Summary
Nrf2 is a master regulator of the antioxidant response. Under basal conditions Nrf2 is polyubiquitinated by the Keap1-Cul3-E3 ligase and degraded by the 26S-proteasome. In response to Nrf2 inducers there is a switch in polyubiquitination from Nrf2 to Keap1. Currently, regulation of the Nrf2-Keap1 pathway by ubiquitination is largely understood. However, the mechanism responsible for removal of ubiquitin conjugated to Nrf2 or Keap1 remains unknown. Here we report that the deubiquitinating enzyme, USP15, specifically deubiquitinates Keap1, which suppresses the Nrf2 pathway. We demonstrated that deubiquitinated-Keap1 incorporates into the Keap1-Cul3-E3 ligase complex more efficiently, enhancing the complex stability and enzymatic activity. Consequently, there is an increase in Nrf2 protein degradation and a reduction in Nrf2 target gene expression. Furthermore, USP15-siRNA enhances chemoresistance of cells through upregulation of Nrf2. These findings further our understanding of how the Nrf2-Keap1 pathway is regulated, which is imperative in targeting this pathway for chemoprevention or chemotherapy.
Keywords: Nrf2, Keap1, USP15, Cul3, ubiquitination, deubiquitination, antioxidant response, chemoresistance
Introduction
Nrf2 (NF-E2-related factor 2) is a transcription factor that regulates a battery of downstream genes that contain an antioxidant response element (ARE) in their promoters, including (i) intracellular redox-balancing proteins (glutamate cysteine ligase, GCL; heme oxygenase-1, HO-1), (ii) xenobiotic metabolizing enzymes (NAD(P)H quinone oxidoreductase-1, NQO1), and (iii) transporters (multidrug resistance-associated proteins, MRPs). Collectively, these genes function in a vast array of processes to protect against oxidative stress and harmful environmental toxicants and carcinogens. This orchestrated response is the underlying mechanism in Nrf2-mediated cell survival and protection. The Nrf2 pathway plays a protective role in many diseases where oxidative stress is thought to play an essential role in disease onset and progression, including cancer, neurodegenerative diseases, aging-related diseases, cardiovascular diseases, inflammatory diseases, pulmonary fibrosis, acute pulmonary injury, and lupus-like autoimmune nephritis (Hayes and McMahon, 2009; Hayes et al., 2010; Jeong et al., 2006; Jiang et al., 2010b; Kensler et al., 2007; Lau et al., 2008; Motohashi and Yamamoto, 2004; Zhang, 2006). Consequently, it is imperative to understand the basic molecular mechanisms of how Nrf2 is regulated so we can target this pathway to help prevent and treat these diseases.
Under basal conditions, Nrf2 protein levels remain relatively low due to negative regulation by the Cul3-Keap1-E3 ubiquitin ligase complex. Keap1, a bric-a-brac, tramtrack, broad complex (BTB) domain containing protein, binds Nrf2 and targets it for Lys-48 linked poly-ubiquitination and subsequent degradation by the 26S proteasome.
Under oxidative stressed or induced conditions the ability of the Keap1-Cul3 E3-ligase to target Nrf2 for degradation becomes impaired. Although the detailed molecular mechanism underlying this reduced Keap1-Cul3 E- ligase activity is still unclear, we previously found a switch of the polyubiquitin chain from Nrf2 to Keap1 in response to tert-butylhydroquinone (tBHQ) or sulforaphane (SF) treatment (Zhang et al., 2005). Under induced conditions, Nrf2 is stabilized and free Nrf2 translocates into the nucleus and heterodimerizes with its small-Maf binding partner to initiate transcription of ARE-bearing genes (Itoh et al., 1999; Kobayashi and Yamamoto, 2006; Motohashi et al., 2004; Villeneuve et al., 2010; Zhang, 2006; Zhang et al., 2004). Unlike Nrf2 which is conjugated with a Lys-48 linked poly-Ub chain, Keap1 is Lys-63 poly-Ub conjugated (Zhang et al., 2005). The function of Keap1 ubiquitination in response to tBHQ or SF has remained elusive until now. Here we demonstrate the significance of Keap1 ubiquitination/deubiquitination in modulating the Nrf2-dependent antioxidant response.
USP15 is a ubiquitously expressed deubiquitinating enzyme that was first discovered in 1999, and belongs to the UBP/USP (Ub-specific processing protease) family of deubiquitinating enzymes. USP15 contains many domains that are important for its function, including the Cys and His boxes that are present in all members of the UBP/USP family (Kim et al., 2003). Additionally, USP15 contains a Zn-finger that is essential for disassembling poly-Ub chains (Hetfeld et al., 2005). Since its discovery, not much has been revealed about the function of USP15. Most of the information comes from studying UBP12, the S. pombe ortholog of human USP15. UBP12 associates with the COP9 signalosome (CSN) and functions to maintain the stability of cullin ring ligase (CRL) adaptor proteins. The CSN is a conserved protein complex involved in the regulation of the ubiquitin proteasome system (UPS) (Cope and Deshaies, 2003). In addition, UBP12 removes Ub from CRL substrates, including BTB domain containing proteins, and protects CRL components from cellular-depletion by preventing auto-ubiquitination and subsequent degradation, thus, facilitating the function of CRLs (Wee et al., 2005; Wu et al., 2006; Zhou et al., 2003). Schmidt et al. demonstrated the specificity of UBP12 in stabilizing CRL components. They discovered that UBP12 regulates the stability of BTB substrate adaptors in S. pombe; however, it is not a major regulator of F-box substrate adaptors (Schmidt et al., 2009). More importantly, it has been shown that USP15 performs similar functions as UBP12. USP15 prevents autoubiquitination and degradation of CRL components, including the E3 ligase component, Rbx1 (Hetfeld et al., 2005; Huang et al., 2009). USP15 deubiquitinates IκBα and promotes its re-accumulation after TNF-α-induced degradation, leading to reduced NF-κB activity (Schweitzer et al., 2007). Recently, It is reported that USP15 can deubiquitinate receptor-activated SMADS (R-SMADS) and deubiquitination is critical for promoter recognition by SMAD complex (Eichhorn et al., 2012; Inui et al., 2011).
Our current study is the first report to describe Keap1 as a direct substrate for USP15 and demonstrate the importance of Keap1 ubiquitination status in the formation of an active Keap1-Cul3 E3 ubiquitin ligase and in regulating steady-state levels of Nrf2. More importantly, this report links downregulation of USP15 to paclitaxel resistance in cells containing a tightly regulated Nrf2-Keap1 axis.
Results
USP15 inhibits Nrf2 protein levels and expression of its target genes
To test if USP15 regulates the Nrf2-dependent pathway, immunoblot analysis was performed to determine protein expression of Nrf2 and its downstream genes. MDA-MB-231 cells were transiently transfected with an empty vector or an expression plasmid containing Myc-USP15. Myc-USP15 was shown to decrease endogenous Nrf2, NQO1, and HO-1 protein levels when compared to control, with no change in Keap1 (Figure 1A). Furthermore, siRNA directed against USP15 significantly knocked down endogenous USP15 protein levels and increased Nrf2 protein expression. As expected, USP15-siRNA resulted in a significant increase in the protein expression of NQO1 and HO-1, indicative of activation of the Nrf2-dependent antioxidant response (Figure 1B). To test the importance of the Zn-finger in the function of USP15, one of the four zinc-coordinating cysteine residues was mutated (C783A). Our results demonstrate that the Myc-USP15-C783A mutant lost its ability to inhibit Nrf2 (Figure 1C).
Figure 1. USP15 inhibits Nrf2 protein levels and expression of its target genes.
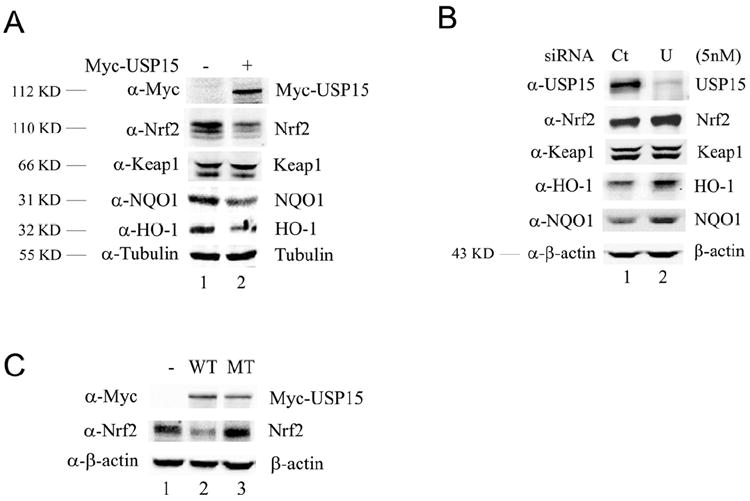
(A) MDA-MB-231 cells were transfected with an empty vector or Myc-USP15 expression vector. (B) MDA-MB-231 cells were transfected with 5 nM of Control-siRNA (Ct) or USP15-siRNA (U). (C) MDA-MB-231 cells were transfected with an expression vector for empty vector (-), Myc-USP15 (WT), or Myc-USP15-C783A (MT). Cell lysates were collected at 48 h post-transfection and subjected to immunoblot analyses using the indicated antibodies. The molecular weight each protein runs as is listed to the left of the blot.
USP15 inhibits the transcriptional activity of Nrf2 and the Nrf2-dependent antioxidant response
To investigate the effect of USP15 on Nrf2 transcriptional activity we conducted a NQO1-ARE-dependent firefly luciferase reporter gene assay. Myc-USP15 inhibited the activity of Nrf2 in a concentration-dependent manner (Figure 2A). Next, we examined the inhibitory effect of Myc-USP15 on Nrf2 transcriptional activity under both basal and induced conditions. As expected, tBHQ induced Nrf2 transcriptional activity when compared to control (Figure 2B, black bars). Interestingly, even though overexpression of Myc-USP15 impaired Nrf2 transcriptional activity, induction of Nrf2-target genes by tBHQ still produced a similar fold increase but from a lower baseline (Figure 2B, light grey bars). Conversely, the inactive Myc-USP15-C783A mutant was unable to inhibit Nrf2-dependent transcriptional activity under basal or induced conditions (Figure 2B, dark grey bars). Next, we wanted to verify that the observed decrease in Nrf2 protein expression and activity in response to ectopic expression of Myc-USP15 was not a result of inhibition of Nrf2 transcription. Real-time RT-PCR was performed using mRNA extracted from cells transiently transfected with Myc-USP15 or USP15-siRNA, which resulted in a 180-fold increase and a 63% reduction in USP15 mRNA expression, respectively (Figure 2C, USP15 panels). Upon transfection of Myc-USP15 or USP15-siRNA, Nrf2 mRNA levels remained unchanged (Figure 2C). mRNA levels of several Nrf2 downstream genes were also investigated. Overall, we observed a similar trend for all genes except AKR1C1 and AKR1B10; Myc-USP15 decreased the mRNA levels of Nrf2-downstream genes, whereas USP15-siRNA resulted in an increase (Figure 2C). Interestingly, AKR1C1 and AKR1B10, which are strongly regulated by Nrf2 (MacLeod et al., 2009; O’Connor et al., 1999), were induced to the greatest extent by USP15-siRNA; however, Myc-USP15 was unable to decrease their mRNA levels (Figure 2C). This may be due to already low basal levels of AKR1C1 and AKR1B10 in MDA-MB-231 cells, therefore Myc-USP15 could not decrease their expression further. These results indicate that USP15 may inhibit Nrf2 protein expression and thus, its activity, by increasing Nrf2 protein degradation. The functional consequence of USP15 in inhibiting the overall Nrf2-mediated response is demonstrated in Figure 2. Following ectopic expression of USP15 a significant increase in ROS was observed (Figure 2D). Taken together, these results further demonstrate that USP15 negatively regulates the Nrf2 antioxidant response.
Figure 2. USP15 inhibits the transcriptional activity of Nrf2 and the Nrf2-dependent antioxidant response.
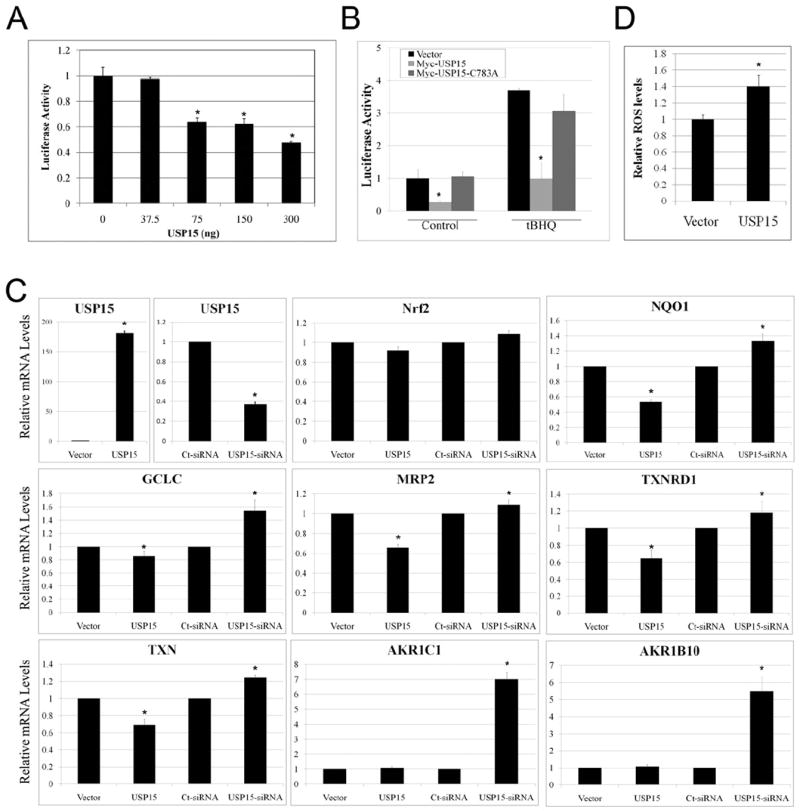
(A) Different amounts of Myc-USP15 expression plasmid were transfected into MDA-MB-231 cells, along with expression plasmids for NQO1-ARE promoter-firefly luciferase and TK-renilla luciferase as an internal control. Both firefly (F) and renilla (R) activity was measured and results are presented as F/R luciferase activity. (B) MDA-MB-231 cells were transfected with expression plasmids for empty vector, Myc-USP15, or Myc-USP15-C783A, along with expression plasmids for NQO1-ARE promoter-firefly luciferase and TK-renilla luciferase. Cells at 32 h post-transfection were left untreated or treated with tBHQ (50 μM) for 16 h prior to measuring luciferase activity. Results were normalized to empty vector transfected control which was set to 1. (C) MDA-MB-231 cells were transfected with either empty vector, Myc-USP15, Control-siRNA (Ct-siRNA), or USP15-siRNA. 48 h post-transfection (Vector and Myc-USP15) or 72 h post-transfection with siRNA, mRNA was extracted and real-time RT-PCR was performed. Values were normalized to GAPDH and controls were set equal to 1. (D) MDA-MB-231 cells were transfected with empty vector (Vector) or Myc-USP15 (USP15). 48 h post-transfection, cells were incubated with CM-H2DCFDA and fluorescence was measured. Data is presented as mean ± SD.
USP15 deubiquitinates Keap1
To elucidate the mechanism responsible for USP15-dependent negative regulation of the Nrf2 level, we further investigated the deubiquitinating properties of USP15. Previously, we reported that endogenous Keap1 is ubiquitinated under basal conditions and ubiquitination of Keap1 is markedly increased whereas ubiquitination of Nrf2 is decreased upon tBHQ treatment (Zhang et al., 2005). First, we confirmed the finding that tBHQ enhanced ubiquitination of Keap1 (Figure 3A). Next, we used an in vivo ubiquitination assay and determined that overexpression of Myc-USP15 led to a decrease in ubiquitinated Keap1 (Figure 3B). To verify this was not an artifact due to overexpression of Myc-USP15, we used siRNA to knock-down endogenous levels of USP15, which resulted in an increase in ubiquitinated-Keap1 in the absence and presence of Nrf2 (Figure 3C). These results suggest that Keap1 is a substrate for USP15. In addition, the specificity of USP15 for Keap1 was demonstrated by an in vitro deubiquitination assay. Ubiquitinated Keap1 was pulled down from cells co-transfected with CBD-Keap1 and HA-Ub and treated with tBHQ. In parallel, ubiquitinated Nrf2 was immunoprecipitated from cells co-transfected with Nrf2 and HA-Ub and treated with MG132. After washing, half of the ubiquitinated Keap1 or Nrf2 lysate was incubated with BSA and half was incubated with purified His-USP15 protein, followed by immunoblot analysis with an anti-HA antibody. His-USP15 was able to deubiquitinate CBD-Keap1, but not Nrf2 (Figure 3D). Since USP15 is known to stabilize components of CRLs and Nrf2 is normally ubiquitinated by the Cul3-Keap1-E3 ubiquitin ligase and degraded by 26S proteasome, we explored the effect of USP15-mediated deubiquitination of Keap1 on Keap1-Cul3 or Keap1-Nrf2 complex formation. First, we generated HA-Cul3-[35S] and Nrf2-[35S] using in vitro transcription/translation, then incubated them with ubiquitinated-Keap1 or deubiquitinated-Keap1 generated in the same way as described in Figure 3D. Autoradiography revealed that deubiquitinated-Keap1 (Figure 3E, lane 2) more readily forms a complex with HA-Cul3-[35S] than does ubiquitinated-Keap1 (-His-USP15, Figure 3E, lane 1). Moreover, the ubiquitination status of Keap1 did not alter its binding to Nrf2-[35S] (Figure 3E, lanes 3-4). Consequently, we hypothesized that deubiquitinated-Keap1 is the form capable of interacting with Cul3 and forming an active Cul3-Keap1-E3 ligase complex, resulting in increased degradation of Nrf2.
Figure 3. USP15 deubiquitinates Keap1.
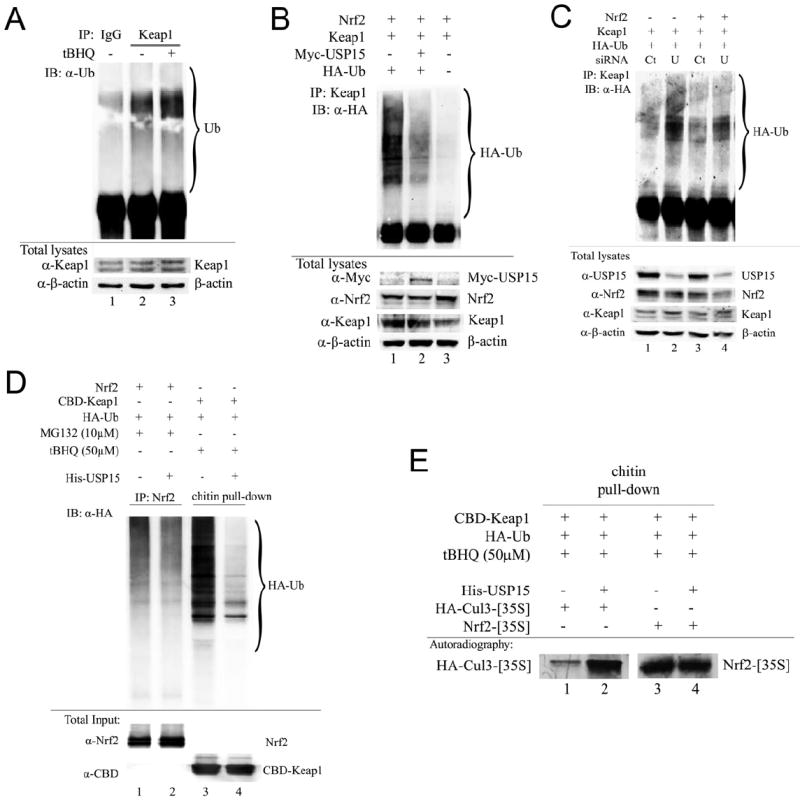
(A) MDA-MB-231 cells were left untreated or treated with 50 μM tBHQ for 4 h. Endogenous Keap1 was immunoprecipitated using an anti-Keap1 antibody and ubiquitinated Keap1 was detected by immunoblot analysis using an anti-Ub antibody. (B,C) MDA-MB-231 cells were transfected with the indicated plasmids and 5 nM control-siRNA (Ct) or USP15-siRNA (U) for 72 h. The Keap1-containing complexes were immunoprecipitated with anti-Keap1 antibodies followed by immunoblot analysis with an anti-HA antibody for detection of ubiquitin-conjugated Keap1. (D, E) Cells were transfected with the indicated plasmids for 48 h followed by treatment with either tBHQ or MG132 for 4 h prior to cell lysis. Nrf2- or CBD-Keap1-containing complexes were precipitated with anti-Nrf2/protein A beads or chitin beads, respectively, followed by incubation with BSA or His-USP15. After washing, half of the sample was eluted in sample buffer and subjected to immunoblot analysis, (E) while the other half was further incubated with HA-Cul3-[35S] or Nrf2-[35S] followed by analysis using autoradiography.
Keap1-K39R, a mutant with a major ubiquitin-accepting lysine residue substituted, is more active in targeting Nrf2 for degradation under induced conditions
Next, we attempted to make a Keap1 mutant that is active in targeting Nrf2 for degradation under basal conditions but is impaired in accepting polyubiquitin chain in response to tBHQ. We hypothesized that such a Keap1 mutant should be more active in forming a complex with Cul3 and therefore more effective in targeting Nrf2 for degradation under induced conditions. In order to identify the ubiquitin-accepting residues we generated several Keap1-mutants containing lysine to arginine mutations in the Nt+BTB, Linker or Kelch+Ct domain (Figure 4A). MDA-MB-231 cells were transiently transfected with Nrf2 expressing vector along with plasmids containing either Keap1-wild type (Keap1-WT) or each of the Keap1 mutants. The Nrf2 protein levels were significantly increased after tBHQ treatment for Keap1-WT or any mutants in the Linker or Kelch+Ct domain, but only increased slightly when cells were cotransfected with the Keap1 mutant containing lysine substitution in the Nt+BTB domain (Figure 4B). The results suggest that the lysine residues located in the Nt+BTB domain are important for Keap1 ubiquitination. Accordingly, several mutations were generated and tested for their abilities in targeting Nrf2 degradation under induced conditions. Any Keap1 mutants with K39 substituted were still capable of suppressing Nrf2 under the tBHQ-induced condition (Figure 4C). Ubiquitination assay showed that the relative ubiquitination level of Keap1 was reduced for the Keap1-K39R mutant compared to Keap1-WT, suggesting that lysine-39 in the N+BTB domain is a major ubiquitin-accepting residue (Figure 4D). Interestingly, ubiquitination of Keap1-K39R still increased after tBHQ treatment, suggesting that lysine-39 is not the only ubiquitin-accepting residue in Keap1. This is consistent with a recent report indicating that lysine-615 of Keap1 was ubiquitinated using a proteomic approach (Ooi et al., 2011). Together, these results indicate that deubiquitinated Keap1 is more active in targeting Nrf2 for degradation. Therefore, it is likely that USP15 generates more active Keap1-Cul3 E3 ligase complexes by deubiquitinating Keap1.
Figure 4. The lysine 39 is a major ubiquitin-accepting residue for Keap1.
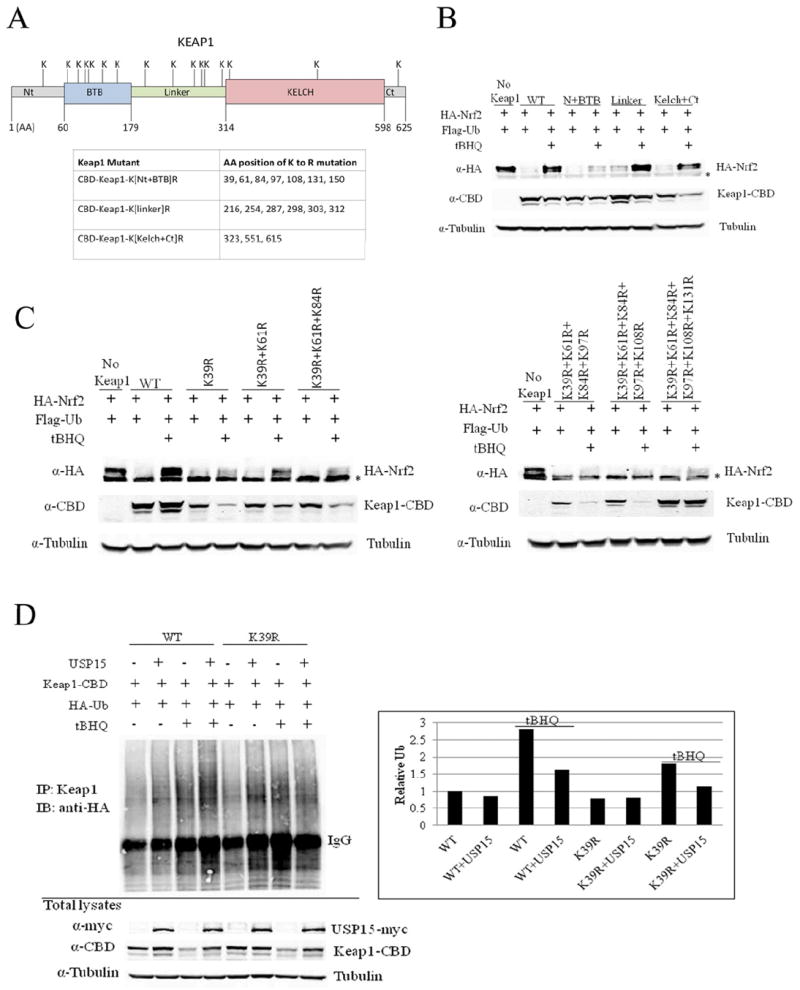
(A) Several Keap1-mutants were constructed: CBD-Keap1-K[Nt+BTB]R; CBD-Keap1-K[linker]R; CBD-Keap1-K[Kelch+Ct]R. The amino acids (AA) mutated are indicated in the table. Nt=N-terminal; Ct=C-terminal. (B) And (C) MDA-MB-231 cells were transfected with the indicated plasmids for 24 h then left untreated or treated with tBHQ (50 μM) for 16 h. Cell lysates were collected and subjected to immunoblot analysis using the indicated antibodies. (D) MDA-MB-231 cells were transfected with the indicated plasmids for 24 h then left untreated or treated with tBHQ (50 μM) for 16 h. Keap1-containing complexes were immunoprecipitated with a Keap1 antibody followed by immunoblot analysis with an HA antibody for detection of ubiquitin-conjugated Keap1. Normalized Keap1 ubiquitin levels (with the Keap1 amount in the total lysates) are shown in the right panel. *, nonspecific band
USP15 increases the degradation of Nrf2 by stabilizing the Cul3-Keap1-E3 ubiquitin ligase complex
Accordingly, we explored the effect of USP15 on the Keap1-Cul3 E3 ligase complex formation in vivo. Consistent with our in vitro results, co-immunoprecipitation analysis revealed that in the presence of Myc-USP15 there was an increase in interaction between Keap1 and Cul3 (Figure 5A). Moreover, USP15-siRNA resulted in a marked decrease in Keap1-Cul3 protein complexes (Figure 5B). These results suggest that deubiquitinated Keap1 binds Cul3 more tightly than ubiquitinated Keap1. To further illustrate this, we also utilized ts20 cells to determine if deubiquitinated-Keap1 is the active form that binds better in the Cul3-Keap1-E3 ligase complex. ts20 cells were derived from the Chinese hamster cell line, E36 (wild-type). They have a temperature-sensitive lesion in the E1 enzyme of the ubiquitination pathway. When ts20 cells, but not E36 cells, are incubated at the non-permissive temperature (39°C) the E1 enzyme is impaired and the process of ubiquitination becomes compromised (Kulka et al., 1988). Indeed, we saw a significant decrease in total ubiquitinated proteins as well as ubiquitinated-Keap1 in ts20 cells at 39°C when compared to the permissive temperature (30°C) (Figure 5C). Next, the effect of ubiquitination status on formation of the Cul3-Keap1-E3 ligase complex was examined. Our results demonstrate that when ubiquitination was inhibited in ts20 cells at 39°C, there was an overwhelming increase in Keap1-Cul3 protein complexes (Figure 5D). In contrast, there was no change in complex formation when E36 cells were incubated at 39°C. It is interesting to note that the total Keap1 protein extracted by RIPA buffer decreased slightly when the ts20 cells were switched to 39°C (Figure 5D, CBD-Keap1 panel). Since USP15 was capable of stabilizing the Cul3-Keap1-E3 ligase in vitro (Figure 3E) and in vivo (Figure 5A-D), we wanted to investigate whether this led to an increase in the Keap1-Cul3 E3 ligase activity and Nrf2 protein degradation. Therefore, the effect of USP15 on the half-life of endogenous Nrf2 protein was measured. Immunoblot analysis revealed that cells transiently transfected with Myc-USP15 had a significant increase in Nrf2 protein degradation when compared to vector transfected cells (Figure 5E). Results from this experiment were quantified and presented in the lower panel. Following ectopic expression of Myc-USP15, the half-life of Nrf2 decreased from 40 to 16 minutes (Figure 5E). Next, we investigated the effect of USP15 on Nrf2 ubiquitination status using an in vivo ubiquitination assay. Our results showed that overexpression of Myc-USP15 led to increased Nrf2 ubiquitination (Figure 5F). Taken together these results demonstrate the mechanism by which USP15 leads to decreased Nrf2 protein levels: USP15 is able to stabilize the Cul3-Keap1-E3 ligase complex through deubiquitination of Keap1, resulting in increased E3 ligase activity and ubiquitination of Nrf2, which ultimately leads to degradation of the Nrf2 protein.
Figure 5. USP15 increases the degradation of Nrf2 by stabilizing the Cul3-Keap1-E3 ubiquitin ligase complex.
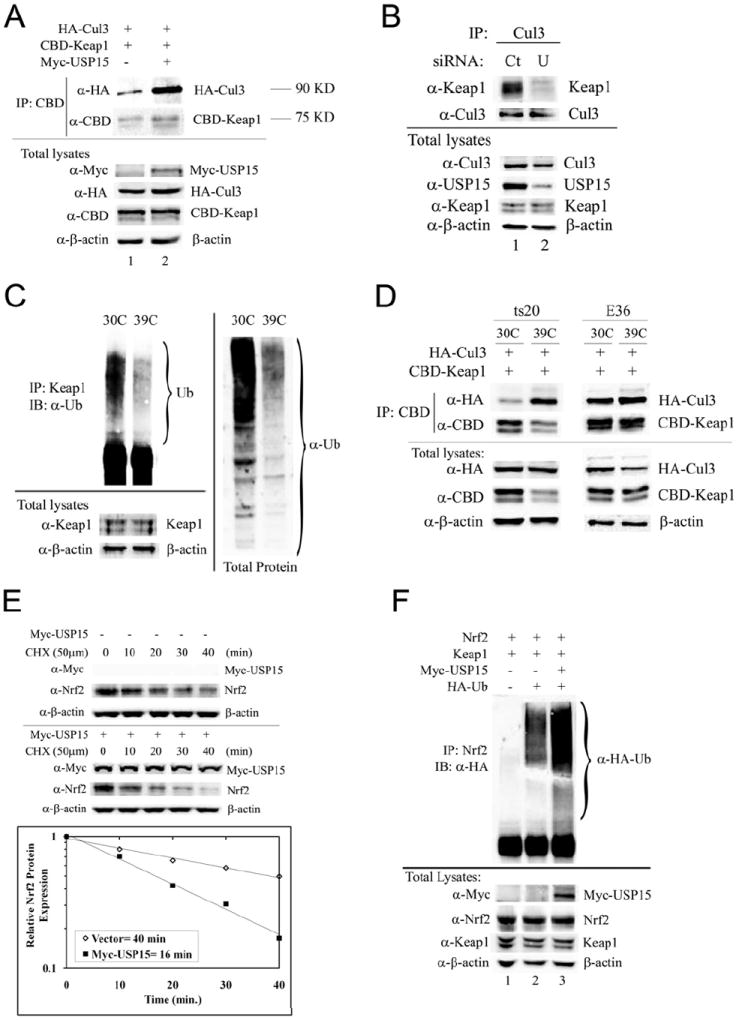
(A) MDA-MB-231 cells were transfected with the indicated plasmids for 48 h. The Keap1-containing complexes were pulled-down with chitin beads and analyzed by immunoblot with the indicated antibodies. The molecular weights HA-Cul3 and CBD-Keap1 run at are listed to the right of the blots. (B) MDA-MB-231 cells were transfected with siRNA (5 nM) for 72 h. The Cul3-containing complexes were immunoprecipitated with anti-Cul3 antibodies and analyzed by immunoblot with the indicated antibodies. (C) Endogenous Keap1 immunoprecipitated with an anti-Keap1 antibody (left panel) or total proteins (right panel) were analyzed by immunoblot using anti-Ub to detect ubiquitinated Keap1 or ubiquitination levels of all proteins. (D) ts20 or E36 cells were transfected with the indicated plasmids for 48 h. Keap1-containing complexes were immunoprecipitated with chitin beads and analyzed by immunoblot with the indicated antibodies for detection of Cul3 or Keap1. (E) MDA-MB-231 cells were transfected with an empty vector or an expression plasmid for Myc-USP15. 48 h post-transfection cells were treated with cycloheximide (CHX) for the indicated time points then cell lysates were analyzed by immunoblot using the indicated antibodies. The semi-log graph represents a quantitative analysis of the western blots. Nrf2 protein expression was normalized to β-actin and the control group was set as 1. Overexpression of USP15 reduced the half-life of Nrf2 from 40 min to 16 min. (F) MDA-MB-231 cells were transfected with the indicated plasmids followed by treatment with MG132 (10μM) for 4 h. The Nrf2-containing complexes were immunoprecipitated with an anti-Nrf2 antibody and analyzed by immunoblot analysis for detection of ubiquitin-conjugated Nrf2. Ct= control; U= USP15.
The effect of USP15 on Nrf2 protein stability is mediated through the Neh2 degron and is dependent on Keap1
The N-terminal region of Nrf2 is termed the Neh2 domain, or redox-sensitive degron (McMahon et al., 2004), which contains seven ubiquitin-accepting lysine residues (Zhang et al., 2004). The Neh2 domain confers negative regulation through interaction with Keap1, resulting in ubiquitination and subsequent degradation by the Cul3-Keap1-E3 ligase complex and the 26S proteasome (Zhang et al., 2004). The Neh6 domain of Nrf2 is recognized as the redox-insensitive degron, which is phosphorylated by GSK3-β, resulting in degradation of Nrf2 by the SCF/β-TrCP-E3 ligase complex, independent of Keap1 (Chowdhry et al., 2012; Rada et al., 2011). To further support our finding that USP15 regulates Nrf2 protein stability in a Keap1-dependent (Neh2 degron), but not Keap1-independent manner (Neh6 degron), we examined the effect of USP15 on Nrf2 in the presence or absence of Keap1, as well as its effect on several Nrf2 mutants. We found that USP15 is unable to inhibit Nrf2 protein levels in the absence of Keap1 (Figure 6A, lanes 3-4). In addition, USP15 is unable to inhibit Nrf2 protein levels when the seven lysine residues in the Neh2 domain required for ubiquitination by the Keap1-Cul3-E3 ligase complex are mutated (Nrf2-K7) (Figure 6B, lanes 3-4), or when the Neh2 domain is deleted (Figure 6B, lanes 5-6). Conversely, when the Neh6 domain of Nrf2 is deleted, USP15 is still able to inhibit Nrf2 protein expression (Figure 6B, lanes 7-8). These results demonstrate that the effect of USP15 on Nrf2 protein stability is Keap1-dependent and mediated through the Neh2 degron. This result is consistent with our model elucidating that negative regulation of Nrf2 by USP15 is through Keap1 deubiquitination to generate more active Keap1-Cul3-E3 ligase, thus enhancing Nrf2 degradation.
Figure 6. The effect of USP15 on Nrf2 protein stability is mediated through the Neh2 degron and is dependent on Keap1.
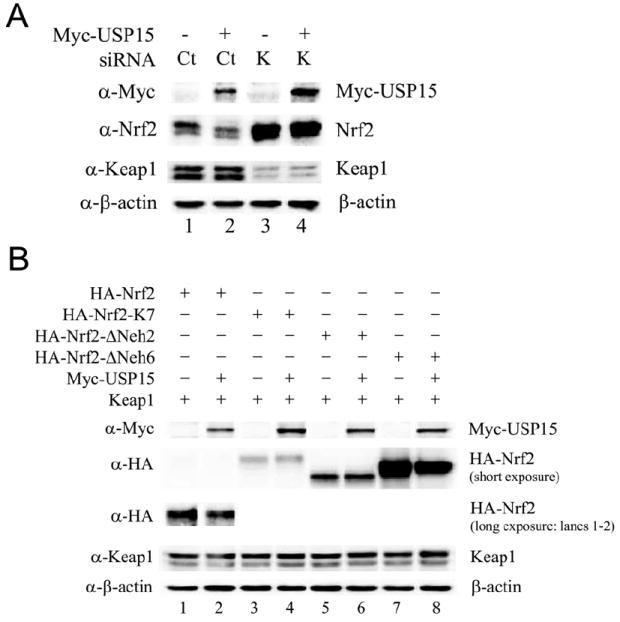
(A) MDA-MB-231 cells were transfected with empty vector or Myc-USP15 along with 5nM Control-siRNA (Ct) or Keap1-siRNA (K) for 72 h. Cell lysates were collected and subjected to immunoblot analysis using the indicated antibodies. (B) MDA-MB-231 cells were transfected with the indicated plasmids. Cell lysates were collected 48 h post-transfection and subjected to immunoblot analysis using the indicated antibodies.
USP15 –siRNA causes paclitaxel resistance through upregulation of Nrf2
Nrf2 has long been regarded as a “good” transcription factor, which is upregulated by many chemopreventive agents. However, recently the “dark-side” of Nrf2 has emerged. High protein expression of Nrf2 in cancer cells confers resistance to chemotherapeutic drugs (Jiang et al., 2010a; Lau et al., 2008; Padmanabhan et al., 2006; Ren et al., 2011; Singh et al., 2006; Wang et al., 2008). Furthermore, decreased expression of USP15 was also shown to cause paclitaxel resistance (Xu et al., 2009). Here, we link paclitaxel resistance in cells with impaired USP15 expression to high expression of Nrf2 in a Keap1-dependent manner. To prove this notion, two stable Spec2 (endometrial serous carcinoma-derived) cell lines were used, one expressing an empty vector (Spec2-Vector) and the other expressing CBD-Keap1 (Spec2-Keap1). Due to low levels of Keap1 in Spec2 cells, when compared to other endometrial cancer cells, Nrf2 is able to escape Keap1-dependent degradation by the 26S proteasome, resulting in high basal levels of Nrf2 (Jiang et al., 2010a). By stably reintroducing Keap1, Spec2-Keap1 cells have lower basal levels of Nrf2 when compared to Spec2-Vector cells. Using the MTT cell viability assay, we examined the effect of USP15 expression on resistance to paclitaxel treatment. As expected, Spec2-Keap1 cells were more sensitive to paclitaxel treatment than Spec2-Vector cells, due to their limited expression of Nrf2 (Figure 7A). Next, we compared the effects of USP15-siRNA and Control-siRNA on paclitaxel treatment in both cell lines. Our results demonstrate that USP15-siRNA had no effect on paclitaxel toxicity when compared to Control-siRNA in Spec2-Vector cells (Figure 7B). Conversely, USP15-siRNA caused resistance to paclitaxel in Spec2-Keap1 cells (Figure 7C). The reason that we did not observe any paclitaxel resistance in Spec2-Vector cells is that USP15-siRNA is unable to upregulate Nrf2 further since the basal level of Nrf2 is already high. Aliquots of cells used for the MTT assay were collected and further analyzed to verify knockdown of USP15 in response to USP15-siRNA and expression of other proteins (Figure 7D). Despite significant inhibition of USP15 protein expression, we found that USP15-siRNA had no effect on expression of Nrf2 or NQO1 in Spec2-Vector cells. Conversely, USP15-siRNA led to an increase in Nrf2 and NQO1 protein levels in Spec2-Keap1 cells (Figure 7D). To confirm that paclitaxel resistance observed when USP15 was knocked-down depends on the regulation of Nrf2, we repeated the experiment using MDA-MB-231 cells stably transfected with an empty vector (231-Vector) or HA-Nrf2 (231-Nrf2). As expected, 231-Vector cells were more sensitive to paclitaxel treatment than 231-Nrf2 cells (Figure 7E). Since basal levels of Nrf2 are low in 231-Vector cells, USP15-siRNA was able to upregulate Nrf2 (1.4-fold) and NQO1 (1.6-fold), causing resistance to paclitaxel (Figure 7F and 7H). Conversely, USP15-siRNA had no effect on paclitaxel resistance in 231-Nrf2 cells due to high basal levels of Nrf2 (Figure 7G). Moreover, USP15-siRNA did not have a significant effect on Nrf2 or NQO1 protein expression in 231-Nrf2 cells (Figure 7H). Taken together these results demonstrate that USP15-siRNA causes paclitaxel resistance through upregulation of Nrf2.
Figure 7. USP15-siRNA causes paclitaxel resistance through upregulation of Nrf2.
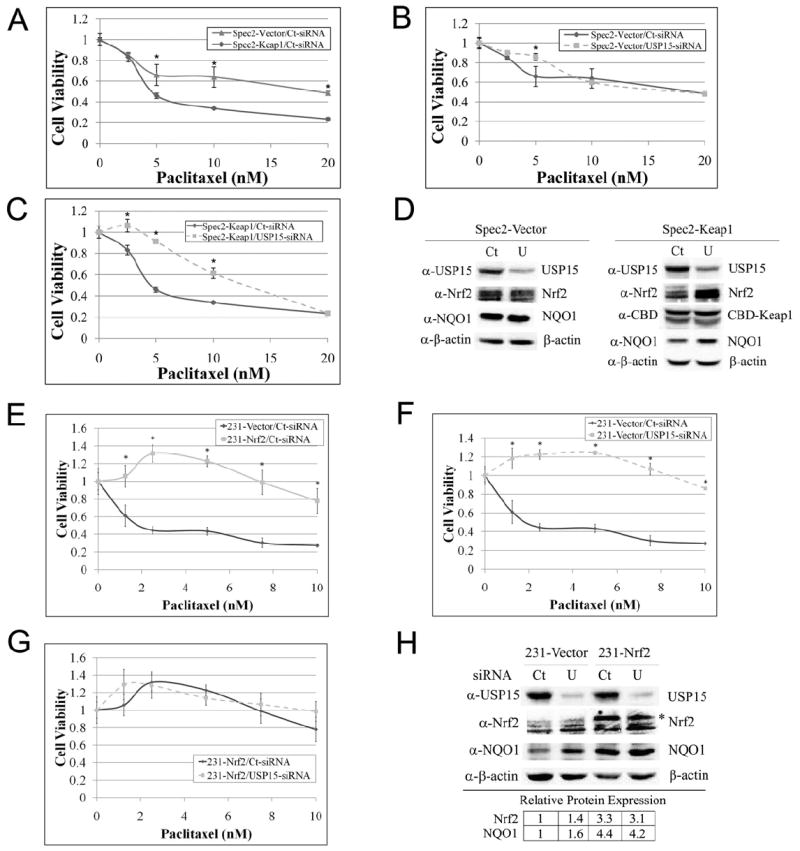
(A-C, E-G) Spec2-Vector, Spec2-Keap1, 231-Vector, or 231-Nrf2 cells were transfected with either Control-siRNA or USP15-siRNA (5 nM), followed by treatment with paclitaxel for 48 h at the indicated doses. Cell viability was measured using the MTT assay at 72 h post-transfection. Data is presented as mean ± SD. (D, H). An aliquot of cells used for the MTT assay were collected and further analyzed by immunoblot using the indicated antibodies. Relative protein expression was quantified in (H) and is presented in the lower table. Ct=Control-siRNA; U=USP15-siRNA; *= HA-Nrf2
Discussion
In this report, we identified USP15 as a negative regulator of the Nrf2-Keap1 pathway and elucidated the molecular mechanisms of regulation. USP15 is able to stabilize the Cul3-Keap1-E3 ligase complex through deubiquitination of Keap1, resulting in increased E3 ligase activity and ubiquitination of Nrf2, which ultimately leads to degradation of the Nrf2 protein. Additionally, we demonstrated the importance of the Nrf2-Keap1 pathway in USP15-dependent paclitaxel chemoresistance.
The S. pombe ortholog of USP15, UBP12, is best known for deubiquitinating E3-ligase components through association with the CSN and preventing autocatalytic destruction (Wee et al., 2005; Zhou et al., 2003). UBP12 has been shown to bind CSN1, CSN3, CSN5, and CSN7 in S. pombe. In addition, it was estimated that 50% of UBP12 co-fractionated with the CSN subunits using gel filtration analysis, suggesting a physical interaction between UBP12 and the CSN complex in S. pombe (Zhou et al., 2003). Moreover, it was shown that USP15 co-purified with subunits of the CSN complex in human erythrocytes (Hetfeld et al., 2005). In this study, we performed in vitro and in vivo binding assays and found that USP15 was not associated with Keap1 or Nrf2 but weakly associated with Cul3 in vivo (data not shown). We believe that the interaction between USP15 and the Keap1-Cul3-E3 ligase complex is most likely mediated by the CSN complex and is therefore difficult to detect due to the dynamic interaction between the E3 ligase and the CSN complex.
Human USP15 has recently been shown to function similarly to UBP12: The CSN and USP15 control the Wnt/β-catenin signaling pathway through deneddylation and USP15-dependent stabilization of adenomatous polyposis coli (Huang et al., 2009); USP15 and CSN5 are required for processing poly-ubiquitinated substrates bound to p97/VCP, a homohexameric ATPase that forms an ATP-dependent supercomplex resembling the proteasome-regulatory particle (Cayli et al., 2009); USP15 stabilizes human papillomavirus type 16 E6 protein stability (Vos et al., 2009); USP15 destabilizes the microtubule end-binding protein 1, a substrate of the UPS, further verifying that USP15 protects CRL components, resulting in increased E3-ligase activity and degradation of their substrates (Peth et al., 2007). In a similar fashion, we showed that USP15 increased the activity of the Cul3-Keap1-E3 ligase complex, resulting in increased degradation of Nrf2. Since ubiquitination of Keap1 does not target Keap1 for degradation, deubiquitination by USP15 did not alter Keap1 protein expression, but it did stabilize the Cul3-Keap1-E3 ligase and increase its activity (Figure 3E, 4 and 5).
Deubiquitinating enzymes play a role in a wide array of cellular processes including, signal transduction, protein degradation, transcriptional regulation, cell cycle regulation, and DNA repair. As a result, they have also been linked to many human diseases, including cancer (Reyes-Turcu et al., 2009). To date, a link between the deubiquitinating enzyme, USP15, and cancer has not been established. Nonetheless, USP15 was shown to have varying activity in many different cancer cell lines, including cervical (HeLa), colon (CoLo), lung (U1906), brain (SH-SY-5Y), kidney (HEK293), and many hematopoietic cell lines (Ovaa et al., 2004), as well as many additional cervical cancer cell lines (Rolen et al., 2006). Despite the fact that USP15 has not been linked directly to cancer, it has been shown to play a role in chemoresistance. USP15 showed a decrease in gene expression in docetaxel-resistant gastric cancer cells when compared to parental cells using microarray analysis. These results were validated using real-time RT-PCR. Further analysis was conducted in 11 cancer cell lines of the digestive system and again, USP15 was significantly correlated to docetaxel sensitivity (Xie et al., 2010). Additionally, paclitaxel-resistant human ovarian cancer samples have lower levels of USP15 mRNA than paclitaxel-sensitive samples. Xu et al. also demonstrated that USP15-siRNA led to chemoresistance in a HeLa cell based model (Xu et al., 2009). Based on the link between Nrf2 and chemoresistance (Jiang et al., 2010a; Ohta et al., 2008; Shibata et al., 2008; Wang et al., 2008), we wanted to demonstrate that USP15-siRNA regulates paclitaxel-chemoresistance in part due to its ability to upregulate Nrf2 protein expression through suppression of Keap1 deubiquitination. Our results demonstrated that USP15-siRNA led to paclitaxel resistance in Spec2-Keap1 and 231-Vector cells. However, USP15-siRNA had no effect in Spec2-Vector cells or in 231-Nrf2 cells (Figure 7), where basal Nrf2 protein levels are high. Taken together, these results demonstrate that activation of the Nrf2-Keap1 pathway may be responsible for paclitaxel chemoresistance observed in cancer cells with decreased expression of USP15.
The Nrf2-Keap1 antioxidant response pathway plays an important role in chemoprevention and cancer therapy. Consequently, tight regulation is imperative to prevent the onset and progression of cancer. Mutations in Nrf2 or Keap1 can lead to constitutive activation of Nrf2, a consequence which has been observed in many cancer tumors and cancer cell lines. Tight regulation of the Nrf2-Keap1 pathway is maintained by the Cul3-Keap1-E3 ligase complex. To date, the mechanisms involved in maintaining low basal levels of Nrf2 through ubiquitination are very well characterized. In this report, we identified a novel mechanism of Nrf2 regulation by USP15. This is the first report to demonstrate the role of deubiquitination in regulating the Nrf2-Keap1 pathway. The function, targets, and regulation of numerous deubiquitinating enzymes still remain unknown. Thus, any information we can unravel will help us understand the important roles that deubiquitinating enzymes play in many cellular processes. Moreover, we determined the role of Keap1-ubiquitination in regulating Nrf2. These findings further our understanding of how the Nrf2-Keap1 pathway is regulated, which is imperative in targeting this pathway for chemoprevention or chemotherapy. Our results represent a potentially significant discovery because our data suggest that agents that antagonize USP15 could be therapeutically useful to prevent disease onset and progression through activation of Nrf2.
Experimental Procedures
Recombinant DNA
The Keap1 mutants: CBD-Keap1-K[Nt+BTB]R; CBD-Keap1-K[linker]R and CBD-Keap1-K[Kelch+Ct]R were generated using site-directed mutagenesis. For others, see previous publications (Hetfeld et al., 2005; Sun et al., 2009; Zhang and Hannink, 2003).
Chemicals, Cell Culture, and Transfection
All chemicals were purchased from Sigma Chemical Co. (St. Louis, MO, USA). For cell culture and transfection, see our previous publications (Jiang et al., 2010a; Wang et al., 2008)
Immunoblot, Immunoprecipitation, and Antibodies
Immunoblot and immunoprecipitation were performed and analyzed as previously described (Lau et al., 2010). All the antibodies used were from Santa Cruz Biotechnology (Santa Cruz, CA, USA) except anti-CBD, which was purchased from New England Biolabs (Ipswich, MA, USA).
Luciferase Reporter Gene Assay
The luciferase reporter gene assay was performed as described in previous publication (Lau et al., 2010) and activity was measured using the dual-luciferase reporter assay system according to manufacturer’s instructions (Promega, Madison, WI, USA).
Protein Half-Life Analysis
Cells were transiently transfected with either empty-vector or Myc-USP15. 48 h post-transfection cells were treated with cycloheximide (CHX, 50 μM) for the indicated time points. Cell lysate was subjected to immunoblot analysis. Quantification and analysis is described as previous (Ren et al., 2011).
mRNA Extraction and qRTPCR
Total mRNA was extracted from cells using TRIZOL reagent (Invitrogen), and equal amounts of RNA were reverse-transcripted to cDNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Indianapolis, IN, USA). The Taqman probes and primers were reported previously (Lau et al., 2010). The Taqman probe used for hUSP15 was #9 and the forward and reverse primers were as follows: hUSP15: forward gacccattgataactctggacttc, reverse ccaattcatcaataaggtgttcc. The real-time PCR reaction was performed as previously described (Lau et al., 2010). The data is expressed as relative mRNA levels and is normalized to GAPDH.
in vivo Ubiquitination Assay
The in vivo ubiqitination assay was performed as previously described (Lau et al., 2010).
in vitro Deubiquitination Assay
Ubiquitinated Keap1 or Nrf2 were generated in cells. Cells were transfected with CBD-Keap1 or Nrf2 plus HA-Ub for 48 h then treated with tBHQ (for Keap1) or MG132 (for Nrf2) for 4 h. Cells were lysed in RIPA buffer containing 1 mM DTT, 1 mM PMSF, and a protease inhibitor cocktail, and incubated with chitin beads (for Keap1) or protein A beads with an Nrf2 antibody (for Nrf2) overnight. Immunoprecipitated complexes were washed, and half of the ubiquitinated Keap1 or Nrf2 lysate was incubated with 1 μg BSA and half was incubated with purified His-USP15 protein (Enzo Life Sciences), and then washed with RIPA buffer before elution in sample buffer. Samples were then subjected to immunoblot analysis. In Figure 3E, following the second wash the samples were incubated with HA-Cul3-[35S] or Nrf2-[35S] and then washed with RIPA buffer 3X before immunoprecipitated complexes were eluted, and subjected to SDS-PAGE and autoradiography.
MTT Assay
Cell viability was measured using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. MTT analysis was performed as previously described (Wang et al., 2007).
ROS Detection
ROS analysis was performed as previously described using CM-H2DCFDA to measure fluorescence (Chen et al., 2009).
Soluble and insoluble fractionation
Cells were lysed in a soluble fraction extraction buffer [50 mM Tris-HCL (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM DTT, 1 mM PMSF, and a protease inhibitor cocktail]. Cells were incubated on ice for 30 min then centrifuged at 12,000 rpm for 10 minutes at 4°C. The supernatant was collected as the soluble fraction. Next, the pellet was washed twice with extraction buffer, then 1X sample buffer was added and the pellet was sonicated (insoluble fraction). Samples were boiled then subjected to immunoblot analysis.
Statistical Analysis
Experiments were conducted in triplicate, and data are shown as mean ± SD. Statistical analysis was performed using two-tailed Student’s t-tests to compare means. Significance was set at p ≤ 0.05. In Figure 2B data were analyzed employing one-way analysis of variance (ANOVA) with Tukey’s post hoc test using the Prism 4.0 software. Differences were considered significant at p ≤ 0.05.
Highlights.
USP15 inhibits Nrf2-mediated antioxidant response via increased Nrf2 degradation.
Keap1 is a direct substrate for the deubiquitinating enzyme, USP15.
Deubiquitination of Keap1 stabilizes the Cul3-Keap1-E3 ligase complex.
USP15-siRNA enhances chemoresistance of cells through upregulation of Nrf2.
Acknowledgments
We would like to thank Dr. Wolfgang Dubiel for his generous contribution of the USP15 constructs. This work was supported by the following grants: NIEHS 2R01 ES015010 and NCI R01 CA154377 to DDZ; ES007091 and ES016652 to NFV, and A center grant P30ES006694.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cayli S, Klug J, Chapiro J, Frohlich S, Krasteva G, Orel L, Meinhardt A. COP9 signalosome interacts ATP-dependently with p97/valosin-containing protein (VCP) and controls the ubiquitination status of proteins bound to p97/VCP. J Biol Chem. 2009;284:34944–34953. doi: 10.1074/jbc.M109.037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Sun Z, Wang XJ, Jiang T, Huang Z, Fang D, Zhang DD. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell. 2009;34:663–673. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, Hayes JD. Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene. 2012 doi: 10.1038/onc.2012.388. in-press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope GA, Deshaies RJ. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell. 2003;114:663–671. doi: 10.1016/s0092-8674(03)00722-0. [DOI] [PubMed] [Google Scholar]
- Eichhorn PJ, Rodon L, Gonzalez-Junca A, Dirac A, Gili M, Martinez-Saez E, Aura C, Barba I, Peg V, Prat A, et al. USP15 stabilizes TGF-beta receptor I and promotes oncogenesis through the activation of TGF-beta signaling in glioblastoma. Nat Med. 2012;18:429–435. doi: 10.1038/nm.2619. [DOI] [PubMed] [Google Scholar]
- Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Hayes JD, McMahon M, Chowdhry S, Dinkova-Kostova AT. Cancer chemoprevention mechanisms mediated through the keap1-nrf2 pathway. Antioxid Redox Signal. 2010;13:1713–1748. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- Hetfeld BK, Helfrich A, Kapelari B, Scheel H, Hofmann K, Guterman A, Glickman M, Schade R, Kloetzel PM, Dubiel W. The zinc finger of the CSN-associated deubiquitinating enzyme USP15 is essential to rescue the E3 ligase Rbx1. Curr Biol. 2005;15:1217–1221. doi: 10.1016/j.cub.2005.05.059. [DOI] [PubMed] [Google Scholar]
- Huang X, Langelotz C, Hetfeld-Pechoc BK, Schwenk W, Dubiel W. The COP9 signalosome mediates beta-catenin degradation by deneddylation and blocks adenomatous polyposis coli destruction via USP15. J Mol Biol. 2009;391:691–702. doi: 10.1016/j.jmb.2009.06.066. [DOI] [PubMed] [Google Scholar]
- Inui M, Manfrin A, Mamidi A, Martello G, Morsut L, Soligo S, Enzo E, Moro S, Polo S, Dupont S, et al. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat Cell Biol. 2011;13:1368–1375. doi: 10.1038/ncb2346. [DOI] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong WS, Jun M, Kong AN. Nrf2: a potential molecular target for cancer chemoprevention by natural compounds. Antioxid Redox Signal. 2006;8:99–106. doi: 10.1089/ars.2006.8.99. [DOI] [PubMed] [Google Scholar]
- Jiang T, Chen N, Zhao F, Wang XJ, Kong B, Zheng W, Zhang DD. High levels of Nrf2 determine chemoresistance in type II endometrial cancer. Cancer Res. 2010a;70:5486–5496. doi: 10.1158/0008-5472.CAN-10-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, Huang Z, Lin Y, Zhang Z, Fang D, Zhang DD. The protective role of Nrf2 in streptozotocin-induced diabetic nephropathy. Diabetes. 2010b;59:850–860. doi: 10.2337/db09-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- Kim JH, Park KC, Chung SS, Bang O, Chung CH. Deubiquitinating enzymes as cellular regulators. J Biochem. 2003;134:9–18. doi: 10.1093/jb/mvg107. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enzyme Regul. 2006;46:113–140. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Kulka RG, Raboy B, Schuster R, Parag HA, Diamond G, Ciechanover A, Marcus M. A Chinese hamster cell cycle mutant arrested at G2 phase has a temperature-sensitive ubiquitin-activating enzyme, E1. J Biol Chem. 1988;263:15726–15731. [PubMed] [Google Scholar]
- Lau A, Villeneuve NF, Sun Z, Wong PK, Zhang DD. Dual roles of Nrf2 in cancer. Pharmacol Res. 2008;58:262–270. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod AK, McMahon M, Plummer SM, Higgins LG, Penning TM, Igarashi K, Hayes JD. Characterization of the cancer chemopreventive NRF2-dependent gene battery in human keratinocytes: demonstration that the KEAP1-NRF2 pathway, and not the BACH1-NRF2 pathway, controls cytoprotection against electrophiles as well as redox-cycling compounds. Carcinogenesis. 2009;30:1571–1580. doi: 10.1093/carcin/bgp176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J Biol Chem. 2004;279:31556–31567. doi: 10.1074/jbc.M403061200. [DOI] [PubMed] [Google Scholar]
- Motohashi H, Katsuoka F, Engel JD, Yamamoto M. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proc Natl Acad Sci U S A. 2004;101:6379–6384. doi: 10.1073/pnas.0305902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- O’Connor T, Ireland LS, Harrison DJ, Hayes JD. Major differences exist in the function and tissue-specific expression of human aflatoxin B1 aldehyde reductase and the principal human aldo-keto reductase AKR1 family members. Biochem J. 1999;343(Pt 2):487–504. [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, Suzuki T, Kobayashi A, Yokota J, Sakiyama T, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- Ooi A, Wong JC, Petillo D, Roossien D, Perrier-Trudova V, Whitten D, Min BW, Tan MH, Zhang Z, Yang XJ, et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell. 2011;20:511–523. doi: 10.1016/j.ccr.2011.08.024. [DOI] [PubMed] [Google Scholar]
- Ovaa H, Kessler BM, Rolen U, Galardy PJ, Ploegh HL, Masucci MG. Activity-based ubiquitin-specific protease (USP) profiling of virus-infected and malignant human cells. Proc Natl Acad Sci U S A. 2004;101:2253–2258. doi: 10.1073/pnas.0308411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, Yamamoto M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Peth A, Boettcher JP, Dubiel W. Ubiquitin-dependent proteolysis of the microtubule end-binding protein 1, EB1, is controlled by the COP9 signalosome: possible consequences for microtubule filament stability. J Mol Biol. 2007;368:550–563. doi: 10.1016/j.jmb.2007.02.052. [DOI] [PubMed] [Google Scholar]
- Rada P, Rojo AI, Chowdhry S, McMahon M, Hayes JD, Cuadrado A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol. 2011;31:1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Villeneuve NF, Jiang T, Wu T, Lau A, Toppin HA, Zhang DD. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc Natl Acad Sci U S A. 2011;108:1433–1438. doi: 10.1073/pnas.1014275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolen U, Kobzeva V, Gasparjan N, Ovaa H, Winberg G, Kisseljov F, Masucci MG. Activity profiling of deubiquitinating enzymes in cervical carcinoma biopsies and cell lines. Mol Carcinog. 2006;45:260–269. doi: 10.1002/mc.20177. [DOI] [PubMed] [Google Scholar]
- Schmidt MW, McQuary PR, Wee S, Hofmann K, Wolf DA. F-box-directed CRL complex assembly and regulation by the CSN and CAND1. Mol Cell. 2009;35:586–597. doi: 10.1016/j.molcel.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer K, Bozko PM, Dubiel W, Naumann M. CSN controls NF-kappaB by deubiquitinylation of IkappaBalpha. Embo J. 2007;26:1532–1541. doi: 10.1038/sj.emboj.7601600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, Yamamoto M, Hirohashi S. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 2008;135:1358–1368. 1368 e1351–1354. doi: 10.1053/j.gastro.2008.06.082. [DOI] [PubMed] [Google Scholar]
- Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Chin YE, Zhang DD. Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response. Mol Cell Biol. 2009;29:2658–2672. doi: 10.1128/MCB.01639-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve NF, Lau A, Zhang DD. Regulation of the nrf2-keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases. Antioxid Redox Signal. 2010;13:1699–1712. doi: 10.1089/ars.2010.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos RM, Altreuter J, White EA, Howley PM. The ubiquitin-specific peptidase USP15 regulates human papillomavirus type 16 E6 protein stability. J Virol. 2009;83:8885–8892. doi: 10.1128/JVI.00605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Sun Z, Chen W, Eblin KE, Gandolfi JA, Zhang DD. Nrf2 protects human bladder urothelial cells from arsenite and monomethylarsonous acid toxicity. Toxicol Appl Pharmacol. 2007;225:206–213. doi: 10.1016/j.taap.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, Chen W, Yi X, Zheng W, Wondrak GT, et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee S, Geyer RK, Toda T, Wolf DA. CSN facilitates Cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat Cell Biol. 2005;7:387–391. doi: 10.1038/ncb1241. [DOI] [PubMed] [Google Scholar]
- Wu JT, Chan YR, Chien CT. Protection of cullin-RING E3 ligases by CSN-UBP12. Trends Cell Biol. 2006;16:362–369. doi: 10.1016/j.tcb.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Xie L, Wei J, Qian X, Chen G, Yu L, Ding Y, Liu B. CXCR4, a potential predictive marker for docetaxel sensitivity in gastric cancer. Anticancer Res. 2010;30:2209–2216. [PubMed] [Google Scholar]
- Xu M, Takanashi M, Oikawa K, Tanaka M, Nishi H, Isaka K, Kudo M, Kuroda M. USP15 plays an essential role for caspase-3 activation during Paclitaxel-induced apoptosis. Biochem Biophys Res Commun. 2009;388:366–371. doi: 10.1016/j.bbrc.2009.08.015. [DOI] [PubMed] [Google Scholar]
- Zhang DD. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev. 2006;38:769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
- Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Sun Z, Habib GM, Lieberman MW, Hannink M. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J Biol Chem. 2005;280:30091–30099. doi: 10.1074/jbc.M501279200. [DOI] [PubMed] [Google Scholar]
- Zhou C, Wee S, Rhee E, Naumann M, Dubiel W, Wolf DA. Fission yeast COP9/signalosome suppresses cullin activity through recruitment of the deubiquitylating enzyme Ubp12p. Mol Cell. 2003;11:927–938. doi: 10.1016/s1097-2765(03)00136-9. [DOI] [PubMed] [Google Scholar]


