Abstract
Studies on hydration are important for better understanding of structure and function of nucleic acids. We compared the hydration of self-complementary DNA, RNA and 2′-O-methyl (2′-OMe) oligonucleotides GCGAAUUCGC, (UA)6 and (CG)3 using the osmotic stressing method. The number of water molecules released upon melting of oligonucleotide duplexes, ΔnW, was calculated from the dependence of melting temperature on water activity and the enthalpy, both measured with UV thermal melting experiments. The water activity was changed by addition of ethylene glycol, glycerol and acetamide as small organic co-solutes. The ΔnW was 3–4 for RNA duplexes and 2–3 for DNA and 2′-OMe duplexes. Thus, the RNA duplexes were hydrated more than the DNA and the 2′-OMe oligonucleotide duplexes by approximately one to two water molecules depending on the sequence. Consistent with previous studies, GC base pairs were hydrated more than AU pairs in RNA, whereas in DNA and 2′-OMe oligonucleotides the difference in hydration between these two base pairs was relatively small. Our data suggest that the better hydration of RNA contributes to the increased enthalpic stability of RNA duplexes compared with DNA duplexes.
INTRODUCTION
Understanding the hydration of biopolymers is important in revealing the most basic mechanisms of biological processes. Recent studies have shown that water is an important contributor to both the affinity and specificity of protein–protein, protein–DNA and drug–DNA interactions (1,2). Water is an integral part of most protein–protein and protein–DNA interfaces, which usually contain at least the same number of water-mediated interactions as direct hydrogen bonds and salt bridges (3). Crystal structure of an early assembly of the signal recognition particle revealed a protein–RNA interface with a complex network of highly ordered water molecules (4). Qu and Chaires found that water was an important participant in the formation of certain DNA intercalation complexes (5,6). Binding of some intercalators is accompanied by the uptake of as many as 30 water molecules. These studies strongly suggest that there is a fine interplay between molecular recognition and hydration of biopolymers.
Our knowledge of nucleic acid hydration is shaped mostly by atomic resolution crystal structures that identify exact sites occupied by water molecules in the immediate hydration layers (7–9). NMR (10–12) and molecular modeling (13–15) studies provide further information on dynamics of water molecules occupying these sites. Although these methods have greatly advanced our understanding of hydration phenomenon, the subtle connection between biopolymer hydration and molecular recognition is still not completely understood.
As a part of our program on design of synthetic biopolymer mimics, we were interested in the relationship between chemical structure and nucleic acid hydration, thermal stability and conformation. Herein we used the osmotic stressing method (16,17) as a straightforward assay to compare the hydration of DNA, RNA and 2′-O-methyl (2′-OMe) modified oligonucleotides. Any equilibria that involve changes in the number of water molecules associated with the biopolymer (e.g. dissociation of a DNA double helix) are sensitive to changes in water activity. Water activity can in turn be manipulated by addition of low molecular weight co-solutes. The osmotic stressing method monitors the depression of DNA melting temperature upon addition of small organic molecules (ethylene glycol, glycerol, etc.) (16). Small molecules in solution are excluded from the immediate vicinity of the biopolymer which leads to preferential hydration of its surface. The osmotic stressing method assumes that the effect of small molecules is to change the water activity only, the excluded co-solutes do not directly interact with the biopolymer. A straightforward calculation gives the number of water molecules that are uniquely bound to the double helix, but released upon melting (17). The method is experimentally simple, fast and does not require sophisticated equipment.
Using the osmotic stressing method, Spink and Chaires (17) found that about four water molecules per base pair were released upon melting of long DNA duplexes [Escherichia coli DNA and poly(dA)-poly(dT)]. Herein we used this methodology to detect differences in hydration of short DNA, RNA and 2′-OMe oligonucleotides. We found that formation of RNA duplexes involved one to two more water molecules (depending on the sequence) than formation of DNA and 2′-OMe duplexes, supporting the general notion that RNA is hydrated more than DNA. Our data support previous suggestions that the increased hydration of RNA contributes to the increased thermal stability of RNA duplexes compared to DNA duplexes (18).
MATERIALS AND METHODS
Oligonucleotides
Oligodeoxyribonucleotides were purchased crude (desalted) from Sigma-Genosys and purified using reverse-phase HPLC. For sample preparation, the crude oligonuclotide was dissolved in water (0.1 ml) and loaded on a Discovery® DSC-18 solid phase extraction tube (1 ml, 100 mg, Supelco). The oligonucleotides were eluted with water (1.5 ml), freeze-dried, dissolved in water (0.1 ml) and filtered through Millex-GN 0.20 µm Nylon filter (4 mm, Millipore). The purification was done on Shimadzu VP gradient HPLC system using a Discovery® BIO Wide Pore C18 column (10 × 250 mm, 5 µm) kept at 50°C using a linear gradient of acetonitrile (2–13% in 15 min) in 50 mM triethylammonium acetate buffer (pH 6.5) with a flow rate of 5 ml/min. The fractions containing oligodeoxyribonucleotides were freeze-dried, dissolved in water and freeze-dried three times to remove residual buffer salts.
Oligoribonucleotides and 2′-OMe oligonucleotides were purchased from MWG-Biotech AG as HPLC-purified material. The quality was checked using reverse-phase HPLC on a Discovery® HS C18 column (4.6 × 250 mm, 3 µm) kept at 50°C using a linear gradient of acetonitrile (2–30% in 30 min) in 50 mM triethylammonium acetate buffer (pH 6.5) with a flow rate of 1 ml/min.
UV meltings
Melting of each oligonucleotide (2 µM) was done in 10 mM sodium cacodylate, 0.1 mM EDTA, and 300 mM NaCl in the presence of 0, 5, 10, 15 and 20% w/v of each of the four osmolytes in Table 2. Oligonucleotide concentrations were determined using data (OD per nmol) provided by suppliers (Sigma-Genosys and MWG-Biotech AG). Absorbance versus temperature profiles were measured at 260 nm (Table 1, 1 and 2) or 280 nm (Table 1, 3 and 4) on a Varian Bio 100 spectrometer equipped with a six-position Peltier temperature controller. The temperature was increased at different rates ranging from 0.1 to 0.5°C/min. Five samples were measured concurrently in the double-beam mode. To increase the accuracy of measurements the sixth position was used to record the temperature data points using a temperature probe directly in a cuvette. At temperatures below 15°C the sample compartment was flushed with dry nitrogen gas.
Table 2. Values of ΔnW (per base pair).
| Sequence | Oligonucleotide | ΔnW | ΔnW | ΔnW | ΔnW |
|---|---|---|---|---|---|
| Ethylene glycol | Glycerol | Acetamide | Sucrose | ||
| 1a | d(GCGAAUUCGC) | 2.7 ± 0.6 | 2.3 ± 0.8 | 3.8 ± 0.9 | 8.1 ± 3.2 |
| 1b | r(GCGAAUUCGC) | 4.4 ± 0.6 | 3.4 ± 0.9 | 6.1 ± 0.6 | 9.0 ± 3.0 |
| 1c | 2′-OMe(GCGAAUUCGC) | 3.1 ± 0.5 | 2.8 ± 0.7 | 3.5 ± 0.5 | 7.9 ± 2.0 |
| 1d | d(GCGAATTCGC) | 4.1 ± 0.8 | 2.5 ± 0.8 | 5.2 ± 1.0 | 8.6 ± 2.9 |
| 2a | d(UA)6 | 1.8 ± 0.5 | 2.2 ± 0.6 | 4.3 ± 0.5 | ndb |
| 2b | r(UA)6 | 3.0 ± 0.5 | 3.3 ± 0.5 | 5.6 ± 0.5 | ndb |
| 2c | 2′-OMe(UA)6 | 2.1 ± 0.5 | 2.6 ± 0.5 | 4.5 ± 0.4 | ndb |
| 2d | d(TA)6 | 1.8 ± 0.6 | 2.2 ± 0.7 | 3.2 ± 0.5 | ndb |
| 3a | d(CG)3 | 2.4 ± 0.5 | 2.6 ± 0.5 | 4.3 ± 0.5 | ndb |
| 3b | r(CG)3 | 4.3 ± 0.7 | 4.3 ± 0.8 | 5.8 ± 0.9 | ndb |
| 3c | 2′-OMe(CG)3 | 2.6 ± 0.7 | 2.6 ± 1.2 | 3.1 ± 0.6 | ndb |
| 4b | r(CGCAAAUUUGCG) | 1.9a | 1.5a | 3.6a | ndb |
| 4d | d(CGCAAATTTGCG) | 2.3a | 1.8a | 3.7a | ndb |
aThese values may contain significant systematic errors due to discrepancies in –ΔH values (see Table 1).
bNot determined.
Table 1. Melting temperatures (tm) and thermodynamic parameters (ΔH and ΔS) for oligonucleotide duplexes.
| Sequence | Oligonucleotide | tma | –ΔHa | –ΔHa | –ΔHb | –ΔSa |
|---|---|---|---|---|---|---|
| (van’t Hoff) | (van’t Hoff) | [δα/δ(1/Tm) versus Tm] | (1/Tm versus lnC) | (van’t Hoff) | ||
| °C | kcal/mol | kcal/mol | kcal/mol | eu | ||
| 1a | d(GCGAAUUCGC) | 42.6 ± 0.6 | 63.8 ± 4.8 | 63.0 ± 6.8 | 62.9 ± 8.1 | 176 ± 15 |
| 1b | r(GCGAAUUCGC) | 53.8 ± 0.5 | 96.4 ± 5.8 | 94.1 ± 4.7 | 114.3 ± 6.9 | 269 ± 17 |
| 1c | 2′-OMe(GCGAAUUCGC) | 62.6 ± 0.5 | 88.5 ± 3.1 | 87.6 ± 4.6 | 97.8 ± 3.9 | 239 ± 7 |
| 1d | d(GCGAATTCGC) | 47.9 ± 0.6 | 75.8 ± 3.9 | 70.1 ± 5.8 | 61.7 ± 3.9 | 210 ± 12 |
| 2a | d(UA)6 | 18.2 ± 0.5 | 56.6 ± 3.3 | 51.6 ± 3.0 | 67.8 ± 5.3 | 169 ± 12 |
| 2b | r(UA)6 | 30.0 ± 0.3 | 70.3 ± 4.0 | 82.4 ± 4.2 | 86.2 ± 5.7 | 206 ± 13 |
| 2c | 2′-OMe(UA)6 | 42.2 ± 0.2 | 67.4 ± 3.8 | 95.3 ± 7.1 | 97.0 ± 9.4 | 188 ± 12 |
| 2d | d(TA)6 | 22.9 ± 0.7 | 41.4 ± 4.1 | 39.5 ± 3.3 | 62.9 ± 4.7 | 113 ± 14 |
| 3a | d(CG)3 | 39.2 ± 0.4 | 60.2 ± 3.3 | 59.2 ± 3.3 | 57.4 ± 4.2 | 168 ± 13 |
| 3b | r(CG)3 | 43.0 ± 0.3 | 63.9 ± 2.5 | 57.7 ± 3.7 | 58.7 ± 4.4 | 176 ± 8 |
| 3c | 2′-OMe(CG)3 | 46.6 ± 0.5 | 59.8 ± 2.3 | 55.3 ± 3.6 | 53.0 ± 5.0 | 161 ± 7 |
| 4b | r(CGCAAAUUUGCG) | 59.5 ± 0.7 | 63.9 ± 6.3 | 51.2 ± 6.2 | 133 ± 46 | 166 ± 19 |
| 4d | d(CGCAAATTTGCG) | 56.6 ± 0.7 | 57.1 ± 2.7 | 53.1 ± 5.6 | 97 ± 18 | 147 ± 8 |
aStandard deviations are given.
bError estimates from plots are given.
Determination of melting temperatures and thermodynamic parameters
The melting temperatures and thermodynamic parameters were obtained using Varian Cary software (version 2.00). The experimental absorbance versus temperature curves were converted into a fraction of strands remaining hybridized (α) versus temperature curves by fitting the melting profile to a two-state transition model, with linearly sloping lower and upper base lines. The melting temperatures (tm in °C) were obtained directly from the temperature at α = 0.5. The final tm was an approximation of, usually, five to eight measurements (Table 1, column 3). The thermodynamic parameters were determined using three different methods (19) as described below.
(i) From the van’t Hoff plot of lnK versus 1/Tm (Table 1, columns 4 and 7). For a bimolecular transition of self-complementary strands the equilibrium constant K = α/[2C(1–α)2] where C is the total strand concentration (C = 2 × 10–6 M). The van’t Hoff plot (lnK versus 1/Tm in K) is linear with –ΔH/R as the slope and ΔS/R as the intercept (R is the universal gas constant 1.986 cal/mol/K). All fitting and calculation operations were done using Varian Cary software (version 2.00) using settings for a bimolecular transition of self-complementary strands. The final –ΔH and –ΔS is the average of at least 10 measurements (five for 2c, Table 1, columns 4 and 7). Full experimental data are given in Supplementary Material Tables S1–S11, S15 and S16.
(ii) From the upper half-width at the half-height of differentiated melting curve (Table 1, column 5). The fraction of strands remaining hybridized (α) versus temperature curves were converted into differentiated melting curves [δα/δ(Tm–1) versus Tm] using Varian Cary software (version 2.00). The upper half-width of the differentiated melting curve at the half-height is inversely proportional to the van’t Hoff transition enthalpy; for a bimolecular transition ΔH = 4.38/(Tmax–1 – T2–1) where Tmax is the temperature at the maximum and T2 is the upper temperature at one-half of [δα/δ(Tm–1)] (19,20). The final –ΔH is the average of at least 10 measurements (five for 2c, Table 1, column 5). Full experimental data are given in Supplementary Material Tables S1–S11, S15 and S16.
(iii) From the concentration dependence of melting temperature (Table 1, column 6). Melting experiments were done in pure buffer over a concentration range of 1–64 µM of oligonucleotides. For a bimolecular association of self-complementary strands 1/Tm = (R/ΔH)lnC + ΔS/ΔH where R is the universal gas constant (1.986 cal/mol/K) and C is the total strand concentration (1 × 10–6–64 × 10–6 M) (19). The plot of 1/Tm versus lnC is linear with R/ΔH as the slope (Supplementary Material, Figs S12–S14). The final –ΔH was obtained by linear fitting using KaleidaGraph software (Version 3.51) with at least 99% confidence level (except 98.5% for 1b, Table 1, column 6). Uncertainties in the slopes of the 1/Tm versus lnC plots were estimated as described below in ‘Error analysis’ (see also Supplementary Material, Fig. S18). Full experimental data are given in Supplementary Material Tables S12–S14.
Calculation of ΔnW
The changes in the number of water molecules associated with the melting process ΔnW were determined as described by Spink and Chaires (17): ΔnW = (–ΔH/nR)[d(Tm–1)/d(lnaw)], where –ΔH is the enthalpy determined from the upper half-width at the half-height of differentiated melting curve in pure buffer, n is the number of base pairs in the duplex and R is the universal gas constant (1.986 cal/mol/K). The experimentally determined values of water activity (lnaw) at given co-solute concentrations were provided by Professors Spink and Chaires. The slope of the plot of reciprocal temperature (in K) of melting versus the logarithm of water activity (lnaw) at different concentrations (0, 5, 10, 15 and 20%) of small co-solutes gave the value of d(Tm–1)/d(lnaw). The final ΔnW were obtained by linear fitting using KaleidaGraph software (version 3.51) with a confidence level usually better than 98%. The plots are given in Supplementary Material, Figs S1–S11, S15 and S16.
Error analysis
The uncertainties in the final ΔnW (Table 2) were estimated from standard deviations of experimental melting temperatures and ΔH according to standard procedures (21). Uncertainties in the slopes of the 1/Tm versus lnaw plots [σd(Tm–1)/d(lnaw)] were estimated by constructing alternative linear fits using error bars (standard deviations of 1/Tm) as the data points. Two alternative plots were obtained by linear fits through 1/Tm plus or minus standard deviation at 0 and 20% co-solute, e.g. 1/Tm plus standard deviation at 0% co-solute and 1/Tm minus standard deviation at 20% co-solute gave one alternative whereas the 1/Tm minus standard deviation at 0% co-solute and the 1/Tm plus standard deviation at 20% co-solute gave the other alternative. The deviations of both alternative plots from the original plot were averaged. If necessary, to maintain the alternative plots inside the limits of error bars of the original plot, error bars at 5 or 15% co-solute were also included to construct the alternative plots (see Supplementary Material Fig. S18 for an example). The final uncertainty in ΔnW was calculated as σΔnW = ΔnW[(σΔH/ΔH)2 + (σslope/slope)2]0.5 where σΔH is the standard deviation and slope = d(Tm–1)/d(lnaw) (21). For detailed input data and results of calculations see Supplementary Material, Table S18.
RESULTS
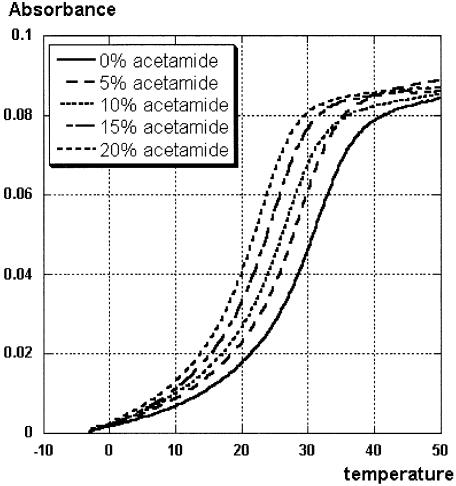
At the outset of our osmotic stressing study we chose four short self-complementary model sequences 1–4 (Table 1) that (i) had been used in previous structural and hydration studies, (ii) had convenient melting temperatures of 20°C < tm < 60°C and (iii) would be relatively easy to prepare as modified analogs in future studies. Model sequence GCGAAUUCGC (1) was an analog of the Dickerson–Drew dodecamer shortened by one base pair at each terminus (22,23). To study the hydration of U-A and C-G base pairs separately we chose (UA)6 (2) and (CG)3 (3), which have been previously studied by NMR (24), crystallography (25,26) and molecular modeling (14,15). Osmotic stressing of CGCAAAUUUGCG (4) (22,23) was also attempted but the studies were discontinued after we found large discrepancies in –ΔH determined by various methods (Table 1). For each model sequence we compared the hydration of 2′-deoxy (a), ribo (b) and 2′-OMe (c) variants. Where appropriate, the 2′-deoxy thymidine variants (d) were also included in the comparison to study the effect of the C5 methyl group. Figure 1 shows typical melting curves and Table 1 presents melting temperatures (tm) in pure buffer and thermodynamic parameters (–ΔH and –ΔS) obtained using three different methods (19). Full experimental data are included in Supplementary Material.
Figure 1.
Changes in thermal stability of r(UA)6 2b with increasing concentration of acetamide.
Enthalpy values obtained from the van’t Hoff plots (Table 1, column 4) and from the differentiated melting curves (Table 1, column 5) were similar (within 10% error limits) with some deviation found for 2b, 2c and 4d. It is possible that the sequence 2 formed a minor amount of imperfect 10 bp duplexes with UA overhangs in equilibrium with the expected perfect duplexes. The fact that the deviations (Table 1, column 4 versus column 5) were observed for the thermally more stable ribo 2b and 2′-OMe 2c oligonucleotides was consistent with this notion: the imperfect oligodeoxyribonucleotide duplexes of 2a and 2d would probably have too small thermal stability. The imperfect duplexes should melt at lower temperatures distorting the lower base lines of melting curves and introducing an error in –ΔH values obtained from the van’t Hoff plots. Enthalpy calculations from the upper half-width at the half-height of differentiated melting curve are relatively insensitive to the choice of the lower base line (19,20) and, therefore, will provide better estimates for –ΔH, especially in the case of 2.
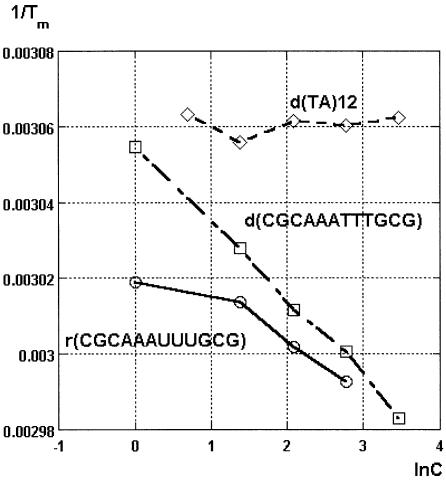
Enthalpy values obtained from the concentration dependence of melting temperature (Table 1, column 6) were generally in accord with the values obtained from the van’t Hoff plots (Table 1, column 4) and from the differentiated melting curves (Table 1, column 5); however, substantial deviations were observed for 1b–d, 2a, 2d and in particular for 4b and 4d. Although more studies are needed to establish the origin of these deviations, some suggestions can be made. The self-complementary sequences may form either bimolecular duplexes or monomolecular hairpin structures. The increase in melting temperature at higher concentration of oligonucleotides (see Supplementary Material Tables S12–S14 and Figs S12–S14) confirmed that the major species under our experimental conditions were duplexes and not hairpins. However, we cannot exclude the possibility that minor amounts of hairpins were present in the melting equilibria, which may be responsible for higher –ΔH values obtained from the concentration dependence of melting temperatures. Future studies are required to test this hypothesis, which is supported by the observation that the –ΔH deviations (Table 1, column 6 versus columns 4 and 5) were negligible for the short sequence 3 that was unlikely to form any hairpins but became severe for 4b and 4d (Table 1). The plot of 1/Tm versus lnC for oligoribonucleotide 4b was not linear at low concentrations (<2 µM), indicating a possible substantial presence of hairpin structures in equilibrium with the duplex (Fig. 2). An osmotic stressing study of d(TA)12 was attempted to explore the impact of the duplex length on ΔnW. However, the thermal stability of d(TA)12 was not concentration dependent, indicating that the major species had a hairpin rather than a duplex structure under our experimental conditions (Fig. 2). Interestingly, a recent comprehensive study on stability of DNA hairpins did not find this motif among unusually stable loops (27). Based on the above analysis, we decided to use the enthalpies obtained from the upper half-width at the half-height of the differentiated melting curve (Table 1, column 5) as the most reliable values for calculation of ΔnW.
Figure 2.
Dependence of reciprocal melting temperatures (1/Tm) on the logarithm of oligonucleotide concentration (lnC) for 4b r(CGCAAA UUUGCG), 4d d(CGCAAATTTTGCG) and d(TA)12.
Consistent with a previous osmotic stressing study by Spink and Chaires (17), we observed a good correlation of ΔnW for ethylene glycol and glycerol, whereas acetamide gave somewhat higher ΔnW values (Table 2). Sucrose gave too high values for model 1, possibly because of some specific solute–oligonucleotide interactions (17) and was excluded from data analysis and further experiments on sequences 2–4. Despite the differences in absolute numbers of ΔnW obtained with different co-solutes (Table 2), comparisons within each series of co-solute were consistent with the general trend discussed below. With ethylene glycol and glycerol we found that upon melting of the DNA and 2′-OMe oligonucleotides, two to three water molecules were released, on average, per base pair. In contrast, melting of RNA sequences consistently released a higher number of water molecules: approximately four from r(GCGAAUUCGC) (1b) and r(CG)3 (3b) and approximately three from r(UA)6 (2b). The only significant deviations from the general trend was 1d in ethylene glycol, which showed a higher ΔnW than expected. The same trend of similar ΔnW released for both DNA and 2′-OMe oligonucleotides and higher ΔnW for RNA oligonucleotides was also observed with acetamide although the data were somewhat less consistent. In accord with the previous studies (14,15,17), we observed little sequence dependency of ΔnW for DNA (cf. 2a, 2d and 3a) and 2′-OMe models (cf. 2c and 3c), whereas the RNA sequences released more water molecules from r(CG)3 (3a) than from r(UA)6 (2a). With a few exceptions (1 in ethylene glycol and in acetamide), we did not observe significant differences between dT- and dU-containing sequences. The ΔnW values for model sequences 4b and 4d were also calculated (Table 2) but were not included in the above analysis because the large discrepancies in –ΔH (Table 1) may cause significant systematic errors.
Accuracy of the melting temperatures is a major concern in UV thermal melting experiments. Sequences 1 and 2 gave good quality melting curves at 260 nm (Fig. 1). However, melting curves of 3 and 4 had non-linear upper base lines at 260 nm; this problem was eliminated by running the melting of 3 and 4 at 280 nm. Most of our measurements fit within ±0.5°C of the averaged melting temperatures (tm). Occasionally, we observed more significant deviations within ±1°C (see Supplementary Material). The final result, the ΔnW per base pair released upon melting, was obtained using plots of reciprocal melting temperature versus the logarithm of water activity (see Materials and Methods). Because the plots were linear fits of five melting temperatures, which in turn were averages of several measurements (usually five to eight), the errors of UV thermal melting experiments were even more efficiently averaged out in the final result. The estimated uncertainties in ΔnW (Table 2), being mostly ±0.5 to ±0.9 water molecules, and the consistency of data among several co-solutes in several model sequences confirm that the detected differences of one to two water molecules are meaningful.
DISCUSSION
Water is an integral part of the nucleic acid structure (13,28,29). Gravimetric, spectroscopic and recent molecular modeling studies suggest that ∼10 water molecules per nucleotide bind directly to B-DNA as the first layer of hydration (30–32). Crystal structures of B-DNA show extensive hydration of the negative phosphate oxygens and a well-ordered water structure as a spine of hydration in the narrow minor groove of adjacent A-T base pairs (33,34) or as a ribbon of two side-by-side water molecules in the wider minor groove of adjacent C-G base pairs (35,36). The major groove of B-DNA is equally well hydrated, but the arrangement of water molecules is irregular. In A-DNA, the situation appears to be reversed: the negative phosphate oxygens are again extensively hydrated, but the narrow and deep major groove now has a well-ordered network of water molecules arranged as fused pentagons, whereas the shallow and wide minor groove is hydrophobic and not as well hydrated (37,38). Z-DNA also displays a spine of hydration running deep in the minor groove and connecting O2 atoms of cytidines (39).
The hydration of phosphates (cones of water) in B-DNA is isolated, whereas in A-DNA and Z-DNA the hydration of phosphates overlap (e.g. a single water molecule bridges two adjacent phosphates) (39,40). It has been suggested that the more economic hydration of phosphates in A and Z conformations (41,42) and the loss of the spine of hydration in A-DNA (43) are responsible for the transition of B-DNA into A or Z conformations when the water activity is lowered (9). Because of the polar 2′-OH, hydration of A-RNA is dramatically changed and displays conserved regular arrangements of water molecules in both grooves (18). The major groove contains regular pentagons formed by four water molecules and the ionic phosphate oxygen. Across the minor groove, tandem water molecules link the 2′-OH groups of adjacent nucleotides. The 2′-OH is suggested to stabilize the RNA duplex by rigidifying the A-type conformation and by linking backbone and base hydration across the strands (18). Thus, the greater thermal stability of RNA duplexes (due to greater stabilization enthalpy) compared with DNA duplexes is suggested to result from the 2′-OH in RNA acting both as a scaffold for the water network in the minor groove and as a site of extensive individual hydration (18). Our results in Table 2 fully support this notion. Melting of RNA duplexes (b) generally released more water molecules than melting of DNA duplexes (a): about two more water molecules from GCGAAUUCGC (1) and (CG)3 (3) and about one more from (UA)6 (2). In general, greater thermal stability of RNA compared to DNA correlated with a favorable enthalpy term (–ΔH, Table 1) and with more water molecules released upon melting (ΔnW, Table 2). An exception was the short model sequence (CG)3 (3) for which the differences in thermal stability and enthalpy for DNA and RNA were relatively small. Because of large discrepancies in –ΔH we did not include model 4 in this analysis. Comparison of 1a and 1d suggested that dT-containing DNA sequences were hydrated better than dU sequences, however, comparison of 2a and 2d did not support this trend. The different behavior of models 1 and 2 may be related to the relatively higher stability of the spine of hydration in (AATT) tracts (as present in 1) compared with (TTAA) and (ATAT) sequences (as present in 2) (11,12). More osmotic stressing studies on sequences modeling different AT motifs are necessary to confirm the increased hydration of the (AATT) motif compared to the (AAUU) motif.
In contrast to two to three water molecules per base pair released from short DNA duplexes in our study, Spink and Chaires (17) found that, on average, four water molecules were released upon melting of E.coli DNA and poly(dA)-poly(dT). Because we are comparing relative differences in ΔnW caused by chemical modifications (2′-H versus 2′-OH versus 2′-OMe), the discrepancy in both studies should not undermine our conclusions, which are not based on the absolute numbers. The discrepancies may be related to the different sizes of nucleic acid duplexes used in both studies. The absolute numbers of ΔnW may be lower for short oligonucleotides due to structural disorder phenomena at the termini of duplexes (the end effect). The experimentally determined ΔnW may also deviate from the true absolute numbers if the co-solutes are not completely excluded from the vicinity of nucleic acids that may occur to different extents for long and short duplexes. Courtenay et al. (44) found that several small solutes (including glycerol) are not completely excluded from the vicinity of proteins, causing an underestimate of the ΔnW in osmotic stressing experiments. Similar studies have not been done for nucleic acids. Our preliminary attempts to investigate the effect of helix length on ΔnW were not successful due to the preference for hairpin formation in longer sequences (see Results). The larger values of –ΔH calculated from the concentration dependence of melting temperatures for some duplexes (Table 1, column 6) might correlate with an underestimate in the ΔnW, but the numbers were significant only in the most severe case of 4b, where the calculated values clearly contrasted with the expected three to four water molecules. Even 4d having major discrepancies in –ΔH values (Table 1), gave ΔnW values only slightly lower than the related sequence 1d (Table 2). Therefore, the much smaller deviations observed for 1b–d, 2a and 2d are not likely to be a source of major errors.
Recent molecular dynamics simulations (14) show that hydration sites around the r(G-C), r(A-U), d(G-C) and d(A-T) are occupied by an average of 21.9, 21.0, 20.1 and 19.8 solvent molecules, respectively. Our data on 2 and 3 (Table 2) were consistent with the above study. The two more water molecules released upon melting of r(CG)3 (3b) compared to d(CG)3 (3a) correlated well with the different hydration of r(G-C) and d(G-C) base pairs (21.9–20.1 = 1.8 water molecules) observed in the molecular dynamics simulation (14). The same correlation was observed for model 2: 21.0–19.8 = 1.2 correlated well with approximately one more water molecule released upon melting of r(UA)6 (2b), as compared to d(TA)6 (2d) and d(UA)6 (2a). Molecular dynamics also show greater sequence dependence of hydration for RNA than for DNA: r(G-C) is hydrated more than r(A-U) (21.9–21.0 = 0.9), whereas the difference between d(G-C) and d(A-T) is relatively small (20.1–19.8 = 0.3). The same trend was observed in Table 2 with ethylene glycol and glycerol, whereas the data with acetamide were not conclusive (cf. 2d with 3a and 2b with 3b). The consistency of our data with previous studies suggests that short oligonucleotides (1–3) are viable model systems for osmotic stressing studies of nucleic acid hydration.
A recent molecular dynamics study (15) of (CG)12 shows that methylation of 2′-OH increases hydrophobicity of the shallow minor groove resulting in a net lower number of water molecules contacting the (G-C) base pairs (∼0.6 less water molecules upon methylation). For model 3 (Table 2) we found that approximately two less water molecules were released upon melting of 2′-OMe(CG)3 (3c) than from r(CG)3 (3b). For models 1 and 2, the difference of water molecules released was somewhat smaller: on average the 2′-OMe duplexes released one less water molecule than the RNA duplexes. This result was consistent with 0.6 less water molecules found in the molecular dynamics study (15) and suggests a net dehydration of the 2′-OMe duplexes as compared to RNA. However, molecular dynamics also show that methylation increases ordering and residence time of the remaining water molecules in the shallow minor groove, which in turn stabilizes the overall hydration in both grooves and rigidifies the duplex (15).
In conclusion, short oligonucleotide duplexes (6–12 bp) could be used in osmotic stressing experiments to detect meaningful differences in nucleic acid hydration. Our data supported the notion that RNA duplexes were hydrated more than DNA duplexes which contributed to the higher thermal stability of RNA. The difference found using osmotic stressing was one to two water molecules per base pair depending on the sequence. Our data on short RNA and DNA oligonucleotides were consistent with the molecular dynamics studies on the same sequences. Synthetic chemistry can be used to modulate hydrophobicity, conformation and hydrogen bonding properties of small oligonucleotides creating novel and intriguing model systems for osmotic stressing studies on nucleic acid hydration. Work along these lines is currently in progress in our laboratory.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Professors Spink and Chaires for providing their experimentally determined values of water activity (lnaw) for the various co-solute concentrations used in this study. We also thank Professors Spink, Chairs and Westhof for helpful discussions. This work was supported by the donors of The Petroleum Research Fund, administered by the American Chemical Society (37599-AC1), Northeastern University and NSF Louis Stokes Alliance for Minority Participation Program.
REFERENCES
- 1.Moravek Z., Neidle,S. and Schneider,B. (2002) Protein and drug interactions in the minor groove of DNA. Nucleic Acids Res., 30, 1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reddy C., Das,A. and Jayaram,B. (2001) Do water molecules mediate protein–DNA recognition? J. Mol. Biol., 314, 619–632. [DOI] [PubMed] [Google Scholar]
- 3.Janin J. (1999) Wet and dry interfaces: the role of solvent in protein–protein and protein–DNA recognition. Structure, 7, R277–R279. [DOI] [PubMed] [Google Scholar]
- 4.Wild K., Sinning,I. and Cusack,S. (2001) Crystal structure of an early protein–RNA assembly complex of the signal recognition particle. Science, 294, 598–601. [DOI] [PubMed] [Google Scholar]
- 5.Qu X. and Chaires,J.B. (1999) Contrasting hydration changes for ethidium and daunomycin binding to DNA. J. Am. Chem. Soc., 121, 2649–2650. [Google Scholar]
- 6.Qu X. and Chaires,J.B. (2001) Hydration changes for DNA intercalation reactions. J. Am. Chem. Soc., 123, 1–7. [DOI] [PubMed] [Google Scholar]
- 7.Berman H.M. (1997) Crystal studies of B-DNA: the answers and the questions. Biopolymers, 44, 23–44. [DOI] [PubMed] [Google Scholar]
- 8.Wahl M.C. and Sundaralingam,M. (1997) Crystal structures of A-DNA duplexes. Biopolymers, 44, 45–63. [DOI] [PubMed] [Google Scholar]
- 9.Schneider B., Patel,K. and Berman,H.M. (1998) Hydration of the phosphate group in double-helical DNA. Biophys. J., 75, 2422–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liepinsh E., Otting,G. and Wüthrich,K. (1992) NMR observation of individual molecules of hydration water bound to DNA duplexes: direct evidence for a spine of hydration water present in aqueous solution. Nucleic Acids Res., 20, 6549–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liepinsh E., Leupin,W. and Otting,G. (1994) Hydration of DNA in aqueous solution: NMR evidence for a kinetic destabilization of the minor groove hydration of d-(TTAA)2 versus d-(AATT)2 segments. Nucleic Acids Res., 22, 2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johannesson H. and Halle,B. (1998) Minor groove hydration of DNA in solution: dependence on base composition and sequence. J. Am. Chem. Soc., 120, 6859–6870. [Google Scholar]
- 13.Makarov V., Pettitt,B.M. and Feig,M. (2002) Solvation and hydration of proteins and nucleic acids: a theoretical view of simulation and experiment. Acc. Chem. Res., 35, 376–384. [DOI] [PubMed] [Google Scholar]
- 14.Auffinger P. and Westhof,E. (2001) Water and ion binding around r(UpA)12 and d(TpA)12 oligomers—comparison with RNA and DNA (CpG)12 duplexes. J. Mol. Biol., 305, 1057–1072. [DOI] [PubMed] [Google Scholar]
- 15.Auffinger P. and Westhof,E. (2001) Hydrophobic groups stabilize the hydration shell of 2′-O-methylated RNA duplexes. Angew. Chem. Int. Ed. Engl., 40, 4648–4650. [DOI] [PubMed] [Google Scholar]
- 16.Parsegian V.A., Rand,R.P. and Rau,D.C. (1995) Macromolecules and water: probing with osmotic stress. Methods Enzymol., 259, 43–94. [DOI] [PubMed] [Google Scholar]
- 17.Spink C.H. and Chaires,J.B. (1999) Effects of hydration, ion release, and excluded volume on the melting of triplex and duplex DNA. Biochemistry, 38, 496–508. [DOI] [PubMed] [Google Scholar]
- 18.Egli M., Portmann,S. and Usman,N. (1996) RNA hydration: a detailed look. Biochemistry, 35, 8489–8494. [DOI] [PubMed] [Google Scholar]
- 19.Breslauer K.J. (1995) Extracting thermodynamic data from equilibrium melting curves for oligonucleotide order-disorder transitions. Methods Enzymol., 259, 221–242. [DOI] [PubMed] [Google Scholar]
- 20.Gralla J. and Crothers,D.M. (1973) Free energy of imperfect nucleic acid helixes. III. Small internal loops resulting from mismatches. J. Mol. Biol., 78, 301–319. [DOI] [PubMed] [Google Scholar]
- 21.Bewington P.R. and Robinson,D.K. (1992) Data reduction and error analysis for the physical sciences, 2nd Edn. WCB/McGraw-Hill, New York, NY, USA. [Google Scholar]
- 22.Soler-Lopez M., Malinina,L., Liu,J., Huynh-Dinh,T. and Subirana,J.A. (1999) Water and ions in a high resolution structure of B-DNA. J. Biol. Chem., 274, 23683–23686. [DOI] [PubMed] [Google Scholar]
- 23.Soler-Lopez M., Malinina,L. and Subirana,J.A. (2000) Solvent organization in an oligonucleotide crystal. The structure of d(GCGAATTCG)2 at atomic resolution. J. Biol. Chem., 275, 23034–23044. [DOI] [PubMed] [Google Scholar]
- 24.Popenda M., Biala,E., Milecki,J. and Adamiak,R.W. (1997) Solution structure of RNA duplexes containing alternating CG base pairs: NMR study of r(CGCGCG)2 and 2′-O-Me(CGCGCG)2 under low salt conditions. Nucleic Acids Res., 25, 4589–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adamiak D.A., Milecki,J., Popenda,M., Adamiak,R.W., Dauter,Z. and Rypniewski,W.R. (1997) Crystal structure of 2′-O-Me(CGCGCG)2, an RNA duplex at 1.30 Å resolution. Hydration pattern of 2′-O-methylated RNA. Nucleic Acids Res., 25, 4599–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adamiak D.A., Rypniewski,W.R., Milecki,J. and Adamiak,R.W. (2001) The 1.19 Å X-ray structure of 2′-O-Me(CGCGCG)2 duplex shows dehydrated RNA with 2-methyl-2,4-pentanediol in the minor groove. Nucleic Acids Res., 29, 4144–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakano M., Moody,E.M., Liang,J. and Bevilacqua,P.C. (2002) Selection for thermodynamically stable DNA tetraloops using temperature gradient gel electrophoresis reveals four motifs: d(cGNNAg), d(cGNABg), d(cCNNGg), and d(gCNNGc). Biochemistry, 41, 14281–14292. [DOI] [PubMed] [Google Scholar]
- 28.Westhof E. (1988) Water: an integral part of nucleic acid structure. Annu. Rev. Biophys. Biophys. Chem., 17, 125–144. [DOI] [PubMed] [Google Scholar]
- 29.Berman H.M. and Schneider,B. (1999) Nucleic acid hydration. In Neidle,S. (ed.), Oxford Handbook of Nucleic Acid Structure. Oxford University Press, New York, pp. 295–312. [Google Scholar]
- 30.Falk M., Hartman,K.A.,Jr and Lord,R.C. (1962) Hydration of deoxyribonucleic acid. I. A gravimetric study. J. Am. Chem. Soc., 84, 3843–3846. [Google Scholar]
- 31.Falk M., Hartman,K.A.,Jr and Lord,R.C. (1963) Hydration of deoxyribonucleic acid (DNA). II. An infrared study. J. Am. Chem. Soc., 85, 387–391. [Google Scholar]
- 32.Feig M. and Pettitt,B.M. (2000) A molecular simulation picture of DNA hydration around A- and B-DNA. Biopolymers, 48, 199–209. [DOI] [PubMed] [Google Scholar]
- 33.Drew H.R., Wing,R.M., Takano,T., Broka,C., Tanaka,S., Itakura,K. and Dickerson,R.E. (1981) Structure of a B-DNA dodecamer. I. Conformation and dynamics. Proc. Natl Acad. Sci. USA, 78, 2179–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drew H.R. and Dickerson,R.E. (1981) Structure of a B-DNA dodecamer. III. Geometry of hydration. J. Mol. Biol., 151, 535–556. [DOI] [PubMed] [Google Scholar]
- 35.Prive G.G., Yanagi,K. and Dickerson,R.E. (1991) Structure of the B-DNA decamer C-C-A-A-C-G-T-T-G-G and comparison with isomorphous decamers C-C-A-A-G-A-T-T-G-G and C-C-A-G-G-C-C-T-G-G. J. Mol. Biol., 217, 177–199. [DOI] [PubMed] [Google Scholar]
- 36.Tereshko V., Minasov,G. and Egli,M. (1999) A ‘Hydrat-ion’ spine in a B-DNA minor groove. J. Am. Chem. Soc., 121, 3590–3595. [Google Scholar]
- 37.Conner B.N., Yoon,C., Dickerson,J.L. and Dickerson,R.E. (1984) Helix geometry and hydration in an A-DNA tetramer: IC-C-G-G. J. Mol. Biol., 174, 663–695. [DOI] [PubMed] [Google Scholar]
- 38.Kennard O., Cruse,W.B.T., Nachman,J., Prange,T., Shakked,Z. and Rabinovich,D. (1986) Ordered water structure in an A-DNA octamer at 1.7 Å resolution. J. Biomol. Struct. Dyn., 3, 623–647. [DOI] [PubMed] [Google Scholar]
- 39.Chevrier B., Dock,A.C., Hartmann,B., Leng,M., Moras,D., Thuong,M.T. and Westhof,E. (1986) Solvation of the left-handed hexamer d(5BrC-G-5BrC-G-5BrC-G) in crystals grown at two temperatures. J. Mol. Biol., 188, 707–719. [DOI] [PubMed] [Google Scholar]
- 40.Egli M., Tereshko,V., Teplova,M., Minasov,G., Joachimiak,A., Sanishvili,R., Weeks,C.M., Miller,R., Maier,M.A., An,H. et al. (2000) X-ray crystallographic analysis of the hydration of A- and B-form DNA at atomic resolution. Biopolymers, 48, 234–252. [DOI] [PubMed] [Google Scholar]
- 41.Saenger W., Hunter,W.N. and Kennard,O. (1986) DNA conformation is determined by economics in the hydration of phosphate groups. Nature, 324, 385–388. [DOI] [PubMed] [Google Scholar]
- 42.Ho P.S. and Mooers,B.H.M. (1997) Z-DNA crystallography. Biopolymers, 44, 65–90. [DOI] [PubMed] [Google Scholar]
- 43.Kopka M.L., Fratini,A.V., Drew,H.R. and Dickerson,R.E. (1983) Ordered water structure around a B-DNA dodecamer. A quantitative study. J. Mol. Biol., 163, 129–146. [DOI] [PubMed] [Google Scholar]
- 44.Courtenay E.S., Capp,M.W., Anderson,C.F. and Record,M.T.,Jr (2000) Vapor pressure osmometry studies of osmolyte–protein interactions: implications for the action of osmoprotectants in vivo and for the interpretation of ‘osmotic stress’ experiments in vitro. Biochemistry, 39, 4455–4471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.