Abstract
Optimization of small interfering RNAs (siRNAs) is important in RNA interference (RNAi)-based therapeutic development. Some specific chemical modifications can control which siRNA strand is selected by the RNA-induced silencing complex (RISC) for gene silencing. Intended strand selection will increase potency and reduce off-target effects from the unintended strand. Sometimes, blocking RISC loading of the unintended strand leads to improved intended strand-silencing potency, but the generality of this phenomenon is unclear. Specifically, unlocked nucleic acid (UNA) modification of the 5′ end of canonical (i.e., 19+2) siRNAs abrogates gene silencing of the modified strand, but the fate and potency of the unmodified strand has not been investigated. Here, we show that 5′ UNA-modified siRNAs show improved silencing potency of the unmodified strand. We harness this advantageous property in a therapeutic context, where a limited target region in a conserved HIV 5′ long terminal repeat U5 region would otherwise yield siRNAs with undesired strand selection properties and poor silencing. Applying 5′ UNA modification to the unintended sense (S) strand of these otherwise poorly targeted siRNAs dramatically improves on-target silencing by the intended antisense (AS) strand in pNL4-3.luciferase studies. This study highlights the utility of 5′ UNA siRNA modification in therapeutic contexts where siRNA sequence selection is constrained.
Keywords: RNAi trigger, siRNA, strand selection, unlocked nucleic acid (UNA)
Introduction
Small interfering RNA (siRNA)–based drugs are emerging as a new platform technology that provides an alternative to small molecule or antibody therapeutics. In contrast to small molecule drugs, the sequence-specific basis of RNA interference (RNAi) gene silencing facilitates rational design of novel inhibitors. RNAi has the power to target any mRNA and to discriminate against different alleles1,2 and alternatively spliced isoforms3 of protein-coding mRNAs. RNAi can also target mRNAs of proteins that have been designated “undruggable” by traditional drug discovery strategies. Upon internalization into cells, the double stranded siRNA is bound and processed by the RNAi machinery, with one strand eventually being chosen and incorporated into the RNA-induced silencing complex (RISC) to mediate target mRNA destruction. The other strand is excluded from the RISC. When designing siRNAs, the strand that will lead to silencing of the intended target mRNA is termed the siRNA antisense (AS) strand and the other strand is termed the siRNA sense (S) strand.
Strand selection is purportedly due to the thermodynamic stability of the 5′ end of an siRNA strand; specifically, the strand with a relatively less stable 5′ ends is preferentially selected.4,5 Optimal siRNAs are biased toward selection and usage of the AS strand. Sub-optimal siRNAs may perform poorly due to significant selection of the undesired S strand. The ability to optimize and control siRNA strand selection, therefore, is an important consideration in the development of efficacious siRNA-based therapeutics. Preferential selection of the AS strand by the cellular RNAi machinery will improve efficacy, reduce effective doses, and mitigate S strand-mediated off-target effects. To achieve preferential strand selection, several structural and chemical modifications to the canonical siRNA molecule have been described, including the development of asymmetric duplexes,6,7 internally destabilized duplexes,8 and 5′O-methyl modifications.9
Wengel and colleagues described a 2′,3′-seco-RNA chemical modification which they termed unlocked nucleic acid (UNA). The UNA modification thermodynamically destabilizes RNA duplexes10 yet preserves the A-form helix of double stranded RNA.11 Several siRNA studies also led researchers to understand the biological consequences12 or other advantageous properties (e.g., reduced seed-based off-target effects)13,14 of UNA substitution at various siRNA positions. For example, placing UNAs at the 2 nucleotide (nt) 3′ overhang position confers increased nuclease stability.15 Furthermore, placement of a UNA at the first (i.e., 5′ terminal) or second position of one strand of an siRNA impairs the gene-silencing ability of the modified strand;12,14 we define this effect as the “strand-blocking” property of the 5′ UNA modification.
Another strategy previously used to influence siRNA strand selection is to transform canonical siRNAs into Dicer substrate interfering RNAs (dsiRNAs).16 The dsiRNA format is a slightly longer asymmetric duplex RNA compared with a canonical siRNA. dsiRNAs are designed with functional polarity, such that Dicer preferentially binds and processes the dsiRNA from one end. As a result, dsiRNAs can promote strand selection towards the AS strand compared with the parent canonical siRNA.17 However, to date, the efficacy of dsiRNA strand biasing has not been tested in situations where the siRNA generated by Dicer cleavage has strongly unfavorable thermodynamics.
Canonical siRNAs are designed with 21 nt of perfectly complementary pairing with target mRNAs to trigger RNAi. When using siRNA as a laboratory tool to silence a full-length target mRNA (i.e., hundreds to thousands of nucleotides long), many possible siRNAs can be generated for the target mRNA. Computational siRNA design algorithms can assist in identifying, but not perfectly predict, highly functional siRNAs from poorly functional siRNAs based on siRNA 5′ end thermodynamics calculations. For some therapeutic applications (e.g., targeting SNPs, alternatively spliced isoforms with small unique exons, species-specific mRNAs in xenograft models, or short highly conserved regions of viral transcripts), the number of possible siRNAs that can be designed may be limited. For example, in human immunodeficiency virus (HIV), the U5 non-coding leader stem region in the long terminal repeat, which is important for viral replication,18 is conserved amongst many different HIV strains but is only ~29 nucleotides in size. Thus, only ten canonical 21mer siRNAs can possibly be designed to target this region, and it may be that none of the possible siRNAs possess favorable end thermodynamics and potency.
Here, we sought to extend the strand-blocking properties of a 5′ UNA modification to other siRNA sequences. In addition, we use cell-based assays to demonstrate the abrogation of RISC loading as a consequence of 5′ UNA modification. Lastly, we rationally apply the 5′ UNA modification to transform a poor siRNA and dsiRNA targeting a small conserved region of the HIV transcript into an improved siRNA to attenuate HIV.
Results
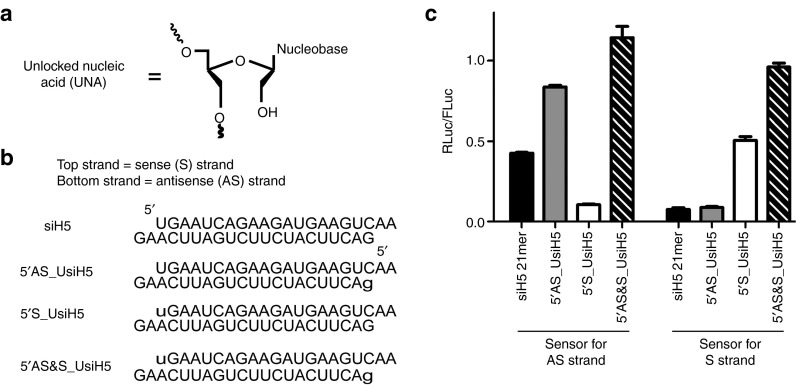
Previously, we demonstrated that an siRNA targeting hnRNPH site 5, termed siH5, had unfavorable AS strand selection properties.19 Poor siRNAs are predicted to have relatively less stable 5′ S strand ΔG compared with 5′ AS strand ΔG, exhibited by a large negative ΔΔG value using our prediction tool.20 Indeed, thermodynamic stability calculations of the 5′ ends of the siH5 AS and S strands predict that the S strand would be selected more by the RISC compared with the AS strand.4,5 To investigate whether the strand-blocking properties of 5′ UNA (Figure 1a) modification is generally applicable to other siRNAs, we conducted dual luciferase assays using unmodified (siH5) and AS-, S-, or both strands 5′ UNA-modified siH5 (5′ AS_UsiH5, 5′ S_UsiH5, 5′ AS&S_UsiH5, respectively) (Figure 1b). Our assay used luciferase sensor plasmids that separately indicate the level of gene silencing conferred by the AS or S strand. The firefly luciferase (FLuc) expression serves as an internal control—a normalized RLuc/FLuc value of 1 indicates no gene silencing. Confirming previous results, siH5 demonstrated relatively strong undesired silencing by the S strand compared with the AS strand (Figure 1c). The incorporation of a UNA at the 5′ end of the S or AS strand, however, impaired silencing activity of the modified strand (Figure 1c). When 5′ UNA modifications were applied on both the AS and S strands, no gene silencing was observed for either strand. Furthermore, consistent with a previous report,14 applying the 5′ UNA modification to the S strand not only retained but improved the silencing potency of the AS strands. These results support the general applicability of 5′ UNA modification to block silencing by the modified strand and concomitantly improve the silencing ability of the unmodified strand.
Figure 1.
Design and gene silencing of unmodified and modified siH5. (a) Schematic of the structure of an unlocked nucleic acid. (b) Sequences of the unmodified siH5 (top), antisense- (5′ AS_), sense- (5′ S), and both (5′ AS&S_) 5′ UNA-modified sequences. Lowercase letters indicates UNA-modified ribonucleotide. (c) Dual luciferase gene-silencing assay, performed at 2 nmol/l. n = 4 in duplicate, mean ± SEM.
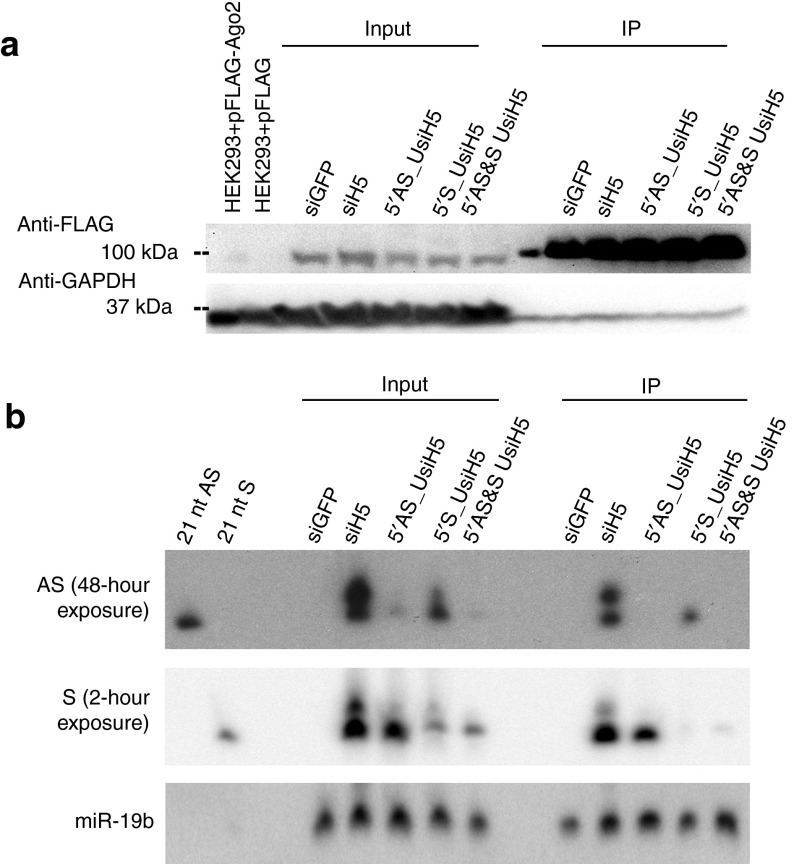
5′ UNA modification was shown to block monophosphorylation of 5′ OH synthetic siRNAs,12 a necessary step for siRNAs to be loaded into the RISC to execute gene silencing. This previous result was exemplified via a cell-free assay, where the lack of phosphorylation of the AS strand prevents AS strand loading into Argonaute 2 (Ago2), the core protein of the RISC. We wished to determine if this was also the case in a cell-based assay, where components are present at physiologically relevant concentrations. We transfected unmodified and UNA-modified siH5 into HEK293 cells stably expressing FLAGHA-tagged Ago2, and conducted small RNA immunoprecipitation assays followed by northern blot. Immunoblotting was performed to confirm successful Ago2 immunoprecipitation (Figure 2a). For unmodified siH5, detectable levels of both AS and S strands were observed in the Ago2-immunoprecipitated samples. Considering the exposure time needed to detect the AS strand, however, we infer that the S strand was more abundant, consistent with the stronger gene-silencing performance of the S strand (Figure 2b). On the basis of the results of our gene-silencing studies, we predict that impairing RISC binding to the S strand with 5′ UNA would result in an increase in relative loading of the otherwise weakly loading AS strand. Indeed, when the 5′ end of the S strand was UNA modified, we did not observe immunoprecipitated S strand but do observe a robust signal for the AS strand. Hence, we observe a concomitant increase in the relative amount of immunoprecipitated AS strand versus S strand. Reciprocally, when the AS strand was modified at the 5′ position, the AS strand was not immunoprecipitated, consistent with a previous report.12 When both the AS and S strand are 5′ UNA-modified, neither strand was appreciably loaded into Ago2. As expected for a single 5′ chemically modified base substitution, the basis of the improved RISC loading is not due to increased nuclease resistance (Supplementary Figure S1). Hence, we demonstrate in a cell-based assay that 5′ UNA-modified strands load very poorly into Ago2, and that the strength of weakly binding unmodified strands can be improved by applying the 5′ UNA modification to the opposite strand.
Figure 2.
The strands with 5′ UNA modifications are not loaded into RISC. The 5′ UNA modification was applied to the AS, S, or both strands of the siH5 sequence. (a) Immunoblot confirming successful immunoprecipitation of FLAGHA-Ago2. HEK293+pFLAGHA-Ago2 samples are from HEK293 cells transiently transfected with a plasmid encoding FLAGHA-Ago2 as a positive control. HEK293+pFLAGHA serves as a negative control. (b) Northern blot of input and small RNAs immunoprecipitated from FLAGHA-Ago2. The 21 nt AS or S strands serve as size markers.
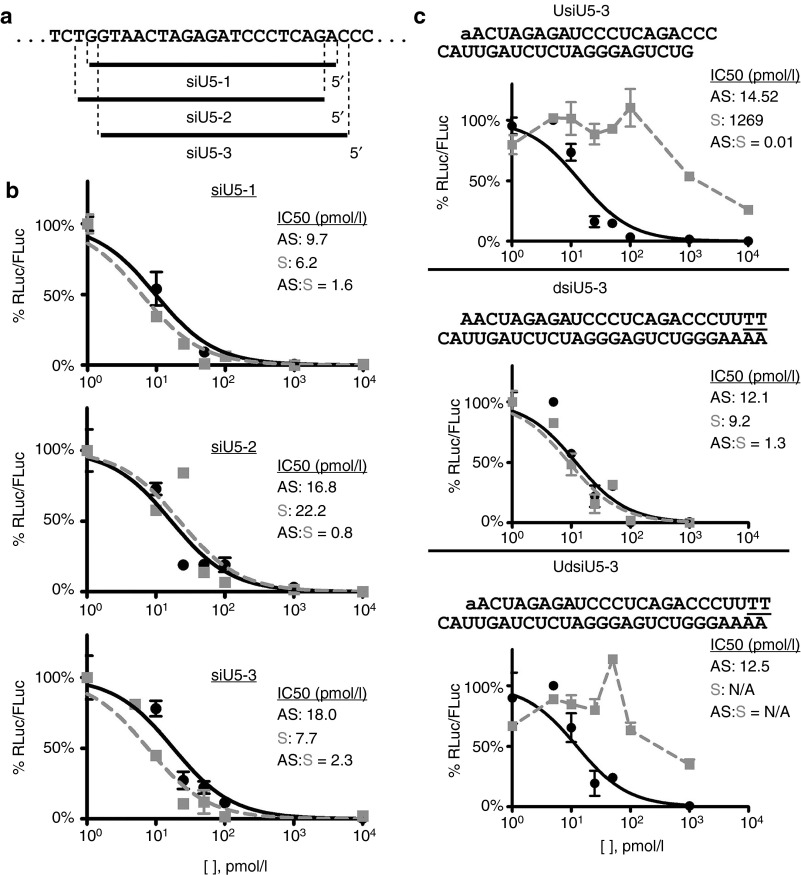
On the basis of the literature as well as our own gene silencing and small RNA immunoprecipitation data, we were intrigued that 5′ UNA modification not only blocks silencing by the modified strand but also improves the silencing potency of the unmodified strand. We reasoned that this property would be useful in situations where the choice of possible siRNAs against an mRNA target is small and limited to siRNAs with poor AS strand gene-silencing potency. This is the case for a highly conserved region the U5 region of HIV (Figure 3). In the HIV 5′ long terminal repeat, the U5 sequence is a good target for anti-HIV therapeutic siRNAs because it is present in both unspliced and spliced transcripts of HIV and because it is highly conserved. Using a psiCHECK-based reporter plasmid spanning a 27 nt segment (i.e., where only eight siRNAs are possible) within the conserved U5 region, we screened three siRNAs (siU5-1, -2, and -3) targeting the conserved HIV U5 region (Figure 4a). In all three cases, gene-silencing ability was observed for both strands (Figure 4b). Consistent with its predicted poor AS strand thermodynamic properties (Table 1),20 based on IC50 calculations, siU5-3 demonstrated the most pronounced difference between AS and S strand silencing (e.g., the largest AS:S ratio of IC50 values) (Figure 4b). An AS:S ratio >1 indicate relatively strong undesired silencing by the S strand, whereas an AS:S ratio <1 indicate relatively strong desired silencing by the AS strand. Therefore, we chose siU5-3 as the parent siRNA on which to apply the 5′ UNA modification to improve AS strand potency. Upon applying the 5′ UNA modification to the S strand of siU5-3 (UsiU5-3), the potency of siU5-3′s AS strand modestly improved (Figure 4c), but S strand silencing was considerably impaired (Figure 4c), yielding a AS:S ratio markedly <1. Inspection of previous 5′ UNA-modified siRNA studies also show that the UNA modification does not always dramatically improve performance of the unmodified strand, particularly at saturating concentrations or when the silencing potency of the unmodified strand is already considerably strong.14 Nonetheless, the ability to block the use of the S strand is still advantageous in regards to RISC competition and siRNA efficiency. As another variation, we transformed siU5-3 into a dsiRNA and a 5′ S strand UNA-modified dsiRNA (dsiU5-3 and UdsiU5-3, respectively (Figure 4c)). The dsiRNA format can partially improve AS strand potency (e.g., dsiRNAs decrease the AS:S ratio relative to the AS:S ratio of the parent siRNA),17 but the dsiRNA format alone may not be able to improve dsiRNAs with highly unfavorable thermodynamics in the proximity of the processed 5′ end of the AS strand. Indeed, the 5′ end of the predicted processed AS strand from dsiU5-3 contains two adjacent guanines, which is highly unfavorable for strand selection. Therefore, as an orthogonal strategy, we were curious whether the 5′ UNA S strand modification could further improve AS strand potency of UdsiU5-3. Compared with the unmodified siU5-3, the potency of the dsiU5-3 AS strand improved modestly, exemplified by the a decrease in the AS:S ratio. The AS:S ratio, however, remains >1, indicating that the dsiU5-3 S strand remained considerably functional. This scenario demonstrates a situation where the dsiRNA format alone was only partially able overcome strongly unfavorable strand selection. Meanwhile, the AS strand of UdsiU5-3 showed improved potency, relative to the canonical siU5-3, with a concomitant impairment of S strand silencing (Figure 4c). This synergistic effect of combining the 5′ UNA modification with dsiRNAs was confirmed using another parent siRNA (Supplementary Figure S2), demonstrating that this combined design can be generalized to other sequences. These screening assays support the generality of the strand blocking properties of a 5′ UNA in another siRNA sequence context, and show that 5′ UNA modification can be used synergistically with dsiRNAs to further improve AS strand potency and dissuade S strand function for a restricted and poor siRNA.
Figure 3.

The targeted region of the HIV 5′ long terminal repeat (LTR) is highly conserved. The conserved sequence of 5′ LTR sequences are taken from 612 different strains of HIV. The height of the font in the weblogo correlates to the degree of sequence conservation amongst strains.
Figure 4.
The 5′ UNA sense strand modification improves strand selection for U5-targeted siRNA and dsiRNA. (a) Design of three siRNAs within U5 reporter target sequence. The black bar indicates the span of the AS strand for each of the indicated siRNAs. (b) Dual luciferase gene-silencing potency of U5-targeted siRNAs. (c) Schematic (shown above each graph) and dual luciferase assays gene-silencing performance of 5′ UNA siU5-3 (UsiU5-3), unmodified dsiU5-3, and 5′ UNA dsiU5-3 (UdsiU5-3). IC50 values for each strand and AS:S IC50 ratios are shown in the insets. n ≥ 3, in duplicate, mean ± SEM.
Table 1. Design and thermodynamic properties of siRNAs targeting the conserved U5 region of the HIV long terminal repeat (LTR).

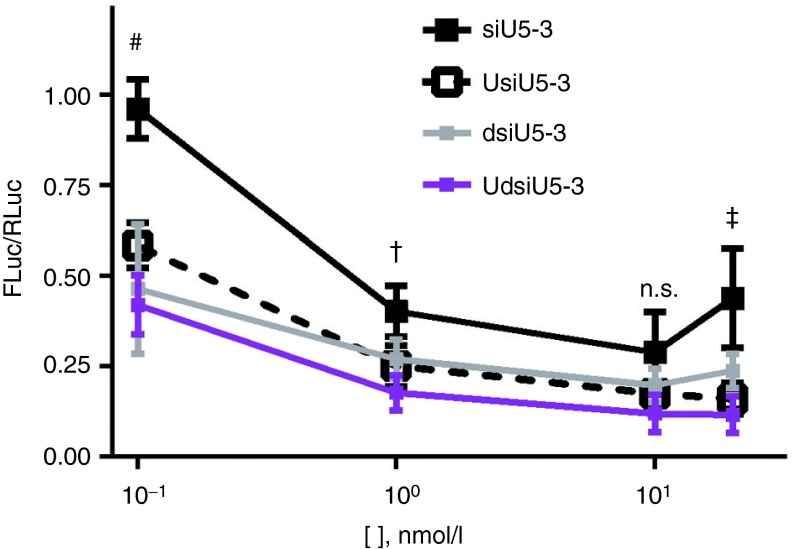
To test if 5′ UNA S strand modification would improve a poor siRNA in a more relevant culture assay, silencing studies were performed using the pNL4-3.luciferase gene-silencing system. This assay is not the same as the 3′UTR 27 nt target site psiCHECK-based assay; the pNL4-3.luciferase plasmid contains the HIV 5′ and 3′ long terminal repeats and 7 of the 9 HIV protein coding genes (lacks rev and nef), with a Firefly luciferase gene inserted in the coding region near the nef gene locus. Therefore, this reporter system more closely simulates an HIV treatment model in culture compared with the psiCHECK-based assay. Transfecting siU5-3 and its derivatives mentioned above, we observed improvements in the 5′ UNA-modified derivatives in two different concentration contexts. First, at low concentrations, siU5-3 was not active in gene silencing, yet UsiU5-3 achieved >40% knockdown (Figure 5). The dsiU5-3 achieved >50% gene silencing, and the UdsiU5-3 achieved the greatest level of silencing out of all four derivatives. At higher concentrations, siU5-3 appeared to reach a threshold of ~70% gene silencing, and dsiU5-3 reached a threshold of ~80%. Strikingly, the 5′ UNA-modified derivatives surpass this threshold, as UsiU5-3 achieved >80% silencing. Furthermore, UdsiU5-3 continued to perform the best, achieving ~90% silencing. These results demonstrate the advantageous properties of a 5′ UNA siRNA modification in a therapeutically relevant assay.
Figure 5.
The 5′ UNA-modified siRNA and dsiRNA silence gene expression more potently in pNL4-3.luciferase assays.The indicated siRNAs were cotransfected with pNL4-3.luciferase and normalization control Renilla luciferase plasmid for 24 hours. n = 3, in duplicate, mean ± SEM. #P < 0.05 between siU5-3 versus all other variants. †P < 0.05 between siU5-3 versus UdsiU5-3. ‡P < 0.05 between siU5-3 versus UsiU5-3 and UdsiU5-3. Statistics performed using two-way analysis of variance followed by multiple comparisons test (Sidak test). n.s., no statistical significance between siU5-3 versus other variants.
Discussion
Previous UNA studies were seminal in exemplifying important biological properties of UNA-modified siRNAs,12,14 but the generality of these observations were not clear. We wished to rationally apply 5′ UNA-modified siRNAs for a specific therapeutic goal, but we first wanted to clarify some of the biological properties. To attribute any phenotypic differences solely to the UNA modification, we maintain the canonical structure of our 5′ UNA-modified siRNAs. Our initial studies, using different siRNA sequences, clarified that the 5′ UNA modification severely impairs, but does not completely inhibit, gene silencing by the modified strand. We also performed Ago2 immunoprecipitation followed by small RNA northern blotting, and observed that 5′ UNA-modified siRNAs do not load into Ago2, consistent with a previous report.12 Our method is cell based (rather than cell lysate based) and thus occurs at physiological conditions. Furthermore, we detected the small RNA strands via northern blot (rather than Taqman qPCR). This latter point is an important technical distinction; many forms of RNA undergo untemplated 3′ additions, including small RNAs. Taqman qPCR primers cannot recognize 3′ tailed targeted species, and therefore identification and quantification of small RNA strands may not be accurate. Northern blotting, however, can identify tailed species for quantification. Indeed, in some of the lanes of Figure 2b, we detected a second larger band that would be consistent with an siRNA with untemplated 3′ additions.
Numerous chemical and structural modifications to siRNAs have been reported, each claiming to improve the efficacy of siRNA in regards to number of different considerations (improved nuclease resistance, improved AS strand selection, decreased off-target effects, etc). Uniquely, we have integrated the basic science knowledge about the 5′ UNA modification as a design feature to improve poorly performing siRNAs for a specific therapeutic context. Therapeutic 5′ UNA-modified siRNAs have been reported,21 but used a non-canonical siRNA structure (a 22mer annealed to a 21mer with a mismatch at the 5′ nucleotide of the S strand) and appeared to select the siRNA sequence based on a broad screen targeting a full mRNA without knowledge of the performance of the corresponding unmodified siRNA. For our study, using an HIV infection model, we maintain the canonical siRNA structure and make sequence- and structure-matched 5′ UNA-modified siRNAs. In a case where choice of target sequence was severely limited by sequence constraints, we rationally modify a poor siRNA with the 5′ UNA modification to improve gene-silencing potency. This possibility of target sequence constraint is a consideration not only for combating viral disease but also cancer and diseases arising from allele- or exon-specific transcripts. Furthermore, we show that the 5′ UNA can be applied to dsiRNAs, and the combination of these two orthogonal modifications achieved the largest enhancement of on-target gene silencing. Because we have excluded the undesired S strand from the RISC, 5′ UNA-modified (d)siRNAs would have the additional benefit of reducing off-target effects mediated by the S strand.
Materials and methods
RNA and UNA oligonucleotides. The siRNAs and dsiRNAs used in this study were purchased as single stranded oligonucleotides. To form siRNAs and dsiRNAs, complementary oligonucleotides were mixed in equimolar ratios, heated to 95 °C, and allowed to cool slowly to room temperature. Unmodified RNA oligonucleotides were synthesized and RNase-free HPLC purified by Integrated DNA Technologies (Coralville, IA). UNA-modified RNA oligonucleotides were synthesized by TriLink Biotechnologies (San Diego, CA).
Reporter plasmids. The reporter sequences described below were cloned into the multiple cloning site of Renilla luciferase (RLuc) of psiCHECK2.1 (Promega, Madison, WI) and used for IC50 determination. A 247 nt segment, both in a forward and reverse-complement orientation, of the coding region of the human HNRNPH gene was used for studying siH5-based siRNAs.17 A 27 nt segment, both in the forward and reverse-complement orientation, of the HIV U5 region was used for studying siU5-based siRNAs. For HIV assays, the pNL4-3.luciferase plasmid22 was cotransfected with an internal control Renilla luciferase-expressing plasmid (pRSV-RLuc) in which the cytomegalovirus promoter of pRL-CMV (Promega) was replaced with a rouse sarcoma virus promoter.
Cells. For dual luciferase assay studies, HEK293 cells (ATCC, Manassas, VA) were used. For small RNA immunoprecipitation assays, HEK293 cells stably expressing doubly tagged FLAGHA human Argonaute 223 were used.
Transfections and small RNA immunoprecipitation. Strand-specific dual luciferase assays (Promega) were conducted as described previously.19
For small RNA immunoprecipitation studies, HEK293 cells stably expressing FLAGHA-tagged Ago2 (HEK293+FLAGHA-Ago2) were transfected with 10 nmol/l of the indicated siRNAs using 10 µl RNAiMAX (Invitrogen, Life Technologies, Grand Island, NY) in 10 cm dishes. After 24 hours, cells were washed once with ice cold PBS and collected and lysed (50 mmol/l Tris, 137 mmol/l NaCl, 1% Triton X-100, pH 8) for 15 minutes on ice. Lysates were cleared via centrifugation, and only the soluble supernatant was used for immunoprecipitation. Input RNA (extracted by RNA STAT-60; Amsbio, Lake Forest, CA) and protein was purified from a portion of the lysate before immunoprecipitation. FLAGHA-Ago2 was immunoprecipitated by incubating 2.5 mg lysate with 20 µl anti-FLAGHA M2 beads (Sigma, St Louis, MO) overnight at 4 °C and washing three times with lysis buffer. After the final wash, 3 µl of the pellet was added to protein loading dye for immunoblot analysis (rabbit anti-FLAG antibody; Cell Signaling, Danvers, MA). Denaturing RNA loading dye was added to the remaining pellet, boiled for 10 minutes, and used for northern blotting. For input protein, ~30 µg was used. For input RNA, 25 µg was used. Small RNAs were detected using 5′ 32P-labeled DNA probes fully complementary to the RNA sequence. Signal was detected via film.
We conducted pNL4-3.luciferase assays as described previously.24 Statistics were performed using GraphPad Prism (GraphPad Software, La Jolla, CA).
Nuclease stability assays. In a 100 µl volume, 3.5 µg of siRNA was incubated with 50% (v/v) human serum diluted in DMEM at 37 °C. At the indicated time points, a 10 µl aliquot was taken, added to 10 µl 2× denaturing RNA loading dye and snap-frozen on dry ice and stored at −80 °C until all time points were taken. A 15% denaturing PAGE was performed, followed by ethidium bromide staining.
HIV sequence analysis. The 612 strains of HIV used for sequence analysis were obtained from the Los Alamos National Lab HIV Database (http://www.hiv.lanl.gov/content/index). Sequence conservation analysis was performed via CLC Bio Main Workbench.
SUPPLEMENTARY MATERIAL Figure S1. The single 5′ UNA modification does not dramatically alter the stability of the siRNAs. Figure S2. The combined dsiRNA and 5′ UNA strand-blocking modification is generalizable to other parent si- and dsiRNA sequences.
Acknowledgments
We thank members of the Rossi laboratory for helpful discussion, Lisa Scherer (City of Hope) for the psiCHECK-U5 sensor plasmids, and Haitang Li (City of Hope) of the Rossi laboratory for helpful discussion and technical assistance regarding HIV-based assays. We also thank the H.N. and Frances C Berger Foundation Fellowship and the Helen and Morgan Chu Fellowship for their support of N.M.S. This work was supported by the United States National Institute of Health (HL074704 to J.J.R.). J.J.R. is a cofounder and scientific advisory board member of Dicerna Pharmaceuticals, a company employing therapeutic dsiRNAs. J.R.E.P. and A.P.M. are employees of TriLink Biotechnologies.
Supplementary Material
References
- Takahashi M, Watanabe S, Murata M, Furuya H, Kanazawa I, Wada K, et al. Tailor-made RNAi knockdown against triplet repeat disease-causing alleles. Proc Natl Acad Sci USA. 2010;107:21731–21736. doi: 10.1073/pnas.1012153107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Yñigo-Mojado L, Martín-Ruíz I, Sutherland JD. Efficient allele-specific targeting of LRRK2 R1441 mutations mediated by RNAi. PLoS ONE. 2011;6:e21352. doi: 10.1371/journal.pone.0021352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MS, Sharp PA. Pyruvate kinase M2-specific siRNA induces apoptosis and tumor regression. J Exp Med. 2012;209:217–224. doi: 10.1084/jem.20111487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Sun X, Rogoff HA, Li CJ. Asymmetric RNA duplexes mediate RNA interference in mammalian cells. Nat Biotechnol. 2008;26:1379–1382. doi: 10.1038/nbt.1512. [DOI] [PubMed] [Google Scholar]
- Chang CI, Yoo JW, Hong SW, Lee SE, Kang HS, Sun X, et al. Asymmetric shorter-duplex siRNA structures trigger efficient gene silencing with reduced nonspecific effects. Mol Ther. 2009;17:725–732. doi: 10.1038/mt.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Ma H, Ye C, Ramirez D, Chen S, Montoya J, et al. Improved siRNA/shRNA functionality by mismatched duplex. PLoS ONE. 2011;6:e28580. doi: 10.1371/journal.pone.0028580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Weinmann L, Gaidatzis D, Pei Y, Zavolan M, Tuschl T, et al. Strand-specific 5'-O-methylation of siRNA duplexes controls guide strand selection and targeting specificity. RNA. 2008;14:263–274. doi: 10.1261/rna.789808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langkjaer N, Pasternak A, Wengel J. UNA (unlocked nucleic acid): a flexible RNA mimic that allows engineering of nucleic acid duplex stability. Bioorg Med Chem. 2009;17:5420–5425. doi: 10.1016/j.bmc.2009.06.045. [DOI] [PubMed] [Google Scholar]
- Pasternak A, Wengel J. Thermodynamics of RNA duplexes modified with unlocked nucleic acid nucleotides. Nucleic Acids Res. 2010;38:6697–6706. doi: 10.1093/nar/gkq561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenski DM, Cooper AJ, Li JJ, Willingham AT, Haringsma HJ, Young TA, et al. Analysis of acyclic nucleoside modifications in siRNAs finds sensitivity at position 1 that is restored by 5'-terminal phosphorylation both in vitro and in vivo. Nucleic Acids Res. 2010;38:660–671. doi: 10.1093/nar/gkp913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramsen JB, Pakula MM, Hansen TB, Bus C, Langkjær N, Odadzic D, et al. A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res. 2010;38:5761–5773. doi: 10.1093/nar/gkq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaish N, Chen F, Seth S, Fosnaugh K, Liu Y, Adami R, et al. Improved specificity of gene silencing by siRNAs containing unlocked nucleobase analogs. Nucleic Acids Res. 2011;39:1823–1832. doi: 10.1093/nar/gkq961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen MB, Pakula MM, Gao S, Fluiter K, Mook OR, Baas F, et al. Utilization of unlocked nucleic acid (UNA) to enhance siRNA performance in vitro and in vivo. Mol Biosyst. 2010;6:862–870. doi: 10.1039/b918869j. [DOI] [PubMed] [Google Scholar]
- Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- Rose SD, Kim DH, Amarzguioui M, Heidel JD, Collingwood MA, Davis ME, et al. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005;33:4140–4156. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerens N, Groot F, Berkhout B. Stabilization of the U5-leader stem in the HIV-1 RNA genome affects initiation and elongation of reverse transcription. Nucleic Acids Res. 2000;28:4130–4137. doi: 10.1093/nar/28.21.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K, Amarzguioui M, Kim DH, Alluin J, Heale B, Song MS, et al. A role for human Dicer in pre-RISC loading of siRNAs. Nucleic Acids Res. 2011;39:1510–1525. doi: 10.1093/nar/gkq846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heale BS, Soifer HS, Bowers C, Rossi JJ. siRNA target site secondary structure predictions using local stable substructures. Nucleic Acids Res. 2005;33:e30. doi: 10.1093/nar/gni026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth S, Matsui Y, Fosnaugh K, Liu Y, Vaish N, Adami R, et al. RNAi-based therapeutics targeting survivin and PLK1 for treatment of bladder cancer. Mol Ther. 2011;19:928–935. doi: 10.1038/mt.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BK, Saksela K, Andino R, Baltimore D. Distinct modes of human immunodeficiency virus type 1 proviral latency revealed by superinfection of nonproductively infected cell lines with recombinant luciferase-encoding viruses. J Virol. 1994;68:654–660. doi: 10.1128/jvi.68.2.654-660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Zhou J, Li H, Li S, Zaia J, Rossi JJ. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol Ther. 2008;16:1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.