Abstract
Embryonic rhabdomyosarcoma (ERMS) is the most common soft-tissue tumor in children. Here, we report the identification of the minor groove DNA-binding factor high mobility group AT-hook 2 (HMGA2) as a driver of ERMS development. HMGA2 was highly expressed in normal myoblasts and ERMS cells, where its expression was essential to maintain cell proliferation, survival in vitro, and tumor outgrowth in vivo. Mechanistic investigations revealed that upregulation of the insulin–like growth factor (IGF) mRNA-binding protein IGF2BP2 was critical for HMGA2 action. In particular, IGF2BP2 was essential for mRNA and protein stability of NRAS, a frequently mutated gene in ERMS. shRNA-mediated attenuation of NRAS or pharmacologic inhibition of the MAP-ERK kinase (MEK)/extracellular signal-regulated kinase (ERK) effector pathway showed that NRAS and NRAS-mediated signaling was required for tumor maintenance. Taken together, these findings implicate the HMGA2–IGFBP2–NRAS signaling pathway as a critical oncogenic driver in ERMS.
Introduction
Rhabdomyosarcoma is the most common soft-tissue sarcoma in children. It displays features of immature skeletal muscle, including the existence of myoblast-like cells, and the expression of myogenic factors such as MyoD and Desmin (1). There are 2 major subtypes of rhabdomyosarcoma, embryonal rhabdomyosarcoma (ERMS) and alveolar rhabdomyosarcoma (ARMS; ref. 2). The ERMS subtype is more common than ARMS, and accounts for about 70% of total rhabdomyosarcoma cases. ERMS and ARMS were originally distinguished by pathologists according to their morphologies. However, it is now clear that these 2 rhabdomyosarcoma subtypes have different genetic mutation signatures. ARMS is characterized by either a t(2;13) or a t(1;13) chromosomal translocation, which generates PAX3–FOXO1 or PAX7–FOXO1 fusion proteins, respectively (3). In contrast, ERMS tumors frequently have mutations in components of the RAS pathway, such as NRAS mutations, HRAS mutations, or NF1 deletions (4). Expression of RAS mutants in muscle progenitors is sufficient to induce an ERMS-like phenotype in mice (5), suggesting that dysregulation of the RAS pathway may be a key event presaging a muscle stem cell to an ERMS fate.
High mobility group AT-hook 2 (HMGA2) is a DNA-binding protein that is highly expressed during embryonic development, but turned off in most adult tissues (6). Interestingly, HMGA2 is frequently reactivated in various cancers. High HMGA2 levels have been associated with increased metastasis and poor prognosis in settings of cancer (7–10). Even though the expression pattern of HMGA2 implies that this gene may play an important role in oncologic transformation, only limited studies have been reported regarding the molecular mechanisms downstream of HMGA2 in cancer cells (8, 11–14).
IGF2 mRNA-binding protein 2 (IGF2BP2), also known as IMP2, was found to be an HMGA2 target gene during early embryonic development (15). IGF2BP2 belongs to a family of RNA-binding proteins that contain 6 characteristics of RNA-binding modules, including 2 N-terminal RNA recognition motifs (RRM1 and RRM2) and 4 C-terminal hnRNP K-homology (KH1–KH4) domains (16). Its homolog, IGF2BP1/IMP1, has been shown to be essential for normal embryonic growth and development (17–19). In contrast, little is known about the function of IGF2BP2 in either development or cancer, other than the widely studied association between its genetic variance and the risk of type II diabetes (16).
Materials and Methods
Animal and xenograft experiments
All procedures were carried out in accordance with the standards of the U.S. Department of Health and Human Services, and were approved by the Novartis Animal Care and Use Committee. RD (a specific rhabodomyosarcoma cell line) cells were transduced with inducible shRNA constructs. Stable shRNA/luciferase-producing clones were selected in media containing 0.5 μg/mL puromycin (Invitrogen). Xenografts were generated by injecting 3 × 106 cells subcutaneously into 8- to 10-week-old nude mice (Taconic). Once tumors grew to sizes between approximately 200 and 400 mm3, mice were treated with 2 mg/mL doxcycline (Sigma) in 5% sucrose drinking water or 5% sucrose drinking water alone for 21 days. Tumor sizes were measured twice a week.
Cell lines and tissue culture
RD and TE617 rhabdomyosarcoma cell lines were purchased from American Type Culture Collection (ATCC). Cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) plus 10% FBS. Human skeletal muscle myoblasts (HSMM) were obtained from Lonza (#CC-2580) and cultured with Clonetics Skeletal Muscle Myoblast Cell System media and additives (#CC-3245 and #CC-5034).
Overexpression and shRNA knockdown experiments
For the overexpression experiments, murine HMGA2 was subcloned from a cDNA library (Open Biosystems #6332337) into the pLenti6/V5-DEST lentiviral expression vector. Human IGF2BP2 (Origene, # SC324486) was subcloned using a similar strategy. Lentivirus particles were produced by ViraPower Lentiviral Packaging Mix (Sigma, # SHP001) in 293FT cells, filtered, and used to infect myoblast cells. For knockdown of HMGA2 or IGF2BP2, lentiviral-based MISSION shRNA constructs (Sigma) against human HMGA2 or IGF2BP2 genes were purchased. Lentivirus was produced by ViraPower Lentiviral Packaging Mix in 293FT cells, filtered and used to infect cells. The efficacy of (shRNA)-mediated knockdown was examined by real time (RT-PCR) and Western blots. A nontargeting (NT) shRNA construct was always used as a negative control (Sigma, #SHC202).
RNA-binding protein immunoprecipitation
RNA-binding protein immunoprecipitation (RBP-IP) was carried out using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) and an IGF2BP2 antibody (Millipore, # 03–251). Briefly, rhabdomyosarcoma cells were lysed in radioimmunoprecipitation assay buffer; immunoprecipitations were carried out using antibodies against IGF2BP2 or normal mouse IgG, with protein A magnetic beads. Magnetic bead–bound complexes were immobilized with a magnet, and unbound materials were rinsed off with washing buffer. RNAs coimmunoprecipitated with IGF2BP2 antibody or IgG were eluted, reverse transcribed, and analyzed by quantitative real-time PCR.
mRNA half-life analysis
RD cells infected with NT-shRNA, IGF2BP2-shRNA, or HMGA2-shRNA were treated with 5 μg/mL actinomycin D (Sigma). Following the addition of actinomycin D, cells were kept in normal culture condition for various periods of time before total RNA was extracted and measured by RT-PCR.
Luciferase assay
NRAS-3′UTR-luciferase construct was a gift from Dr. Frank Slack (Addgene, # 13797). NRAS-3′UTR-Luciferase, Renilla, and IGF2BP2 plasmids were transfected into 293 cells seeded in 96-well plates using FUGENE 6 (Roche) transfection reagent following the manufacturer's protocol. Forty-eight hours later, the cells were lysed and luciferase assays were conducted using a Dual-Luciferase Reporter Assay System (Promega) on a luminometer. Transfection of each combination was carried out in triplicate in each assay. Luciferase readings were taken as singlets. Ratios of Renilla luciferase readings to Firefly luciferase readings were taken for each experiment and triplicates were averaged.
Immunofluorescence and immunohistochemistry
For immunofluorescence, cells grown in chamber slides were fixed by 4% paraformaldehyde for 15 minutes at room temperature and permeabilized by 0.5% Triton X-100. Samples were then stained with primary antibody for 2 hours and with secondary antibody for 30 minutes at room temperature. Nuclei were labeled with 4′, 6-diamidino-2-phenylindole (DAPI). Primary antibodies used were HMGA2 (Cell Signaling, #5269) and MHC (Millipore, #05–833). Secondary antibodies used are goat-anti-rabbit IgG Alexa Fluor 488 (Invitrogen) and goat-anti-mouse IgG Alexa Fluor 594 (Invitrogen). For immunohistochemistry, the paraffin-embedded sections were cleared and the sections were incubated with 0.1% Pronase (Roche #165 921) in 0.1% CaCl2 pH 7.8. at 37° C for 10 minutes. They were blocked with 3% H2O2 in Tris-buffered saline (TBS) for 10 minutes. After washing, they were further blocked with 10% normal rabbit serum for 10 minutes at room temperature and incubated with HMGA2 antibody (Cell Signaling, 1:100) for 2 hours at room temperature, then with biotinylated goat–antimouse at 1/100 for 30 minutes at room temperature, and finally with Strep–ABC complex (Dako) at 1/100 for 30 minutes at room temperature. The sections were developed with an AEC substrate kit (vector lab) at room temperature for 20 minutes, counter-stained with hematoxylin, and after drying were mounted with DAKO aqueous mount (Dako, 003181).
Results
HMGA2 is highly expressed in human myoblasts and ERMS cells
Given that rhabdomyosarcoma cells share many characteristics with undifferentiated myoblasts, such as expression of muscle lineage markers, and a proliferation-competent phenotype, we asked whether genes controlling early myogenesis might also contribute to the transformed phenotype seen in rhabdomyosarcoma. To address this question, we first identified a group of genes that were highly expressed in proliferating myoblasts, but not in differentiated myofibers. HMGA2 was a top candidate because it was highly expressed in undifferentiated HSMMs, rapidly downregulated during differentiation, and became essentially undetectable in mature muscle fibers (Fig. 1A and B). We next examined whether HMGA2 was also highly expressed in rhabdomyosarcoma. Comprehensive database analyses confirmed that HMGA2 mRNA was absent in all adult tissues examined but reactivated in rhabdomyosarcoma samples (Fig. 1C), particularly in those of the ERMS subtype (Fig. 1D). In contrast, while HMGA1 shares significant homology with HMGA2, HMGA1 mRNA remained detectable in adult tissues and its level was below average in rhabdomyosarcoma samples (Supplementary Fig. S1).
Figure 1.
HMGA2 mRNA and HMGA2 protein are highly expressed in myoblasts and embryonic rhabdomyosarcoma cells. A, HMGA2 mRNA levels gradually drop during human myoblast differentiation. Eighty percent of confluent HSMMs were cultured in differentiation media for 4 days. mRNA samples were harvested at days 0, 3, and 4. RT-PCR was then conducted to quantify the level of HMGA2 mRNA. Data were normalized to GAPDH and 18S mRNAs. *, P < 0.05 versus mRNA level at day 0. B, HMGA2 expression is inversely correlated with myoblast terminal differentiation. Human myoblasts were grown to more than 80% confluence in proliferation media, and switched to differentiation media for 2 days. Cells were then fixed and stained for HMGA2 and myosin heavy chain (MHC). At this stage, 30% to 40% cells became terminally differentiated and expressed MHC (scale bar, 50 μm). Interestingly, HMGA2 and MHC expression were mutually exclusive. MHC-positive cells are negative for HMGA2 [white arrow, HMGA2(+)/MHC(−) nuclei; yellow arrow, HMGA2 (−)/MHC(+) nuclei]. C, HMGA2 mRNA is absent in adult human tissues but reactivated in various cancer cells. Rhabdomyosarcoma (RMS) shows the highest average expression level. ES, embryonic stem cells. D, HMGA2 mRNA is highly expressed in normal human myoblasts and rhabdomyosarcoma cells of the ERMS subtype. *, P < 0.05, Student t test. E, HMGA2 protein is selectively expressed in ERMS cell lines, whereas HMGA1 is ubiquitously expressed. N, nuclei; C, cytoplasmic. F HMGA2 protein is localized to the nuclei of normal human myoblasts and rhabdomyosarcoma cells. Scale bar, 25 μm. G HMGA2 is widely expressed in primary human ERMS specimens. Scale bar, 50 μm.
To investigate whether the HMGA2 protein level was also high in ERMS, protein samples were harvested from several cancer or immortalized cell lines, including the ERMS-type RD and TE617 cell lines. RD and TE617 cells expressed much higher levels of HMGA2 in comparison with other cancer/immortal cell lines (Fig. 1E). Therefore, HMGA2 is not a general cell proliferation marker; rather, it is selectively activated in ERMS. Similar to its mRNA profile, HMGA1 was ubiquitously expressed in all cell lines examined. Consistent with HMGA2 as a DNA-binding protein, nuclei versus cytoplasm was fractionated, and lysates run on a Western blot analysis have shown that HMGA2 protein was exclusively localized to the nuclei of ERMS cells (Fig. 1E); this was confirmed by immunofluorescence staining (Fig. 1F). Immunohistochemistry also confirmed that HMGA2 was clearly evident in primary ERMS samples (Fig. 1G).
HMGA2 is indispensable for ERMS survival and growth in vitro
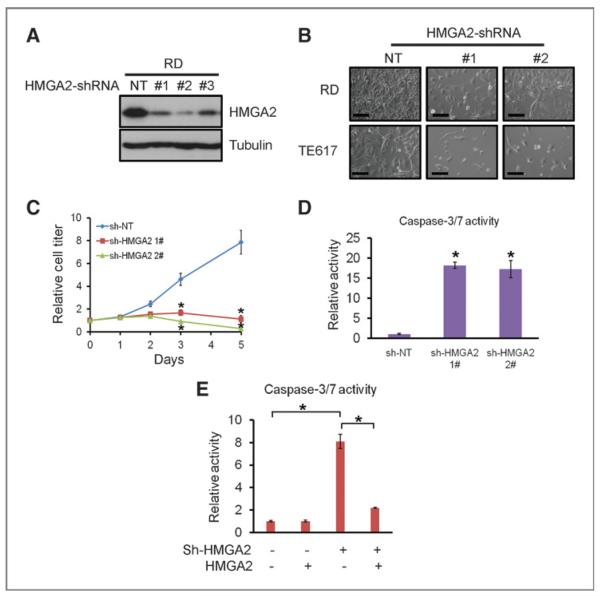
The unique and strong expression of HMGA2 in ERMS prompted the query as to whether it is required for ERMS proliferation or survival. Lentiviral-based HMGA2 shRNAs achieved more than 80% knockdown of HMGA2 protein, in comparison with a NT shRNA control (Fig. 2A). Knockdown of HMGA2 in ERMS cell lines resulted in a strong inhibition of cell proliferation (Fig. 2B and C). Caspase-3/7 activity increased 15- to 20-fold after HMGA2 knockdown (Fig. 2D), indicating that loss of HMGA2 induced apoptosis in rhabdomyosarcoma cells.
Figure 2.
HMGA2 is required for embryonic rhabdomyosarcoma growth in vitro. A, knockdown of HMGA2 in RD cells using lentiviral-based shRNAs. NT shRNA control or 3 independent shRNAs (1#, 2#, 3#) targeting HMGA2 were used. All 3 HMGA2 shRNAs significantly reduced HMGA2 levels, in comparison to the NT shRNA. B, knockdown of HMGA2 in ERMS cells reduces proliferation and induces cell death. Images were taken 96 hours after infection. Scale bar, 100 μm. C, knockdown of HMGA2 in ERMS cells reduces proliferation, measured by cell-titer assays. *, P < 0.05, Student t test. D, knockdown of HMGA2 in ERMS cells induces apoptosis, measured by activated caspase 3/7 activity. *, P < 0.05, Student t test, sh-HMGA2 sample versus sh-NT sample. E, overexpression of HMGA2 significantly rescues the apoptosis effect of HMGA2 shRNA. *, P < 0.05, Student t test.
To examine the specificity of HMGA2 shRNA constructs, and make sure the observed phenotypes were not due to off-target effects, we conducted rescue experiments; overexpression of murine HMGA2 protein largely rescued the inhibitory effect of human HMGA2 shRNAs on rhabdomyosarcoma cells (Supplementary Fig. S2). Expression of murine HMGA2 also significantly reduced apoptosis in HMGA2 shRNA-treated cells (Fig. 2E). In addition, we introduced HMGA2 shRNAs into the MCF7 breast cancer cell line, which does not express endogenous HMGA2 (Fig. 1E). HMGA2 shRNAs had minimal growth inhibitory effects on MCF7 cells (data not shown). These data together suggest that the phenotypes observed in rhabdomyosarcoma cells largely resulted from on-target effects of HMGA2 shRNAs.
HMGA2 is crucial for ERMS tumorigenesis in vivo
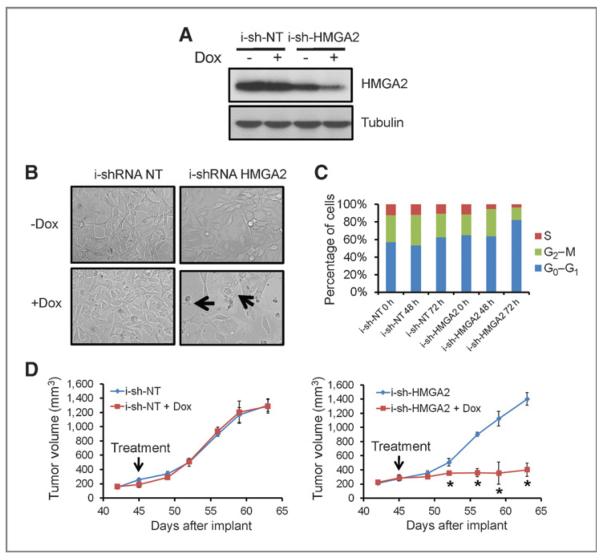
To investigate the effect of targeting HMGA2 in vivo, NT and HMGA2 shRNA hairpins were subcloned into a doxycyclineinducible lentiviral construct. Without doxycycline treatment, cell lines with NT shRNA or HMGA2 shRNA grew at similar rates. Upon doxycycline treatment, HMGA2 shRNA successfully reduced HMGA2 mRNA and protein levels by more than 70% (Fig. 3A). Similar to constitutively active shRNAs, inducible HMGA2 shRNA inhibited cell growth and increased cell death of ERMS cells in vitro (Fig. 3B). Inducible HMGA2 shRNAs gradually led to a G0–G1 cell-cycle arrest in ERMS cells (Fig. 3C). The inducible HMGA2 shRNAs also caused a decrease in proliferation, as shown by a more than 80% reduction in cell number that stained positive for Ki67 (Supplementary Fig. S3A).
Figure 3.
HMGA2 is required for rhabdomyosarcoma growth in vivo. A, doxycycline (Dox)-inducible shRNA successfully reduced HMGA2 protein levels. B, inducible shRNA against HMGA2 caused doxycycline-dependent cell growth inhibition and cell death. The inhibition/death phenotypes were observed in sh-HMGA2–infected cells but not inducible sh-NT–infected cells. Arrows indicate dying cells. C, knockdown of HMGA2 in ERMS cells reduces the time cells spend in S-phase and causes G0–G1 arrest. Doxycycline treatment led to a gradual decrease of cells in S-phase and accumulation of cells in G0–G1 phase. D, doxycycline-inducible shRNA shows that HMGA2 is required for rhabdomyosarcoma growth in vivo. *, P < 0.05, Student t test. +Dox group versus −Dox group at the same time point.
To examine whether HMGA2 is essential for ERMS tumor growth and maintenance in vivo, RD cells infected with NT or HMGA2-inducible shRNA were transplanted into nude mice. Cells grew at similar rates in vivo without doxycycline treatment. When the average tumor size reached 300 to 400 mm3, 2 mg/mL doxycycline was added to the drinking water. HMGA2 shRNA, but not NT shRNA, significantly blocked further tumor growth and caused tumor regression in some cases (Fig. 3D)—showing that HMGA2 is required for ERMS proliferation and maintenance in vivo.
Even though HMGA2 inhibition significantly reduced tumor growth, eventually, resistant tumors arose in the HMGA2 shRNA group. This could be either due to inefficiency in the maintenance of the expression knockdown by HMGA2 shRNA or as a result of an activation of compensatory pathways. To determine which of these possibilities were operative, resistant tumors were harvested from the HMGA2 inducible-shRNA group and stained for HMGA2 protein. All resistant tumors showed strong staining for HMGA2 (Supplementary Fig. S3B), indicating these tumors came from clones that failed to turn off HMGA2 upon doxycycline treatment (though still one might argue that a secondary mechanism stabilized residual HMGA2 protein, so that mechanism is not excluded).
HMGA2 regulates IGF2BP2 in rhabdomyosarcoma
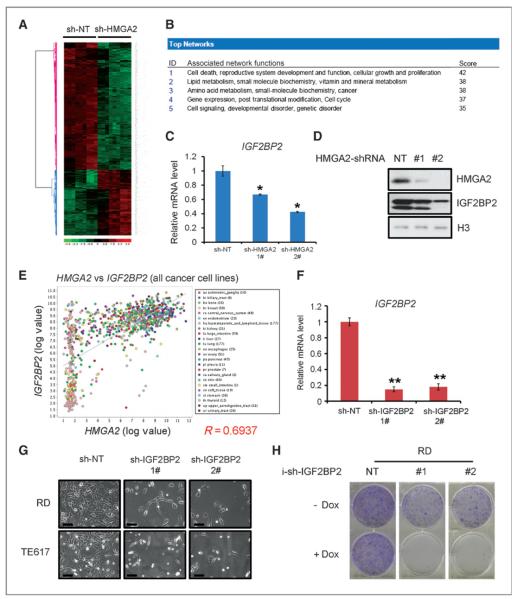
To better understand the mechanism by which HMGA2 mediates ERMS oncogenesis, microarray analyses were conducted to compare the mRNA profiles of RD cells infected with NT or HMGA2 shRNAs at several time points. Five hundred and twenty five genes were significantly downregulated and 242 genes were significantly upregulated in HMGA2 shRNA-infected samples (Fig. 4A). Consistent with the strong cytostatic and cytotoxic phenotype associated with HMGA2 knockdown, the most downregulated gene networks were involved in cell death, growth, and proliferation (Fig. 4B and Supplementary Table S1). As a transcriptional coregulator, HMGA2 may regulate multiple pathways. Our approach was to identify distinct pathways that HMGA2 regulates in ERMS, and then determine the pathway's requirement for the transformed phenotype. IGF2BP2, a known target of HMGA2 during embryonic development (15), was identified as one of the top genes downregulated by HMGA2 shRNAs (Supplementary Table S1). We confirmed that IGF2BP2 mRNAs and protein were both reduced upon HMGA2 knockdown in RD cells (Fig. 4C and D). Consistent with the possibility that IGF2BP2 is a direct target of HMGA2 in rhabdomyosarcoma, HMGA2 and IGF2BP2 mRNA levels were positively correlated in 19 soft-tissue sarcoma cell lines including rhabdomyosarcoma lines (Supplementary Fig. S4). To examine whether HMGA2 may regulate IGF2BP2 in other tumor cells, the analysis was extended to 1,000 cancer cell lines from various tissues. HMGA2 and IGF2BP2 mRNAs indeed showed significant correlation even in this highly heterogeneous cancer group (Fig. 4E), strongly suggesting that the regulation of IGF2BP2 by HMGA2 may be conserved in multiple cancer types. Because HMGA2 was reported to enhance E2F activity (8) and as the E2F pathway is strongly associated with cell-cycle regulation, we examined whether E2F pathway was regulated by HMGA2 shRNAs in ERMS. Gene set enrichment assay indicated that E2F pathway was significantly downregulated 72 hours, but not 48 or 96 hours after HMGA2 knockdown (Supplementary Fig. S5A). Quantitative RT-PCR confirmed that the mRNA levels of multiple E2F targets, including CDC25A, CDC6, CDk1, CyclinA2, and TK1 were reduced at the 72-hour time point (Supplementary Fig. S5B). When HMGA2 was knocked down for 72 hours, E2F reporter activity was significantly reduced by about 40% (Supplementary Fig. S5C). These data together indicate that HMGA2 is a positive regulator of E2F pathway in ERMS, which is not terribly surprising given the perturbation of proliferation; however, given the rather modest changes, and the further data showing a requirement for the HMGA2/IGF2BP2 pathway, the changes in E2F do not seem to be primary in terms of mechanism downstream of HMGA2.
Figure 4.
HMGA2 regulates IGF2BP2, which is necessary for ERMS growth. A, heatmap of microarray on HMGA2 knockdown samples illustrates HMGA2-regulated genes in rhabdomyosarcoma cells. B, genes significantly downregulated by HMGA2 knockdown are enriched for those controlling cell death, growth, and proliferation, including NRAS. Gene sets were analyzed by IPA software. C, knockdown of HMGA2 reduces IGF2BP2 mRNA. *, P < 0.05, Student t test, sh-HMGA2 sample versus sh-NT sample. D, knockdown of HMGA2 reduces IGF2BP2 protein. E, HMGA2 and IGF2BP2 mRNA levels are correlated in cancer cell lines. F, knockdown of IGF2BP2 using lentiviral-based shRNAs. **, P < 0.01, Student t test, sh-IGF2BP2 sample versus sh-NT sample. G, knockdown of IGF2BP2 in ERMS cells reduces proliferation and induces cell death. Images were taken 96 hours after infection. Scale bar, 100 μm. H, knockdown of IGF2BP2 using doxycycline-inducible shRNAs in rhabdomyosarcoma cells inhibits colony formation.
IGF2BP2 is crucial for ERMS survival and growth
The biologic roles of IGF2BP2 in cancer were unknown. To determine whether IGF2BP2 is required for ERMS growth, shRNAs against IGF2BP2 were used to inhibit its expression (Fig. 4F). Two shRNAs successfully knocked down IGF2BP2 mRNA levels by more than 80%. Similar to that of HMGA2, knockdown of IGF2BP2 in ERMS cells inhibited cell growth and led to cell death (Fig. 4G). IGF2BP2 shRNAs also reduced colony formation in vitro (Fig. 4H). To determine whether IGF2BP2 is a critical functional target of HMGA2 in ERMS, we overexpressed IGF2BP2 in an HMGA2 knockdown background. Overexpression of IGF2BP2 partially rescued the proliferation deficiency caused by HMGA2 knockdown in ERMS cells (Supplementary Fig. S5D). Therefore, IGF2BP2 is important for ERMS growth and represents a key target downstream of HMGA2 in ERMS cells.
IGF2BP2 binds to a set of mRNAs, including NRAS, and regulates NRAS mRNA stability and protein production
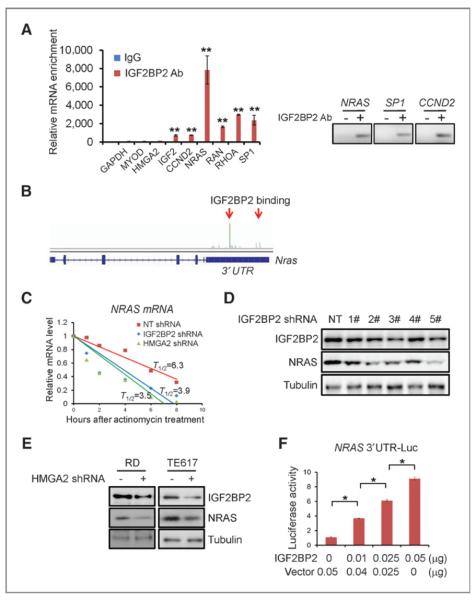
It was next of interest to understand the mechanism by which IGF2BP2 regulates proliferation and survival in ERMS cells. IGF2BP family members have been shown to perturb stability and translation efficiency of the mRNAs they bind to (16, 20–22). Therefore, we first tried to identify mRNAs bound by IGF2BP2 in ERMS cells. A previous study identified top RNAs, including mRNAs, tRNAs, rRNAs, and microRNAs, bound by overexpressed IGF2BP2 in HEK293 cells (23). We reanalyzed the data and noticed that the top mRNAs bound by IGF2BP2 were enriched for those encoding proteins involved in regulating cell proliferation and migration, most notably NRAS, but also including CCND2 and RHOA (Supplementary Table S2). The reason NRAS was of particular interest is that this gene is one of the most frequently mutated genes in ERMS (4, 24–26), including in the RD cell line, which was used for most of these experiments; so this finding would help explain the mechanism by which expression of a causative oncogene is maintained in ERMS. To confirm whether endogenous IGF2BP2 also bound to NRAS, CCND2, and RHOA mRNAs in ERMS cells, RBP-IP was carried out. A specific IGF2BP2 antibody was used to pull down endogenous IGF2BP2 protein, and many of the top mRNA candidates, including NRAS, RAN, RHOA, and SP1, were coimmunoprecipitated with IGF2BP2 protein. The IGF2BP2 antibody did not pull down GAPDH, MYOD, or HMGA2 mRNAs (Fig. 5A). Through analyzing RNA-sequencing data, we found that IGF2BP2 strongly bound to the 3′UTR of NRAS mRNA (Fig. 5B). Because the NRAS 3′UTR is a crucial element targeted by multiple microRNAs (27, 28), and involved in regulating NRAS mRNA stability, this finding raised the possibility that IGF2BP2 could directly regulate NRAS mRNA stability and protein production. Indeed, when IGF2BP2 or HMGA2 was knocked down in ERMS cells, NRAS mRNA half-life was reduced from 6.3 hours to 3.9 hours and 3.5 hours, respectively (Fig. 5C). Knockdown of either HMGA2 or IGF2BP2 also reduced NRAS protein in ERMS cells (Fig. 5D and E). To determine whether IGF2BP2 regulates NRAS mRNA stability through binding to its 3′UTR, HEK293 cells were cotransfected with a luciferase-NRAS 3′UTR fusion construct (27) and an IGF2BP2 expressing construct. IGF2BP2 overexpression caused a dose-dependent increase of luciferase activity (Fig. 5F), indicating that IGF2BP2 indeed was able to bind to the 3′UTR of NRAS mRNA and increase its mRNA stability and thus NRAS protein production.
Figure 5.
IGF2BP2 binds to mRNAs and regulates multiple oncogenic genes, including NRAS. A, IGF2BP2 binds to mRNAs of Igf2, Ccnd2, NRas, Ran, RhoA, and Sp1, but not GAPDH, MYOD, and HMGA2. **, P < 0.01, Student t test, IGF2BP2 antibody versus IgG control. B, IGF2BP2 binds to the 3′UTR of NRAS mRNA. C, IGF2BP2 regulates the stability of NRAS mRNA. After IGF2BP2 shRNA treatment, NRAS mRNA half-life (T1/2) is reduced from 6.3 to 3.9 hours. HMGA2 shRNA reduces half-life to 3.5 hours. D, knockdown of IGF2BP2 reduces NRAS protein levels. Five different shRNAs were used to knock down IGF2BP2; NRAS protein levels correlate well with IGF2BP2 knockdown efficacy. E, knockdown of HMGA2 reduces the NRAS protein level in ERMS cells. F, IGF2BP2 stabilizes NRAS-3′UTR-luciferase protein. Luciferase assays show a dose-dependent increase of luciferase activity with IGF2BP2 overexpression. *, P < 0.01, Student t test.
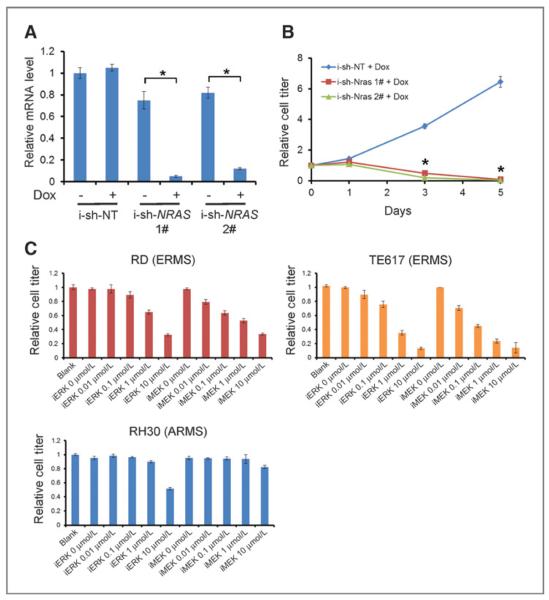
The proliferation of ERMS cells with NRAS mutations is dependent on NRAS level and activity
The proliferation of cancer cells containing mutated onco-genes is usually dependent on the mutated protein products, a phenotype termed oncogene addiction (29). To determine whether the regulation of NRAS protein levels by IGF2BP2 is required in ERMS with NRAS mutations, NRAS was knocked down in RD cells by 2 independent shRNAs (Fig. 6A). The proliferation of RD cells was significantly reduced upon NRAS knockdown (Fig. 6B). In addition, RD and TE617 cells were much more sensitive to MEK and ERK inhibitors AZD6244 and VX-11e, compared with the ARMS cell line RH30, which has a wild-type NRAS gene (Fig. 6C). Therefore, like many cancers with NRAS mutations, ERMS cells are highly dependent on the hyperactivated NRAS-MEK-ERK-MAPK pathway for their growth and survival. The HMGA2–IGF2BP2 pathway thus comprises a novel and important node maintaining NRAS protein production via mRNA stabilization
Figure 6.
Growth of ERMS cells is dependent on NRAS protein and downstream pathway. A, successful knockdown of NRAS mRNA by inducible shRNAs. *, P < 0.05, Student t test. B, knockdown of NRAS in RD cells reduces proliferation. *, P < 0.05, Student t test. NRAS shRNA versus NT shRNA control. C, RD and TE617 ERMS cell lines are more sensitive to ERK inhibitor (VX-11e) and MEK inhibitor (AZD6244) than ARMS cell line RH30.
Discussion
The proliferation and subsequent differentiation of skeletal muscle stem cells into myofibers are essential steps for muscle development, regeneration, repair, and maintenance. Failure of terminal differentiation is a hallmark of the muscle-lineage tumor RMS. We reasoned that it might be helpful to understand the molecular mechanisms controlling early myogenesis, to gain an understanding of what might go wrong in the switch to the transformed phenotype seen in RMS. Toward this goal, in a distinct study (29), we tried to identify genes that are enriched in proliferating human myoblasts during the normal differentiation process, and in this study asked which of these were also highly expressed in RMS cells. HMGA2 conforms to this profile only in the ERMS, as opposed to the ARMS type of the disease. HMGA2 is more than a simple marker of the ERMS subtype of disease; using both gain-of-function and loss-of-function assays, we identified HMGA2 as a required gene, mediating proliferation and survival of ERMS cells both in vitro and in vivo. In a parallel study, we have also shown that HMGA2 is an important factor regulating normal mouse myoblast growth and myogenesis—in normal myoblasts, HMGA2 must be downregulated in order for the differentiation process to take place (30). These studies together help to explain why the pathologic upregulation and maintenance of HMGA2 expression may presage oncogenic transformation—the differentiation stage is thereby blocked.
HMGA2 was previously shown to enhance E2F activity and promote cell proliferation in pituitary adenomas (8). We confirmed that many E2F targets were affected by HMGA2 shRNAs in ERMS. However, the regulation of the E2F pathway by HMGA2 in ERMS is relatively modest, and may not be able to explain all the phenotypes we observed upon HMGA2 knockdown. Given that any mechanism that results in a perturbation of the cell cycle might in some way perturb E2F pathway members, it seemed that a more unbiased approach to HMGA2 was in order. Using microarray and expression correlation analyses, we identified IGF2BP2 as a top regulated gene by HMGA2 in cancers.
To determine whether HMGA2 is necessary or sufficient to mediate oncogenesis in ERMS cells, the downstream pathway was further studied. Functional experiments further show the importance of IGF2BP2 activation downstream of HMGA2 upregulation for ERMS proliferation and survival. IGF2BP2 is a known target for HMGA2 during embryonic development (15, 31), and has been reported to be highly expressed in some cancers (32, 33). However, its potential role in tumorigenesis has not been elucidated. The data presented in the current study, together with a second report, which showed that IGF2BP2 regulates a glioma cancer stem cell metabolism (34), strongly suggest that the HMGA2–IGF2BP2 pathway plays a required role to maintain proliferation and survival, perhaps by blocking differentiation mechanisms, in particular settings of cancer.
In ERMS cells, IGF2BP2 was in turn found in this study to regulate the mRNA stability of NRAS, an oncogene frequently mutated in ERMS but not ARMS, providing a plausible explanation as to the terminal step in maintaining the transformed phenotype upon HMGA2–IGF2BP2 activation. In a mutated NRAS background, ERMS cells with high HMGA2 expression may have a growth advantage due to their ability to maintain high NRAS level and activity and therefore be selected for during early tumorigenesis. It is worth noting that HMGA2 expression is dependent on the RAS pathway in some contexts (11). Therefore, there seems to be a positive feedback loop of RAS–HMGA2–IGF2BP2, in turn stimulating HMGA2/IGFBP2/NRAS/MEK/ERK signaling in ERMS cells here.
NRAS is a well-known oncogene that is frequently mutated in many tumors, including ERMS. Cells with NRAS mutations are usually dependent on this oncogene for survival (35, 36) and we have confirmed that ERMS cell lines are also dependent on the mutated NRAS protein. Knockdown of NRAS inhibited ERMS cell proliferation and decreased survival. Furthermore, a negative effect on cell number was observed with MEK and ERK pharmacologic inhibitors, showing that in ERMS cells NRAS is operating through the well-studied RAS/MEK/ERK pathway. Although NRAS mutations have been extensively studied, little attention has been paid to how the NRAS protein is maintained. Our study identifies new layers of regulation of NRAS, in this case, at the step of maintaining the half-life of its mRNA, resulting in higher protein levels. Because the development of RAS inhibitors has been largely unsuccessful so far, the HMGA2–IGF2BP2 axis may provide a novel target for future drug development to treat this childhood cancer.
Supplementary Material
Acknowledgments
The authors thank Mark Fishman and Brian Richardson for their support and Robin Ge for excellent advice and assistance.
Grant Support This work was supported in part by R01 CA138265 (D.G. Kirsch).
Footnotes
Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
Authors' Contributions Conception and design: Z. Li, D.J. Glass
Development of methodology: Z. Li, D.J. Glass
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): Z. Li, K. Ramanujan
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Z. Li, Y. Zhang, K. Ramanujan, D.G. Kirsch, D.J. Glass
Writing, review, and/or revision of the manuscript: Z. Li, D.G. Kirsch, D.J. Glass
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): Z. Li, Y. Ma
Study supervision: Z. Li, D.J. Glass
References
- 1.Bizer LS. Rhabdomyosarcoma. Am J Surg. 1980;140:687–91. doi: 10.1016/0002-9610(80)90059-8. [DOI] [PubMed] [Google Scholar]
- 2.Davicioni E, Anderson MJ, Finckenstein FG, Lynch JC, Qualman SJ, Shimada H, et al. Molecular classification of rhabdomyosarcoma—genotypic and phenotypic determinants of diagnosis: a report from the Children's Oncology Group. Am J Pathol. 2009;174:550–64. doi: 10.2353/ajpath.2009.080631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xia SJ, Pressey JG, Barr FG. Molecular pathogenesis of rhabdomyosarcoma. Cancer Biol Ther. 2002;1:97–104. doi: 10.4161/cbt.51. [DOI] [PubMed] [Google Scholar]
- 4.Martinelli S, McDowell HP, Vigne SD, Kokai G, Uccini S, Tartaglia M, et al. RAS signaling dysregulation in human embryonal rhabdomyosarcoma. Genes Chromosomes Cancer. 2009;48:975–82. doi: 10.1002/gcc.20702. [DOI] [PubMed] [Google Scholar]
- 5.Rubin BP, Nishijo K, Chen HI, Yi X, Schuetze DP, Pal R, et al. Evidence for an unanticipated relationship between undifferentiated pleomorphic sarcoma and embryonal rhabdomyosarcoma. Cancer cell. 2011;19:177–91. doi: 10.1016/j.ccr.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou X, Chada K. HMGI family proteins: architectural transcription factors in mammalian development and cancer. Keio J Med. 1998;47:73–7. doi: 10.2302/kjm.47.73. [DOI] [PubMed] [Google Scholar]
- 7.Cleynen I, Van de Ven WJ. The HMGA proteins: a myriad of functions (Review) Int J Oncol. 2008;32:289–305. [PubMed] [Google Scholar]
- 8.Fedele M, Visone R, De Martino I, Troncone G, Palmieri D, Battista S, et al. HMGA2 induces pituitary tumorigenesis by enhancing E2F1 activity. Cancer Cell. 2006;9:459–71. doi: 10.1016/j.ccr.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 9.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and HMGA2 enhances oncogenic transformation. Science. 2007;315:1576–9. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–30. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe S, Ueda Y, Akaboshi S, Hino Y, Sekita Y, Nakao M. HMGA2 maintains oncogenic RAS-induced epithelial-mesenchymal transition in human pancreatic cancer cells. Am J Pathol. 2009;174:854–68. doi: 10.2353/ajpath.2009.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fedele M, Fusco A. HMGA and cancer. Biochim Biophys Acta. 2010;1799:48–54. doi: 10.1016/j.bbagrm.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Thuault S, Tan EJ, Peinado H, Cano A, Heldin CH, Moustakas A. HMGA2 and Smads co-regulate SNAIL1 expression during induction of epithelial-to-mesenchymal transition. J Biol Chem. 2008;283:33437–46. doi: 10.1074/jbc.M802016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li AY, Lin HH, Kuo CY, Shih HM, Wang CC, Yen Y, et al. High-mobility group A2 protein modulates hTERT transcription to promote tumorigenesis. Mol Cell Biol. 2011;31:2605–17. doi: 10.1128/MCB.05447-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brants JR, Ayoubi TA, Chada K, Marchal K, Van de Ven WJ, Petit MM. Differential regulation of the insulin-like growth factor II mRNA-binding protein genes by architectural transcription factor HMGA2. FEBS Lett. 2004;569:277–83. doi: 10.1016/j.febslet.2004.05.075. [DOI] [PubMed] [Google Scholar]
- 16.Christiansen J, Kolte AM, Hansen TO, Nielsen FC. IGF2 mRNA-binding protein 2: biological function and putative role in type 2 diabetes. J Mol Endocrinol. 2009;43:187–95. doi: 10.1677/JME-09-0016. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19:1262–70. doi: 10.1128/mcb.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen J, Adolph SK, Rajpert-De Meyts E, Lykke-Andersen J, Koch G, Christiansen J, et al. Nuclear transit of human zipcode-binding protein IMP1. Biochem J. 2003;376:383–91. doi: 10.1042/BJ20030943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen TV, Hammer NA, Nielsen J, Madsen M, Dalbaeck C, Wewer UM, et al. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol. 2004;24:4448–64. doi: 10.1128/MCB.24.10.4448-4464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaeffer V, Hansen KM, Morris DR, Leboeuf RC, Abrass CK. RNA-binding protein IGF2BP2/IMP2 is required for laminin-beta2 mRNA translation and is modulated by glucose concentration. Am J Physiol Renal Physiol. 2012;303:F75–82. doi: 10.1152/ajprenal.00185.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai N, Rapley J, Angel M, Yanik MF, Blower MD, Avruch J. mTOR phosphorylates IMP2 to promote IGF2 mRNA translation by internal ribosomal entry. Genes Dev. 2011;25:1159–72. doi: 10.1101/gad.2042311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leeds P, Kren BT, Boylan JM, Betz NA, Steer CJ, Gruppuso PA, et al. Developmental regulation of CRD-BP, an RNA-binding protein that stabilizes c-myc mRNA in vitro. Oncogene. 1997;14:1279–86. doi: 10.1038/sj.onc.1201093. [DOI] [PubMed] [Google Scholar]
- 23.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaaf G, Hamdi M, Zwijnenburg D, Lakeman A, Geerts D, Versteeg R, et al. Silencing of SPRY1 triggers complete regression of rhabdomyosarcoma tumors carrying a mutated RAS gene. Cancer Res. 2010;70:762–71. doi: 10.1158/0008-5472.CAN-09-2532. [DOI] [PubMed] [Google Scholar]
- 25.Stratton MR, Fisher C, Gusterson BA, Cooper CS. Detection of point mutations in N-ras and K-ras genes of human embryonal rhabdomyosarcomas using oligonucleotide probes and the polymerase chain reaction. Cancer Res. 1989;49:6324–7. [PubMed] [Google Scholar]
- 26.Paulson V, Chandler G, Rakheja D, Galindo RL, Wilson K, Amatruda JF, et al. High-resolution array CGH identifies common mechanisms that drive embryonal rhabdomyosarcoma pathogenesis. Genes Chromosomes Cancer. 2011;50:397–408. doi: 10.1002/gcc.20864. [DOI] [PubMed] [Google Scholar]
- 27.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Luo XJ, Xiong AW, Zhang ZD, Yue S, Zhu MS, et al. MicroRNA-214 promotes myogenic differentiation by facilitating exit from mitosis via down-regulation of proto-oncogene N-ras. J Biol Chem. 2010;285:26599–607. doi: 10.1074/jbc.M110.115824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma SV, Settleman J. Oncogene addiction: setting the stage for molecularly targeted cancer therapy. Genes Dev. 2007;21:3214–31. doi: 10.1101/gad.1609907. [DOI] [PubMed] [Google Scholar]
- 30.Li Z, Gilbert JA, Zhang Y, Zhang M, Qiu Q, Ramanujan K, et al. An HMGA2-IGF2BP2 axis regulates myoblast proliferation and myogenesis. Dev Cell. 2012;23:1176–88. doi: 10.1016/j.devcel.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cleynen I, Brants JR, Peeters K, Deckers R, Debiec-Rychter M, Sciot R, et al. HMGA2 regulates transcription of the Imp2 gene via an intronic regulatory element in cooperation with nuclear factor-kappaB. Mol Cancer Res. 2007;5:363–72. doi: 10.1158/1541-7786.MCR-06-0331. [DOI] [PubMed] [Google Scholar]
- 32.Himoto T, Kuriyama S, Zhang JY, Chan EK, Nishioka M, Tan EM. Significance of autoantibodies against insulin-like growth factor II mRNA-binding proteins in patients with hepatocellular carcinoma. Int J Oncol. 2005;26:311–7. [PubMed] [Google Scholar]
- 33.Zhang L, Liu Y, Hao S, Woda BA, Lu D. IMP2 expression distinguishes endometrioid from serous endometrial adenocarcinomas. Am J Surg Pathol. 2011;35:868–72. doi: 10.1097/PAS.0b013e318219c6f9. [DOI] [PubMed] [Google Scholar]
- 34.Janiszewska M, Suva ML, Riggi N, Houtkooper RH, Auwerx J, Clement-Schatlo V, et al. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev. 2012;26:1926–44. doi: 10.1101/gad.188292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelleher FC, McArthur GA. Targeting NRAS in melanoma. Cancer J. 2012;18:132–6. doi: 10.1097/PPO.0b013e31824ba4df. [DOI] [PubMed] [Google Scholar]
- 36.Jaiswal BS, Janakiraman V, Kljavin NM, Eastham-Anderson J, Cupp JE, Liang Y, et al. Combined targeting of BRAF and CRAF or BRAF and PI3K effector pathways is required for efficacy in NRAS mutant tumors. PLoS ONE. 2009;4:e5717. doi: 10.1371/journal.pone.0005717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.