into similarly digested pRS316 (Sikorski and Hieter, 1989) to create pRS316-SPO21. The oligonucleotides ANO183 (5′CTT GTT GAG CTC CTG GTT GGT TGG AAG C) and ANO184 (5′CTT GTT CTC GAG TCA ATC GAT GGA GTG CGG AAC) were then used to reamplify SPO21 and introduce a ClaI site just before the stop codon of SPO21. This PCR fragment was then digested with SpeI and ClaI and the 850-bp fragment carrying the 3′ end of the SPO21 gene isolated. Plasmid pGFP-C-FUS (Niedenthal et al., 1996) was digested with ClaI and KpnI, and the 800-bp fragment bearing GFP and the CYC1 terminator was isolated. Finally, plasmid pRS316-SPO21 was digested with SpeI and KpnI, and the 4.5-kb backbone containing the promoter and 5′ end of the SPO21 gene was isolated. These three fragments were ligated to produce pRS316-SPO21::GFP2. SPO21::GFP2 differs from the spo21::GFP allele only in the linker sequence between the SPO21 and GFP coding regions. In spo21::GFP the amino acid sequence RIPGLIN links the last amino acid residue of Spo21p to the second amino acid of GFP, whereas in SPO21::GFP2, the terminal K residue is removed and the sequence DTVD connects Spo21p to the first Met residue of GFP.
Electron Microscopy
For examination of spindle pole bodies, cells were prepared essentially as described (Byers and Goetsch, 1991). Sporulating cells were harvested by centrifugation, washed once in water, and then incubated 5 min in pretreatment buffer (0.2 M Tris, pH 9.4, 20 mM EDTA, pH 8.0, 0.1 M 2-mercaptoethanol, 1 M NaCl). Samples were then washed two times in 0.7 M sorbitol, resuspended in 0.7 M sorbitol, and fixed by addition of glutaraldehyde to a final concentration of 2.5%. After overnight incubation at 4°C, samples were pelleted and resuspended in phosphate citrate buffer (0.17 M KH2PO4, 30 mM sodium citrate), zymolyase (ICN Pharmaceuticals, Aurora, OH) was added to a final concentration of 0.5 μg/ml, and samples were incubated at 37°C for 3 h. Cells were then washed twice with 0.1 M NaOAc, stained in 2% OsO4 for 15 min, washed again in distilled water, and stained in 1% uranyl acetate for 1 h. Finally, the samples were dehydrated by a series of ethanol washes and embedded in Spurr resin for sectioning and analysis. Samples were analyzed with the use of a JEOL 1200EX (JEOL USA, Peabody, MA) transmission electron microscope at the Stony Brook University Microscopy Imaging Center.
To visualize meiosis II SPBs, cells were prepared for EM from time points in which, as judged by 4′,6′-diamidino-2-phenylindole (DAPI) staining, >60% of the cells were in meiosis II. In such cultures, >80% of the SPBs present should be meiosis II SPBs. At least 10 SPBs were examined in each strain (AN120, AN180, and AN161).
For examination of the prospore membranes, cells were stained in KMnO4 as described previously (Neiman, 1998).
Indirect Immunofluorescence
Immunofluorescence studies were performed essentially as described (Neiman et al., 1997), except that cells containing SPO21::GFP2 were fixed in 3.7% formaldehyde for only 5 min to preserve GFP fluorescence. Affinity-purified anti-Ssop, anti-Sncp (Rossi et al., 1997), and anti-Spr3p (Fares et al., 1996) antibodies were used at 1:1 dilution. Antitubulin antibodies were provided by N. Hollingsworth (SUNY, Stony Brook, NY). The secondary antibodies used were goat anti-rabbit coupled to Cy3 (Cappel Laboratories, Malvern, PA), goat anti-rat coupled to rhodamine (Cappel Laboratories), and goat anti-rabbit coupled to Alexa 488 (Molecular Probes, Eugene, OR). Immunofluorescence images were generated with the use of a Zeiss Axioskop and a 300T CCD camera (Dage MTI, Michigan City, IN) or a SPOT camera (Diagnostic Instruments, Sterling Heights, MI) and NIH Image 6.1 and prepared with the use of Adobe Photoshop 5.0 (Adobe Software, San Jose, CA).
RESULTS
spo21Δ Mutants Do Not Form Prospore Membranes
A sporulation defect in diploids deleted for open reading frame YOL091w (now designated SPO21) was originally noted as part of the EUROFAN systematic knockout project (Pearson et al., 1998). Subsequently, transcription of SPO21 was shown by microarray analysis to be induced during midsporulation (Chu et al., 1998). The protein encoded by SPO21 has a region of predicted coiled coil, but no obvious homologues in the GenBank database. However, careful examination of the sequence revealed a region of moderate homology (23% identity and 48% similarity >300 amino acids) to the spindle pole body component Spc72p (Figure 1). In both proteins, this region of homology includes ∼200 amino acids predicted by the Coils2 algorithm (Lupas et al., 1991) to be coiled coil as well as 120 residues to the amino terminal side of the coiled coil.
Figure 1.
Alignment of Spo21p and Spc72p sequences. The alignment was prepared with the use of the BLAST 2 algorithm (Tatusova and Madden, 1999). The residues at the amino terminal ends of the predicted coiled-coil regions, Arg283 of Spo21p and His359 of Spc72p, are highlighted.
Given these observations, we decided to more carefully examine the sporulation defect associated with mutation of the SPO21 gene. A deletion of SPO21 was constructed in the efficiently sporulating SK1 strain background (Kane and Roth, 1974). Consistent with previous work, diploids homozygous for spo21Δ fail to sporulate. A meiotic time course in the spo21Δ strain was performed to determine where in the sporulation process spo21Δ mutants were defective. The DNA-binding dye DAPI was used to follow the progress of the meiotic divisions by fluorescence microscopy. The spo21Δ strains proceeded through the two meiotic divisions with kinetics comparable to an isogenic wild-type control and produced tetranucleate cells with high efficiency (Figure 2). However, spores were never seen, indicating that spo21Δ is defective in some aspect of spore formation. At later time points, in the spo21Δ strain, but not in the wild type, the proportion of tetranucleate cells declined, and a new class of cells appeared in which DAPI staining was fragmented. This nuclear fragmentation phenotype has been reported in several mutants defective in packaging nuclei into spores (Rose et al., 1995; Nag et al., 1997; Neiman, 1998).
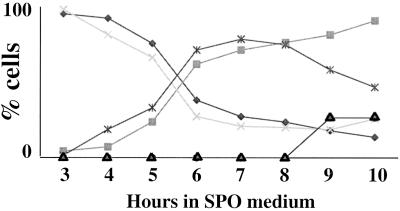
Figure 2.
spo21 mutants show no defect in meiotic progression. Strains AN120 and AN180 were transferred to 2% KOAc, and at indicated times samples were removed and fixed, and progression through meiosis was examined by DAPI staining. ×, AN120, mononucleate cells; ♦, AN180 mononucleate cells; ▪, AN120 binucleate and tetranucleate cells; ∗, AN180 binucleate and tetranucleate cells; ▴, AN180 cells displaying nuclear fragmentation.
To examine the nature of the spore formation defect, spo21Δ mutants were sporulated and stained for immunofluorescence studies with antibodies to three different prospore membrane associated markers; Sso1/2p, Snc1/2p, and Spr3p (Neiman, 1998; Rudge et al., 1998). No localized staining of any of the antibodies was detected in the spo21Δ diploid, indicating a complete failure in prospore membrane formation (Figure 3). A similar absence of prospore membranes has previously been seen in mutants that interfere with vesicular trafficking such as sec4 and spo14 (Neiman, 1998; Rudge et al., 1998), suggesting a role for SPO21 in secretory pathway function during sporulation. Alternatively, a failure to form prospore membranes might result from spindle pole body defects (Davidow et al., 1980).
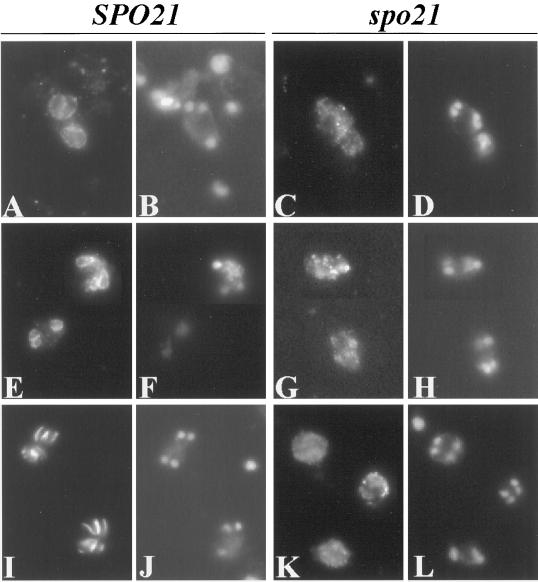
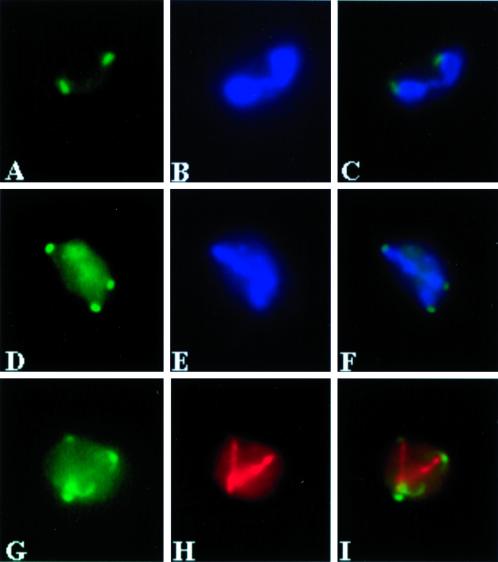
Figure 3.
No prospore membranes are formed in a spo21Δ mutant. Strains AN120 (SPO21/SPO21) and AN180 (spo21Δ/spo21Δ) were transferred to 2% KOAc and analyzed by indirect immunofluorescence as described in MATERIALS AND METHODS. (A, E, and I) AN120 stained with anti-Ssop, anti-Sncp, and anti-Spr3p antibodies, respectively. (B, F, and J) DAPI staining of the corresponding cells in A, E, and I. (C, G, and K) AN180 stained with anti-Ssop, anti-Sncp, and anti-Spr3p antibodies, respectively. (D, H, and L) DAPI staining of the corresponding cells in C, G, and K, demonstrating that the cells analyzed have undergone meiosis II. Images in E, F, G, H, K, and L are composites.
SPO21 Dosage Determines the Number of Spores Formed
Analysis of a partially functional allele of SPO21, generated in an effort to produce a Spo21-GFP fusion for localization studies, revealed that the pattern of spore formation is highly sensitive to the dosage of functional SPO21 (Table 1). A diploid carrying one copy of the spo21Δ allele and one copy of the spo21::GFP allele failed to sporulate, indicating that the spo21::GFP allele is largely nonfunctional. However, a strain homozygous for spo21::GFP did sporulate. Strikingly, this strain did not form tetrads but rather almost exclusively formed dyads.
Table 1.
Effects of SPO21 gene dosage on ascus formation
| Strain | Relevant genotype | Distribution of
ascal types (%)
|
||||
|---|---|---|---|---|---|---|
| % Asci | monad | dyad | triad | tetrad | ||
| AN180 | spo21Δ/spo21Δ | 0 | ||||
| AN231 | spo21∷GFP/spo21Δ | 0 | ||||
| AN230* | spo21∷GFP/spo21∷GFP | 12.6 | 0 | 97 | 3 | 0 |
| AN178* | SPO21/spo21Δ | 83 | 8 | 57 | 0 | 35 |
| AN229* | SPO21/spo21∷GFP | 79 | 1 | 47 | 0 | 52 |
| AN120 | SPO21/SPO21 | 90 | 0 | 19 | 1 | 80 |
Strains of the indicated genotype were scored for sporulation by light microscopy. For comparison of ascal types, at least 500 individual cells were scored.
The number of dyads and tetrads observed in strains AN230, AN178, and AN229 are all significantly different (χ2, p < 0.001) from that in AN120.
The SPO21/spo21::GFP heterozygote strain forms more dyads than wild type (Table 1). This observation could indicate a dominant negative effect of the spo21::GFP allele. However, dyad formation is also elevated in a strain heterozygous for a deletion of SPO21 (SPO21/spo21Δ). This result indicates that a single copy of SPO21 is insufficient to support full tetrad formation. Thus, the residual dyads formed in the SPO21/spo21::GFP strain are not due to a novel activity of Spo21-GFP protein but rather to an insufficiency of SPO21.
These data demonstrate a direct relationship between spore formation and SPO21 dosage. As SPO21 dosage decreases, more dyads and fewer tetrads are formed until exclusively dyads are formed (spo21::GFP/spo21::GFP). At even lower levels of SPO21 (spo21::GFP/spo21Δ) spore formation is blocked.
Nonsister Dyads Are Formed in the spo21::GFP Strain
If the dyads formed in the AN230 were NSDs, it would implicate SPO21 in spindle pole body function. To determine the nature of the dyads formed, the spo21::GFP/ spo21::GFP strain was examined in two ways. First, dissections were performed, and the segregation of the centromere-linked markers ARG4, RME1, and MAT was followed. If diploid (or aneuploid) spores are formed, then a significant fraction of the spores should carry both MATa and MATα information and be nonmaters. For >200 spore colonies scored, all mated as either MATa or MATα. Therefore, it is likely that these dyads contain haploid spores. Consistent with this idea, DAPI staining of spo21::GFP/spo21::GFP cells indicates that the cells complete meiosis and form 4 daughter nuclei even though only 2 spores are formed. For all three loci, a strong predominance of +/− dyads was observed (Table 2). For a marker completely linked to a centromere, all NSDs will be of the +/− class. However, if recombination occurs between the marker and the centromere, then +/+ or −/− class dyads can result from packaging of nonsister nuclei. For each of the markers examined, the observed number of +/+ and −/− class dyads was in good agreement with the expected frequencies for NSDs, based on the reported map distances between ARG4, RME1, MAT, and their respective centromeres. The observed distributions are also significantly different (χ2, p < 0.05) from the expected distribution from random packaging of haploid nuclei. These data indicate that the dyads formed in the spo21::GFP/spo21::GFP strain are probably NSDs.
Table 2.
Segregation of centromere-linked markers in spo21∷GFP/spo21∷GFP dyads
| Ascal Type | Locus
|
Random | ||
|---|---|---|---|---|
| ARG4 | RME1 | MAT | ||
| +/+ | 7 (3.8) | 3 (3.3) | 7 (7.7) | 16.5 |
| +/− | 88 (92.4) | 93 (93.4) | 86 (84.6) | 67 |
| −/− | 5 (3.8) | 4 (3.3) | 7 (7.7) | 16.5 |
The observed distributions for markers are shown. Numbers in parentheses are the expected distributions of each marker in NSDs based on the established map distances between each marker and the centromere (data from the Saccharomyces Genome Database http://genome-www.stanford.edu/Saccharomyces/). The expected distribution of ascal types for random packaging of haploid nuclei is also shown. The observed distributions are significantly different (χ2, p < 0.05) from random.
As a second method to analyze the dyads, sporulating cells of the spo21::GFP/spo21::GFP strain were fixed and stained with antitubulin antibodies to visualize the meiotic spindles and anti-Ssop to visualize the prospore membranes. Cells in meiosis II, defined by the presence of 2 meiotic spindles, never contained more than 2 prospore membranes. When 2 prospore membranes were evident, each was associated with a different meiotic spindle (Figure 4). These data not only indicate that the spores that develop from these prospore membranes are nonsisters, but that the NSDs in this strain are caused, as expected, by the failure of 2 of the 4 spindle poles to initiate formation of a prospore membrane. Taken together with the gene dosage results above, these data suggest that SPO21 is required for SPBs to initiate prospore membrane formation and, further, that the level of SPO21 determines how many SPBs become competent to do so.
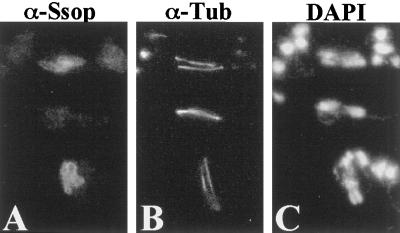
Figure 4.
Prospore membranes form on opposite spindles in a spo21::GFP strain. Strain AN230 (spo21::GFP/spo21::GFP) was transferred to 2% KOAc and analyzed by fluorescence microscopy as described in MATERIALS AND METHODS. (A) Anti-Ssop staining. (B) Antitubulin staining of the cells in A. (C) DAPI staining of the same cells.
SPB Outer Plaques Are Improperly Formed in spo21Δ Mutants
The results described strongly implicate SPO21 as a component or regulator of the meiotic spindle pole body. To determine if mutation of SPO21 alters SPB modification, strains AN120 (SPO21/SPO21) and AN180 (spo21Δ/spo21Δ) strain were transferred to KOAc medium. Cells were fixed and prepared for electron microscopy at time points in which, as judged by DAPI staining, >60% of cells were in meiosis II. At these times ≥75% of the SPBs present in the culture should be from cells in meiosis II. Fifteen SPBs were examined in each strain. All of the SPBs seen in the wild-type strain had a characteristic meiosis II morphology (Moens and Rapport, 1971; Byers, 1981), with a well-defined bilayer structure connected to the central plaque by amorphous material (Figure 5A). By contrast, all the SPBs in the spo21Δ strain displayed similar abnormalities in the outer plaque (Figure 5, B and C). The well-defined bilayer was absent, but amorphous material extending from the cytoplasmic surface of the central plaque was still evident. This observation suggests that if Spo21p is a component of the outer plaque, it might reside in the bilayer structure, the region of the outer plaque that contacts the prospore membrane.
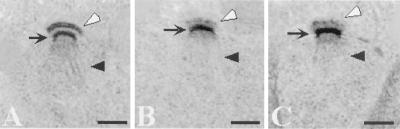
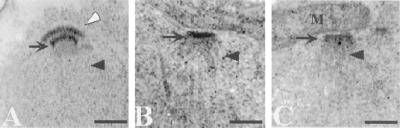
Figure 5.
SPO21 is required for proper modification of the outer plaque. Strains AN120 (SPO21/SPO21) and AN180 (spo21Δ/spo21Δ) were transferred to 2% KOAc and prepared for transmission EM as described in MATERIALS AND METHODS. (A) Wild-type meiosis II SPB. (B and C) Meiosis II SPBs in the spo21 mutant. Clear arrowhead, outer plaque; arrow, central plaque; dark arrowhead, spindle microtubules. Scale bar, 200 nm.
Spo21-GFP Localizes to the Meiotic Spindle Pole
To generate a functional, tagged allele of SPO21 for localization studies, a second SPO21-GFP fusion was constructed by conventional subcloning. When introduced into strain AN180 on a CEN vector, this second fusion, SPO21::GFP2, produced 30% sporulation, and 63% of the asci formed were tetrads. By comparison, the wild-type gene on the same CEN plasmid gave 50% sporulation with 78% of the asci tetrads. Thus, the SPO21::GFP2 fusion has nearly wild-type function. When the plasmid bearing this new fusion allele was introduced into strain AN230 (spo21::GFP/spo21::GFP), sporulation efficiency and the frequency of tetrads were comparable to strain AN120 (SPO21/SPO21). Therefore, AN230 carrying pRS316-SPO21::GFP2 was used to examine the localization of Spo21-GFP.
In sporulating cells, Spo21-GFP fluorescence was first visible after an ∼4-h incubation in sporulation medium and appeared as one or two discrete dots located near the periphery of the nucleus (Figure 6). In cells judged by DAPI staining to be in meiosis I, one or two Spo21-GFP foci were visible (Figure 6A). In meiosis II cells, three or four dots were present (Figure 6D). These Spo21-GFP spots were generally found near the tips of the segregating chromatin (Figure 6, A–F). Taken as a whole, this pattern is suggestive of localization to the two SPBs present during meiosis I and then to the four SPBs present in meiosis II. Consistent with this interpretation, labeling with antitubulin antibodies demonstrates that Spo21-GFP is located at the ends of the spindle (Figure 6, G–I). A similar pattern of localization was observed in strain AN180 (spo21Δ/spo21Δ) carrying pRS316-SPO21::GFP2 as the only source of Spo21p, indicating that the functional Spo21-GFP protein is localized at the SPBs.
Figure 6.
Spo21-GFP localizes to the spindle pole. Strain AN230 (spo21::GFP/spo21::GFP) carrying pRS316-SPO21::GFP2 was transferred to 2% KOAc and analyzed by fluorescence microscopy as described in MATERIALS AND METHODS. (A) Spo21-GFP in a meiosis I cell. (B) DAPI staining of the same cell in A. (C) Merged image of A and B showing localization of Spo21-GFP to the periphery of the nucleus. (D) Spo21-GFP foci in a meiosis II cell. (E) DAPI staining of the same cell in D. (F) Merged image of D and E showing Spo21-GFP near the leading edge of the segregating chromatin. (G) A meiosis II cell showing four Spo21-GFP foci. (H) Antitubulin staining of the same cell in G. (I) Merged image of G and H, demonstrating the Spo21-GFP foci are at the spindle poles.
Spo21p Localizes to the Poles of Opposite Spindles during NSD Formation
Wild-type strains can be induced to form NSDs instead of tetrads by transfer to water after a brief incubation in sporulation medium (Srivastava et al., 1983). This “interrupted sporulation” protocol was performed on AN230 carrying pRS316-SPO21::GFP2. Under these conditions, Spo21-GFP again initially appeared as two spots, but the subsequent appearance of cells containing four dots was reduced from 25% of cells to 4%. Labeling of these cells with antitubulin antibodies confirmed that the Spo21-GFP foci are located at the spindle poles and that the two spots, when present, are always on opposite spindles (Figure 7). A similar localization of Spo21-GFP to opposite spindles was seen when AN230 (spo21::GFP/spo21::GFP) was used to produce dyads under normal sporulation conditions (Bajgies and Neiman, unpublished observations). These observations support the inference that NSD formation is caused by the failure of Spo21p to localize to 2 of the 4 spindle poles.
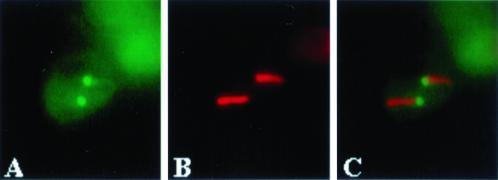
Figure 7.
Spo21p is found on the poles of opposite spindles in during NSD formation. Strain AN230 (spo21:: GFP/spo21::GFP) carrying pRS316-SPO21::GFP2 was subjected to interrupted sporulation and analyzed by fluorescence microscopy as described in MATERIALS AND METHODS. (A) Localization of Spo21-GFP. (B) Antitubulin staining of the cell in A. (C) Merged image of A and B showing localization of Spo21-GFP to the poles of opposite spindles.
The spo21 Sporulation Phenotype Is Distinct from cnm67
To determine if the effects of the spo21Δ mutation on prospore membrane formation are a more general consequence of perturbation of the outer plaque, we examined the sporulation phenotypes of cells lacking CNM67, which encodes a mitotic outer plaque component also required for sporulation (Brachat et al., 1998). A diploid strain homozygous for a deletion of CNM67 was transferred to sporulation medium, and the structure of the outer plaques of cells in meiosis II was examined by electron microscopy (Figure 8). Similar to what has been seen in mitotic cells (Brachat et al., 1998), the meiotic SPBs completely lacked outer plaque structures in the cnm67Δ mutant. The effect is somewhat more severe than spo21Δ in that the residual outer plaque material present in spo21Δ mutants is absent in cnm67Δ. These data suggest that Cnm67p is a component of the meiotic as well as the mitotic outer plaque.
Figure 8.
Outer plaques are absent in the cnm67 mutant. Strains AN120 (SPO21/SPO21) and AN161 (cnm67Δ/cnm67Δ) were transferred to 2% KOAc and prepared for transmission EM as described in MATERIALS AND METHODS. (A) Wild-type meiosis II SPB. (B and C) Meiosis II SPBs in the cnm67 mutant. Clear arrowhead, outer plaque; arrow, central plaque; dark arrowhead, spindle microtubules. M, mitochondrion; Scale bar, 200 nm.
To examine prospore membrane formation, the cnm67Δ/cnm67Δ mutant strain was sporulated and stained with antibodies to the Ssop and Sncp (Figure 9, A and C). In contrast to the spo21Δ mutant, a significant fraction of the cnm67Δ cells (10–20%) display intracellular rings consistent with prospore membrane localization of Ssop and Sncp. Unlike the situation in wild-type cells, these membranes are not associated with nuclei. EM studies of the cnm67Δ/cnm67Δ mutant also reveal prospore membranes, identifiable both by their shape and the characteristic knob structure at the membrane lip (Moens, 1971), that are apparently unassociated with nuclei (Figure 9F). These results distinguish the phenotypes of cnm67Δ and spo21Δ mutants and indicate that it is not the absence of a normal outer plaque per se that causes the failure of the spo21Δ mutant to form prospore membranes. Rather, there is a specific requirement for SPO21 for membrane formation.
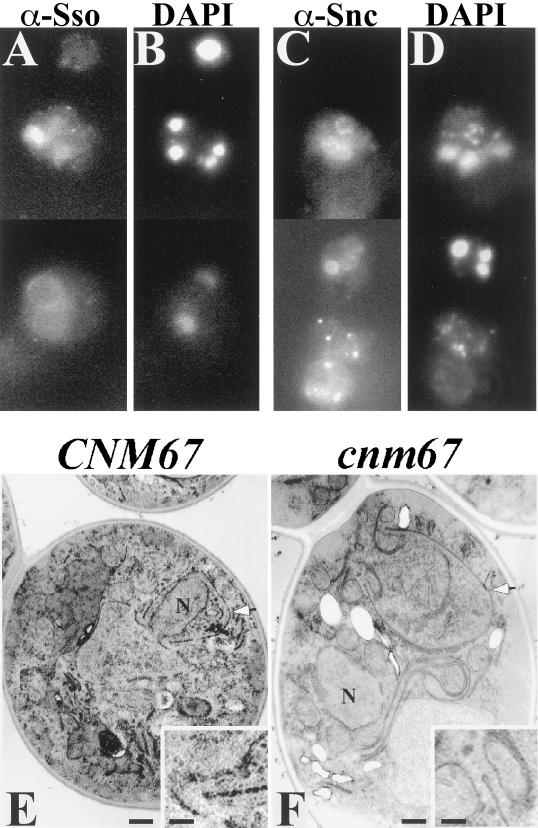
Figure 9.
Prospore membranes form in abnormal locations in a cnm67 mutant. Strain AN161 (cnm67Δ/cnm67Δ) was transferred to 2% KOAc and prepared for immunofluorescence and transmission EM as described in MATERIALS AND METHODS. (A) Anti-Ssop staining. (B) DAPI staining of cells in A. (C) Anti-Sncp staining. (D) DAPI staining of cells in C. (A–D) Composite images. (E) Transmission EM of a wild-type cell prepared by permanganate staining shows a prospore membrane engulfing a nuclear lobe (arrowhead). (F) A cnm67 mutant prepared similarly to the cell in E displays a prospore membrane (arrowhead) unassociated with nucleus (N). Insets: Higher magnification showing the characteristic knob on the end of the prospore membranes in E and F. Scale bars: large panels, 500 nm; insets, 150 nm.
Prospore Membrane Formation in cnm67 Strains Requires SPO21
If SPO21 is directly required for prospore membrane formation, then the residual prospore membranes formed in the cnm67 mutant should still depend on SPO21 function. To test this possibility, a spo21 cnm67 double mutant strain was constructed. This strain (AN254) was sporulated and prospore membrane formation was monitored by staining with anti-Sso antibodies (Table 3). A significant fraction (36%) of the cnm67 cells contain prospore membranes. The cnm67 spo21 double mutant, however, completely lacks these membranes. This demonstrates that SPO21 is required for prospore membrane formation even in the absence of obvious outer plaque structures.
Table 3.
Prospore membrane formation in spo21, cnm67, and spo21 cnm67 strains
| Strain | Relevant genotype† | No. of
prospore membranes observed (%)*
|
||||
|---|---|---|---|---|---|---|
| 4 | 3 | 2 | 1 | 0 | ||
| AN120 | SPO21 CNM67 | 75 | 3 | 3 | 0 | 19 |
| AN180 | spo21Δ | 0 | 0 | 0 | 0 | 100 |
| AN161 | cnm67Δ | 16 | 1 | 19 | 0 | 64 |
| AN254 | spo21Δ cnm67Δ | 0 | 0 | 0 | 0 | 100 |
Cells displaying tetranucleate DAPI staining were scored for the number of prospore membranes observed when counterstained with the anti-Sso antibody. At least 135 cells in each culture were counted.
DISCUSSION
SPO21 Encodes a Component of the Meiosis-specific Outer Plaque Necessary for Prospore Membrane Formation
Rearrangement of the secretory pathway is a common feature of cellular differentiation. In many instances this rearrangement involves the de novo formation of novel intracellular compartments (Flucher et al., 1991; Staehelin and Hepler, 1996). Prospore membrane formation provides an excellent model system for understanding how new membrane compartments are created. Although it has been known for some time that the meiosis-specific modification of the outer plaque of the SPB is important for initiating prospore membrane formation (Davidow et al., 1980; Okamoto and Iino, 1982), proteins specific to this structure have not been identified. Our observations that Spo21-GFP localizes to the spindle pole and that the outermost layer of the outer plaque is defective in spo21 mutants suggest that SPO21 encodes a component of the meiosis-specific SPB outer plaque. During the course of this work, an independent study (Knop and Strasser, 2000) identified Spo21p (called Mpc70p) and a second protein, Mpc54p as meiosis-specific components of the SPB. Importantly, immuno-EM localization of Mpc70p/Spo21p indicates that it is localized to the outermost layer of the outer plaque, consistent with a role in the formation of the prospore membrane.
Knop and Strasser (2000) also report that the appearance of Mpc70p/Spo21p at the spindle pole coincides with the disappearance of γ-tubulin complex binding protein Spc72p. In the vegetative outer plaque, Spc72p is found on the outermost surface (Adams and Kilmartin, 1999), analogous to the position Spo21p on the meiotic plaque (Knop and Strasser, 2000). Replacement of Spc72p with Spo21p could therefore account for the change in function of the outer plaque from microtubule nucleation to membrane attachment. In this light, the homology between the Spo21 and Spc72 proteins (Figure 1) may represent a common binding site for a component of the outer plaque present in both vegetative and meiotic cells such as Nud1p or Cnm67p (Brachat et al., 1998; Wigge et al., 1998; Knop and Strasser, 2000).
The finding that prospore membrane formation occurs in the cnm67Δ strain despite the absence of outer plaque structures indicates that the lack of prospore membranes in the spo21Δ mutant is not simply a consequence of a defective outer plaque. Rather, these data suggest a direct role for Spo21p in membrane biogenesis. The prospore membrane is proposed to form by the coalescence of secretory vesicles (Neiman, 1998). The initiating event in this process may be the homotypic fusion of two secretory vesicles with each other to produce the nascent prospore membrane. Factors such as Spo21p might be required to promote this specialized fusion event.
The Cnm67 protein binds to the central plaque component Spc42p (Adams and Kilmartin, 1999); thus, Cnm67p is likely present in the amorphous material connecting the meiotic outer plaque to the central plaque. Deletion of CNM67 might, therefore, lead to the release of an outer plaque complex containing Spo21p into the cytoplasm. Prospore membrane formation initiating on these “free” Spo21p complexes would lead to the generation of prospore membranes separated from the cell nucleus, as observed. This hypothesis is also consistent with the requirement of SPO21 for prospore membrane formation in the cnm67 mutant (Table 3). It should be noted, however, that we have not yet been able to see a discrete localization for Spo21-GFP in the cnm67 mutant (Bajgier and Neiman, unpublished observations).
SPO21 Is a Potential Target for Regulation of the SPB
A functional role for SPO21 in initiating membrane formation is also consistent with the SPO21 gene dosage effects. The sensitivity of the cell to SPO21 dosage is highlighted by the observation that the SPO21/spo21Δ strain shows a significant shift to dyad formation. A haplo-insufficient phenotype has also been reported for another SPB component, NDC1, during mitotic growth (Chial et al., 1999).
The fact that decreased dosage of SPO21 causes NSD formation, as opposed to randomly packaged spores as seen for other mutants that form dyads (Esposito et al., 1974; Uno et al., 1985) suggests that the cell is responding in a regulated manner to decreased SPO21 dosage. A regulatory mechanism is also indicated by the ability of the cell to control the number of spores formed, depending on the level of available nutrients. If the available carbon source is depleted early in sporulation only two SPBs will be modified, and cells will form NSDs instead of tetrads (Davidow et al., 1980; Srivastava et al., 1983).
The observation that Spo21p is localized to both spindle poles during meiosis I suggests that the assembly of expanded outer plaques on the 2 daughter SPBs formed at the onset of meiosis II occurs after assembly of the outer plaques on the mother SPBs. Consistent with this hypothesis, previous EM studies demonstrate an accumulation of electron-dense material on the outer plaque late in meiosis I (Moens and Rapport, 1971). When cells were triggered to form NSDs by interrupted sporulation, the initial localization to meiosis I SPBs occurred normally, but Spo21p was usually present on only 2 of the 4 meiosis II SPBs. A simple explanation for these observations is that during NSD formation, Spo21 fails to localize to the 2 new daughter SPBs formed at the start of meiosis II. Thus, each spindle would have at one end a modified, mother SPB and an unmodified daughter SPB. In this model, when SPB duplication occurs in the absence of nutrients, environmental signals block the assembly of Spo21p onto the outer plaque of the daughter SPBs formed at meiosis II.
Presumably, there exists an intracellular signaling pathway that monitors carbon source availability and regulates the decision to modify the daughter spindle poles. A signaling pathway mediated by the phosphatidylinositol kinase related genes TOR1 and TOR2 is involved in nutrient sensing in vegetative yeast cells (Thomas and Hall, 1997; Cutler et al., 1999). The immunosuppressive drug rapamycin inhibits signaling through the Tor kinases (Thomas and Hall, 1997; Cutler et al., 1999). Interestingly, treatment of sporulating cells with rapamycin leads to the accumulation of haploid dyads (Zheng and Schreiber, 1997). Although it has not been reported whether the dyads formed under these conditions are NSDs, this result raises the possibility that the Tor signaling pathway is responsible for mediating the decision to form dyads or tetrads. We propose that SPO21 expression or Spo21p activity is a target of the hypothetical signaling pathway, possibly the Tor pathway, which mediates the formation of NSDs.
Future studies will be needed to determine both whether SPO21 is regulated in response to nutrient depletion and how Spo21p contributes to the coalescence of vesicles initiating membrane formation. Thus, identification of SPO21 serves as an important first step toward understanding both the molecular mechanisms by which the SPB outer plaque contributes to membrane formation and how formation of the outer plaque is regulated in response to environmental cues.
ACKNOWLEDGMENTS
The authors thank N. Hollingsworth and R. Sternglanz for helpful discussion, N. Hollingsworth for comments on the manuscript, and R. Sternglanz for material support in the early stages of this work. The authors are also grateful to G. Rudomen for assistance with the electron microscopy, N. Hollingsworth, J. Pringle (University of North Carolina, Chapel Hill), and P. Brennwald (Cornell Medical College, New York) for antibodies, and M. Knop (Max Planck Institute, Martinsried) for communication of results before publication. This work was supported by National Institutes of Health grant GM62184 to A.M.N.
Abbreviations used:
- NSD
non-sister dyad
- SPB
spindle pole body
- DAPI
4′,6′-diamidino-2-phenylindole
REFERENCES
- Adams IR, Kilmartin JV. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J Cell Biol. 1999;145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvan P, Castle D. Sorting and storage during secretory granule biogenesis: looking backward and looking forward. Biochem J. 1998;332:593–610. doi: 10.1042/bj3320593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachat A, Kilmartin JV, Wach A, Philippsen P. Saccharomyces cerevisiae cells with defective spindle pole body outer plaques accomplish nuclear migration via half-bridge-organized microtubules. Mol Biol Cell. 1998;9:977–991. doi: 10.1091/mbc.9.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne RD, Morgan A. Regulated exocytosis. Biochem J. 1993;293:305–316. doi: 10.1042/bj2930305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B. Cytology of the yeast life cycle. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1981. pp. 59–96. [Google Scholar]
- Byers B, Goetsch L. Preparation of yeast cells for thin-section electron microscopy. Methods Enzymol. 1991;194:602–608. doi: 10.1016/0076-6879(91)94044-d. [DOI] [PubMed] [Google Scholar]
- Chial HJ, Giddings TH, Jr, Siewert EA, Hoyt MA, Winey M. Altered dosage of the Saccharomyces cerevisiae spindle pole body duplication, NDC1, leads to aneuploidy and polyploidy. Proc Natl Acad Sci USA. 1999;96:10200–10205. doi: 10.1073/pnas.96.18.10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Cutler NS, Heitman J, Cardenas ME. TOR kinase homologs function in a signal transduction pathway that is conserved from yeast to mammals. Mol Cell Endocrinol. 1999;155:135–142. doi: 10.1016/s0303-7207(99)00121-5. [DOI] [PubMed] [Google Scholar]
- Davidow LS, Goetsch L, Byers B. Preferential occurrence of nonsister spores in two-spored asci of Saccharomyces cerevisiae: evidence for regulation of spore-wall formation by the spindle pole body. Genetics. 1980;94:581–595. doi: 10.1093/genetics/94.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito MS, Esposito RE, Moens PB. Genetic analysis of two spored asci produced by the spo3 mutant of. Saccharomyces. Mol Gen Genet. 1974;135:91–95. doi: 10.1007/BF00264777. [DOI] [PubMed] [Google Scholar]
- Fares H, Goetsch L, Pringle JR. Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. J Cell Biol. 1996;132:399–411. doi: 10.1083/jcb.132.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flucher BE, Terasaki M, Chin HM, Beeler TJ, Daniels MP. Biogenesis of transverse tubules in skeletal muscle in vitro. Dev Biol. 1991;145:77–90. doi: 10.1016/0012-1606(91)90214-n. [DOI] [PubMed] [Google Scholar]
- Geissler S, Pereira G, Spang A, Knop M, Soues S, Kilmartin J, Schiebel E. The spindle pole body component Spc98p interacts with the gamma-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Jones AL, Fawcett DW. Hypertrophy of the agranular endoplasmic reticulum in hamster liver induced by phenobarbital (with a review on the functions of this organelle in liver) J Histochem Cytochem. 1966;14:215–232. doi: 10.1177/14.3.215. [DOI] [PubMed] [Google Scholar]
- Kane SM, Roth R. Carbohydrate metabolism during ascospore development in yeast. J Bacteriol. 1974;118:8–14. doi: 10.1128/jb.118.1.8-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Receptors determine the cellular localization of a gamma-tubulin complex and thereby the site of microtubule formation. EMBO J. 1998;17:3952–3967. doi: 10.1093/emboj/17.14.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Strasser K. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 2000;19:3657–3667. doi: 10.1093/emboj/19.14.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Lynn RR, Magee PT. Development of the spore wall during ascospore formation in Saccharomyces cerevisiae. J Cell Biol. 1970;44:688–692. doi: 10.1083/jcb.44.3.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall LG, Jeng RL, Mulholland J, Stearns T. Analysis of Tub4p, a yeast gamma-tubulin-like protein: implications for microtubule-organizing center function. J Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB. Fine structure of ascospore development in the yeast Saccharomyces cerevisiae. Can J Microbiol. 1971;17:507–510. doi: 10.1139/m71-084. [DOI] [PubMed] [Google Scholar]
- Moens PB, Rapport E. Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae (Hansen) J Cell Biol. 1971;50:344–361. doi: 10.1083/jcb.50.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag DK, Koonce MP, Axelrod J. SSP1, a gene necessary for proper completion of meiotic divisions and spore formation in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:7029–7039. doi: 10.1128/mcb.17.12.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J Cell Biol. 1998;140:29–37. doi: 10.1083/jcb.140.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM, Katz L, Brennwald PJ. Identification of domains required for developmentally regulated SNARE function. Genetics. 2000;155:1643–1655. doi: 10.1093/genetics/155.4.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM, Mhaiskar V, Manus V, Galibert F, Dean N. Saccharomyces cerevisiae HOC1, a suppressor of pkc1, encodes a putative glycosyltransferase. Genetics. 1997;145:637–645. doi: 10.1093/genetics/145.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenthal RK, Riles L, Johnston M, Hegemann J. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Iino T. Selective abortion of two nonsister nuclei in a developing ascus of the hfd-1 mutant in Saccharomyces cerevisiae. Genetics. 1981;99:197–209. doi: 10.1093/genetics/99.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S, Iino T. Genetic block of outer plaque morphogenesis at the second meiotic division in an hfd1–1 mutant of Saccharomyces cerevisiae. J Gen Microbiol. 1982;128:1309–1317. doi: 10.1099/00221287-128-6-1309. [DOI] [PubMed] [Google Scholar]
- Pearson BM, Hernando Y, Schweizer M. Construction of PCR-ligated long flanking homology cassettes for use in the functional analysis of six unknown open reading frames from the left and right arms of Saccharomyces cerevisiae chromosome. XV. Yeast. 1998;14:391–399. doi: 10.1002/(SICI)1097-0061(19980315)14:4<391::AID-YEA235>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Rose K, Rudge SA, Frohman MA, Engebrecht J. Phospholipase D signaling is essential for meiosis. Proc Natl Acad Sci USA. 1995;92:12151–12155. doi: 10.1073/pnas.92.26.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1990. [Google Scholar]
- Rossi G, Salminen A, Rice LM, Brunger AT, Brennwald P. Analysis of a yeast SNARE complex reveals remarkable similarity to the neuronal SNARE complex and a novel function for the C terminus of the SNAP-25 homolog, Sec9. J Biol Chem. 1997;272:16610–16617. doi: 10.1074/jbc.272.26.16610. [DOI] [PubMed] [Google Scholar]
- Rudge SA, Morris AJ, Engebrecht J. Relocalization of phospholipase D activity mediates membrane formation during meiosis. J Cell Biol. 1998;140:81–90. doi: 10.1083/jcb.140.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel SG, Snyder M. A highly divergent gamma-tubulin gene is essential for cell growth and proper microtubule organization in Saccharomyces cerevisiae. J Cell Biol. 1995;131:1775–1788. doi: 10.1083/jcb.131.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Geissler S, Grein K, Schiebel E. gamma-Tubulin-like Tub4p of Saccharomyces cerevisiae is associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J Cell Biol. 1996;134:429–441. doi: 10.1083/jcb.134.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spritz RA. Multi-organellar disorders of pigmentation: intracellular traffic jams in mammals, flies and yeast. Trends Genet. 1999;15:337–340. doi: 10.1016/s0168-9525(99)01785-0. [DOI] [PubMed] [Google Scholar]
- Srivastava PK, Harashima S, Oshima Y. Two-spored asci produced by interrupted sporulation: a novel approach to linkage analysis in yeast. Mol Gen Genet. 1983;191:165–166. doi: 10.1007/BF00330906. [DOI] [PubMed] [Google Scholar]
- Staehelin LA, Hepler PK. Cytokinesis in higher plants. Cell. 1996;84:821–824. doi: 10.1016/s0092-8674(00)81060-0. [DOI] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Thomas G, Hall MN. TOR signaling and control of cell growth. Curr Opin Cell Biol. 1997;9:782–787. doi: 10.1016/s0955-0674(97)80078-6. [DOI] [PubMed] [Google Scholar]
- Uno I, Matsumoto K, Hirata A, Ishikawa T. Outer plaque assembly and spore encapsulation are defective during sporulation of adenylate cyclase-deficient mutants of Saccharomyces cerevisiae. J Cell Biol. 1985;100:1854–1862. doi: 10.1083/jcb.100.6.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Jensen ON, Holmes S, Soues S, Mann M, Kilmartin JV. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol. 1998;141:967–977. doi: 10.1083/jcb.141.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XF, Schreiber SL. Target of rapamycin proteins and their kinase activities are required for meiosis. Proc Natl Acad Sci USA. 1997;94:3070–3075. doi: 10.1073/pnas.94.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]