Abstract
Purpose
To identify regional network covariance patterns of gray matter associated with Alzheimer’s disease (AD) and to further evaluate its replicability and stability.
Materials and Methods
This study applied a multivariate analytic approach based on scaled subprofile modeling (SSM) to structural MRI data from 19 patients with AD and 19 healthy controls (HC). We further applied the derived covariance patterns to examine the replicability and stability of AD-associated covariance patterns in an independent dataset [13 AD and 14 HC] acquired with a different scanner.
Results
The AD-associated covariance patterns identified from SSM combined principal components mainly involved the temporal lobe and parietal lobe. The expression of covariance patterns was significantly higher in AD patients than HC (t(36) = 5.84, p= 5.75E–7) and predicted the AD/HC group membership (84% sensitivity and 90% specificity). In replicability evaluation, the expression of the forward applied covariance patterns was still statistically significant and had acceptable discriminability (69% sensitivity and 71% specificity).
Conclusion
AD patients showed regional gray matter alterations in a reliable covariance manner. The results suggest that SSM has utility for characterizing covariant features, therefore, can assist us with further understanding covariance patterns of gray matter in AD based on the view of the network.
Keywords: multivariate analysis, scaled subprofile model, Alzheimer’s disease, structural MRI, voxel-based morphometry
INTRODUCTION
Alzheimer’s disease (AD) is a neurodegenerative disorder. It is typically characterized by progressive memory impairment, other associated cognitive decline, amyloid plaques, neurofibrillary tangles in multiple brain regions and regional and global brain volume atrophy (1–4). Structural magnetic resonance imaging (MRI) has long been used in the study of /AD, providing remarkable opportunities to investigate the morphological abnormalities in brain anatomy. Using different analytic approaches and measurement indices, a large number of structural MRI studies have demonstrated abnormal changes in gray matter (5–9). These studies consistently confirm that regional brain tissue atrophy starts at the medial temporal structures, including the bilateral hippocampus and entorhinal cortex, and subsequently extends to the posterior cingulate gyrus, medial thalamus and the parietal and frontal lobes with the advancement of the disease.
Measures from structural MRI have been proposed to serve as AD biomarkers (10,11). Global and/or regional brain morphometry information provided by structural MRI is abundant and could be used for the early diagnosis and quantitative evaluation of the disease progression of AD (12). The brain anatomic structure is a highly complex network system with different regions that are morphometrically interconnected and influenced by each other, and these interconnections are affected by aging, disease and other factors (6,13,14). Untill now, few structural MRI studies have identified AD-related anatomical changes based on the view of structural networks. Alterations of the covarying pattern in the regional brain networks, however, could play a vital role in understanding the patterns and features of AD pathology.
To date, numerous structural MRI studies have applied univariate analysis methods, such as manually traced or automatically defined regions of interest (ROI), or voxel-based morphometry (VBM) to investigate gray matter abnormalities in AD (5,7). Voxel-based or ROI-based univariate analysis approaches can only examine differences in peak voxels and provide local information but not the inter-regional covarying relationship of gray matter volumes among different brain regions. In contrast, multivariate analysis methods, such as scaled subprofile modeling (SSM) (15), view various brain regions as inter-related and coordinated nodes of an integrated network. SSM establishes a global index (subject score) that integrates the interregional covarying information over the entire brain volume on a voxel-by-voxel basis and that, at the same time, is free of multiple comparisons typically associated with univariate approaches (15,16). Thus SSM may capture the subtle changes and has greater sensitivity in examining the regional effects in brain tissue components (13). SSM has been widely and successfully used in brain functional imaging studies (17,18), and recently in structural MRI studies (13,19–21).
In the current study, we applied the SSM multivariate analytic approach to structural MRI data from patients with AD and healthy controls (HC). We aimed to identify regional network covariance patterns of structural MRI gray matter associated with AD and to further evaluate the replicability and stability of AD-associated covariance patterns in an independent dataset acquired with a different scanner.
MATERIALS AND METHODS
Participants And MRI Acquisition
Participants were recruited from two hospitals, Beijing Tiantan Hospital (Group 1) and Xuanwu Hospital of Capital Medical University (Group 2), Beijing, China, respectively.
In Group 1, there were 19 patients with AD [7 males and 12 females; ages between 55 and 81 years old (mean±SD: 65.2±8.3); mean Mini-Mental State Examination (MMSE): 11.7, range: 0–23] and 19 HC [12 males and 7 females; ages between 57 and 79 years old (67.8±6.9); mean MMSE: 29, range: 27–30]. The diagnosis of probable AD was made according to the International Statistical Classification of Diseases and Related Health Problems 10th Revision (ICD-10) criteria for dementia. The severity of cognitive impairment was assessed with the MMSE. HC had no cognitive complaints and did not have neurological or psychiatric disorders. The AD group did not differ significantly from HC in sex ratio (χ2(1) = 2.63, p= 0.105) or age (t(36) = 1.04, p= 0.110). MRI was performed with a 3.0T Siemens Trio Tim scanner. For each subject, whole-brain T1-weighted, sagittally oriented 3D anatomical imaging data were acquired using a magnetization-prepared rapid-acquisition gradient echo (MPRAGE) sequence (TR=2100 ms, TE=3.25 ms, TI=1200 ms, flip angle=10°, field of view=256×256 mm2, matrix size=256×256 and voxel size=1×1×1 mm3).
For Group 2, the detailed demographic information on participants and scanning parameters has been reported in our previous study (22). Thirteen patients with AD [6 males and 7 females; ages between 58 and 81 years old (72.1±6.5); mean MMSE: 18.5, range: 12–23] and 14 HC [6 males and 8 females; ages between 61 and 82 years old (70.4±3.5); mean MMSE: 28.5, range: 27–29] were included. Volumetric T1-weighted 3D MPRAGE anatomical MRI data were acquired on a 3.0T Siemens Trio Tim scanner for all subjects (TR=1900 ms, TE=2.2 ms, TI=900 ms, flip angle=9°, field of view=224×256 mm2, matrix size=448×512 and voxel size=0.5×0.5×1 mm3).
This current study was approved by the local Medical Research Ethics Committee. Written informed consent was obtained from all participants.
Image Preprocessing
The spatial preprocessing of structural MRI data was performed using SPM8 (http://www.fil.ion.ucl.ac.uk/spm) with a DARTEL-based VBM protocol (23,24). DARTEL, standing for “Diffeomorphic Anatomical Registration using Exponential Lie Algebra”, is a recently introduced algorithm for diffeomorphic image registration, running as a toolbox in SPM8. DARTEL utilizes a single constant velocity field to generate diffeomorphic and invertible deformations. The template creation and image registration are iteratively implemented and the structural MRI data for each subject are warped to a new template in each iteration. The first step of the DARTEL-based VBM procedure was to segment the native space images into rigid-body aligned gray and white matter images of the subjects (rigid-body aligned to the SPM8 default tissue probability maps). The second step imported gray matter images to create specific deformation fields by aligning gray matter to a gray matter template using the DARTEL registration algorithm. The third step used flow fields to separately warp the native gray matter images to the final template space and further Montreal Neurological Institute (MNI) space. In addition, the registered gray matter partitions were multiplied by the Jacobian determinants from the deformations to preserve the total amount of tissue in the native spaces. Finally, the gray matter maps for all subjects were smoothed with a 10 mm full width at half maximum (FWHM) Gaussian kernel.
SSM Analysis
Multivariate SSM analysis was performed for the DARTEL-based VBM processed gray matter volume images using the SSMPCA toolbox in MATLAB (http://www.feinsteinneuroscience.org). SSM was based on a modified principal component analysis (PCA), including 1) log-transformation, the initial data matrix (subjects by voxels) was logarithmically transformed; 2) global normalization, the voxel mean across the whole brain was substracted from the log-transformed data for each subject, and then the mean across subjects was subtracted to produce the subject residual profile (SRP) matrix, and 3) PCA analysis, a subject-by-subject covariance matrix was computed from the SRP matrix, and subsequently, PCA was performed to acquire subject scaling factors (SSF) (15,16). The images of both AD patients and healthy control subjects from Group 1 were included to produce a series of principle components (covariance patterns) and their corresponding SSF (subject scores). The SSF value indicates the relative degree of similarity of each subject’s gray matter map to the corresponding regional covariance patterns.
Akaike’s information criterion (AIC) was used to determine the optimal number of principal components (25). The set of principal components (SSF values) with the lowest AIC value was entered into a multiple linear regression model to predict AD/HC group membership. Covariance patterns that were significant as AD/HC predictors were identified with p ≤ 0.05. Based on the multiple regression results, the AD-related covariance pattern was created as a linear combination. Thus-generated covariance pattern was converted to a z-score map standardized by the standard deviation. Voxels with z-scores threshold of z ≥ |1.65| (p ≤ 0.05) were visually examined and used to reflect the related brain regions involved in the covariance network.
Application of the Covarying Patterns to an Independent Dataset: Replicability Evaluation
The covarying patterns that were first identified with SSM analysis based on data from Group 1 was forward applied to the subjects in Group 2 to evaluate the replicability and stability of AD-associated covarying patterns. The forward application is the procedure of calculating subject scores of Group 2 based on the derived covariance patterns from Group 1. These subject scores were mathematically implemented as the topographic profile rating (TPR) scores for subjects in Group 2. TPR scores, representing the degree to which the derived covariance patterns were manifest in an independent dataset, were obtained by multiplying the subject residual profile vector by the previously derived pattern vector (i.e. the dot product of these two vectors) (16). TPR scores were z-transformed to determine the sensitivity and specificity of discrimination classification for subjects in Group 2.
RESULTS
Generation of AD-Related Covariance Patterns
The optimal number of principal components was 10 according to AIC. The regression model including the SSF for the first 10 components significantly predicted the AD/HC group membership (F(10,27) = 4.42, p= 9.94E–4), and the first component was the most significant (t(27) = 3.89, p= 5.91E - 4). A linear combination of the first four significant components, accounting for 49% of the total variance, was the best discriminating pattern (F(4,33) = 7.82, p=1.51E – 4). The expression of covariance patterns was significantly higher in AD patients than in the HC group (t(36) = 5.84, p= 5.75E–7) (Fig. 1a). The receiver operating curve (ROC) analysis demonstrated discrimination classification with 84% sensitivity and 90% specificity (area under curve = 0.889, SE = 0.059, p= 4.10E–5) (Fig. 1b). Moreover, the expression of covariance patterns was significantly correlated with the MMSE scores in AD patients (r(19) =−0.479, p= 0.038).
Figure 1.
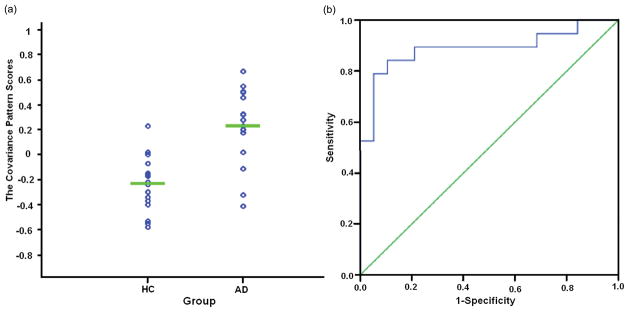
The statistical results based on AD-associated covariance pattern scores in Group 1. (a) The scatterplot of covariance pattern scores. The covariance pattern scores were calculated from the linear combination of the four significant principal components in 19 AD patients and 19 HC. The expression of covariance patterns was significantly higher in AD patients than in the HC group. (b) ROC curve of discriminability from the expression of covariance patterns.
Negative weights in the covariance pattern map represent reduced regional gray matter volumes in AD patients compared with HC, and vice versa. The gray matter voxel-based volume decreased mainly in the middle/inferior temporal gyrus, hippocampus, parahippocampal gyrus, inferior parietal lobule, putamen, caudate nucleus, insula, middle/superior frontal gyrus, and middle occipital gyrus and was relatively preserved mainly in the calcarine sulcus, postcentral gyrus, supplementary motor area, paracentral lobule, lingual gyrus and precentral gyrus (Fig. 2). Table 1 shows specific brain areas, Talairach coordinates and z values.
Figure 2.
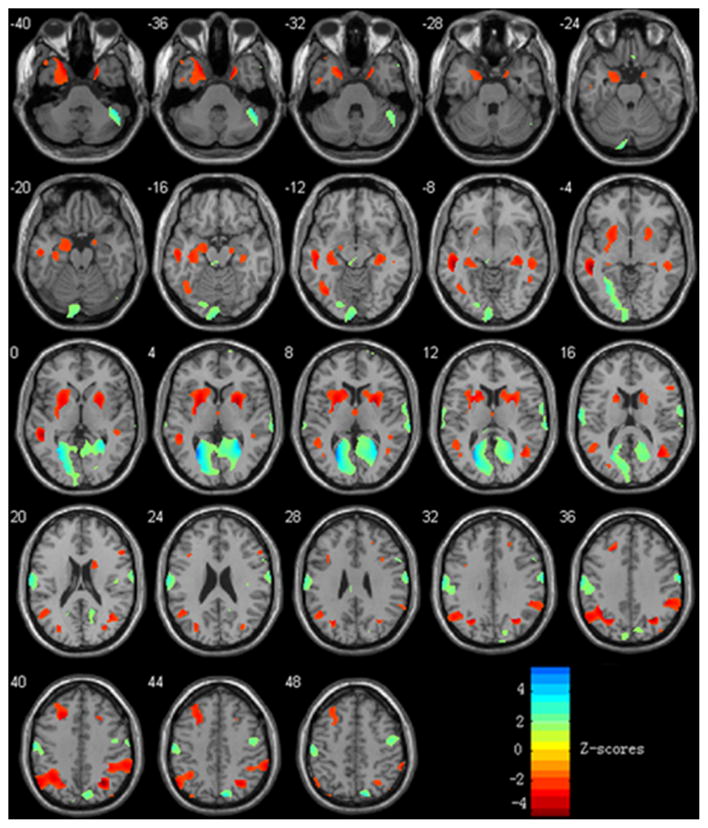
Covariance pattern map in gray matter volume in AD patients compared with HC in Group 1. Negative weights (“hot” colors) in the map represent reduced regional gray matter volumes in AD patients compared with HC, and positive weighs (“cold” colors) illustrate increases. The color bar represents the z scores, and the left of the plane is the left of the brain.
Table 1.
Specific brain areas in the covariance pattern map in AD patients compared to HC.
| Brain regions | Peak coordinates | z* | Cluster size (mm3) | ||
|---|---|---|---|---|---|
| Minima | |||||
|
| |||||
| L Middle Temporal Gyrus | −50 | −34 | −2 | −4.02 | 6,429 |
| R Middle Temporal Gyrus | 41 | −59 | 18 | −2.83 | 2,987 |
| L Middle Temporal Pole | −20 | 8 | −32 | −3.62 | 1,742 |
| L Inferior Temporal Gyrus | −41 | −63 | −6 | −2.15 | 965 |
| L Hippocampus | −34 | −20 | −12 | −2.26 | 3,061 |
| R Hippocampus | 35 | −26 | −5 | −2.38 | 1,495 |
| L Parahippocampal Gyrus | −19 | 4 | −27 | −2.51 | 1,573 |
| R Parahippocampal Gyrus | 21 | 4 | −31 | −2.62 | 749 |
| L Inferior Parietal Lobule | −32 | −54 | 37 | −3.50 | 6,379 |
| R Inferior Parietal Lobule | 30 | −57 | 38 | −4.24 | 5,755 |
| L Putamen | −23 | 3 | 5 | −3.32 | 4,273 |
| R Putamen | 26 | 4 | 6 | −3.93 | 2,879 |
| L Caudate Nucleus | −13 | 16 | 10 | −2.56 | 1,350 |
| R Caudate Nucleus | 15 | 12 | 16 | −2.23 | 1,458 |
| L Insula | −28 | 13 | 9 | −2.26 | 1,013 |
| R Insula | 32 | 13 | 4 | −2.23 | 665 |
| L Middle Frontal Gyrus | −23 | 25 | 34 | −3.19 | 2,612 |
| L Superior Frontal Gyrus | −22 | 25 | 34 | −3.25 | 709 |
| L Middle Occipital Gyrus | −26 | −61 | 34 | −3.34 | 1,455 |
| R Middle Occipital Gyrus | 30 | −58 | 38 | −4.36 | 1,090 |
|
| |||||
| Maxima | |||||
|
| |||||
| L Calcarine Sulcus | −26 | −63 | 10 | 5.55 | 9,383 |
| R Calcarine Sulcus | 29 | −51 | 8 | 4.49 | 5,832 |
| L Postcentral Gyrus | −66 | −10 | 19 | 3.08 | 7,530 |
| R Supplementary Motor Area | 12 | −4 | 69 | 3.04 | 5,758 |
| L Paracentral Lobule | −7 | −17 | 53 | 3.33 | 4,030 |
| L Lingual Gyrus | −22 | −63 | 5 | 3.76 | 4,010 |
| R Precentral Gyrus | 24 | −10 | 68 | 3.87 | 3,486 |
z*: p ≤ 0.05; L: left; R: right; coordinates in Talairach space.
Replicability Evaluation
For Group 2, the expression of the forward applied covariance patterns was significantly higher in AD patients than in the HC group (t(25) = 2.55, p= 0.009) (Fig. 3a). The ROC analysis demonstrated discrimination classification with 69% sensitivity and 71% specificity (area under curve = 0.747, SE = 0.096, p= 0.029) (Fig. 3b).
Figure 3.
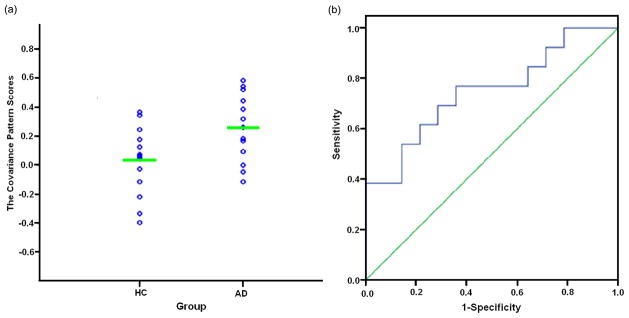
The statistical results of replicability evaluation according to the forward application in Group 2. (a) The scatterplot of AD-associated covariance pattern scores. (b) ROC curve of discriminability for the forward application.
DISCUSSION
The present study performed SSM multivariate analysis of structural MRI data to capture AD-related regional network covariance patterns. AD patients showed the distributed effects, such as gray matter volume decreases over multiple brain regions in a covarying manner. The regions with gray matter volume decreases in AD patients mainly included the middle temporal gyrus, hippocampus, parahippocampal gyrus, and inferior parietal lobule (Fig. 2). Additionally, relatively preserved regions were also observed and included mainly the calcarine sulcus, postcentral/precentral gyrus, supplementary motor area and paracentral lobule (Fig. 2). More importantly, the forward application of the derived pattern expression established in Group 1 confirms the replicability and stability of AD-associated covariance patterns in an independent sample.
AD-associated covariance pattern scores from SSM combined principal components were significantly higher in AD patients than in HC in both Group 1 (Fig. 1a) and Group 2 (Fig. 3a). Moreover, ROC analysis demonstrated discrimination classification with 84% sensitivity and 90% specificity in the pattern generation group (Fig. 1b), and it was still statistically significant and consistent but with somewhat lower sensitivity and specificity in the independent group (Fig. 3a and 3b). These statistical results suggest that the multivariate SSM technique could differentiate AD patients from healthy control subjects, as we expected.
The SSM findings of the interrelated gray matter volume reductions in AD patients from this current investigation are generally consistent with previous structural MRI studies using univariate voxel-based analysis methods (5,7,22). In particular, SSM detected the typically affected brain regions, such as the middle temporal gyrus, hippocampus, parahippocampal gyrus and subcortical nuclei, including the putamen and caudate nucleus in AD patients. Volume decreases in the hippocampus and parahippocampal gyrus are considered as the most valuable structural MRI biomarkers of AD pathology for prediction and early diagnosis (10,11,26). In addition, SSM also found that gray matter volume reductions within the parietal lobe involved the inferior parietal lobule. The pathological alteration of the parietal lobe has been of great importance in the development of AD, and the inferior parietal lobule may be more vulnerable than other parietal association areas (27). Together with previous studies, this study demonstrated that SSM detected the covarying patterns relevant to AD and could be used to characterize anatomical abnormalities in AD patients compared with HC.
Functional MRI (fMRI) studies demonstrated selective changes of the functional network in individuals at risk for AD or patients with mild AD (28,29). Greicius et al. found that resting-state default mode network (DMN) activity distinguished AD from healthy aging and confirmed that functional connectivity reflected structural connectivity in DMN (28,30). Seely et al. tested the network degeneration hypothesis in five different neurodegenerative syndromes and suggested that specific neurodegenerative diseases targeted distinct function-structure networks (31). It is also interesting to note that AD patients exhibited covariant volume reductions in gray matter, such as the inferior temporal gyrus, hippocampus, parahippocampal gyrus, inferior parietal lobule and middle/superior frontal gyrus, in DMN. The gray matter volume alterations in covarying patterns indicated that these regions were possibly in the same structural network involved in episodic memory and executive function impairment in AD (1). The anatomical structure serves as the brain function substrate, and changes in functional networks are possibly associated with abnormalities in structural networks (9,29). Therefore, investigations of brain structural networks help to construct a relevant neuroanatomy model and provide theory evidence and basis for studies on functional networks together (31).
This study also found some spare brain regions in which gray matter volumes were relatively preserved in AD patients compared with the normal controls. These regions were mainly in the calcarine sulcus, postcentral/precentral gyrus, supplementary motor area and paracentral lobule. Brain areas with preserved gray matter volumes were also observed in most univariate voxel-based structural MRI studies (7). The existing documents on the whole are in agreement with significant gray matter atrophy in the temporal and parietal lobes with sparing sensor and motor cortices (27,32).
One advantage of the SSM method is that each participant has a subject score representing its contribution to the expression of covariance patterns. This single score can be used free of the concern of using multiple comparisons to characterize AD differences (with regard to normal controls in this study) in the clinical setting. The correlation analysis showed that the subject scores were significantly negatively correlated with the MMSE scores in the AD group. Such negative correlation suggested that greater expression of the covariance patterns was associated with poorer clinical performance in AD patients. To make this subject score objective and reliable, its sensitivity and specificity need to be examined using an independent dataset. In the current study, a forward calculation was performed to evaluate the ability of the identified covariance patterns to differentiate AD patients from controls. Satisfactorily, AD patients in Group 2 demonstrated a significantly greater reflection of the covariance patterns established in Group 1 (Fig. 3a). It should be noted that the number of samples in Group 2 is small, the data were acquired from a different scanner and the subject scores had relatively greater variability. Nevertheless, ROC analysis still indicated discriminability with acceptable sensitivity and specificity (Fig. 3b).
In addition, we also conducted a univariate VBM analysis in Group 1. No voxels survived at the threshold of p ≤ 0.05 when corrected for multiple comparisons. When the significance level was set at uncorrected p ≤ 0.05, the locations of gray matter volume alteration in AD patients compared to HC were similar to the multivariate SSM based findings. The similarity of pattern is often used as a way to validate multivariate techniques such as SSM (13). The univariate approach can only examine differences in peak voxels, while multivariate SSM integrates covariance information and identifies gray matter volume changes across all voxels; moreover, the SSM subject scores do not need multiple comparison correction, while statistics based on the univariate approach do. The SSM method emphasizes the covariance information of imaging data and yields interrelated morphological features and a representation of the defined network patterns, thus SSM may detect the subtle regional alterations and has greater sensitivity (13,19). Our results from both univariate and multivariate methods confirmed this perspective.
The coordinated variations in brain anatomical regions are affected by various factors. In recent years, multivariate analytic techniques aiming at the interrelationship among brain regions, have been increasingly utilized to evaluate neural networks in different populations (6,13,19,31,33). A small number of structural MRI studies using SSM primarily focused on healthy aging (13,20,21). These studies successfully captured age-associated networks of regional covariance in structural MRI data in normal aging and provided an important basis for exploring the interrelationship of gray matter volume in AD in the present study. In contrast to reports on covariance patterns of normal aging, there are relatively few AD-related network studies in structural MRI and the covariance patterns and features of AD pathology are still poorly understood. Using a multivariate small world approach, He et al. found both decreases and increases in cortical thickness intercorrelations in the parietal and temporal cortex implying aberrant changes in cortical morphometry in AD patients (6). Alexander et al. investigated gray matter network associated with risk for AD using SSM in young to middle-aged adults (19). Our current study indicated that gray matter damage affected broad networks in AD.
In conclusion, AD patients showed regional gray matter alterations in a covarying manner. More importantly, the forward application suggests that the identified regional covariance patterns are reliable and generalizable. Although the size of the sample is relatively small in the current study, but is comparable to the other published SSM reports in structural MRI studies (19,20). Our results suggest that multivariate SSM analysis of structural MRI is a potentially suitable approach for identifying network-based AD biomarkers. We believe that network features contribute to explore the underlying architectures of complex brain networks in AD patients. SSM findings in this study can assist us with further understanding covariance patterns of gray matter in AD based on the view of the network.
Acknowledgments
We would like to thank Ms. Jiachao Liu and Ms. Li Liu for assistance with processing data.
Grant Support: This work was supported by the National Key Basic Research Program (973 Program), China (2012CB720704), National Natural Science Foundation (NNSF), China (81000603), Key Program of NNSF, China (60931003), the Funds for International Cooperation and Exchange of NNSF, China (61210001), the Fundamental Research Funds for the Central Universities, China; the National Institute of Mental Health, US (RO1MH57899), the National Institute on Aging, US (9R01AG031581-10, P30AG19610, k23AG24062), and the State of Arizona.
References
- 1.Buckner RL, Snyder AZ, Shannon BJ, et al. Molecular, structural, and functional characterization of Alzheimer’s disease: evidence for a relationship between default activity, amyloid, and memory. J Neurosci. 2005;25:7709–7717. doi: 10.1523/JNEUROSCI.2177-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furst AJ, Rabinovici GD, Rostomian AH, et al. Cognition, glucose metabolism and amyloid burden in Alzheimer’s disease. Neurobiol Aging. 2012;33:215–225. doi: 10.1016/j.neurobiolaging.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nestor PJ, Scheltens P, Hodges JR. Advance in the early detection of Alzheimer’s disease. Nat Rev Neurosci. 2004;5:s34–s41. doi: 10.1038/nrn1433. [DOI] [PubMed] [Google Scholar]
- 4.Petersen RC, Aisen PS, Beckett LA, et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010;74:201–209. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baxter LC, Sparks DL, Johnson SC, et al. Relationship of cognitive measures and gray and white matter in Alzheimer’s disease. J Alzheimers Dis. 2006;9:253–260. doi: 10.3233/jad-2006-9304. [DOI] [PubMed] [Google Scholar]
- 6.He Y, Chen Zh, Evans A. Structural insights into aberrant topological patterns of large-scale cortical networks in Alzheimer’s disease. J Neurosci. 2008;28:4756–4766. doi: 10.1523/JNEUROSCI.0141-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karas GB, Burton EJ, Rombouts SA, et al. A comprehensive study of gray matter loss in patients with Alzheimer’s disease using optimized voxel-based morphometry. NeuroImage. 2003;18:895–907. doi: 10.1016/s1053-8119(03)00041-7. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Wang Y, Wu G, et al. Discriminant analysis of longitudinal cortical thickness changes in Alzheimer’s disease using dynamic and network features. Neurobiol Aging. 2012;33:427.e15–427.e30. doi: 10.1016/j.neurobiolaging.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoub TR, deToledo-Morrell L, Stebbins GT, Leurgans S, Bennett DA, Shah RC. Hippocampal disconnection contributes to memory dysfunction in individuals at risk for Alzheimer’s disease. Proc Natl Acad Sci. 2006;103:10041–10045. doi: 10.1073/pnas.0603414103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ewers M, Sperling RA, Klunk WE, Weiner MW, Hampel H. Neuroimaging markers for the prediction and early diagnosis of Alzheimer’s disease dementia. Trends Neurosci. 2011;34:430–442. doi: 10.1016/j.tins.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hampel H, Frank R, Broich K, et al. Biomarkers for Alzheimer’s disease: academic, industry and regulatory perspectives. Nat Rev Drug Discov. 2010;9:560–574. doi: 10.1038/nrd3115. [DOI] [PubMed] [Google Scholar]
- 12.Vemuri P, Jack CR., Jr Role of structural MRI in Alzheimer’s disease. Alzheimers Res Ther. 2010;2:23–32. doi: 10.1186/alzrt47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brickman AM, Habeck C, Zarahn E, Flynn J, Stern Y. Structural MRI covariance patterns associated with normal aging and neuropsychological functioning. Neurobiol Aging. 2007;28:284–295. doi: 10.1016/j.neurobiolaging.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 14.Mechelli A, Friston KJ, Frackowiak RS, Price CJ. Structural covariance in the Human Cortex. J Neurosci. 2005;25:8303–8310. doi: 10.1523/JNEUROSCI.0357-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moeller JR, Strother SC, Sidtis JJ, Rottenberg DA. Scaled subprofile model: a statistical approach to the analysis of functional patterns in positron emission tomographic data. J Cereb Blood Flow Metab. 1987;7:649–658. doi: 10.1038/jcbfm.1987.118. [DOI] [PubMed] [Google Scholar]
- 16.Spetsieris PG, Eidelberg D. Scaled subprofile modeling of resting state imaging data in Parkinson’s disease: Methodological issues. NeuroImage. 2011;54:2899–2914. doi: 10.1016/j.neuroimage.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Habeck C, Foster NL, Perneczky R, et al. Multivariate and univariate neuroimaging biomarkers of Alzheimer’s disease. NeuroImage. 2008;40:1503–1515. doi: 10.1016/j.neuroimage.2008.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma Y, Tang C, Spetsieris P, Dhawan V, Eidelberg D. Abnormal metabolic network activity in Parkinson’s Disease: test-retest reproducibility. J Cereb Blood Flow Metab. 2007;27:597–605. doi: 10.1038/sj.jcbfm.9600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexander GE, Bergfield KL, Chen K, et al. Gray matter network associated with risk for Alzheimer’s disease in young to middle-aged adults. Neurobiol Aging. 2012;33:2723–2732. doi: 10.1016/j.neurobiolaging.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bergfield KL, Hanson KD, Chen K, et al. Age-related networks of regional covariance in MRI gray matter: reproducible multivariate patterns in healthy aging. NeuroImage. 2010;49:1750–1759. doi: 10.1016/j.neuroimage.2009.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brickman AM, Habeck C, Ramos MA, Scarmeas N, Stern Y. A forward application of age associated gray and white matter networks. Hum Brain Mapp. 2008;29:1139–1146. doi: 10.1002/hbm.20452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo XJ, Wang ZQ, Li KC, et al. Voxel-based assessment of gray and white matter volumes in Alzheimer’s disease. Neurosci Lett. 2010;468:146–150. doi: 10.1016/j.neulet.2009.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashburner J, Friston KJ. Voxel-based morphometry the methods. NeuroImage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 24.Ashburner J. A fast diffeomorphic image registration algorithm. NeuroImage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 25.Akaike H. A new look at statistical model identification. IEEE Trans Automat Contr. 1974;19:716–723. [Google Scholar]
- 26.Caroli A, Frisoni GB. The dynamics of Alzheimer’s disease biomarkers in the Alzheimer’s Disease Neuroimaging Initiative cohort. Neurobiol Aging. 2010;31:1263–1274. doi: 10.1016/j.neurobiolaging.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs HIL, Van Boxtel MP, Jolles J, Verhey FR, Uylings HB. Parietal cortex matters in Alzheimer’s disease: an overview of structural, functional and metabolic findings. Neurosci Biobehav Rev. 2012;36:297–309. doi: 10.1016/j.neubiorev.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Greicius MD, Srivastava G, Reiss AL, Menon V. Default mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI. Proc Natl Acad Sci. 2004;101:4637–4632. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sorg C, Riedl V, Mühlau M, et al. Selective changes of resting-state networks in individuals at risk for Alzheimer’s disease. Proc Natl Acad Sci. 2007;104:18760–18765. doi: 10.1073/pnas.0708803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greicius MD, Supekar K, Menon V, Dougherty RF. Resting-state functional connectivity reflects structural connectivity in the default mode network. Cereb Cortex. 2009;19:72–78. doi: 10.1093/cercor/bhn059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seely WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large-scale human brain networks. Neuron. 2009;62:42–52. doi: 10.1016/j.neuron.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fox NC, Crum WR, Scahill RI, Stevens JM, Janssen JC, Rossor MN. Imaging of onset and progression of Alzheimer’s disease with voxel-compression mapping of serial magnetic resonance images. Lancet. 2001;358:201–205. doi: 10.1016/S0140-6736(01)05408-3. [DOI] [PubMed] [Google Scholar]
- 33.Xu L, Groth KM, Pearlson G, Schretlen DJ, Calhoun VD. Sourec-based morphometry: the use of independent component analysis to identify gray matter differences with application to schizophrenia. Hum Brain Mapp. 2009;30:711–724. doi: 10.1002/hbm.20540. [DOI] [PMC free article] [PubMed] [Google Scholar]


