Abstract
Using circular dichroism spectroscopy, UV absorption spectroscopy and polyacrylamide gel electrophoresis, we studied conformational properties of guanine-rich DNA strands of the fragile X chromosome repeats d(GGC)n, d(GCG)n and d(CGG)n, with n = 2, 4, 8 and 16. These strands are generally considered in the literature to form guanine tetraplexes responsible for the repeat expansion. However, we show in this paper that the repeats are reluctant to form tetraplexes. At physiological concentrations of either Na+ or K+ ions, the hexamers and dodecamers associate to form homoduplexes and the longer repeats generate homoduplexes and hairpins. The tetraplexes are rarely observed being relatively most stable with d(GGC)n and least stable with d(GCG)n. The tetraplexes are exclusively formed in the presence of K+ ions, at salt concentrations higher than physiological, more easily at higher than physiological temperatures, and they arise with extremely long kinetics (even days). Moreover, the capability to form tetraplexes sharply diminishes with the oligonucleotide length. These facts make the concept of the tetraplex appearance in this motif in vivo very improbable. Rather, a hairpin of the fragile X repeats, whose stability increases with the repeat length, is the probable structure responsible for the repeat expansion in genomes.
INTRODUCTION
The fragile X chromosome, the most common form of inherited mental retardation in humans (reviewed in 1), is associated with expansion of a d(CGG)n trinucleotide repeat in the 5′-untranslated region of the FMR1 gene (reviewed in 2,3). Whereas in normal individuals the number of d(CGG) repeats ranges between 6 and 54, in phenotypically normal ‘premutation’ carriers the number of repeats is increased to >50–200 and in individuals affected with fragile X syndrome the number is expanded to >200–2000 (4). It was proposed (5,6) that the formation and stability of d(CGG)n intrastrand folded structures might promote expansion during DNA replication, and cause pausing of DNA polymerase inside the trinucleotide sequences both in vitro (6,7) and in vivo (8). It was reported (reviewed in 9,10) that d(GGC)n chains (n = 5–11) formed hairpins and antiparallel homoduplexes in the presence of sodium cations (11,12). The same structures were also observed in the case of d(GCG)n chains in the presence of potassium cations (13). Mitas et al. (14) reported formation of highly thermostable hairpins of d(CGG)15 in the presence of K+ ions. Usdin and Woodford (6) observed that repeated d(CGG)n sequences formed series of barriers hindering DNA synthesis in vitro, in the case where the number n of the trinucleotide repeats was 13 at least. The authors concluded that a monomolecular tetraplex, rather than a hairpin, stood behind the stop of the DNA synthesis. The four-stranded tetraplex as a minor conformation of short d(CGG)n (n = 4, 5, 7) sequences was observed by Fry and Loeb (5), Chen et al. (11), and also by ourselves (15). However, it follows from our results that the d(CGG)n stretches form tetraplexes unwillingly. On the other hand, Fry and colleagues (16–19) claimed that d(CGG)n (n = 7, 8) chains formed tetraplexes easily under physiological conditions, and even in the absence of K+ ions. It is thus obvious that the literature data on guanine tetraplexes of d(CGG)n [or d(GCG)n, d(GGC)n] are ambiguous.
In this paper we deal in detail with the conditions of guanine tetraplex formation by d(CGG)n chains (n = 2, 4, 8, 16) and the permuted triplet repeats d(GCG)n and d(GGC)n. We show that these sequences preferentially adopt homoduplexes or hairpins, whereas their transition into a tetraplex is a difficult, long-lasting process. The tetraplex formation is remarkably influenced by the base permutation in the repeated triplet and, further on, by the chain length, DNA concentration, type and concentration of cations, and temperature. We have observed a tetraplex of the repeat only in the presence K+ ions, and at their concentrations higher than physiological. The willingness to form tetraplexes distinctly decreased with the repeat length. Contrary to the literature data (16–20) we have never observed tetraplexes of G-rich fragile X chromosome repeat in the presence of sodium cations.
MATERIALS AND METHODS
The oligonucleotides were synthesized and purified by the Laboratory of Plant Molecular Physiology, Faculty of Science, Masaryk University (Brno, Czech Republic). The hexamers were also bought from VBC Genomics Bioscience Research (Vienna, Austria). The same results were obtained with the samples from the two sources. The lyophilized oligonucleotides were dissolved in 1 mM Na phosphate and 0.3 mM EDTA, pH 7.4, to give a stock solution concentration of ∼100 OD/ml.
The precise sample concentrations were determined from their absorption measured at 90°C in the above buffer using molar extinction coefficients calculated according to Gray et al. (21). The UV absorption spectra were measured on a UNICAM 5625 UV/VIS spectrometer. Before starting any experiments, the samples of the G-rich oligonucleotides were denatured (for 10 min at 90°C) to remove aggregates. In this way the circular dichroism (CD) spectral changes were reproducible starting from the same initial conditions.
CD spectra were measured using a Jobin–Yvon Mark VI dichrograph in 0.1 cm and, in special cases, in 0.01 cm pathlength Hellma cells, placed in a thermostatted holder. The DNA concentration was chosen to give an absorption of ∼0.6–0.8 at the absorption maximum, which gives an optimum signal-to-noise ratio. CD was expressed as the difference in the molar absorption of the right-handed and left-handed circularly polarized light, Δε, in units of M–1 cm–1. The molarity (M) was related to the nucleoside residues in the DNA samples. Unless stated otherwise, the DNA concentration was ∼0.7 mM.
Salts, i.e. KCl or NaCl, were added directly to the cells. The salt and DNA concentrations were then corrected for the increase of the sample volume. The pH values were checked using a Sentron Titan pH meter and a Sentron Red-Line electrode. The electrode measures pH in volumes as small as 3 µl. The buffers used were sodium or potassium phosphate (pH 7–8) or Britton–Robinson buffer pH 7 (26 mM mixture of boric, phosphoric and acetic acids and 69 mM NaOH), or pH 7.5 (25 mM mixture of the acids and 73 mM NaOH).
Non-denaturing polyacrylamide gel electrophoresis was performed in a thermostatted submersible apparatus (SE-600; Hoefer Scientific, San Francisco, CA). Gels (16%, 29:1 monomer/bis ratio), 14 × 16 × 0.1 cm in size, were run for 20 h at 70 V (∼5 V/cm) and 0°C. Two to three micrograms of DNA (∼10 µl of 0.7 mM DNA) was loaded on gels. The gels were stained with Stains-All (Sigma) and ethidium bromide in selected cases. Densitometry was performed using a Personal Densitometer SI, Model 375-A (Molecular Dynamics, Sunnyvale, CA).
RESULTS
Duplexes and hairpins
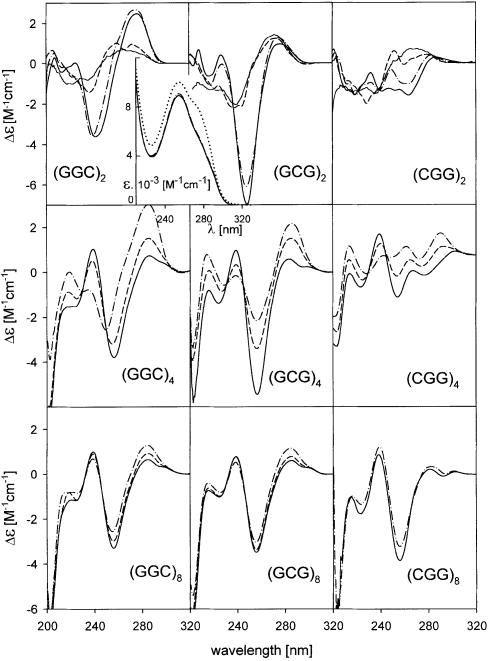
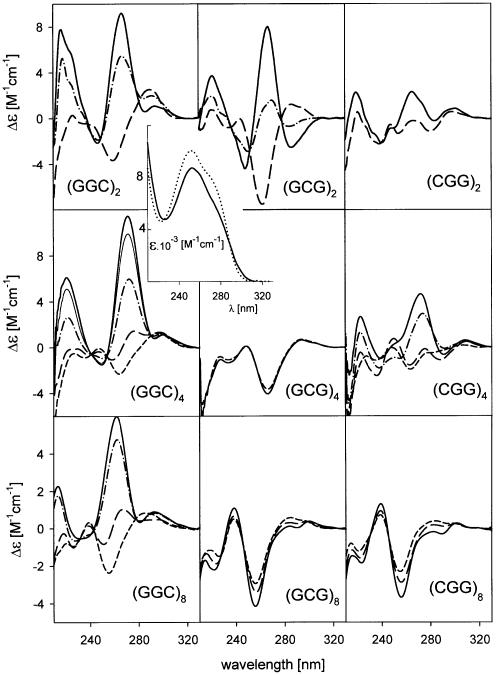
Hexamers. The hexamers d(GGC)2, d(GCG)2 and d(CGG)2 yield mutually distinct CD spectra (Fig. 1, top) at low salt (1 mM Na phosphate and 0.3 mM EDTA) and 0°C, though their theoretical spectra calculated (22) on the basis of the spectra of their constituting mono- and dinucleotides (Fig. 1, top) are very similar. The spectra thus reflect the hexamer conformation. The conformation disappears before room temperature is reached. Then the spectra of the hexamers become similar to those calculated. Increasing ionic strength (Na+ or K+) stabilizes an ordered structure. The spectra do not differ substantially from those measured at low salt, the spectral amplitudes only become more intense (Fig. 1). Also, the UV absorption spectra display the maximum hypochromicity change when the low salt samples are cooled to 0°C (insert in Fig. 1). The ionic strength increase induces small changes. The hexamers migrate through both low and physiological ionic strength gels like the d(GGC)2·d(GCC)2 heteroduplex (Fig. 2A). There is no doubt that these bimolecular structures are homoduplexes. Under all conditions d(GGC)n run slightly slower than the remaining two permuted oligonucleotides. The distinct CD spectra of the particular hexamers indicate that their detailed conformations differ.
Figure 1.
CD spectra of d(GGC)n, d(GCG)n and d(CGG)n in the presence of Na+ ions. (Top) hexamers measured in 1 mM Na phosphate and 0.3 mM EDTA, pH 7, at 22 (dashes) and 0°C (dots and dashes) and in 0.1 M NaCl at 0°C (full line). The thin full lines correspond to the calculated (22) spectra. Insert: UV absorption spectra of the hexamer d(GCG)2 measured under the same conditions as CD spectra and drawn with the same line types. The dotted spectrum corresponds to denatured hexamer measured in 1 mM Na phosphate and 0.3 mM EDTA, pH 7, at 90°C; CD spectra of 12mers (middle) and of 24mers (bottom) in 1 mM Na phosphate and 0.3 mM EDTA, pH 7 (dots and dashes), Na phosphate concentration increased to 10 mM, pH 7.5 (dashes) and with 150 mM NaCl, pH 7.5 (full line). Temperature 0°C.
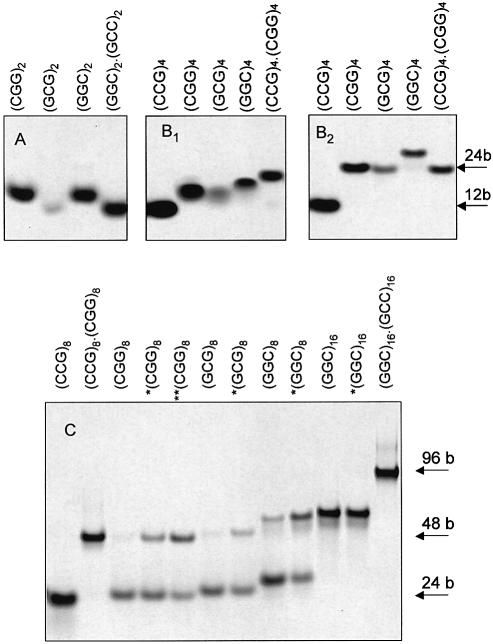
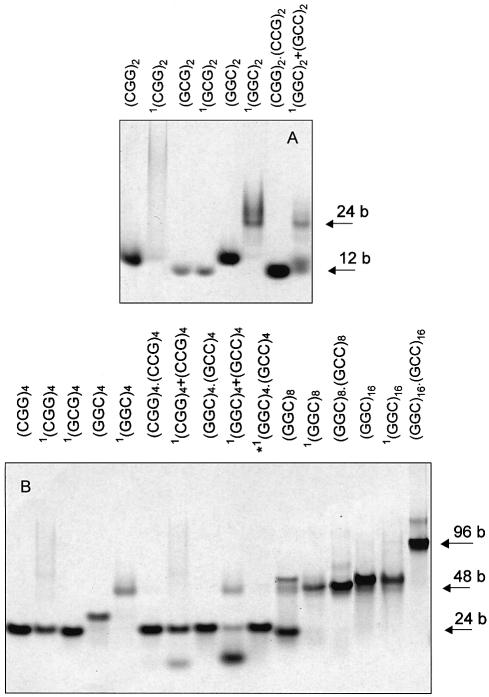
Figure 2.
Polyacrylamide gel electrophoreses of d(GGC)n, d(GCG)n and d(CGG)n in the presence of Na+ ions. (Top) Hexamers (A) and dodecamers (B2) running in Britton–Robinson buffer, pH 7 and 0.1 M NaCl, and dodecamers (B1) running in 1 mM Na phosphate and 0.3 mM EDTA, pH 7. (Bottom) 24mers and 48mers running in Britton–Robinson buffer, pH 7.5 and 0.15 M NaCl. The samples marked with the asterisk were denatured (for 5 min at 90°C) in the electrophoretic buffer prior to loading on the gel. The two asterisks mark the sample which was denatured twice in the electrophoretic buffer and very slowly cooled to room temperature. The gel B1 contains the dodecamer samples corresponding to the CD spectra drawn by dashes and dots (Fig. 1, middle), all the other gels contain samples corresponding to full line spectra in Figure 1. The heteroduplexes and d(CCG)n hairpin stand for 12, 24, 48 and 96 base long markers. All the gels were run at 2°C.
Dodecamers. At low salt (1 mM Na phosphate) pH ∼7 and at 0°C, d(GGC)4, d(GCG)4 and d(CGG)4 migrate between the d(CCG)4 hairpin and the d(CCG)4·d(CGG)4 heteroduplex (Fig. 2B1), i.e. at a position corresponding to denatured single strands. Their CD spectra (Fig. 1, middle), are more similar to those of the denatured structures than in the case of hexamers. The UV absorption spectra of the dodecamers (data not shown) are slightly hypochromic in the low salt, and the hypochromism markedly increases after increasing salt concentration. The absorption spectra in 150 mM Na+ are principally the same as those shown in the insert of Figure 1. Thus, the samples are essentially denatured at a low ionic strength. The increasing ionic strength causes retardation of all the dodecamers (Fig. 2B2) so that they run at the level of the d(CCG)4·d(CGG)4 heteroduplex. Again, d(GGC)4 migrates most slowly. The increasing ionic strength leads to extensive CD spectral changes with all the three 12mers (Fig. 1, middle). The spectra, initially conservative, become, with fast kinetics (not detectable by CD), non-conservative with the main band amplitudes at negative values. The spectra become similar to those of hexamer homoduplexes. They still remain mutually different, though their primary structures differ by two nucleotides only. In the course of the salt dependence the CD spectra of all the three 12mers intersect in two isoelliptic points. The presence of the isoelliptic points testifies that the fast process is a two-state one. Undoubtedly, it is a transition from a partially denatured structure, most probably from a short poorly stable hairpin, to a homoduplex. Further increasing sodium ions above 150 mM concentration does not change the spectra of the 12mers any more (data not shown). Thus, the transition is complete. These results agree well with the literature data indicating that d(GGC)4 (12) and d(CGG)4 (23) form antiparallel homoduplexes in the presence of (0.1–0.2 M) sodium ions.
24mers. The spectra of the three 24mers (Fig. 1, bottom), as could be envisaged, are much more similar than the shorter repeats (but their conformational properties still differ as will be shown below). Increasing ionic strength from 1 mM Na phosphate to 150 mM NaCl at 0°C induces only slight changes in the CD spectra. The oligonucleotides actually form stable single-stranded hairpins at the low ionic strength, which remain stable even after increasing salt concentration to 150 mM (Fig. 2C). The homoduplex population partially appeared only after heating the samples in 150 mM Na+ (at 90°C) (Fig. 2C). The homoduplex was stabilized by further increasing salt. The change in the proportion of the mono- and bimolecular fraction was not much reflected in the CD spectra. An increasing population of duplexes was accompanied by a slight deepening of the negative bands and the positive one around 280 nm [the spectrum of d(CGG)8 after heating is shown at the bottom of Fig. 1]. The change is the same as that caused by further increasing ionic strength. Fry and colleagues (16,18) report that they induce bimolecular tetraplexes in d(CGG)7 and d(CGG)8 in this way. We have studied both d(CGG)8 (Figs 1, bottom and 2C) and d(CGG)7 (Figs 3 and 4) under different conditions including precisely (DNA concentration, temperature and time of sample incubation, etc.) those of Fry and colleagues (16,18) (0.3 M Na+, and even 0.5 M K+ concentrations) but have invariably obtained the same result: the shape of the CD spectrum (Fig. 3A), as well as that of the UV absorption spectrum (Fig. 3, insert) remained principally the same after heating as before heating. The type of CD spectra remained the same in the presence of Na+, K+, Li+ (Fig. 3B), Ca2+ or Mg2+. The same was true with all the three 24mers with permuted triplets. The only consequence of the heating was an increase of the bimolecular fraction (Fig. 4). These results indicate that the structural features of both conformers, i.e. of the mono- as well as the bimolecular one, are in principle the same. The two strictly separated electrophoretic bands of the two conformers (Figs 2 and 4) give evidence of a kinetic barrier between them, in the same way as observed with the stable hairpins and homoduplexes of various oligonucleotides (24). The barrier increases with the length of the molecule. Along with the fact that the same CD and absorption spectrum is displayed in 100–150 mM Na+ by 24-, 12- and even 6mers, the results indicate that the bimolecular structure is a homoduplex. At the same time, the results imply that the homoduplex is antiparallel and that both the hairpin and the homoduplex have the same topology (9,10).
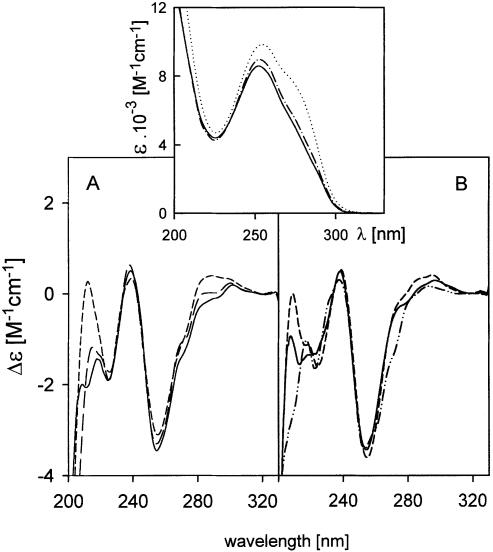
Figure 3.
CD spectra of d(CGG)7 measured under the conditions of Weisman-Shomer et al. (17). (A) 0.15 M NaCl (short dashes) immediately after the addition of salt, and after denaturation (for 5 min at 90°C) (long dashes); 0.3 M NaCl (full line), the spectrum did not change after denaturation and incubation for 24 h at 4°C. (B) 0.5 M KCl measured immediately after the addition of salt (short dashes) and after denaturation (5 min at 90°C) and incubation for 48 h at 4°C (full line); d(CGG)7 in 0.3 M LiCl (two dots and dash). The spectrum did not change after denaturation and incubation at 4°C for 24 h. All of the spectra were recorded at 0°C, the buffer always contained 10 mM Na phosphate and 0.3 mM EDTA, pH 8. Insert: UV absorption spectra of d(CGG)7 in 1 mM Na phosphate and 0.3 mM EDTA, pH 7 measured at 90 (dots) and 0°C (dots and dashes); in 0.3 M NaCl, measured at 0°C (full line). The spectrum did not change after denaturation and incubation at 4°C for 24 h. This spectrum corresponds to the sample whose CD spectrum is drawn in full line in (A).
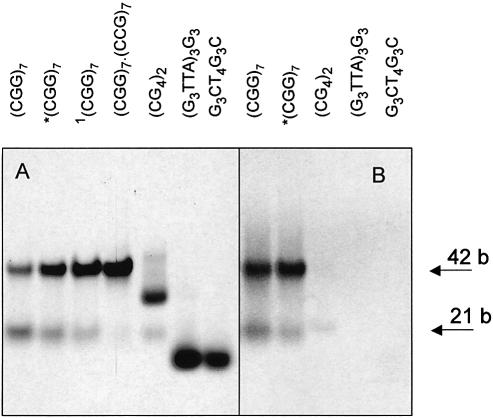
Figure 4.
Polyacrylamide gel electrophoresis of d(CGG)7 running in Britton–Robinson buffer pH 7.5 and 0.15 M NaCl, at 2°C. The samples marked with an asterisk were denatured (for 5 min at 90°C) in the electrophoretic buffer. The sample marked with 1 was transformed into 0.3 M NaCl and then denatured (for 5 min at 90°C). The samples of d(CGG)7 (from left to right), respectively, correspond to short dashes, long dashes and full line spectra in Figure 3A. Heteroduplex d(CGG)7·d(CCG)7 stands for the marker. The gel was cut into two pieces. One part (A) was stained with Stains-All and the other part (B) with ethidium bromide. d(CG4)2, d[(G3TTA)3G3] and d(G3CT4G3C), which form various types of tetraplexes, were used as controls.
The salt-induced CD changes (Fig. 3A and B) are characteristic (25,26) of the winding of DNA double helix due to increasing ionic strength. The resulting spectrum and the course of its appearance do not correspond to tetraplex formation irrespective of whether one has in mind the so-called parallel stranded tetraplex (positive maxima at 260 and 210 nm) or the antiparallel stranded tetraplex (positive maxima at 295 and 210 nm, or at 295, 260 and 210 nm) (27). Also, the UV absorption spectrum does not correspond to tetraplex (28). As a complement evidence testifying against tetraplex, we used staining with ethidium, which does not intercalate into the guanine tetraplex (P. Fojtík, I. Kejnovská and M. Vorlíčková, unpublished results). We cut the gel shown in Figure 4 and stained one part, as usual, with Stains-All (Fig. 4A) and the other with ethidium bromide (Fig. 4B). As can be seen, both the hairpin and homoduplex of the d(CGG)7 stain with ethidium bromide like the normal duplexes containing W-C pairs. We show in Figure 4B the following controls: d(CG4CG4), d[(G3TTA)3G3] and d(G3CT4G3C). Under the conditions of the electrophoresis, d(CG4CG4) forms duplex and two types of ‘parallel’ tetraplex (P. Fojtík, I. Kejnovská and M. Vorlíčková, unpublished results), d[(G3TTA)3G3] an intramolecular tetraplex (29) and d(G3CT4G3C) a bimolecular tetraplex containing GCGC tetrads (30). None of these tetraplexes is visible on the electrophoresis stained with ethidium bromide (Fig. 4B). The only trace of the three samples belongs to d(CG4CG4) duplex.
We also undertook a study of d(GGC)16 in NaCl. In a wide range of salt concentrations and pH values, the CD spectrum of d(GGC)16 was the same as with d(GGC)8. Under all conditions studied (1 mM phosphate–300 mM Na+, pH 4.5–9), including heating to various temperatures, d(GGC)16 co-migrated with the d(GGC)8 duplex. Thus, d(GGC)16 adopted a stable hairpin.
Tetraplexes
In spite of all our efforts, we have never observed a different type of CD with the studied trinucleotide repeats in the presence of sodium ions and neutral pH than those described above. Therefore, we started trying to stabilize tetraplexes in the presence of KCl. However, tetraplexes of all the d(GGC)n and of their permuted analogs appeared very unwillingly. To promote tetraplex formation we used high DNA concentrations (∼0.7 mM in a 0.1 cm, and even ∼7 mM in a sandwich 0.01 cm pathlength cell).
Hexamers. Adding KCl to hexamer solutions induced the same CD (Fig. 5, top) and UV absorption (data not shown) spectra as with NaCl. Thus, the duplex was also induced in KCl immediately after addition of the salt. Electrophoretic migration of the samples (Fig. 6A) also corresponds to the spectroscopic result. The CD spectra start changing (Fig. 5) with very long kinetics [e.g. one half of (GGC)2 is transformed into the tetraplex after 2 days in 0.4 M KCl] only at higher K+ concentrations. The final CD spectra of d(GGC)2 and d(GCG)2 correspond to those of the tetraplex described in the literature (29,31–34). The sample of d(CGG)2 rather non-specifically aggregates, which also follows from its electrophoresis (Fig. 6A). d(GGC)2 yields electrophoretic bands corresponding to four-molecular associates, the tetraplex of d(GCG)2 does not remain stable under the conditions of the electrophoresis (Fig. 6A). The observed tetraplexes are the so-called parallel-stranded tetraplexes {‘so-called’ because we know [P. Fojtík, I. Kejnovská and M. Vorlíčková, unpublished results and Dapic et al. (27)] that the same type of CD spectrum is also observed with antiparallel bimolecular and even intramolecular tetraplexes}. Apart from the main maximum around 260 nm, the spectrum also contains a positive maximum at 210 nm, which is usually present with both parallel- and also antiparallel-stranded tetraplexes. The UV absorption spectrum of the KCl-induced tetraplex [shown with, for example, d(GGC)2 in the insert of Fig. 5] displays a distinct decrease in the absorption maximum and an increase in absorption at long wavelengths. An increase in absorption around 295 nm as compared with the denatured sample was suggested as an easy way to monitor tetraplex formation (28).
Figure 5.
CD spectra of d(GGC)n, d(GCG)n and d(CGG)n in the presence of KCl. KCl concentration and time of incubation increase from dashed to full lines. (Top) d(GGC)2 in 0.1 M KCl (dashes), 2 days after addition of KCl to 0.4 M (dots and dashes) and after 11 days (full line). Insert: UV absorption spectra of (GGC)2: full line corresponds to conditions of the full line CD spectrum, dotted spectrum corresponds to the denatured sample measured in 1 mM Na phosphate and 0.3 mM EDTA, pH 7, at 90°C. d(GCG)2: in 2 M KCl measured immediately (dashes), 2 days (dots and dashes) and 7 days (full line) after adding KCl. d(CGG)2: in 0.4 M KCl measured immediately (dashes) and 5 days (full line) after adding KCl. Buffer 10 mM K phosphate, pH 7.4, temperature 0°C. (Middle) d(GGC)4: in 0.1 M KCl immediately (short dashes) and after 20 h (long dashes), in 0.2 M KCl after a further 20 h (dots and dashes), in 0.3 M KCl after 3 days (thin full line), in 0.4 M KCl after 7 days (full line). d(GCG)4 and d(CGG)4: in 0.1 M KCl immediately (short dashes) and in 0.4 M KCl immediately (long dashes), 2 days (dots and dashes) and 8 days (full line) after increasing the salt concentration. (Bottom) 24mers in 0.1 M KCl (short dashes), 0.3 M KCl, 2 days after incubation for 60 min at 60°C and slow cooling to 37°C (long dashes), 0.5 M KCl, immediately after incubation for 5 min at 90°C and slow cooling to 37°C (dots and dashes) and after a further 20 days (full line). In the middle and bottom panel the buffer was 10 mM K phosphate, pH 7.5, the temperature of the measurement and of the sample incubation was 37°C.
Figure 6.
Polyacrylamide gel electrophoresis of d(GGC)n, d(GCG)n and d(CGG)n running in Britton–Robinson buffer, pH 7 and 0.2 M KCl, temperature 2°C. The unmarked samples were loaded on the gel in the electrophoretic buffer. The samples marked 1 were loaded in 0.4 or 0.5 M KCl in (A) and (B), respectively. With the exception of (GCG)2 (higher KCl concentration was needed to induce CD changes), the samples marked with the index 1 correspond to those whose CD spectra are drawn in Figure 5 with full lines. The heteroduplexes serving as markers were mixed and annealed under low salt conditions. In the neighboring lines there are mixtures of the particular marked samples with their complements. The sample marked with the asterisk was denatured (for 5 min at 90°C) before loading on the gel and slowly cooled.
Dodecamers. The CD spectra of the dodecamers measured at 0°C (or 37°C, Fig. 5, middle) immediately after adding K+ to 0.1 M concentration are the same as in the presence of Na+ ions. They thus correspond to homoduplexes, which is in accord with the electrophoresis (Fig. 6B). The spectra do not change with time. Only the temperature increase to 37°C leads to a very slow increase of the CD positive band at 260 nm reflecting tetraplex formation but only with d(GGC)4 (Fig. 5, middle). With increasing potassium cation concentration the band grows faster and, in 0.4 M KCl, it finally reaches an equilibrium Δε value of ∼12 M–1 cm–1. The UV absorption spectrum of this sample corresponds to that shown with (GGC)2 in the insert of Figure 5. The analogous treatment leads to only partial tetraplex formation with d(CGG)4, whereas d(GCG)4 does not form any tetraplex at all. We observed a tetraplex of d(GCG)4 only at an oligonucleotide concentration higher by an order of magnitude (data not shown). Again, the tetraplex appeared with very slow kinetics and through a two-state mechanism. The tetraplex of d(GGC)4 arose much faster at higher DNA concentrations, whereas d(CGG)4 aggregated under the same conditions. Aggregation usually takes place when the oligonucleotide cannot adopt an ordered conformation in the particular extreme solvent conditions (M. Vorlíčková, personal communication). Aggregation of d(CGG)4 at high potassium concentration was also observed by Sha et al. (35).
The three samples in which the tetraplex was induced by 0.4 M KCl at 0.7 mM DNA concentration and at 37°C were loaded on the gel run in 0.1 M KCl. The electrophoresis (Fig. 6B) shows that d(GGC)4 forms a four-molecular tetraplex under these conditions. The d(GCG)4 strand displays a trace in the position of the d(CCG)4·d(CGG)4 duplex so that, in accord with the spectroscopic results, it remains duplex like in the presence of sodium cations. The d(CGG)4 strand displays the main electrophoretic band also at the position of the heteroduplex, but, in addition, a slower, fuzzy trace can be seen on the gel, which indicates the presence of heavier structures aggregating non-specifically.
24mers. The CD spectra of all the three 24mers in 0.1 M K+ (Fig. 5, bottom) again correspond to those in the presence of Na+, and thus the 24mers form hairpins and partially homoduplexes as follows from the electrophoretic results (Fig. 6B). We saw indications of tetraplex formation only in the case of d(GGC)8, but at 0.3 M K+ only and after an incubation at 60°C for 60 min and then at 37°C for 2 days (Δε at 261 nm reached only 1 M–1 cm–1). Only further increasing the potassium concentration to 0.5 M, along with a temperature increase to 90 °C (for 5 min), led to a more extensive growth of the 261 nm CD maximum at 37°C, while Δε maximally reached the value of 6 M–1 cm–1 (Fig. 5, bottom). The electrophoresis of this sample (Fig. 6B) shows that the bands corresponding to d(GGC)8 hairpin and the homoduplex disappeared and a new band appeared, corresponding to a bimolecular tetraplex. The new band appeared at the very same place as the band of the heteroduplex d(GCC)8·d(GGC)8. The tetraplex thus ran faster than the d(GGC)8 homoduplex, which is usual in the case of hairpin dimers. Increasing DNA concentration by 10 times supported tetraplex formation (data not shown). Seven millimolar d(GGC)8 in 0.5 M KCl displayed (at 37°C after a previous short incubation at 70°C) a slow, two-state transition into tetraplex, while its final equilibrium CD spectrum (obtained after 2 days) reached an amplitude value of 12 M–1 cm–1 at 261 nm and ∼6 M–1 cm–1 at 210 nm (data not shown). The CD spectrum of d(CGG)8 at 7 mM DNA concentration changed only slightly after the same treatment and reached the final 261 nm amplitude value of ∼3.5 M–1 cm–1. The CD spectrum of the remaining d(GCG)8 did not change, the 24mer invariably adopted the hairpin and homoduplex. The CD spectrum of d(GGC)16 was identical to that of d(GGC)8 in NaCl, and it also did not change upon adding K+ and the treatment described above (data not shown). The 48mer always remained a hairpin. It is obvious that the G-rich strand of the fragile X chromosome repeat is not disposed to form tetraplex. The reluctance to tetraplex formation increases (in the range of oligonucleotide lengths studied) with the increasing number of trinucleotide repeats. The tetraplex stability depends on DNA concentration, which is in line with the fact that we have never observed its intramolecular formation.
DISCUSSION
The relevant published studies generally state (5,6,11,16–19,36–38) that the G-rich strand of the fragile X chromosome region folds into guanine tetraplexes, and the tetraplexes were proposed to be responsible for the repeat expansion. In contrast, we show in this report that the guanine-rich strands are very unwilling to fold into guanine tetraplexes. We demonstrate that the guanine tetraplex formation is considerably influenced by the base sequence in the repeated triplet, d(GGC)n and d(GCG)n being the most and the least promotive repeats, respectively. The tetraplexes only appear in the presence of potassium cations, especially at higher than physiological concentrations and at higher than physiological temperatures. The kinetics of the transition into the guanine tetraplex is very slow. The tetraplex is furthermore destabilized with increasing DNA length. The hexamers and dodecamers form four-molecular tetraplexes. Higher potassium cation concentrations are needed to induce the tetraplex with longer chains, and higher temperatures should be used to overcome the transition barrier. Among the present 24mers, d(GGC)8 was the only one providing the tetraplex. The tetraplex was bimolecular. We have never observed monomolecular tetraplexes with the present oligonucleotides. d(GGC)16 was entirely resistant to generate a tetraplex of any kind. In contrast to the literature (16–20), we have observed no tetraplexes at all in the presence of sodium cations at neutral pH.
In the presence of sodium cations, the hexamers are homoduplexes at both low (1 mM Na phosphate) and physiological ionic strengths. Surprisingly, the dodecamers are less ordered than the hexamers at low ionic strength. The UV absorption spectra and the electrophoretic migration (Fig. 2B1) suggest that the dodecamers are hairpins (Fig. 7) whose loops occupy a considerable part of the molecule. The increasing ionic strength transforms the dodecamers into homoduplexes (Fig. 7) through a two-state mechanism (Fig. 1, middle). The CD spectra of the homoduplexes become similar to those of the hexamers. The CD spectra of d(CGG)4 and d(CGG)2 differ from those of the remaining permuted motifs. The difference could be caused by different alignment (9,10) of the strands.
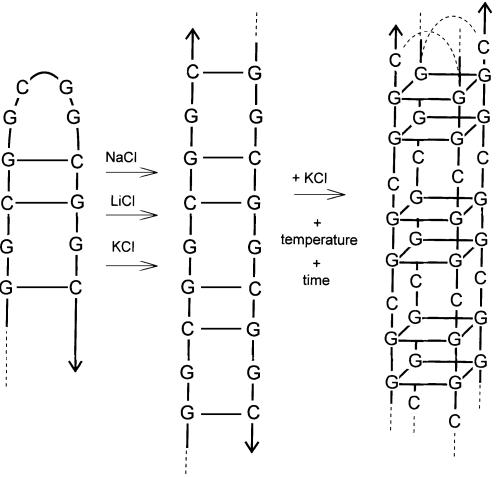
Figure 7.
Sketch of the (GGC)n hairpin (left), duplex (middle) and tetraplex (right). For n ≥ 7 the tetraplex is bimolecular.
The 24mers are stable hairpins at low ionic strength, as follows from the UV absorption spectra and PAGE. The increasing ionic strength causes a partial transition of the hairpins to homoduplexes (Fig. 2C). The transition is promoted by elevated temperatures. The strict electrophoretic bands indicate that the hairpins and homoduplexes are distinct conformers separated by an energy barrier. The barrier is connected with the necessity to break GC pairs within the hairpin and their reunion in the same way between two molecules (see Fig. 7). However, the CD spectra of the hairpins and homoduplexes are almost the same. The CD spectra identity suggests that the hairpin stem has the same conformation as the homoduplex and that the homoduplex is antiparallel (Fig. 7).
CD spectroscopy is useful to map the whole conformational space of the studied DNA molecule (39) because it enables an easy titration of the sample to study the DNA conformational properties in a wide range of conditions. However, we failed to stabilize a tetraplex in the G-rich strand of the fragile X repeat in the presence of sodium cations. A very acid pH was the only exception, which is in line with the results of Chen (36) and our own previous results (15). The hexamers provided tetraplexes around pH 4 through a two-state mechanism and without aggregation. The CD spectra were the same as the CD spectra of tetraplexes induced by KCl, i.e. the positive maxima were located at 260 and 210 nm. The UV absorbance was also enhanced at 295 nm. We think that the K+-induced and acid-induced tetraplexes are the same. According to the suggestion of Chen (36), the tetraplex is formed by a couple of guanine tetrads and by a cytosine tetrad in between (Fig. 7). The duplex–tetraplex transition requires breaking of GC pairs and at least a shift in the strand alignment (possibly even a change in strand polarity). That is why the transition is difficult.
A correct structural interpretation of the observed CD spectra requires a careful analysis of the mechanisms of the appearance of the particular conformers and a search for analogies with related molecules. The observation in the presence of sodium cations of the same and through the same mechanism arising CD spectrum for both long and short molecules, for example, excludes the possibility that the CD spectrum with the dominating negative band at 260 nm corresponds to the antiparallel tetraplex described by Kettani et al. (37). These authors observed, in the presence of NaCl, a tetraplex of d(GCGGTTTGCGG) which was a dimer of two hairpins containing G tetrads at the ends, and GCGC tetrads in between. It is this work that is often cited as a reference for the statement that the G-rich strands of the fragile X chromosome triplet repeats form a tetraplex. First of all, this molecule contains six Gs and only two Cs, which is a substantial difference from the fragile X chromosome motif. Secondly, the Ts generate a loop, which promotes the hairpin. The present molecules lacking the thymine run fail to fold into the hairpin. The shortest molecule we observed to form a stable hairpin was d(GGC)7. Furthermore, we studied d(GCGGTTTGCGG) exactly under the conditions of NMR measurements including the oligonucleotide concentration. Its tetraplex formation was accompanied by an increase of the positive CD bands at 290, 260 and 210 nm. It corresponds to spectra of antiparallel tetraplexes (27), namely of the chair type (M. Vorlíčková, unpublished results). The present spectra of d(GCG)n, d(GGC)n or d(CGG)n in NaCl are entirely different. Another tetraplex of d(G3CT4G3C) also contains GCGC tetrads (30). It displays a CD spectrum (M. Vorlíčková, unpublished results) corresponding to antiparallel tetraplexes of the basket type, the classical representative of which is d(G4T4G4) (40). The formation of these tetraplexes is accompanied not only by a deepening of the negative band around 260 nm, but also by an increase of the positive band at 290 nm and namely of that at 210 nm. The band at 210 nm is significant because it is usually present with tetraplexes, though the long wavelength parts of the CD spectrum are different for various tetraplex types. Not only the CD spectrum but also the UV absorption spectrum of d(CGG)n and their permutations differ from those of tetraplexes. If, in spite of all this, we would admit that the bimolecular structure of the studied repeat in NaCl is a tetraplex (association of two hairpins would result in formation of one G tetrad and two tetrads of GCGC; see Fig. 7), then the hairpin dimerization not including any substantial change in their arrangement (according to no change in the CD and the absorption) should happen fast, without any difficulties. The reverse is true in our case. The presence of an energy barrier gives evidence of a structural rearrangement which we meet with hairpin–duplex transitions of longer DNA molecules (24). Further on, both the hairpin and homoduplex are stained with ethidium bromide (Fig. 4B) in the same way as regular DNA molecules containing W-C pairs. In contrast, the tetraplexes of all the above-mentioned types (tetraplex of GCG2T3GCG2 is not stable under the conditions of the electrophoresis) are not stained with ethidium bromide.
Patel et al. (41) studied the d(TGGCGGC) heptamer, which is another analog of the fragile X sequence. They found that the heptamer was a tetraplex in the presence of potassium cations, whereas it formed a duplex at neutral pH but a tetraplex at acid pH. This observation exactly corresponds to our results with the d(GGCGGC) hexamer. Even the CD spectra that we have obtained with d(TGGCGGC) are identical (data not shown) to the CD spectra of d(GGCGGC). Another paper cited on the fragile X tetraplexes is that of Fry and Loeb (5) where they observed the tetraplexes with short fragments, which is consistent with our observations.
It follows from our unpublished observations and is also obvious from Figure 6 that the fragile X tetraplex does not bind the complementary sequence. Usdin and Woodford demonstrated (6) that the tetraplex of the d(CGG)n repeats blocked DNA synthesis in vitro. The block was only observed in the presence of potassium cations, which is consistent with our results. However, the synthesis was blocked by long d(CGG)n repeats in their experiments, which is a difference from our observations. We do not exactly know which factors contribute to the tetraplex stabilization in their experiments but the repeated cycles of denaturation and annealing are certainly among the factors.
Conformation of the G-rich fragile X motif was studied by means of DNA interaction with dimethylsulphate (DMS) and other agents by several authors. The results of all the studies support our conclusion that the motif does not form tetraplex in the presence of sodium cations. Usdin and Woodford (6) were the only authors who observed a protection of N7 guanines in d(CGG)20 against interaction with DMS, which suggests the presence of all guanines in the tetraplex. This, however, was only true in the presence of potassium. On the contrary, in the absence of potassium, the results of the paper (also following from reaction with bromoacetaldehyde) were consistent with d(CGG)20 hairpin. Mitas et al. (14) showed that no guanine residue in d(CGG)15 was completely protected from methylation in a wide range of conditions (including K+) to rule out tetraplex. Furthermore, their DMS and P1 experiments showed that there was only one loop in the d(CGG)15 structure, again suggesting a single hairpin. Even Nadel et al. found (13) that all the guanine residues in unimolecular structures of d(GCG)n were modified by DMS. In addition, they found by modification with diethylpyrocarbonate that the d(GCG)11 contained only one loop of unpaired bases.
Folding of the d(GGC)n (or its permuted analogs) motifs into tetraplexes may block DNA synthesis in vivo (6). On the other hand, the tetraplex can hardly be the factor standing behind the fragile X repeat expansion. It follows from our results that the tetraplex formation is difficult and its stability considerably decreases with the length of the chain. In fact, we have never observed its intramolecular formation. The hairpin of the repeat, whose stability increases with the repeat length, is a much more suspect candidate to cause fragility of chromosome X.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful to Dr Jaroslav Kypr and Dr Karel Nejedly for critical reading of the manuscript and kind discussions. The study was supported by grants 204/01/0561 and A4004201, awarded to M.V. by the Grant Agency of the Czech Republic and by the Grant Agency of the Academy of Sciences of the Czech Republic, respectively.
REFERENCES
- 1.Ashley C.T.J. and Warren,S.T. (1995) Trinucleotide repeat expansion and human disease. Annu. Rev. Genet., 29, 703–728. [DOI] [PubMed] [Google Scholar]
- 2.Oostra B.A. and Willems,P.J. (1995) A fragile gene. Bioessays, 17, 941–947. [DOI] [PubMed] [Google Scholar]
- 3.Richards R.I. (2001) Dynamic mutations: a decade of unstable expanded repeats in human genetic disease. Hum. Mol. Genet., 10, 2187–2194. [DOI] [PubMed] [Google Scholar]
- 4.Mandel J.L. (1993) Questions of expansion. Nature Genet., 4, 8–9. [DOI] [PubMed] [Google Scholar]
- 5.Fry M. and Loeb,L.A. (1994) The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl Acad. Sci. USA, 91, 4950–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Usdin K. and Woodford,K.J. (1995) CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res., 23, 4202–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang S., Ohshima,K., Shimizu,M., Amirhaeri,S. and Wells,R.D. (1995) Pausing of DNA synthesis in vitro at specific loci in CTG and CGG triplet repeats from human hereditary disease genes. J. Biol. Chem., 270, 27014–27021. [DOI] [PubMed] [Google Scholar]
- 8.Samadashwily G.M., Raca,G. and Mirkin,S.M. (1997) Trinucleotide repeats affect DNA replication in vivo. Nature Genet., 17, 298–304. [DOI] [PubMed] [Google Scholar]
- 9.Darlow J.M. and Leach,D.R.F. (1998) Secondary structures in d(CGG) and d(CCG) repeat tracts. J. Mol. Biol., 275, 3–16. [DOI] [PubMed] [Google Scholar]
- 10.Darlow J.M. and Leach,D.R.F. (1998) Evidence for two preferred hairpin folding patterns in d(CGG).d(CCG) repeat tracts in vivo. J. Mol. Biol., 275, 17–23. [DOI] [PubMed] [Google Scholar]
- 11.Chen X., Mariappan,S.V.S., Catasti,P., Ratliff,R., Moyzis,R.K., Laayoun,A., Smith,S.S., Bradbury,E.M. and Gupta,G. (1995) Hairpins are formed by the single DNA strands of the fragile X triplet repeats: structure and biological implications. Proc. Natl Acad. Sci. USA, 92, 5199–5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mariappan S.V.S., Catasti,P., Chen,X., Ratliff,R., Moyzis,R.K., Bradbury,E.M. and Gupta,G. (1996) Solution structures of the individual single strands of the fragile X DNA triplets (GCC)n.(GGC)n. Nucleic Acids Res., 24, 784–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nadel Y., Weisman-Shomer,P. and Fry,M. (1995) The fragile X syndrome single strand d(CGG)n nucleotide repeats readily fold back to form unimolecular hairpin structures. J. Biol. Chem., 270, 28970–28977. [DOI] [PubMed] [Google Scholar]
- 14.Mitas M., Yu,A., Dill,J. and Haworth,I.S. (1995) The trinucleotide repeat sequence d(CGG)15 forms a heat-stable hairpin containing Gsyn.Ganti base pairs. Biochemistry, 34, 12803–12811. [DOI] [PubMed] [Google Scholar]
- 15.Vorlickova M., Zimulova,M., Kovanda,J., Fojtik,P. and Kypr,J. (1998) Conformational properties of DNA dodecamers containing four tandem repeats of the CNG triplets. Nucleic Acids Res., 26, 2679–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fry M. and Loeb,L.A. (1999) Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem., 274, 12797–12802. [DOI] [PubMed] [Google Scholar]
- 17.Weisman-Shomer P., Naot,Y. and Fry,M. (2000) Tetrahelical forms of the fragile X syndrome expanded sequence d(CGG)(n) are destabilized by two heterogeneous nuclear ribonucleoprotein-related telomeric DNA-binding proteins. J. Biol. Chem., 275, 2231–2238. [DOI] [PubMed] [Google Scholar]
- 18.Weisman-Shomer P., Cohen,E. and Fry,M. (2000) Interruption of the fragile X syndrome expanded sequence d(CGG)(n) by interspersed d(AGG) trinucleotides diminishes the formation and stability of d(CGG)(n) tetrahelical structures. Nucleic Acids Res., 28, 1535–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weisman-Shomer P., Cohen,E., Hershco,I., Khateb,S., Wolfovitz-Barchad,O., Hurley,L.H. and Fry,M. (2003) The cationic porphyrin TMPyP4 destabilizes the tetraplex form of the fragile X syndrome expanded sequence d(CGG)n. Nucleic Acids Res., 31, 3963–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uliel L., Weisman-Shomer,P., Oren-Jazan,H., Newcomb,T., Loeb,L.A. and Fry,M. (2000) Human Ku antigen tightly binds and stabilizes a tetrahelical form of the fragile X syndrome d(CGG)n expanded sequence. J. Biol. Chem., 275, 33134–33141. [DOI] [PubMed] [Google Scholar]
- 21.Gray D.M., Hung,S.-H. and Johnson,K.H. (1995) Absorption and circular dichroism spectroscopy of nucleic acid duplexes and triplexes. Methods Enzymol., 246, 19–34. [DOI] [PubMed] [Google Scholar]
- 22.Cantor C.R., Warshaw,M.M. and Shapiro,H. (1970) Oligonucleotide interactions. III. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers, 9, 1059–1077. [DOI] [PubMed] [Google Scholar]
- 23.Zheng M., Huang,X., Smith,G.K., Yang,X. and Gao,X. (1996) Genetically unstable CXG repeats are structurally dynamic and have a high propensity for folding. An NMR and UV spectroscopic study. J. Mol. Biol., 264, 323–336. [DOI] [PubMed] [Google Scholar]
- 24.Vorlickova M., Kejnovska,I., Tumova,M. and Kypr,J. (2001) Conformational properties of DNA fragments containing (GAC) trinucleotide repeats associated with skeletal displasias. Eur. Biophys. J., 30, 179–185. [DOI] [PubMed] [Google Scholar]
- 25.Studdert D.S., Patroni,M. and Davis,R.C. (1972) Circular dichroism of DNA: temperature and salt dependence. Biopolymers, 11, 761–779. [DOI] [PubMed] [Google Scholar]
- 26.Ivanov V.I., Minchenkova,L.E., Schyolkina,A.K. and Poletayev,A.I. (1973) Different conformations of double-stranded nucleic acid in solution as revealed by circular dichroism. Biopolymers, 12, 89–110. [DOI] [PubMed] [Google Scholar]
- 27.Dapic V., Abdomerovic,V., Marrington,R., Peberdy,J., Rodger,A., Trent,J.O. and Bates,P.J. (2003) Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res., 31, 2097–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mergny J.L., Phan,A.T. and Lacroix,L. (1998) Following G-quartet formation by UV-spectroscopy. FEBS Lett., 435, 74–78. [DOI] [PubMed] [Google Scholar]
- 29.Balagurumoorthy P., Brahmachari,S.K., Mohanty,D., Bansal,M. and Sasisekharan,V. (1992) Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res., 20, 4061–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kettani A., Bouaziz,S., Gorin,A., Zhao,H., Jones,R.A. and Patel,D.J. (1998) Solution structure of a Na cation stabilized DNA quadruplex containing G·G·G·G and G·C·G·C tetrads formed by G-G-G-C repeats observed in adeno-associated viral DNA. J. Mol. Biol., 282, 619–636. [DOI] [PubMed] [Google Scholar]
- 31.Gray D.M. and Bollum,F.J. (1974) A circular dichroism study of poly dG, poly dC and poly dG:dC. Biopolymers, 13, 2087–2102. [DOI] [PubMed] [Google Scholar]
- 32.Guo Q., Lu,M. and Kallenbach,N.R. (1993) Effect of thymine tract length on the structure and stability of model telomeric sequences. Biochemistry, 32, 3596–3603. [DOI] [PubMed] [Google Scholar]
- 33.Penazova H. and Vorlickova,M. (1997) Guanine tetraplex formation by short DNA fragments containing runs of guanine and cytosine. Biophys. J., 73, 2054–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porumb H., Monnot,M. and Fermandjian,S. (2002) Circular dichroism signatures of features simultaneously present in structured guanine-rich oligonucleotides: a combined spectroscopic and electrophoretic approach. Electrophoresis, 23, 1013–1020. [DOI] [PubMed] [Google Scholar]
- 35.Sha F., Mu,R., Henderson,D. and Chen,F.M. (1999) Self-aggregation of DNA oligomers with XGG trinucleotide repeats: kinetic and atomic force microscopy measurements. Biophys. J., 77, 410–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen F.-M. (1995) Acid-facilitated supramolecular assembly of G-quadruplexes in d(CGG)4. J. Biol. Chem., 270, 23090–23096. [DOI] [PubMed] [Google Scholar]
- 37.Kettani A., Kumar,R.A. and Patel,D.J. (1995) Solution structure of a DNA quadruplex containing the fragile-X syndrome triplet repeat. J. Mol. Biol., 254, 638–656. [DOI] [PubMed] [Google Scholar]
- 38.Usdin K. (1998) NGG-triplet repeats form similar intrastrand structures: implications for the triplet expansion diseases. Nucleic Acids Res., 26, 4078–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vorlickova M. (1995) Conformational transitions of alternating purine-pyrimidine DNAs in perchlorate ethanol solutions. Biophys. J., 69, 2033–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith F.W. and Feigon,J. (1993) Strand orientation in the DNA quadruplex formed from the Oxytricha telomere repeat oligonucleotide d(G4T4G4) in solution. Biochemistry, 32, 8682–8692. [DOI] [PubMed] [Google Scholar]
- 41.Patel P.K., Bhavesh,N.S. and Hosur,R.V. (2000) Cation-dependent conformational switches in d-TGGCGGC containing two triplet repeats of fragile X syndrome: NMR observations. Biochem. Biophys. Res. Commun., 278, 833–838. [DOI] [PubMed] [Google Scholar]