Summary
The neuronal growth cone, a highly motile structure at the tip of neuronal processes, is an excellent model system for studying directional cell movements. While biochemical and genetic approaches unveiled molecular interactions between ligand, receptor, signaling and cytoskeleton-associated proteins controlling axonal growth and guidance, in vitro live cell imaging has emerged as a crucial approach for dissecting cellular mechanisms of growth cone motility and guidance. Important insights into these mechanisms have been gained from studies using the large growth cones elaborated by Aplysia californica neurons, an outstanding model system for live cell imaging for a number of reasons. Identified neurons can be isolated and imaged at room temperature. Aplysia growth cones are 5–10 times larger than growth cones from other species, making them suitable for quantitative high-resolution imaging of cytoskeletal protein dynamics and biophysical approaches. Lastly, protein, RNA, fluorescent probes and small molecules can be microinjected into the neuronal cell body for localization and functional studies. The following chapter describes culturing of Aplysia bag cell neurons, live cell imaging of neuronal growth cones using differential interference contrast and fluorescent speckle microscopy as well as the restrained bead interaction assay to induce adhesion-mediated growth cone guidance in vitro.
Keywords: Neuronal growth cone, Aplysia californica, cell motility, cytoskeletal dynamics, live cell imaging, differential interference contrast, fluorescent speckle microscopy, restrained bead interaction
1. Introduction
Neuronal growth cones are highly motile structures at the tip of neuronal processes that detect molecular information in the environment via cell surface receptors during both development and regeneration. Various signaling cascades transduce this information from the plasma membrane to the underlying actin and microtubule cytoskeleton to affect the dynamic properties of these structures and ultimately lead to changes in speed and direction of growth cone movement (1–3). In vitro experiments using cultured neurons from various species have contributed significantly to our understanding of the molecular and cellular mechanisms of neuronal growth cone motility and guidance. The benefits of in vitro experiments include (1) working with an identified cell type, (2) the possibility of manipulating specific neurons in a temporarily and spatially defined manner, and (3) the capability of observing cell motility and protein dynamics by live cell imaging without the need for confocal or multi-photon fluorescence microscopy. Commonly used neuronal cell culture systems for growth cone studies include: retinal ganglion cells and spinal cord neurons from Xenopus embryos (4, 5), dorsal root ganglion neurons from chicken embryos (6), superior cervical ganglion neurons from rat and mouse embryos (7), hippocampal neurons from rat and mouse embryos (8), and bag cell neurons from adult Aplysia californica (9, 10).
Aplysia growth cones are 5–10 times larger than typical vertebrate growth cones, making them an excellent model system for high-resolution live cell imaging of protein dynamics, particularly of cytoskeletal proteins through the use of quantitative fluorescent speckle microscopy (FSM) (11–13). Other benefits of this system include the purity of neuronal cultures as well as the fact that neurons can be cultured and imaged without any serum or growth factors at room temperature. The large size of the growth cones and the room temperature compatibility make these cells suitable for biophysical manipulations, e.g. by micropipettes (14) and atomic force microscopy (15). Lastly, the large cell bodies of Aplysia neurons (50–100 µm) allow microinjection of various probes, including protein, RNA, fluorescent probes and small molecules. Thus, Aplysia growth cones represent one of the best model systems for studying growth cone motility and guidance in vitro.
To study directional growth cone movements various approaches of presenting soluble and substrate-bound guidance cues in a gradient or patterned fashion have been developed in recent years. A commonly used method of applying a gradient of a soluble factor is the so-called “pipette-gradient” assay, which involves pulsed ejections of diffusible small molecules or proteins from micropipette in a defined angle to the advance axis of the growth cone (16, 17). Different techniques have been developed to produce gradients of substrate-bound proteins, including filter application (18), diffuse printing (19), microcontact printing (20), microfluidics (21), and photoimmobilization (22). In vitro growth cone turning responses have also been studied with patterned substrates, using a variety of approaches to create stripe patterns of two different substrates, including microcontact printing (23), microfluidics (24), and silicon masks (25, 26). We have developed an assay referred to as “restrained bead interaction (RBI)” assay to investigate adhesion-mediated growth cone responses (14). This assay involves microbeads coated with adhesion proteins manipulated with a micropipette in order to study the transition of growth cone adhesion from one substrate to another. The large Aplysia growth cones are particularly suitable for this RBI assay. This chapter discusses preparation of cultured Aplysia bag cell neurons, live cell imaging of growth cone motility and cytoskeletal dynamics using differential interference contrast (DIC) and fluorescent speckle microscopy (FSM) of actin and microtubule dynamics, as well setting up the restrained bead interaction assay.
2. Materials
2.1. Aplysia bag cell neuronal culture
Leibovitz's L15 cell culture medium supplemented with artificial seawater (L15-ASW): L15 (powder, containing glutamine, Invitrogen #41300-039), pH 7.9, including 400 mM NaCl, 27 mM MgSO4, 28 mM MgCl2, 9 mM CaCl2, 4 mM L-glutamine, 50 µg/ml gentamicin, 5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES). Osmolarity should be 950–1000 mmol/kg. Filter through 0.22 µm sterile filter. Store at 4° C for up to 1 month (see Note 1).
Dispase II solution: 10 mg dispase II (Roche # 04942078001) dissolved in 900 µl L15-ASW plus 100 µl sterile H2O ultrapure. Prepare freshly right before dissection of ganglia. 1 ml of dispase II solution is sufficient for the digestion of 1–3 ganglia.
Magnesium chloride solution: 0.5 M MgCl2, prepared in H2O ultrapure. Store at 4° C.
6 N HCl for acid-cleaning of coverslips. To prepare mix 1 volume H2O ultrapure with 1 volume of 36% HCl.
60 ml syringe with Luer-lok tip (BD), 18.5 gauge sterile hypodermic needle, 2 inch long.
Microdissecting scissors: Vannas Spring Scissor, 8.5 cm long, 7 mm blade (WPI #500086); regular dissecting scissors; 2 pair of forceps: Dumont Tweezers #5 – 11cm long (WPI #500341).
Poly-L-lysine (10×): 200 µg/ml Poly-L-lysine (70–150 kD; Sigma #P-6282) stock solution is prepared in sterile H2O ultrapure. Store 1 ml aliquots at −20° C.
Aplysia californica, adult, 100–200 g, from Marinus Scientific or the National Resource for Aplysia at the University of Miami.
Coverglass: #1.5, 22×22 mm, acid-cleaned and stored in 100% ethanol.
35 mm Petri dishes.
Water bath; laminar flow bench; dissecting microscope (e.g. Leica MZ 9.5); 14° C incubator.
2.2. Live cell imaging using differential interference contrast (DIC) and fluorescent speckle microscopy (FSM)
Artificial seawater (ASW): 400 mM NaCl, 10 mM KCl, 55 mM MgCl2, 10 mM CaCl2, 15 mM HEPES, pH 7.8. Osmolarity should be 950–1000 mmol/kg. Filter through 0.45 µm sterile filter. Store at 4° C.
Imaging medium: ASW supplemented with 2 mg/ml BSA, 1 mg/ml L-carnosine and 25 mM vitamin E. Filter through 0.22 mm sterile filter. Prepare freshly on the day of the experiment.
Injection buffer: 100 mM 1,4-Piperazinediethanesulfonic acid (PIPES) pH 7.0, 1 mM MgCl2, 1 mM ethylene glycol tetraacetic acid (EGTA). Filter through 0.22 mm sterile filter. Store at 4° C.
G-buffer: 5 mM Tris pH 8.1, 0.2 mM CaCl2, 0.2 mM dithiothreitol, 0.2 mM ATP, 10% (w/v) sucrose. Filter through 0.22 mm sterile filter. Prepare freshly for dilution of G-actin.
Rhodamine tubulin (for microinjection): 1 mg/ml rhodamine-labeled bovine tubulin (Cytoskeleton, Inc, old Cat. # TL331M; new Cat. # TL590M) prepared in injection buffer, store at −80° C (see Note 2).
Alexa Fluor 488-phalloidin (for microinjection): 20 µM Alexa Fluor 488-phalloidin (Invitrogen # A12379) freshly prepared in injection buffer. Therefore, transfer a small amount (10 µl) of phalloidin dissolved in methanol into microcentrifuge tube, remove the methanol in a speed vacuum concentrator and reconstitute the phalloidin in injection buffer (see Note 2).
Alexa Fluor 568-G-actin (for microinjection): 1 mg/ml Alexa Fluor 568-G-actin (rabbit muscle actin; Invitrogen # A12374) freshly prepared in G-buffer; store stock aliquots of 7–8 mg/ml at −80° C (see Note 2).
Imaging chamber: We use a custom-made holder for an imaging chamber made by two coverslips separated by plastic spacers (Fig. 1).
Diamond pen; 22×2 mm plastic spacers (cut from 0.020" plastic shims, # SHSP-200 SmallParts, Inc.); nylon grommets (# CGN-250-02 SmallParts, Inc.) used as clips to hold top and bottom part of the holder together; high vacuum grease (Dow Corning); Sparkle glass cleaner; Q-tips.
Microinjection needles: borosilicated glass capillaries (1B100F-4, World Precision Instruments). Microloader tips (Eppendorf #930001007).
Vertical micropipette puller (Narishige PP830).
Micromanipulator: NP-2 patchman (Eppendorf); injection system: FemtoJet (Eppendorf).
Imaging set up: Various combinations of imaging hardware and software are possible. The following set up is used for most of our live cell imaging experiments (Fig. 1F): Eclipse TE2000E2 (Nikon) inverted microscope equipped with 10× objective for phase contrast imaging as well as 40× 1.3 NA oil, 60× 1.4 NA oil and 100× 1.45 NA oil immersion objectives for DIC imaging. Flat top motorized stage H117 ProScan™ (Prior) with linear encoders and joystick control. Electron-multiplying gain charge-coupled device (EMCCD) camera Cascade II (Roper Scientific). X-cite 120 metal halide lamp (EXFO Photonic Solutions, Inc). Lambda 10-3 high speed filter wheels and shutters (Sutter). Filter sets (all from Chroma) for rhodamine-tubulin: TRITC exciter (555/25nm) and emitter (HQ610/60nm) of a FITC/TRITC set; for Alexa Fluor 488-phalloidin: FITC exciter (485/20nm) and emitter (517/30nm) of a FITC/TRITC set; for Alexa Fluor 568-G-actin: TxRed exciter (575/15nm) and emitter (610/40nm) of a FITC/TxRed/Cy5 set. MetaMorph 7.0 software (Universal Imaging). PC with Core 2 Duo processor (3 GHz, Intel).
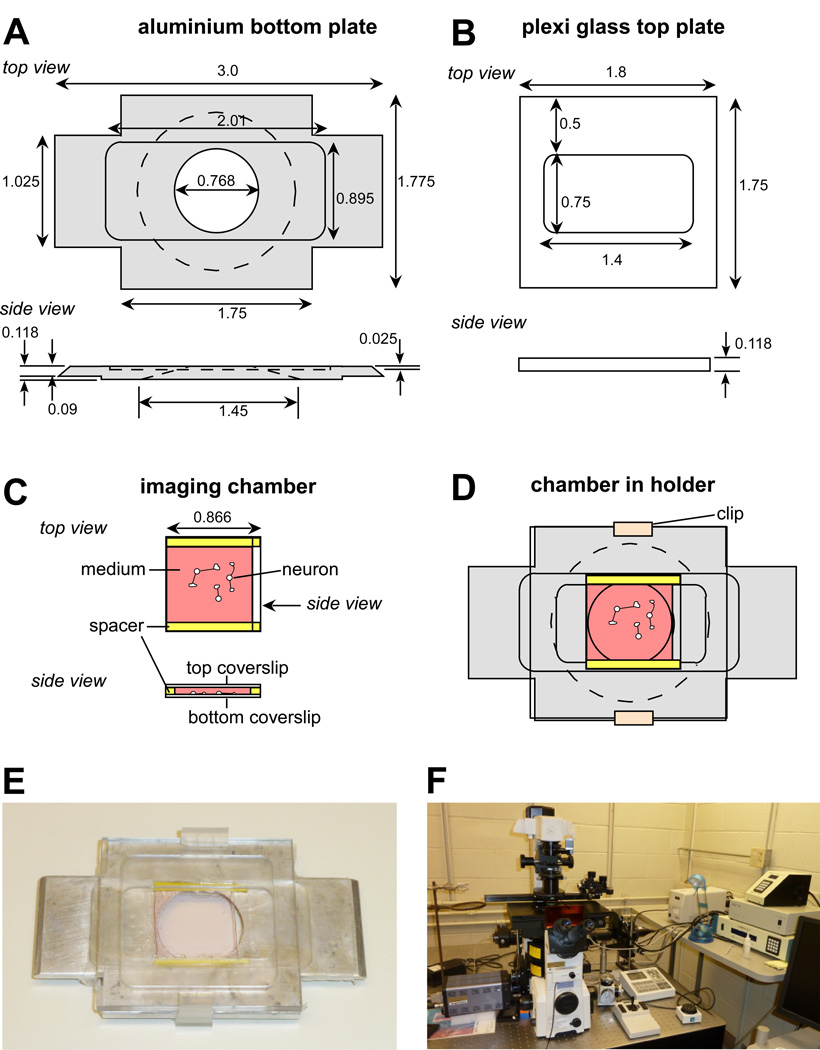
Figure 1. Imaging chamber used for live cell imaging of neuronal growth cones.
(A) Schematic drawing of bottom aluminium plate of custom-made holder used for the imaging chamber described in this chapter. Dimensions are in inches. (B) Plexiglass top plate of custom-made holder. (C) Imaging chamber made by two coverslips separated by two plastic spacers. The bottom coverslip contains the cultured neurons. Left and right side are open for medium exchange. (D) Imaging chamber mounted between bottom and top part held together by plastic clips. (E) Picture of an assembled chamber within the holder. (F) Live cell imaging workstation based on an Eclipse TE2000E2 (Nikon) inverted microscope as described in this article.
2.3. Protein coating of 5 µm silica beads for restrained bead interaction (RBI) assay
NTA silica beads (diameter of 5 µm); 5 % w/v in solution; Sicastar 43-11-503 (Micromod, Germany). Store at 4° C. Never freeze!
0.1 M Ni-sulfate. Store at room temperature (RT).
Phosphate buffered saline (PBS): 137.0 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4. Filter through 0.22 µm sterile filter. Store at 4° C.
6His-tagged recombinant protein used as adhesive substrate for growth cones, e.g. apCAM, 200–400 µg/ml in PBS.
Blocking solution for bead coupling: 5 mg/ml BSA in PBS. Filter through 0.22 µm sterile filter. Store at 4° C.
Refrigerated bench top centrifuge, e.g. Beckman-Coulter AllegraTM X-22R.
End-over end rotator, e.g. Barnstead/Thermolyne Rotisserie Labquake.
2.4. Restrained bead interaction (RBI) assay
L15-ASW containing 5 mg/ml BSA.
Phosphate buffered saline (PBS): 137.0 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4. Filter through 0.22 µm sterile filter. Store at 4° C.
Protein-coated 5 µm silica beads.
5 µl calibrated pipettes (#2-000-001 Drummond Scientific Company).
Vertical micropipette puller (Narishige PP830).
Narishige 3D hydraulic micromanipulator.
Custom-made open imaging chamber with aluminum base and brass inset (Fig. 3).
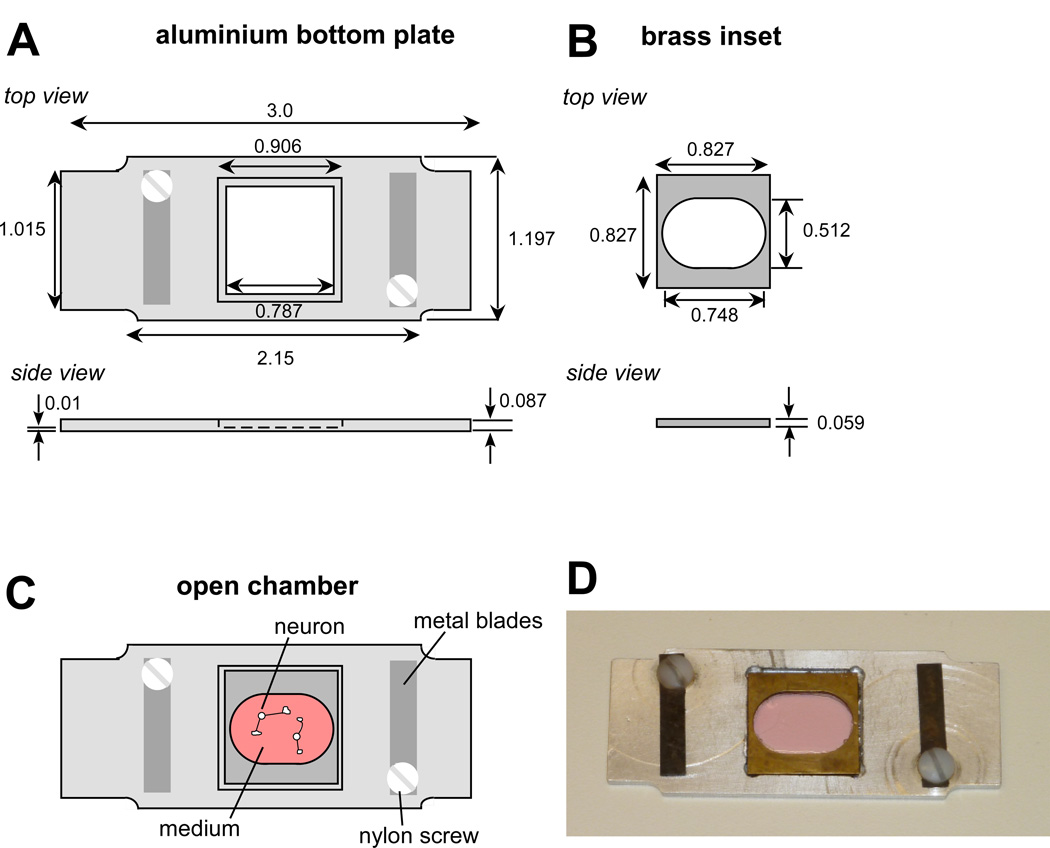
Figure 3. Open imaging chamber used for RBI growth one guidance assay.
(A) Schematic drawing of bottom aluminium plate of custom-made holder used as open imaging chamber. The chamber is open to the top allowing accessing cultured cells via micropipettes. Dimensions are in inches. (B) Brass inset to be mounted on coverslip with cells using high vacuum grease. (C) Assembled open chamber. Brass inset with coverslip attached is positioned into the bottom plate. Metal blades can be used to hold tubing for perfusion of medium. (D) Picture of a fully assembled open chamber.
3. Methods
3.1. Aplysia bag cell neuronal culture
Individual identified Aplysia neurons can be isolated and cultured alone or together with other neurons to form small circuits in vitro, e.g. between sensory and motor neurons (27). The following protocol describes culturing bag cell neurons derived from the abdominal ganglion of adult Aplysia. The main function of the neurosecretory bag cell neurons is to produce an egg laying pro-hormone and related peptides in response to electrical stimulation. We use bag cell neurons to investigate molecular and cellular mechanisms of growth cone motility and guidance; however, other neurons such as sensory neurons derived from the pleural ganglia can be used for this purpose as well. Video protocols for preparing cultures of bag cell (10) and sensory/motor neurons (28) have recently been published in the Journal of Visualized Experiments.
Transfer 10 mg of dispase II into a microcentrifuge tube and add 900 µl L15-ASW and 100 µl sterile H2O ultrapure. Gently mix the solution until the dispase is completely dissolved and keep the solution at RT during dissection.
Fill a 60 ml syringe with 50 ml of 0.5 M MgCl2 and attach an 18.5 gauge needle. Use a styrofoam box cover as dissection area. Sterilize regular dissecting scissors, microdissecting scissors and two forceps with 70% ethanol and let air dry.
Take an adult Aplysia from the tank and inject all of the MgCl2 solution into the body cavity right behind the head. Keep the animal on the styrofoam board during the injection. Gently rub the animal to spread the MgCl2 solution throughout the body. Wait until animal is completely anesthetized and does not show any signs of muscle contractions (1–3 min; see Note 3).
Pin the head and tail of the animal to the dissection board using two needles, head facing to the right.
Take one forceps to lift up the skin and body wall muscle. Open the body wall with regular dissecting scissors without damaging internal organs. Cut the body wall open along the side towards the head and the tail.
Locate the abdominal ganglion towards the mantle and gill. Use Fig.4.4 in (29) as a reference. Use new forceps to lift up the connecting nerves just rostrally to the ganglion and cut the connecting nerves rostrally and caudally to the abdominal ganglion with microdissecting scissors. Be sure not to damage the ganglion. The two bag cell clusters are attached rostrally to the larger hemiganglia (Fig.4.7 in (29)). Residual nervous system tissue can be collected if needed. The residual body of the animal is stored at −20° C before incineration.
Transfer the ganglion into a microcentrifuge tube with 1 ml of dispase II solution. Digest the tissue for 15–16h at 22°C using a temperature-controlled water bath (see Note 4).
On the following day, bag cell plating is performed in a laminar flow bench. Prepare three 35 mm Petri dishes filled with 4 ml of L15-ASW each for dissecting the bag cell clusters. Transfer the digested abdominal ganglion to one of these dishes using either a 20 µl pipette or sterile forceps. Make sure medium temperature has been adjusted to RT.
- Preparing poly-lysine-coated coverslips:
- Use acid-cleaned #1.5 square glass coverslips (22×22 mm; see Note 5 regarding acid-cleaning), stored in 100% EtOH. Take the coverglass out of the storage container using sterile forceps and briefly flame it with a Bunsen burner before placing it onto the edge of an uncovered empty 35 mm Petri dish. Alternatively air dry the coverglass without flaming. Depending on the number of neurons plated per coverglass, 6–12 is a reasonable number coverslips to prepare for bag cells derived from one animal.
- Prepare enough 20 µg/ml poly-L-lysine solution by diluting 200 µg/ml stock solution 10 times with sterile H2O ultrapure. Once the coverslip is dry, place it inside the Petri dish and coat it with 0.5 ml of 20 µg/ml poly-L-lysine solution. Incubate for at least 20 min at RT (see Note 6).
- Wash each coverglass three times with 0.5 ml of sterile H2O ultrapure. Remove solutions with vacuum. After the last wash add 4 ml of L15-ASW into each dish.
Clean microdissecting scissors and two forceps with 70% EtOH. Separate the two bag cell clusters from the rest of the abdominal ganglion while observing them with a dissection microscope. Therefore, first cut the abdominal ganglion into two halves right through the center along the rostral-caudal axis. Then, cut each cluster from the hemiganglion and the residual nerve. Be sure not to damage the clusters with the dissecting tools and to keep the clusters and cells in medium all the time.
Use two forceps to remove the connective tissue sheath from each cluster one at a time. Therefore, hold the cluster with one forceps while pushing against the connective tissue sheath with the second forceps. Try to find an opening where the bag cell neuronal cluster pops out through the connective tissue sheath. Ideally separate the sheath from the cellular cluster as one piece. The bag cell cluster will appear as a “berry-like” structure.
Use a forceps to bend a yellow P20 pipette tip that has been briefly (1–2 sec) warmed up over a Bunsen burner flame. Be careful not to melt the tip. The tip is bent at a 45 degree angle to facilitate the positioning of the tip during cell plating. Use the bent tip to transfer the cluster into a new dish with L15-ASW (see Note 7).
Triturate the cluster with the yellow tip to remove individual bag cells. Therefore, gently suck the cluster into the tip and push it out. Start out with low shear forces to avoid killing
Pick up 3–5 healthy cells with medium and transfer them onto the first poly-lysine-coated coverslip. Avoid picking up dead cells or pieces of extracellular tissue.
Repeat steps 13 and 14 and place about 20–40 live neurons into the center of the coverslip. The number of cells depends on the experiment; thus, plate more for microinjections since some cells will not survive the injections. Pay attention to the positioning of the cells because it will be impossible to image cells that are located close to the edge of the coverslip when using oil immersion objectives and the sample holder described here. Remove extracellular debris and dead cells from the coverslips by picking them up with the pipette or gently blowing them away from healthy cells.
After all cells from one cluster have been plated, start removing the sheath of the second cluster and proceed with cell separation and cell plating as for the first cluster. The whole plating procedure will take between 1h 30 min and 2 h 30 min depending on the number of cells. The size of the bag cell clusters varies between animals. A large cluster contains about 300–400 neurons. A good plating typically results in about 200–300 live neurons from one animal.
After all healthy neurons have been positioned onto the coverslips, keep the culture dishes for at least 2 hours at RT to allow cells to attach. Avoid any potential vibrations during this time. Then, place the dishes into an incubator at 14 °C overnight or until further use. Growth cones develop at the tips of cut neuronal processes or emerge from cell bodies within 4–10 hours after plating. We use the cells within the first 48 h after plating for most experiments, since growth cones get smaller with time in culture.
3.2. Live cell imaging using differential interference contrast (DIC) and fluorescent speckle microscopy (FSM)
3.2.1 DIC imaging
Inspect the cells on the day after cell plating using a tissue culture microscope. Perform a half volume medium change with fresh L15-ASW that has been adjusted to RT. If growth cones have not yet developed during the overnight incubation at 14 °C, one can speed up the process by keeping the cultures at RT for 1–2h.
For live cell imaging, place the coverslip with the cells in either custom-made or commercial imaging chamber, for example the Series 20 platform from Warner Instruments. We use a custom-made chamber system originally designed and developed by Paul Forscher and Stephen Smith at Yale University (9). This system is relatively simple and consists of a sandwich chamber made from two coverslips mounted between an aluminium bottom plate and plexiglass top plate (Fig. 1A–B). This system allows exchange of solutions on the microscope while imaging with both high numerical aperture oil immersion objectives and condenser.
To make the imaging chamber, use a diamond pen and ruler to cut off a two millimeterwide stripe of a clean square coverslip (22×22 mm; Fig. 1C). Cutting off this stripe will facilitate the exchange of solution through the chamber. Fill a 1 ml syringe with high vacuum grease and attach a yellow pipette tip to the syringe for grease application (see Note 8). Apply grease along each of the shorter sides of the cut cover glass. Then, cut two 22×2 mm plastic spacers from 0.020" yellow plastic shims and press the spacers onto the applied grease using the wooden end of two Q-tips.
Apply grease on top of each of the spacers. Using forceps lift up the coverslip/spacer assembly and invert it onto a coverslip with cultured neurons within a Petri dish. Align the inverted coverslip with the one containing the cells and push it down gently using the wooden end of two Q-tips. Make sure the two coverslips stick well together before gently lifting the sandwich out of the medium using forceps (Fig. 1C; see Note 9).
Align the two coverslips before positioning the sandwich over the round hole of the bottom part of the sample holder. The slightly lowered inner portion of the aluminum bottom part is covered with a piece of parafilm with a cut out center hole. Align the sandwich well before mounting the plexiglass top part. Use plastic clips cut from nylon grommets on each side to hold the top and bottom part together (Fig. 1D; see Note 10).
Wipe off any excess medium. Clean each cover glass with a Q-tip soaked with Sparkle glass cleaner. Quickly dry off any of the glass cleaner using a dry Q-tip. Clean glass surfaces are essential to high-quality imaging, particularly using differential interference contrast (DIC) optics.
Mount the chamber holder onto the microscope stage of an inverted microscope. We use an Eclipse TE2000E2 (Nikon; Fig. 1F). Use the 10× phase objective to locate individual bag cells. Since the number of neurons is relatively low (~20–40) compared to other neuronal culture systems, it is helpful to use a motorized stage with memory function to mark positions of individual cells.
After marking the cell body positions, switch the objective to a high resolution DIC oil immersion objective, e.g. 60×. Set up the microscope for DIC imaging and perform Koehler illumination. Search for an appropriate growth cone for imaging.
The optimal camera settings will depend on the type of CCD camera. We use either a Cascade II or a CoolSnap HQ (both from Roper Scientific) and normally select an exposure time of 50 msec for DIC imaging. Timelapse sequences of growth cone dynamics are acquired in 5–10 sec intervals, typically for 10–30 min (Fig. 2A and D). Minimize the light exposure to the amount needed for imaging without negatively impacting growth cone dynamics (see Note 11).
Make sure that the chamber is always filled with medium since evaporation will occur overtime. Exchange with fresh medium (1–3 ml) at least every 30 min by adding medium into one side of the chamber while removing the medium on the other side with a vacuum line and a yellow pipette tip. Pay attention to possible leaks of solution onto the objective.
Using spatial calibrations and image analysis software, analyze image stacks for various parameters of growth cone dynamics, including rates of filopodia and lamellipodia growth (Fig. 2).
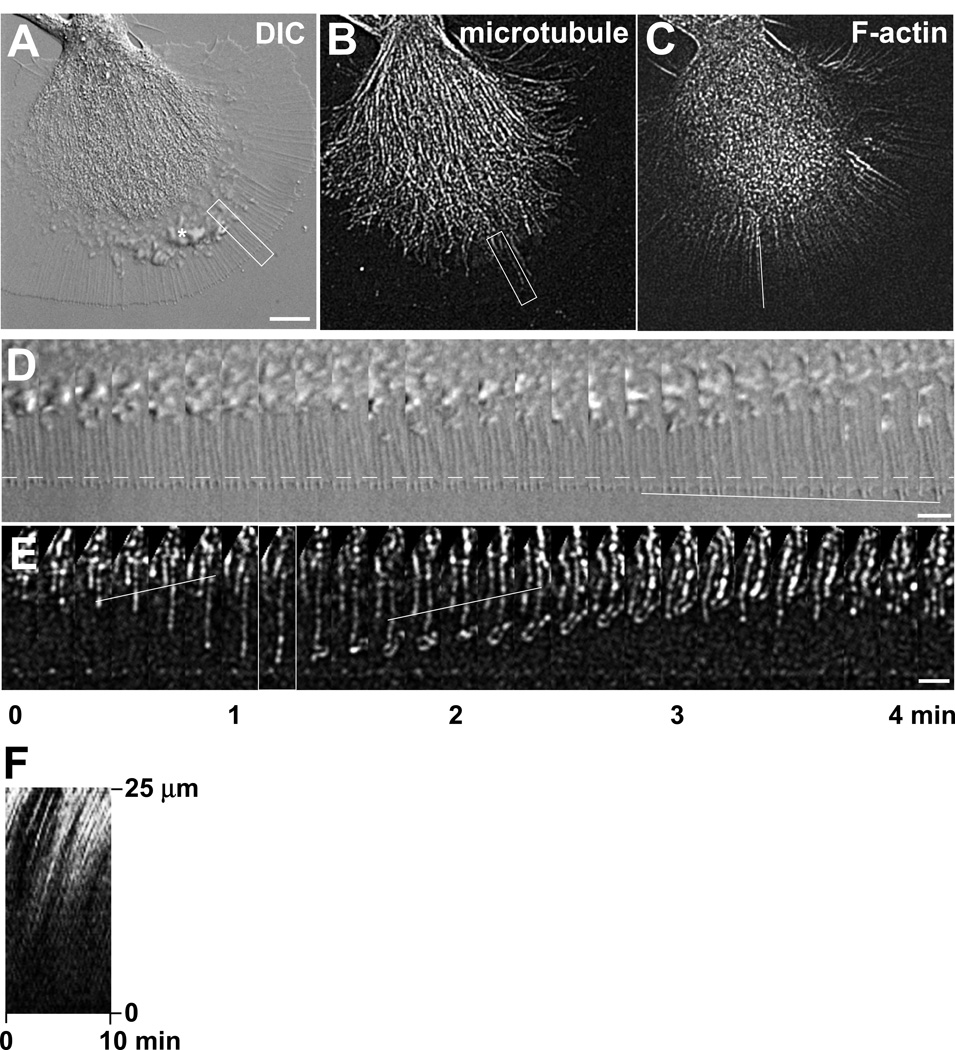
Figure 2. Aplysia bag cell growth cone visualized by DIC and actin/microtubule FSM.
(A) DIC image of live Aplysia bag cell neuronal growth cone on poly-L-lysine. White box (5 by 25 µm) demarks region of interest in the peripheral domain used for time lapse montage shown in (D). (B) Microtubule FSM image of the same growth cone. Rhodamine-labeled tubulin was injected into the neuronal cell body according to the protocol 3.2.2, resulting in a “speckled” labeling of individual microtubules. White box (5 by 25 µm) demarks region of interest different from the one shown in (A) and used for time lapse montage shown in (E). This region contains a dynamic microtubule with fluorescent marks. The corresponding time point shown in image (B) is marked in (E) with a white border. (C) Actin FSM image of the same growth cone using Alexa 488 phalloidin labeling. Note that F-actin-labeling with phalloidin highlights actin bundles in the peripheral and central domain, but not highly dynamic actin structures such as ruffles between the central and peripheral domain (indicated by an asterisk in (A)). White line demarks position of line scan used for kymograph shown in (F). (D) DIC time lapse montage of region of interest indicated in (A) imaged at 10 sec intervals. During the time period of 4 min and 10 sec, the leading edge and filopodia slowly advance from the initial position marked by the dashed line. The solid line demarks the tip position of a filopodium growing at a rate of 1 µm/min. (E) Microtubule FSM time lapse montage of region of interest indicated in (B). The position of the leading edge is at the bottom. This montage shows a fast growing microtubule that loops around and undergoes retrograde flow. The two solid white lines indicate internal reference speckles moving retrogradely. (F) Kymograph of F-actin signals scanned along the white line shown in (C) over a time period of 10 min. The slope of these lines indicates the rate of retrograde actin flow. Scale bars: 10 µm in (A)–(C); 5 µm in (D)–(E).
3.2.2 Actin and microtubule FSM imaging
Pull needles for microinjection of cytoskeletal probes. A tip opening of 1 µm is suitable for injections into bag cell neuronal cell bodies. We use borosilicate glass capillaries and perform a double pull with all weights on the vertical puller PP830 using the heat setting “65” for the first pull and “53” for the second pull. Inspect the pulled needles on a microscope for desired tip shape before storing or using them.
Prepare cytoskeletal probes for microinjection as described in section 2.2 steps 5–7.
Clarify cytoskeletal probes by centrifugation at 10,000 g for 30 minutes at 4°C before microinjection. This reduces the clogging problem when performing microinjections (see Note 12).
Prepare your microinjection set up. We use the FemtoJet system for controlled pressure application. Start up the FemtoJet following the instructions of the manufacturer. Turn on micromanipulator if you use a motorized one. We use an NP-2 patchman. Position the microneedle holder in a 40 degree angle to the stage.
Position the Petri dish containing a coverslip with cultured bag cell neurons onto the stage. Search for position of individual neurons using a 10× phase objective and mark the cell positions.
Using microloader tips, backfill injection needles with a small amount of cytoskeletal probe (see Note 13).
Connect the microneedle to the needle holder. Keep compensation pressure at ~30–40 hPa.
Quickly lower the injection needle into the medium. Position the tip of the needle in the field of view without touching the surface. Switch to a 40× long working distance phase objective for microinjection and position the cell body of the neuron into the center of the field of view.
Check either by phase or fluorescence optics whether there is continuous flow coming out of the pipette. A small “cloud” of fluorescent material should appear in front of the tip. If not the case, increase the compensation pressure or clear out the clog by pressing the “clean” function on the FemtoJet (see Note 12).
Position the needle tip adjacent to the cell body using the fine speed setting of the NP-2 micromanipulator. Inject the cell body by moving the needle tip quickly into an area of cytoplasm avoiding the nucleus. A fast movement increases the chance of actually penetrating the plasma membrane. An injection will result in a visible local expansion of cell volume. Leave needle in cell for ~ 1–3 seconds and then quickly move needle out since a too large injection volume can kill the cell. Inject an estimated volume of no more than 10% of the cell body. Compensation pressure is usually sufficient for injections; if needed use an increased injection pressure.
To make sure the cell body was injected, switch to fluorescent mode using an appropriate fluorescent filter. A clearly injected cell will immediately exhibit fluorescent signals in the cell body that can be seen with the eye piece (see Note 14).
After all cell bodies in one dish are injected, place the dish in a light-protected area for the cells to recover. Once all dishes are injected, carefully exchange half of the L15-ASW medium in each dish. Let the injected cells recover for at least 1 h in the dark before starting the imaging.
Assemble an imaging chamber as described under steps 3–6 of protocol 3.2.1 and exchange the L15-ASW with imaging medium.
Mark cell positions with 10× phase objective before switching to 60× or 100× oil immersion lenses in order to identify suitable growth cones for imaging.
Set up the appropriate illumination and acquisition parameters suitable for the injected fluorescent probes. We use a TRITC exciter (555/25nm) and emitter (HQ610/60nm) of a FITC/TRITC set for rhodamine-tubulin and a FITC exciter (485/20nm) and emitter (517/30nm) for Alexa Fluor 488-phalloidin. Reduce the fluorescence intensity with neutral density filters (see Note 15). The gain setting and exposure time will depend on the camera used. We typically use 500 msec exposure, gain 3 and additional EM gain on the Cascade II camera. Time intervals between images are typically 5–10 sec. Healthy neurons can be imaged up to 1h under such conditions. It is recommended to exchange fresh imaging medium every 30 min, or perfuse with medium throughout the experiment. Make sure the sample remains in focus during timelapse imaging and perform the necessary adjustments.
Image processing and data analysis: Raw images will show labeled microtubules and actin bundles; however, only after appropriate image processing one can clearly see the speckles on the filaments (Fig. 2). We use MetaMorph 7.0 software for image processing, quantitative analysis of microtubule and actin dynamics as well as for making movies and montages. To enhance speckles, microtubule and actin images are processed with spatial filters in the following sequence: (1) Low pass 4 × 4; (2) Laplace 2 edge enhancement; (3) Low pass 3 × 3. Select clearly labeled microtubules or actin bundles for quantitative speckle analysis (see Note 16). Make a montage and movie of the selected filament and make sure that the appropriate spatial calibration is applied to the montage (Fig. 2E). To measure microtubules assembly rate, measure the distance between a newly added speckle at the microtubule plus end and an internal previously added speckle and divide by the time passed between the two speckle additions. To measure translocation rate, measure the distance an internal speckle moves between two time points and divide by the time. Enter the data into an Excel spreadsheet. For final rate calculation we time-weigh individual rate measurements; thus, rates are multiplied by the time during which they are measured, added up and divided by the total observation time. To measure percentage of time spent in one dynamic behavior, add up all the relevant times during which the behavior is observed and divide the sum by the total time. For actin retrograde flow analysis, use the kymograph function of MetaMorph and draw a line along the lines of speckle movements (Fig. 2F). The slope of this line will directly indicate the rate of actin flow. Calculate average values, standard error of the mean and significance values in Excel. To get reliable data, it is advised to analyze at least 30 filaments from 10 different growth cones in 2–3 independent experiments.
3.3. Protein coating of 5 µm silica beads for restrained bead interaction (RBI) assay
The following protocol describes the preparation of microbeads coated with recombinant Aplysia cell adhesion protein, apCAM. These beads can be used to induce adhesion-mediated neuronal growth as described in protocol 3.4. Other adhesion molecules and protein coupling methods can be used as well.
Transfer 0.1 ml of 5 µm NTA silica beads (5% weight/volume stock solution) into a 1.5 ml microcentrifuge tube (see Note 17).
Wash beads 3× with 1 ml sterile H2O ultrapure. Therefore, resuspend beads with wash solution and sediment the beads by centrifugation at 20,000×g, 4°C for 5 min using a refrigerated table-top centrifuge.
Resuspend bead pellet with 0.1 ml of 0.1 M Ni-sulfate and incubate for 30 min at RT by placing tube on rotating device, such as an end-over-end rotator, in order to keep the beads in suspension.
Transfer 20 µl of Ni-NTA beads (at 5% w/v) into a new tube and wash 2× with 1 ml H2O ultrapure as described in step 2.
Wash beads 2× with 1 ml sterile PBS sterile as described in step 2.
Resuspend bead pellet with 100 µl purified 6His-tagged protein, such as apCAM (250–400 µg/ml in PBS), and incubate on rotator overnight at 4° C.
Sediment the beads by centrifugation at 20,000×g, 4° C for 5 min. Transfer supernatant containing free apCAM protein solution into separate tube.
Resuspend bead pellet with 100 µl blocking solution for bead coupling (5 mg/ml BSA in PBS sterile) and incubate for 30 min at RT on rotator.
Spin the bead solution as above, remove free blocking solution and add free apCAM ligand solution back for storage of beads (see Note 18). These apCAM-coupled beads can be used for the RBI assay described in the next section. Beads that were not incubated with apCAM can be used as controls.
3.4. Restrained bead interaction (RBI) assay to study growth cone guidance
Culture Aplysia bag cell neurons on poly-lysine-coated coverslips overnight as described in protocol 3.1.
To prepare apCAM beads for RBI experiments, remove any unbound free ligand from the bead solution. Therefore, pellet the beads at 20,000×g, 4°C for 5 min and transfer free apCAM ligand into separate tube.
Resuspend apCAM bead pellets once with 500 µl sterile PBS and spin at 20,000×g, 4°C for 5 min; then resuspended beads in 100 µl PBS.
Prepare a 1:500 dilution of washed beads in experimental medium, such as L15/ASW 5 mg/ml BSA (in case of DIC imaging) or imaging medium (in case of actin and microtubule FSM imaging) by adding 1 µl beads into 500 µl medium. 50 µl diluted beads are typically sufficient for one open chamber. Spin residual bead stock, remove PBS supernatant and resuspend beads in apCAM solution for storage.
Pull microneedles with short shanks using 5 µl calibrated pipettes. We use a double pull (heating settings 61/54) on the PP830 puller. The tip should be no more than 1 µm wide and closed. If it is open, it can be fire-polished with a microforge.
Identify a coverslip containing several neurons elaborating healthy and large growth cones. We use an open chamber system containing an aluminum base plate in slide format and a brass inset to hold the coverglass (Fig. 3A and B). The design shown in Fig. 3 is slightly different from the original one made by Paul Forscher (Yale University) as it uses square 22×22 mm coverslips as opposed to round coverslips (14). Apply vacuum grease generously to the bottom side of the brass inset (Fig. 3B) and mount the brass inset onto the coverslip with cells within the Petri dish filled with medium. Use forceps to lift the brass inset together with the coverslip and medium out of the dish.
Align the brass inset well with the coverslip, wipe off excess medium on the bottom side and apply some vacuum grease along the edges on the bottom side of the coverslip to attach the inset to the aluminum plate. Position the brass inset together with the cells into the aluminum plate (Fig. 3C and D) and wipe off an extra medium. Clean glass coverslip on the bottom side as described in step 6 of protocol 3.2.1.
Exchange L15-ASW with L15-ASW containing 5 mg/ml BSA (if performing DIC imaging only) or with imaging medium (if performing both DIC and FSM imaging). Mount open chamber onto motorized stage and mark cell positions with 10× phase objective.
Position the microneedle into the center of the field of view with a relatively flat angle (25 degree to stage) using a hydraulic micromanipulator (see Note 19). Avoid touching the surface as the needle may break.
Switch to 60× oil immersion DIC objective and search for large growth cones with distinct cytoplasmic domains that are suitable for the RBI assay.
Add 50 µl of diluted beads into the center of the chamber. Identify a bead in vicinity of the growth cone. Using the micromanipulator, position the bead close to the leading edge of the growth cone. This can be achieved by picking up the bead with the microneedle or by moving it over the coverslip. One can position the bead at various places along the leading edge of the growth cone; however, orienting the bead-growth cone axis perpendicular to the microneedle provides the best control of the bead during the RBI assay (Fig. 4).
Set up the program for time lapse imaging. Using the microneedle, move the bead onto the leading edge of the growth cone (Fig. 4B). If the protein on the bead binds to cell surface receptors that can couple to the actin flow, the bead will undergo retrograde translocation. Position the microneedle tip behind the bead to block its movement and start the time lapse program taking images every 10 sec (see Note 20). Replace medium every 30 min or set up a continuous perfusion system because of evaporation.
During the initial phase of bead restraint, called “latency” phase (Fig. 4B), there are hardly any structural changes that can be observed; during the later phase, termed “interaction” or “traction” phase (Fig. 4C), the central domain including microtubules extend towards the restrained bead (12, 14). This is accompanied by attenuation of retrograde actin flow, tension increase and protrusion of the leading edge specifically along the growth cone-bead axis (Fig. 4C). These cytosolic and cytoskeletal rearrangements mimic events observed during growth cone interactions with cellular targets. Parameters to analyze are the percentage of successful bead interactions and the latency time. A high percentage of interactions and short latency time indicate a favorable growth condition. This assay can be combined with live cell imaging of fluorescently labeled proteins, such as actin/microtubule FSM (12), as well as immunocytochemistry (14).
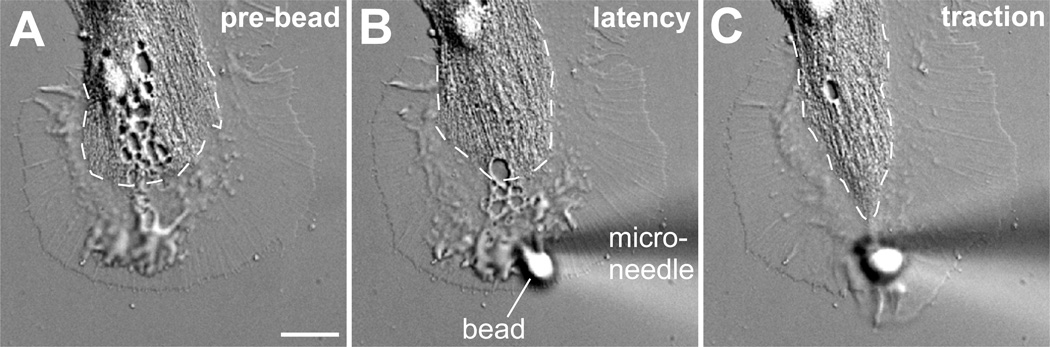
Figure 4. Restrained bead interaction assay.
(A) DIC image of an Aplysia growth cone on poly-lysine substrate. Dashed line indicates the central domain boundary. (B) The same growth cone is shown shortly after an apCAM-coated bead is positioned onto the peripheral domain close to the leading edge and restrained with a microneedle against retrograde translocation. This image is taken at the beginning of latency phase during which the coupling between apCAM and F-actin is weak. (C) As coupling strength increases, the growth cone undergoes rearrangements, such as central domain extension towards the bead and leading edge growth in front of the bead. Scale bar: 10 µm.
Acknowledgements
I would like to thank Aih Cheun Lee for contributing the images for Figures 2 and 4. I am also grateful to Aih Cheun Lee and Yingpei He for excellent suggestions on this chapter. Work in the Suter lab is supported by grants from the NIH (R01 NS049233) and the Bindley Bioscience Center at Purdue University to D.M.S.
Footnotes
Use L15-ASW within a month or add additional glutamine after one month because of its degradation in aqueous solutions.
We have used the cytoskeletal probes mentioned in section 2.2; however, both Cytoskeleton Inc. and Invitrogen sell tubulin, actin and phalloidin conjugated to different fluorochromes as well. Alternatively, tubulin and actin can be purified and labeled in the lab (30). Stock solutions of labeled tubulin and G-actin are prepared in the buffers described in section 2.2., and small aliquots (1–5 µl) are stored at −80°C after drop-freezing in liquid nitrogen. An aliquot is typically used only once but could be refrozen once and used a second time.
Inking and body wall muscle contractions can make the dissection more difficult. Both can be avoided most of the time by gentle handling of the animal and firm injection procedure. Avoid getting ink into the body cavity while dissecting the animal. Make sure to penetrate the needle through the body wall muscle into the cavity without damaging internal organs.
The activity of the dispase, the time of digestion and temperature will determine how effectively the tissue is digested. The goal is to loosen the fibrous connective tissue sheaths without killing many neurons. The optimal amount of enzyme depends on its specific activity and needs to be established for each batch. We use a heating water bath placed into a cold room to maintain 22° C.
Acid cleaning of coverslips can be done ahead of time. Incubate coverslips in 6 N HCl for at least 1 h, followed by thorough washing with H2O ultrapure. Make sure that coverslips do not stick together during the acid-incubation. After extensive washing with water transfer the coverslips with forceps into a new container with 100% EtOH for sterilization and storage at RT.
Poly-L-lysine is a non-physiological adhesive substrate that works excellent for plating Aplysia neurons. Growth cones are fan-shaped and neurites grow at a rate of 5–10 µm/h on this substrate (Fig. 2). Poly-lysine-coated coverslips can also be stored dry.
Keep the bag cell clusters and cells within the medium during the whole dissection procedure. Avoid air bubbles attaching to the cluster as they may lift up the cluster to the medium/air interface which will damage the cells.
To fit the yellow pipette tip onto the 1 ml syringe, use a razor blade to cut off the wider end of the tip. Make sure the tip is tightly attached before filling it with grease.
The assembly needs to be done with the coverslips fully covered with medium. Avoid getting the cells exposed to air. Make sure to press the coverslips firmly against each other without breaking them. It is okay if the vacuum grease is pushed outside the spacers. This indicates a well-sealed chamber.
It is essential that the two coverslips are well aligned with each other as well as with the groove of the aluminum bottom part before the top part is mounted. Uneven positioning will generate local pressure after applying the clips and result in breaking the coverslips.
Light intensity can be controlled by the lamp voltage, neutral density and color filters. For DIC live cell imaging of growth cones, we often use a 590 nm cut off filter to avoid the shorter wavelength light spectrum. Make sure to shut off the illumination between taking images either by using a transillumination shutter or by reducing the lamp voltage.
Clogging of the injection needle is one of the biggest challenges in performing microinjections. Samples should be spun before injection. If clogging persists, try the following: a) spin sample at higher g forces; b) coat needles with hexamethyldisilazane (HMDS, Pierce Chemical) by placing a few drops of volatile HMDS on the bottom of a needle-holding box and allowing the vapor to coat needles overnight in a fume hood; c) clear out clog by increasing the pressure of the microinjection system.
Filling about 1 cm of the narrow microloader tip provides a sufficient amount for injecting several cells. Make sure not to touch the bottom of the tube where protein aggregates could have accumulated. Gently tap needle to remove any potential air bubbles from the solution in the microneedle. Alternative to pulling injection needles: commercial injection capillaries such as Femtotips (Eppendorf). Alternative to microloader tips: long and thin pulled glass capillaries.
The key to successful injections is using minimal pressure to yield a high percentage of injected cells without killing them. As the probability of clogging increases with time, it is important to perform the injections as quickly as possible to avoid the need for continuously increasing the pressure or frequently exchanging the needle.
To minimize phototoxicity use at least one neutral density filter and a minimally-open field diaphragm in the fluorescent light path.
Select clearly labeled filaments for quantitative speckle analysis. For example, an ideal microtubule has clear speckles against background, does not cross over with other microtubules or ruffling structures containing a lot of free tubulin, and preferably has distinct growth/collapse phases.
This protocol describes non-covalent coupling of recombinant 6His-tagged protein to Ni-NTA silica beads. There are many other ways of coupling proteins to different types of microbeads in oriented and non-oriented manner, covalently and non-covalently.
Always store the beads (1% w/v) in ligand solution (such apCAM in PBS) at 4°C. Otherwise the density of the protein on the beads will gradually decrease over time since the 6His-Ni-NTA binding has an on- and off-rate.
We found that a non-motorized, hydraulic micromanipulator works best for well-controlled bead manipulations without any vibrations.
The RBI assay is vibration-sensitive. It is critical to minimize any vibrations by using an optical table, detaching shutters from the microscope body and/or controlling the transillumination via the lamp voltage instead of via a shutter.
References
- 1.Bashaw GJ, Klein R. Signaling from axon guidance receptors. Cold Spring Harb Perspect Biol. 2010;2:a001941. doi: 10.1101/cshperspect.a001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huber AB, Kolodkin AL, Ginty DD, Cloutier JF. Signaling at the growth cone: ligand-receptor complexes and the control of axon growth and guidance. Annu Rev Neurosci. 2003;26:509–563. doi: 10.1146/annurev.neuro.26.010302.081139. [DOI] [PubMed] [Google Scholar]
- 3.Lowery LA, Van Vactor D. The trip of the tip: understanding the growth cone machinery. Nat Rev Mol Cell Biol. 2009;10:332–343. doi: 10.1038/nrm2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gomez TM, Harrigan D, Henley J, Robles E. Working with Xenopus spinal neurons in live cell culture. Methods Cell Biol. 2003;71:129–156. doi: 10.1016/s0091-679x(03)01008-2. [DOI] [PubMed] [Google Scholar]
- 5.Leung KM, Holt CE. Live visualization of protein synthesis in axonal growth cones by microinjection of photoconvertible Kaede into Xenopus embryos. Nat Protoc. 2008;3:1318–1327. doi: 10.1038/nprot.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallo G, Lefcort FB, Letourneau PC. The trkA receptor mediates growth cone turning toward a localized source of nerve growth factor. J Neurosci. 1997;17:5445–5454. doi: 10.1523/JNEUROSCI.17-14-05445.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis AK, Bridgman PC. Nerve growth cone lamellipodia contain two populations of actin filaments that differ in organization and polarity. J. Cell Biol. 1992;119:1219–1243. doi: 10.1083/jcb.119.5.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goslin K, Banker G. Experimental observations on the development of polarity by hippocampal neurons in culture. J Cell Biol. 1989;108:1507–1516. doi: 10.1083/jcb.108.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forscher P, Kaczmarek LK, Buchanan JA, Smith SJ. Cyclic AMP induces changes in distribution and transport of organelles within growth cones of Aplysia bag cell neurons. J Neurosci. 1987;7:3600–3611. doi: 10.1523/JNEUROSCI.07-11-03600.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee AC, Decourt B, Suter D. Neuronal cell cultures from aplysia for high-resolution imaging of growth cones. J Vis Exp. 2008 doi: 10.3791/662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waterman-Storer CM, Desai A, Bulinski JC, Salmon ED. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr Biol. 1998;8:1227–1230. doi: 10.1016/s0960-9822(07)00515-5. [DOI] [PubMed] [Google Scholar]
- 12.Lee AC, Suter DM. Quantitative analysis of microtubule dynamics during adhesion-mediated growth cone guidance. Dev Neurobiol. 2008;68:1363–1377. doi: 10.1002/dneu.20662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaefer AW, Kabir N, Forscher P. Filopodia and actin arcs guide the assembly and transport of two populations of microtubules with unique dynamic parameters in neuronal growth cones. J Cell Biol. 2002;158:139–152. doi: 10.1083/jcb.200203038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suter DM, Errante LD, Belotserkovsky V, Forscher P. The Ig superfamily cell adhesion molecule, apCAM, mediates growth cone steering by substrate-cytoskeletal coupling. J Cell Biol. 1998;141:227–240. doi: 10.1083/jcb.141.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong Y, Lee AC, Suter DM, Lee GU. Topography and nanomechanics of live neuronal growth cones analyzed by atomic force microscopy. Biophys J. 2009;96:5060–5072. doi: 10.1016/j.bpj.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohof AM, Quillan M, Dan Y, Poo MM. Asymmetric modulation of cytosolic cAMP activity induces growth cone turning. J Neurosci. 1992;12:1253–1261. doi: 10.1523/JNEUROSCI.12-04-01253.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng JQ, Felder M, Connor JA, Poo MM. Turning of nerve growth cones induced by neurotransmitters. Nature. 1994;368:140–144. doi: 10.1038/368140a0. [DOI] [PubMed] [Google Scholar]
- 18.Halfter W. The behavior of optic axons on substrate gradients of retinal basal lamina proteins and merosin. J Neurosci. 1996;16:4389–4401. doi: 10.1523/JNEUROSCI.16-14-04389.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mai J, Fok L, Gao H, Zhang X, Poo MM. Axon initiation and growth cone turning on bound protein gradients. J Neurosci. 2009;29:7450–7458. doi: 10.1523/JNEUROSCI.1121-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Philipsborn AC, Lang S, Loeschinger J, Bernard A, David C, Lehnert D, Bonhoeffer F, Bastmeyer M. Growth cone navigation in substrate-bound ephrin gradients. Development. 2006;133:2487–2495. doi: 10.1242/dev.02412. [DOI] [PubMed] [Google Scholar]
- 21.Dertinger SK, Jiang X, Li Z, Murthy VN, Whitesides GM. Gradients of substrate-bound laminin orient axonal specification of neurons. Proc Natl Acad Sci U S A. 2002;99:12542–12547. doi: 10.1073/pnas.192457199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams DN, Kao EY, Hypolite CL, Distefano MD, Hu WS, Letourneau PC. Growth cones turn and migrate up an immobilized gradient of the laminin IKVAV peptide. J Neurobiol. 2005;62:134–147. doi: 10.1002/neu.20075. [DOI] [PubMed] [Google Scholar]
- 23.von Philipsborn AC, Lang S, Bernard A, Loeschinger J, David C, Lehnert D, Bastmeyer M, Bonhoeffer F. Microcontact printing of axon guidance molecules for generation of graded patterns. Nat Protoc. 2006;1:1322–1328. doi: 10.1038/nprot.2006.251. [DOI] [PubMed] [Google Scholar]
- 24.Romanova EV, Fosser KA, Rubakhin SS, Nuzzo RG, Sweedler JV. Engineering the morphology and electrophysiological parameters of cultured neurons by microfluidic surface patterning. FASEB J. 2004;18:1267–1269. doi: 10.1096/fj.03-1368fje. [DOI] [PubMed] [Google Scholar]
- 25.Turney SG, Bridgman PC. Laminin stimulates and guides axonal outgrowth via growth cone myosin II activity. Nat Neurosci. 2005;8:717–719. doi: 10.1038/nn1466. [DOI] [PubMed] [Google Scholar]
- 26.Walter J, Kern-Veits B, Huf J, Stolze B, Bonhoeffer F. Recognition of position-specific properties of tectal cell membranes by retinal axons in vitro. Development. 1987;101:685–696. doi: 10.1242/dev.101.4.685. [DOI] [PubMed] [Google Scholar]
- 27.Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, Bailey CH, Kandel ER. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Y, Wang DO, Martin KC. Preparation of Aplysia sensory-motor neuronal cell cultures. J Vis Exp. 2009 doi: 10.3791/1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kandel ER. Cellular basis of behavior. San Francisco: W. H. Freeman and Company; 1976. [Google Scholar]
- 30.Waterman-Storer C. Fluorescent speckle microscopy (FSM) of microtubules and actin in living cells. Curr Protoc Cell Biol. 2002;Chapter 4(Unit 4):10. doi: 10.1002/0471143030.cb0410s13. [DOI] [PubMed] [Google Scholar]