Abstract
Ligand 12459, a potent G-quadruplex-interacting agent that belongs to the triazine series, was previously shown to downregulate telomerase activity in the human A549 lung carcinoma cell line. We show here that the downregulation of telomerase activity is caused by an alteration of the hTERT splicing pattern induced by 12459, i.e. an almost complete disappearance of the active (+α,+β) transcript and an over-expression of the inactive –β transcript. Spliced intron 6 forming the –β hTERT transcript contained several tracks of G-rich sequences able to form G-quadruplexes. By using a specific PCR-stop assay, we show that 12459 is able to stabilize the formation of these G-quadruplex structures. A549 cell line clones selected for resistance to 12459 have been analyzed for their hTERT splicing pattern. Resistant clones are able to maintain the active hTERT transcript under 12459 treatment, suggesting the appearance of mechanisms able to bypass the 12459-induced splicing alterations. In contrast to 12459, telomestatin and BRACO19, two other G-quadruplex-interacting agents, have no effect on the hTERT splicing pattern in A549 cells, are cytotoxic against the A549-resistant clones and display a lower efficiency to stabilize hTERT G-quadruplexes. These results lead us to propose that 12459 impairs the splicing machinery of hTERT through stabilization of quadruplexes located in the hTERT intron 6. Differences of selectivity between 12459, BRACO19 and telomestatin for these hTERT quadruplexes may be important to explain their respective activity and inactivity against hTERT splicing.
INTRODUCTION
Telomeres are essential to maintain the stability of chromosome ends and are synthesized by a specialized enzyme called telomerase. Telomerase is over-expressed in a large number of tumors and is involved in cell immortalization and tumorigenesis, whereas it is not expressed in most somatic cells (1). Such differential expression provided the initial rationale for the evaluation of telomerase inhibitors as potential anticancer agents (2). Folding of the telomeric G-rich single strand into a quadruplex DNA has been found to inhibit telomerase activity. Stabilization of G-quadruplexes can then be considered an original strategy to achieve antitumor activity (3–6).
The 2,4,6-triamino-1,3,5-triazine derivatives are potent telomerase inhibitors that bind to telomeric G-quadruplex (7). In this series, 12459 (Fig. 1a) is one of the most selective G-quadruplex-interacting compounds that displayed a 25-fold selectivity when telomerase inhibition was compared with the Taq polymerase inhibition by using the TRAP-G4 assay (8). 12459 also presented strong affinities for different G-quadruplex structures when compared with other forms of nucleic acids (L.Guittat and J.L.Mergny, unpublished results). In addition, 12459 is able to induce both telomere shortening and apoptosis in the human lung adenocarcinoma A549 cell line, as a function of its concentration and time exposure (7). 12459 was also shown to downregulate telomerase activity in A549 cells after a short-term exposure to the ligand (7). We present evidence that short-term treatment of A549 cells with 12459 induces a downregulation of telomerase activity caused by an alteration of the hTERT splicing pattern, in which the inactive –β transcript becomes over-expressed. Since the hTERT spliced intron 6 contains several repeated GGG motifs able to form G-quadruplexes and analogous to reported splicing enhancer elements, we have determined that 12459 could stabilize these quadruplexes by using a specific PCR-stop assay and compared its activity with that of two other G-quadruplex ligands, telomestatin and BRACO19. The biological effect of 12459 against hTERT splicing was found to be strongly decreased in A549 clones selected for resistance to 12459. Our data provide evidence of a potential correlation between the ability of these ligands to impair hTERT splicing and their potency to stabilize quadruplexes located in the hTERT intron 6. We propose as a hypothesis that 12459 might modulate hTERT splicing through an interaction with hTERT pre-mRNA.
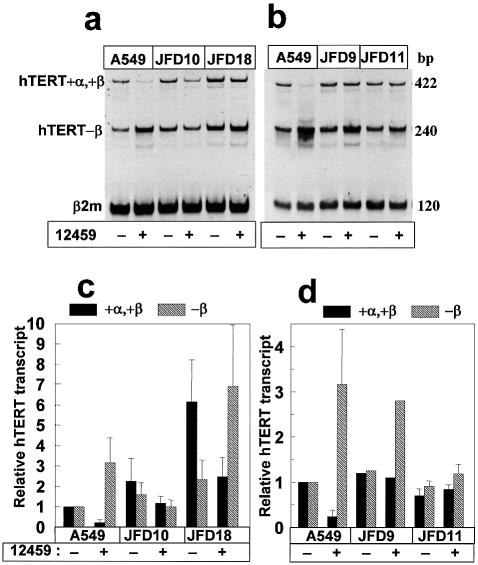
Figure 1.
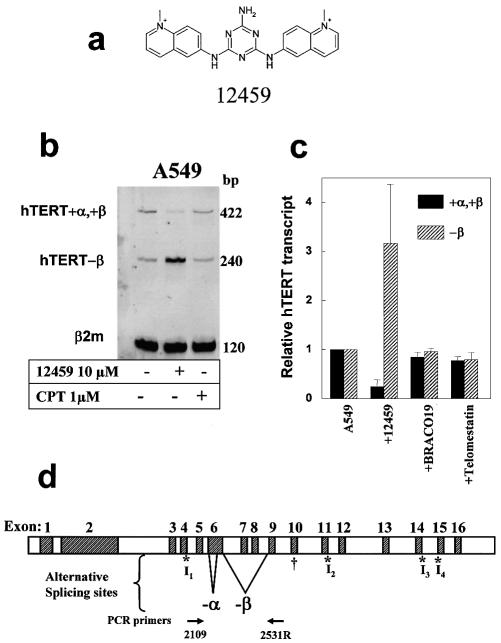
hTERT transcriptional alterations induced by a G-quadruplex ligand in A549 cells. (a) Chemical structure of 12459. (b) Ligand 12459 induced an hTERT splicing pattern alteration. RT–PCR of hTERT in parental A549 cells. As indicated, cells were treated with 10 µM 12459 for 48 h or 1 µM camptothecin (CPT) for 24 h before RNA extraction and RT–PCR analysis according to Yi et al. (14) that allowed the detection of active (+α,+β) and inactive (–β) hTERT transcripts (representative of three independent experiments). β2-Microglobulin transcript (β2m) is used as a control for mRNA expression. The size of the PCR products (bp) is indicated on the right. (c) A549 cells were treated with either 10 µM 12459, telomestatin or BRACO19 for 48 h. Quantification of RT–PCR experiments (from three independent RNA extractions) for the active +α,+β and the inactive –β hTERT transcripts. Data were normalized relative to the β2-microglobulin transcript and to the transcript level obtained in untreated A549 cells. (d) Genomic organization of the hTERT gene. Exons 1–16 and alternative splicing insertions (I1–I4, indicated by asterisks) and deletions (–α and –β, indicated by solid lines) are marked (14). A cross marks the location of the termination codon for the variant containing the β deletion. The location of the PCR primers used to determine the splicing patterns is shown at the bottom.
MATERIALS AND METHODS
Oligonucleotides and compounds
All oligonucleotides were synthesized and purified by Eurogentec, Seraing, Belgium. Triazine derivative 12459 was synthesized according to patent WO 0140218. Telomestatin was purified according to Shin-ya et al. (9). BRACO19 was synthesized according to Gowan et al. (10). Other compounds were commercially available (Sigma). Solutions of compounds were prepared in 10 mM dimethylsulfoxide (DMSO), except telomestatin, which was prepared at 5 mM in MeOH/DMSO (50:50). Further dilutions were made in water.
UV melting experiments
All measurements were performed as described previously (11,12) on G4TERT1, G4TERT2 VNTR6-1 and mutant oligonucleotides (sequences shown in Table 1). Melting profiles were recorded at 295 nm in a 10 mM pH 7.0 sodium cacodylate buffer containing 0.1 M NaCl (or LiCl or KCl) using quartz cuvettes with a 1 cm pathlength.
Table 1. List of the oligomers used in the PCR-stop assay.
| Oligomer name | Sequence |
|---|---|
| G4TERT1 | 5′-GGGGTGAAAGGGGCCCTGGGCTTGGG-3′ |
| G4TERT1mu | 5′-AGGATGAAAGGAGCCCTGAGCTTGGG-3′ |
| TERT1reva | 5′-TCTCGTCTTCCAATCCCA-3′ |
| G4TERT2 | 5′GGGGGCCTTGGGGCTCGGCAGGGGTGAAAGGGG-3′ |
| G4TERT2mu | 5′-GAGAGCCTTGAAGCTCGGCAGGAGTGAAAGGGG-3′ |
| TERT2reva | 5′-ATCGCTTCTCGTCCCCTTTC-3′ |
| VNTR6-1 | 5′-GGGGTAGGTGGGGATCTGTGGGATTGG-3′ |
| VNTR6-1mu | 5′-AGGATAGGTGAAGATCTGTGGGATTGG-3′ |
| VNTR6-1reva | 5′-TCTCGTCTTCCAATCCCA-3′ |
aComplementary oligomers were used with either the corresponding G4 or mutated oligomers.
Underlined text corresponded to sequence complementary to the test oligomer and bold text corresponded to G-repeats. TERT1rev and VNTR6-1rev correspond to the same oligomer sequence.
Cell culture conditions and selection of resistant clones
A549 human cell lung carcinoma cell line was from the American Type Culture Collection. These cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) with Glutamax (Invitrogen), supplemented with 10% fetal calf serum and antibiotics. JFD9, JFD10, JFD11 and JFD18 clones derived from the parental A549 cell line were selected for resistance to 10 µM 12459 after ethyl methanesulfonate (EMS) mutagenesis treatment (13). Clones were cultured in the same medium as the A549 cell line in the absence of 12459.
The MTT survival assay (4 days) in the presence of various cytotoxic compounds was performed in 96-well plates, each point in quadruplicate, as recommended by the manufacturer (Sigma). Due to an interference of 12459 with the coloration induced by MTT, the survival assay for this compound was performed in 6-well plates, each point in triplicate. The number of viable cells was counted after trypan blue dye exclusion in a hematocytometer.
A 48 h survival assay in the presence of 12459, telomestatin and BRACO19 was performed in 25 cm2 culture flasks, in duplicate. Cells were seeded at 75 000 cells and treated 4 h later with various concentrations of compounds. After 48 h, cells were trypsinized and the number of viable cells was counted after trypan blue dye exclusion in a hematocytometer.
For apoptosis determination, A549 cells were seeded on 4-well cover slides and treated with 12459. Cells were washed with phosphate-buffered saline (PBS) and stained with Hoechst 33342 at 1 µg/ml. Cells with apoptotic nuclei were counted in different part of the slide by fluorescence microscopy. Results corresponded to the mean of three separate determinations (±SD) relative to control untreated cells.
RNA preparation and RT–PCR assays
Total RNA was isolated from 1 × 106 cells using Tri-Reagent (Sigma) as recommended by the manufacturer. A 1 µg aliquot of total RNA was reverse transcribed in a 20 µl reaction volume using random hexamers, AMV reverse transcriptase and the reaction buffer provided in the Reverse Transcription kit (Promega). The volume of the sample was adjusted to 200 µl with diethylpyrocarbonate (DEPC)-treated water at the end of the reaction. A 10 µl aliquot of cDNA was used for PCR amplifications. hTERT was amplified using the forward TERT2109 primer (5′-GCCTGAGCTGTACTTTGTCAA-3′) and the reverse TERT2531R primer (5′-AGGCTGCAGAGC AGCGTGGAGAGG-3′) according to Yi et al. (14), with the following cycling conditions: 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 58°C for 30 s and 72°C for 30 s. Amplification of β2-microglobulin was performed as a control using the same PCR conditions with forward primer (5′-ACCCCCACTGAAAAAGATGA-3′) and reverse primer (5′-ATCTTCAAACCTCCATGATG-3′). Amplified products were resolved on 6% non-denaturing polyacrylamide gels in 1× TBE and stained with SYBR Green I (Roche). Quantification was performed by a CDD camera (Bioprint) and a BioCapt software analysis, relative to the signal of β2-microglobulin. Results represent the mean value of three independent RNA extractions.
PCR-stop assay on G-quadruplex oligonucleotides
The stabilization of G-quadruplex structures by specific ligands was investigated by a PCR-stop assay using a test oligonucleotide and a complementary oligonucleotide that partially hybridizes to the last G-repeat of the test oligonucleotide. Sequences of the test oligonucleotides and their corresponding complementary sequences used here are presented in Table 1.
Assays were performed in a final volume of 25 µl, in a 10 mM Tris pH 8.3 buffer with 25 mM LiCl, 25 mM KCl, 1.5 mM Mg(OAc)2, 7.5 pmol of each oligonucleotide, 2.5 U of Taq polymerase and the indicated amount of the ligand. Reaction mixtures were incubated in a thermocycler with the following cycling conditions: 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 58°C for 30 s and 72°C for 30 s. Amplified products were resolved on a 12% non-denaturing polyacrylamide gel in 1× TBE and stained with SYBR Green I (Roche). Fluorescence was scanned with a phosphorimager (Typhoon 9210, Amersham). Results represent the mean ± SD of four independent experiments, except as indicated.
RESULTS
12459 altered hTERT alternative RNA splicing in A549 cells
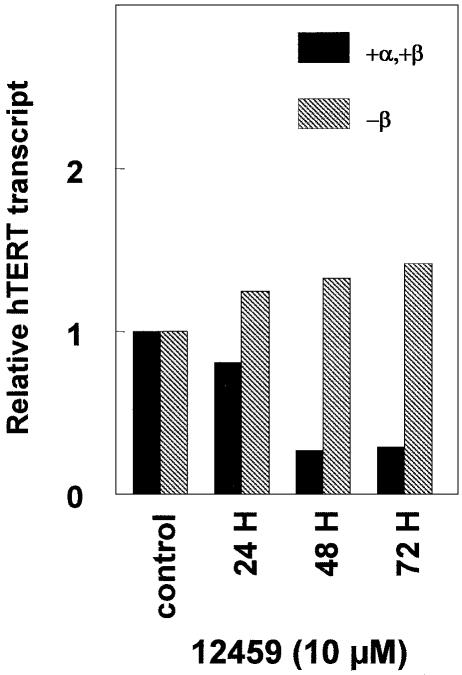
Short-term treatment with 12459 (10 µM, 48 h) was initially found to downregulate telomerase activity in A549 cells extracts, but the precise mechanism of such an effect was not elucidated (7). We examined whether 12459 could alter hTERT transcription in parental A549 cells. The expression of the hTERT transcript was investigated by RT–PCR analysis. hTERT presents a complex splicing pattern that includes an active +α,+β transcript and several inactive species including the major –β, transcript and the minor –α and –α,–β transcripts (Fig. 1d) (14,15). Surprisingly, we have found that 12459 had no effect on the global transcription level of hTERT but markedly altered its splicing pattern, leading to an almost complete disappearance of the active transcript (+α,+β) and to an increase of the inactive one (Fig. 1b). The effect of 12459 on hTERT splicing was detectable after 24 h and became maximal at 48 h (Fig. 2). Further prolongation of the treatment to 72 h did not increase the effect on hTERT splicing (Fig. 2). Such downregulation of the active hTERT transcript would probably account for the short-term downregulation of the telomerase enzymatic activity reported earlier (7). Telomestatin and BRACO19 (10 µM, 48 h), two other potent telomerase inhibitors that were shown to stabilize G-quadruplex (10,16), did not significantly downregulate hTERT transcription nor alter its splicing pattern (Fig. 1c), suggesting that the splicing alteration is specific for the triazine derivative. A possible explanation for the (+α,+β) hTERT transcript downregulation might be the selective death of cells expressing the active transcript or a consequence of the cell growth inhibition induced by 12459. This seems unlikely since the onset of apoptosis induced by 12459 appeared with a delay at 96 h (13) and the kinetics of the splicing alteration reached a plateau at 48 h (Fig. 2). In addition, camptothecin at a concentration and time exposure (1 µM, 24 h) that generated >30% apoptosis did not alter transcription and splicing of hTERT (Fig. 1b). Cell growth experiments (48 h) with 10 µM 12459, telomestatin or BRACO19 produced 52, 62 or 68% growth inhibition, respectively, against A549 cells. Since growth inhibition is similar between 12459 and two other G-quadruplex ligands that did not induce splicing, we concluded that splicing alteration is independent of the antiproliferative effect of these ligands.
Figure 2.
Kinetics of 12459-induced hTERT splicing pattern alteration in A549 cells. Quantification of RT–PCR experiments for the active α,+β and the inactive –β hTERT transcripts. As indicated, cells were treated with 10 µM 12459 for 24, 48 and 72 h before RNA extraction and RT–PCR analysis. Data were normalized relative to the β2-microglobulin transcript and to the transcript level obtained in untreated A549 cells (control).
hTERT intron 6 contains G-rich elements able to form G-quadruplexes in vitro
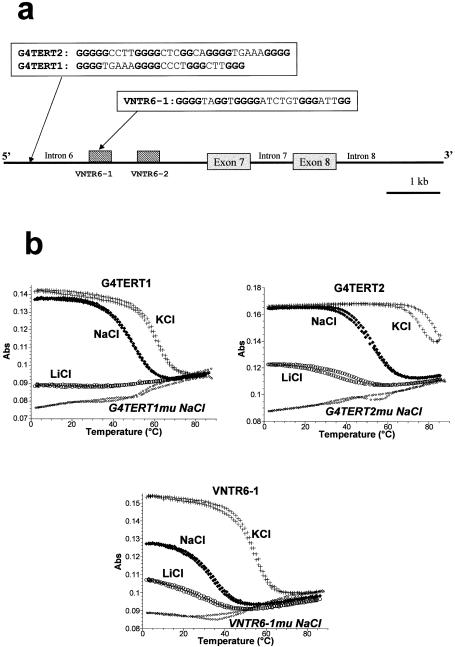
Interestingly, the splicing alteration induced by 12459 might be related to the presence of G-rich elements identified in two regions within intron 6 of the hTERT gene and thought to affect the mRNA splicing of the –β form of hTERT (17). The first element is a stretch of six G-repeats located at the 5′ end of intron 6, and the second is the repeat unit of a 1–2 kb VNTR (variable number of tandem repeat). DNA oligonucleotides synthesized from these sequences (G4TERT1, G4TERT2 and VNTR6-1, see Fig. 3a) displayed the characteristic features of G-quadruplexes as shown by UV absorbance. An inverted transition is observed at 295 nm upon heating; it corresponds to the melting of the quadruplex (12). As expected for these structures, the Tm of the quadruplexes depends on the nature of the monocation added to the buffer. For all oligonucleotides, the highest Tm is obtained with KCl followed by NaCl. G4TERT1, G4TERT2 and VNTR6-1 quadruplexes exhibit a Tm of 61, >75 and 55°C, respectively in KCl, as compared with 49, 52 and 36°C, respectively, in NaCl. Little or no signal is observed in 0.1 M LiCl for the three different oligonucleotides, arguing against the formation of stable quadruplexes under these conditions. Parallel experiments with control oligomers that contains mutations in the guanine repeats were also performed; the results are shown in Figure 3b for oligonucleotides G4TERT1mu, G4TERT2mu and VNTR6-1mu (sequences presented in Table 1) in 0.1 M NaCl. These controls are theoretically unable to form unimolecular G-quadruplexes. In agreement with this assumption, no transition at 295 nm is observed in NaCl (Fig. 3b, diamonds) or KCl (not shown).
Figure 3.
G-quadruplex stabilization of sequences from the hTERT intron 6. (a) Schematic representation of the –β deletion of hTERT that encompassed intron 6, exon 7, intron 7, exon 8 and intron 8 according to Leem et al. (17). G4TERT1 and G4TERT2 are located in the 5′ part of intron 6. VNTR6-1 represented a fragment of a 38 base repeat that extended 1.0–1.8 kb in the mid-part of intron 6. (b) UV melting behavior of G4TERT1, G4TERT2 and VNTR6-1 oligonucleotides and mutated sequences (3 µM) in 10 mM sodium cacodylate buffer (pH 7.0) and 100 mM KCl, NaCl or LiCl, as indicated. Denaturation of the quadruplexes is shown by a decrease in absorbance at 295 nm (12). G4TERT1 in KCl, –0.01 offset; G4TERTa in LiCl, –0.004 offset; G4TERT2 in KCl, –0.01 offset; G4TERT2mu in NaCl, –0.03; G4TERT1mu in NaCl, –0.017.
12459 is more efficient than telomestatin in stabilizing the formation of hTERT intron 6 G-quadruplexes
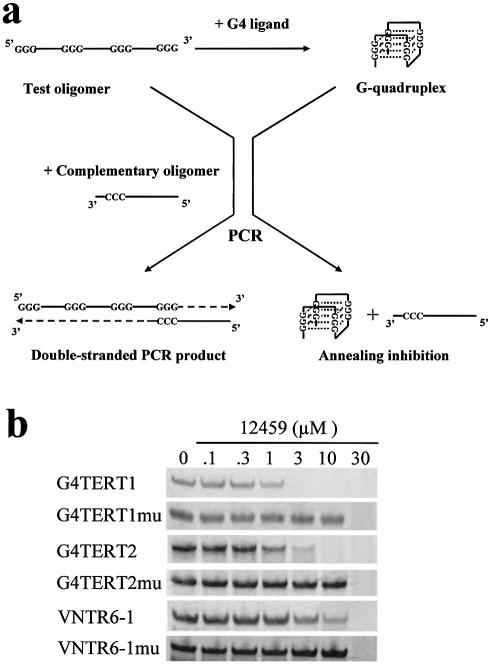
The role of 12459 in the formation of G-quadruplex structures from the hTERT intron 6 was investigated using a PCR-stop assay. In the presence of the ligand, DNA oligomers from hTERT intron 6 sequences were stabilized into G-quadruplex structures that blocked hybridization with a complementary strand overlapping the last G-repeat (Fig. 4a and Table 1). In that case, 5′ to 3′ extension with Taq polymerase was inhibited and the final double-stranded DNA PCR product was not detected. Oligomers G4TERT1, G4TERT2 or VNTR6-1 were incubated with increasing concentrations of 12459 in the presence of their complementary strand for 30 PCR cycles. The final double-stranded DNA product was inhibited in a dose-dependent manner by 12459 (Fig. 4b). The IC50 values which indicate the concentration of 12459 required to achieve 50% inhibition of the reaction were found to be 1.75, 2 and 6.25 µM for G4TERT1, G4TERT2 and VNTR6-1, respectively (Table 2). To further demonstrate that inhibition induced by 12459 was due to G-quadruplex stabilization of the oligomer, parallel experiments with oligomers that contain mutations in the guanine repeats (G4TERT1mu, G4TERT2mu and VNTR6-1mu, see Table 1) were performed. In that case, IC50 was increased to 26, 20 and 20 µM, for G4TERT1mu, G4TERT2mu and VNTR6-1mu, respectively (Table 2), and represented either non-specific Taq polymerase extension inhibition or oligonucleotide hybridization inhibition. IC50 differences for 12459 were observed between the quadruplex-forming oligomers (from 1.75 to 6.25 µM) while non-specific inhibition against all the mutated oligomers remained constant and above 10 µM. PCR inhibition by 12459 was found to be more efficient with G4TERT1 or G4TERT2 oligomers than with VNTR6-1 oligomer.
Figure 4.
Effect of 12459 on the formation of hTERT intron 6 quadruplex by a PCR-stop assay. (a) Principle of the assay. Test oligomer was amplified with a complementary oligomer overlapping the last G-repeat. Taq polymerase extension resulted in the formation of a final double-stranded PCR product. In the presence of a ligand that stabilizes oligomer into a G-quadruplex structure, annealing of oligomer and, therefore, Taq polymerase extension is inhibited. Note that the diagram represents a G-quadruplex formed with three G-quartets, as an example. (b) Effect of increasing concentrations of 12459 on the PCR-stop assay with G-quadruplex-forming oligomers (G4TERT1, G4TERT2 and VNTR6-1) or with control mutated oligomers (G4TERT1mu, G4TERT2mu and VNTR6-1mu). The sequence of oligomers is indicated in Table 1.
Table 2. Effects of 12459, BRACO19, TMPyP4 and telomestatin on the formation of hTERT intron 6 G-quadruplexes measured by the PCR-stop assay.
| IC50 (µM) on test oligomers | ||||||
|---|---|---|---|---|---|---|
| Compound | G4TERT1a | G4TERT1mub | G4TERT2b | G4TERT2mub | VNTR6-1a | VNTR6-1mub |
| 12459 | 1.75 ± 0.37 | 26 | 2 | 20 | 6.25 ± 1.8 | 20 |
| Telomestatin | 5.0 ± 1.7 | 10 | 6 | 10 | 6.25 ± 0.37 | 9 |
| BRACO19 | 5.7 ± 1.1 | 7.7 | >30 | >30 | >30 | >30 |
| TMPyP4 | 1.38 ± 0.28 | 1.39 | 1.8 | 1.8 | 2.37 ± 0.82 | 2.3 |
aMean ± SD of quadruplicates.
bMean of duplicates.
A comparison of 12459 with telomestatin and BRACO19 was made under the same experimental conditions. For telomestatin, inhibition of the double-stranded DNA product was detected with IC50 equal to 5, 6 and 6.25 µM for G4TERT1, G4TERT2 and VNTR6-1, respectively (Table 2). Thus, except for VNTR6-1, telomestatin was found to be less efficient than 12459 in converting these oligomers into a G-quadruplex structure. The non-specific inhibition of the PCR by telomestatin gave an IC50 equal to 10, 10 and 9 µM against G4TERT1mu, G4TERT2mu and VNTR6-1mu, respectively (Table 2). Thus, telomestatin also presented a higher non-specific effect than 12459 in this assay. For BRACO19, inhibition of double-stranded DNA product was only detected for G4TERT1 with an IC50 equal to 5.7 µM, and no inhibition was found up to 30 µM against G4TERT2 and VNTR6-1 (Table 2). Therefore, BRACO19 was also less efficient than 12459 in converting the hTERT sequences into G-quadruplex. Again, the non-specific inhibition for BRACO19 against G4TERT1mu was higher than 12459. TMPyP4 was also evaluated in this assay and presented potent inhibitory properties, with the IC50 ranging from 1.3 to 2.3 µM against these G-quadruplex-forming oligomers. However, control experiments with mutated oligomers indicated identical IC50 values, suggesting that TMPyP4 had no selectivity in this assay, as compared with other G-quadruplex ligands evaluated here (Table 2).
These results provide evidence that 12459 stabilizes the G-quadruplex DNA structures that may be formed with a sequence found in the hTERT intron 6 with a marked preference for the sequence located at the 5′ end of intron 6. In addition, differences in the efficiency for stabilizing this quadruplex were observed between 12459, telomestatin and BRACO19. It is interesting to note that the stabilizing effect of all these compounds was also evaluated on the human telomeric A(GGGTTA)3GGG quadruplex, using a FRET-based melting assay that we have previously described (11). 12459, telomestatin and TmPyP4 (1 µM ligand) stabilized the F21T quadruplex by 8, 17 and 22°C, respectively. This indicates that the stabilization depends on the quadruplex sequence: 12459 which is the least stabilizing compound on the human telomeric motif is the most active compound in the PCR-stop assay.
12459-resistant clones presented resistance to the 12459-induced hTERT downregulation
We have established A549 clones selected for resistance to the short-term effect of 12459 (13). Clones JFD10 and JFD18 displayed hTERT transcript over-expression and an increased telomerase activity leading to an increase in telomere length. Two other 12459-resistant clones, JFD9 and JFD11, did not over-express the hTERT transcript. Telomerase activity was also found to contribute to the resistance to 12459 since transfection of a DN-hTERT cDNA into JFD18 cells restored sensitivity to 12459 (13). To further demonstrate that there is a link between the ability of 12459 to modulate hTERT RNA alternative splicing and the biological effect of 12459 against telomerase activity, we have analyzed 12459-resistant clones for the hTERT splicing pattern alteration induced by 12459. Interestingly, 12459 treatment of the resistant clones JFD10 and JFD18 induced a different hTERT splicing pattern from that of the parental A549: the active (+α,+β) transcript was reproducibly maintained at a level at least comparable with that of untreated parental A549 cells, equal to 1.2-fold for JFD10 and 2.5-fold for JFD18 (Fig. 5a and c). In agreement with this observation, we have previously shown that JFD10 and JFD18 cells were resistant to the downregulation of the telomerase activity induced by 12459 treatment, in contrast to A549-treated cells (13). Analysis of other resistant clones, including JFD9, and JFD11 that did not overexpress hTERT mRNA (13), indicated that the level of the active +α,+β transcript is weakly affected by the 12459 treatment and is maintained at a level comparable with that of untreated parental A549 (Fig. 5b and d). These data suggested a common phenotype aiming at the maintenance of an active hTERT transcript level under treatment with 12459. Resistance to 12459 also produced two phenotypic classes of clones. In the first, active hTERT transcript is upregulated (i.e. JFD10, JFD18, etc.) and presented a qualitative alteration of the hTERT splicing pattern induced by 12459. In the second, active hTERT transcript is normal, as compared with A549 (i.e. JFD9, JFD11, etc.) and presented a qualitative alteration of the effect of 12459 on hTERT splicing. Interestingly, telomestatin, BRACO19 and camptothecin, that did not downregulate hTERT or alter splicing, were not found to be cross-resistant in the JFD18 clone (13). These results support the notion that the 12459-induced splicing alteration of hTERT is a key element in the short-term downregulation of telomerase activity in A549 cells.
Figure 5.
hTERT splicing pattern alteration in 12459-resistant clones. (a) RT–PCR of hTERT in parental A549 and resistant clones (JFD10 and JFD18). As indicated, cell lines were treated with 10 µM 12459 for 48 h before RNA extraction and RT–PCR analysis (representative of four independent experiments). (b) RT–PCR of hTERT in parental A549 and resistant clones (JFD9, JFD11). As indicated, cell lines were treated with 10 µM 12459 for 48 h before RNA extraction and RT–PCR analysis according to Yi et al. (14). The size of the PCR products (bp) is indicated on the right. (c) Quantification of four independent RT–PCR experiments (from independent RNA extractions) for the active +α,+β and the inactive –β hTERT transcripts on A549, JFD10 and JFD18. Cells were treated with 12459 (10 µM, 48 h) as indicated. Data were normalized relative to the β2-microglobulin transcript (β2m). (d) Quantification of the RT–PCR experiments (three independent RNA extractions, except for JFD9 with one single experiment) for the active +α,+β and the inactive –β hTERT transcripts on A549, JFD9 and JFD11. Cells were treated with 12459 (10 µM, 48 h) as indicated. Data were normalized relative to the β2-microglobulin transcript).
DISCUSSION
G-quadruplex structures have been extensively studied in the telomeric single-stranded overhang, and two different forms of telomeric quadruplexes have been structurally characterized (3). G-quadruplexes could also be formed from duplex telomeric DNA under appropriate ionic and pH conditions (18) or in the presence of specific ligands (19). G-quadruplexes were also found in the promoter or regulatory regions of important oncogenes such as c-myc, c-myb, c-Fos and c-ABL (20), and a recent study demonstrated that the c-myc quadruplex formed in the promoter region functions as a transcriptional repressor element (21). c-myc transcription can be inhibited by ligand-mediated G-quadruplex stabilization. Two other regions of the genome are G-rich and have considerable potential to form G-quadruplex DNA: rDNA and mammalian immunoglobulin heavy chain switch regions (20).
We report here that the hTERT intron 6 contains two G-rich sequences able to form G-quadruplexes. 12459 ligand is also capable of promoting the stabilization of these G-quadruplexes, as evidenced by a specific PCR-stop assay. Interestingly, 12459 is unable to downregulate total hTERT transcription, unlike TMPyP4 ligand that blocks c-myc gene transcription, but rather modifies the hTERT alternative RNA splicing in A549 cells, thus suggesting that 12459 affects hTERT mRNA processing rather than hTERT DNA transcription. The effect of 12459 leads to an almost complete downregulation of the active +α,+β hTERT transcript and to an increase of the inactive –β hTERT transcript, explaining the downregulation of telomerase activity previously observed in A549 cells (7,13).
Some genes are known to encode G-rich mRNA capable of forming G-quadruplex RNA. RNA quadruplexes were found in several mRNAs that are bound to the fragile X mental retardation protein that plays a role in the regulation of RNA translation (22). The human fibroblast growth factor-2 internal ribosome entry site (IRES) also contains an intramolecular G-quadruplex motif that contributes to the IRES activity (23). It is thus tempting to speculate that an enhanced stabilization of an intron 6 RNA quadruplex by 12459 could modulate the binding of factors participating in branch site selection. Interestingly, the GGG repeat observed in the 5′ end of hTERT intron 6 (encompassing G4TERT1 and G4TERT2 oligomers) displayed strong similarity to an intronic GGG repeat that was shown to enhance the splicing of an alternative intron of the chicken β-tropomyosin pre-mRNA (24) and in the human growth hormone (25) (Fig. 6). Mutations in these motifs were found to cause exon skipping. A possible explanation for the –βhTERT splicing would be that these G-rich sequences bind to a splicing factor involved in the negative control of the hTERT exon 7 and 8 skipping, during alternative splicing. In that case, the stabilization of this region into G-quadruplex by 12459 would impair the binding of this factor and would provoke the alternative splicing. At the molecular level, G-quadruplex formation might be analogous to a mutation in these motifs to mask or modify protein-binding sites and cause exon skipping. It is interesting to note that hnRNPA1/UP1 that played an important role in RNA metabolism and the control of telomere length is also known to bind to G-quadruplex-forming sequences and to destabilize G-quadruplexes (26,27). On the other hand, we cannot exclude that 12459 induces a direct effect on the splicing machinery, independent from any interaction with hTERT intron 6. A detailed analysis of the action of 12459 by in vitro experiments aiming to reconstitute the splicing of hTERT would be necessary to understand the effect of this ligand.
Figure 6.
Sequence alignment of the G-repeats from the 5′ end of hTERT intron 6 (A) with intronic repeats from the chicken β-tropomyosin pre-mRNA (B) and human growth hormone (C). Oligomers G4TERT1 and G4TERT2 derived from the hTERT intron 6 sequence are indicated by arrows.
Interestingly, two other potent G-quadruplex ligands, BRACO19 and telomestatin, did not alter splicing and transcript levels of hTERT, suggesting that the splicing alteration is specific for the triazine derivative. A recent study with distamycin, a well known minor groove binder of AT-rich DNA tracts, demonstrated that this compound also bind specifically to the terminal quartet of quadruplex DNA (28). Distamycin inhibits G-quadruplex binding by the RGG domain of nucleolin. In contrast, distamycin does not inhibit G-quadruplex DNA binding by BLM helicase or by RBD domains of nucleolin. A recent report from L. Hurley’s group suggested a link between the selectivity of telomestatin for intramolecular G-quadruplex or TMPyP4 for intermolecular G-quadruplex and the ability of these compounds to mediate different biological effects against telomerase-positive or -negative (ALT) tumor cell lines (19). In addition, TMPyP4 was shown to stabilize G-quadruplex from the telomere sequence but to destabilize G′2 quadruplex from the 5′-untranslated region of the first exon of the FMR1 gene (29). These data demonstrated that the mode of interaction of the ligand with the quadruplex or the nature of the quadruplex are critical for biochemical or biological properties of the ligand. Our results indicated that 12459 is ∼3-fold more potent than telomestatin in stabilizing the oligomers G4TERT1 and G4TERT2 that correspond to parts of the GGG repeat from the 5′ end of hTERT intron 6 putatively involved in alternative splicing. In contrast, the oligomer corresponding to the G-rich VNTR6-1 repeat was stabilized with equal efficiency by 12459 and telomestatin but at 3-fold higher IC50. Similarly, BRACO19 displayed important differences compared with 12459 in stabilizing these oligomers into G-quadruplex. On the other hand, 12459, telomestatin and BRACO19 were found to be equally potent in stabilizing the telomeric quadruplex in the TRAP-G4 assay (8) with an IC50 equal to 0.7, 0.65 and 0.8 µM, respectively. Therefore, significant quantitative differences in the stabilization of different G-quadruplex species were found between these three ligands. Since G4TERT1 and G4TERT2 are preferentially stabilized by 12459 as compared with VNTR6-1, and with higher efficiency when compared with telomestatin and BRACO19, this constitutes a basis for a future investigation of the effect of these ligands in the stabilization of an RNA quadruplex in hTERT intron 6 that might explain their biological differences regarding hTERT splicing.
Unlike other G-quadruplex ligands, 12459 may contribute to the impairment of telomerase activity by two different mechanisms. The first corresponded to the induction of quadruplex formation in telomeric overhang repeats that inhibited telomerase activity and therefore telomere stabilization, as reported with different classes of ligands including telomestatin and BRACO19 (3). The second corresponded to the findings of our present study relative to the hTERT active transcript downregulation mediated by the 12459-induced hTERT splicing alteration. This new mechanism was also found to be specific for the triazine derivative. Interestingly, all 12459-resistant clones were also found to be resistant for the effect of 12459 against hTERT splicing through either quantitative or qualitative alterations, suggesting that the maintenance of an active hTERT transcript is a critical determinant in the resistance to this compound. Additional experiments on the JFD18 clone demonstrated that hTERT functions contribute to the resistance to 12459 (13). Furthermore, telomestatin and BRACO19 that did not impair hTERT splicing in A549 cells were not found to be cross-resistant in JFD18 and JFD10 clones presenting quantitative alterations of the hTERT expression and 12459-induced hTERT splicing. A possible explanation for this lack of resistance for telomestatin might be related to the lower efficiency of telomestatin in stabilizing G4TERT1 and/or G4TERT2 quadruplexes and its inability to impair the splicing machinery. However, we could not exclude that telomestatin and 12459 induced specific and unrelated antiproliferative pathways other than telomerase inhibition or hTERT splicing that could explain their resistance differences. Experiments aiming to analyze differences in the splicing machinery between A549 and 12459-resistant clones would provide interesting clues to the answers to these questions.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank J. Tazi, A. Londono-Vallejo and E. Mandine for helpful discussion, and K. Shin-ya for the gift of telomestatin. This work was supported by an Action Concertée Incitative, ‘Molécules et Cibles Thérapeutiques’ grant from the French Ministry of Research and by grants from the Association pour la Recherche sur le Cancer (ARC 4691 to J.F.R., 4321 to J.L.M). T.L. is supported by a grant from l’Association Régionale pour l’Enseignement et la Recherche Scientifique et technologique (ARERS).
REFERENCES
- 1.McEachern M.J., Krauskopf,A. and Blackburn,E.H. (2000) Telomeres and their control. Annu. Rev. Genet., 34, 331–358. [DOI] [PubMed] [Google Scholar]
- 2.Sharma S., Raymond,E., Soda,H., Sun,D., Hilsenbeck,S.G., Sharma,A., Izbicka,E., Windle,B. and Von Hoff,D.D. (1997) Preclinical and clinical strategies for development of telomerase and telomere inhibitors. Ann. Oncol., 8, 1063–1074. [DOI] [PubMed] [Google Scholar]
- 3.Neidle S. and Parkinson,G.N. (2003) The structure of telomeric DNA. Curr. Opin. Struct. Biol., 13, 275–283. [DOI] [PubMed] [Google Scholar]
- 4.Mergny J.L., Riou,J.F., Mailliet,P., Teulade-Fichou,M.P. and Gilson,E. (2002) Natural and pharmacological regulation of telomerase. Nucleic Acids Res., 30, 839–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neidle S. and Parkinson,G. (2002) Telomere maintenance as a target for anticancer drug discovery. Nature Rev. Drug Discov., 1, 383–393. [DOI] [PubMed] [Google Scholar]
- 6.Rezler E.M., Bearss,D.J. and Hurley,L.H. (2002) Telomeres and telomerases as drug targets. Curr. Opin. Pharmacol., 2, 415–423. [DOI] [PubMed] [Google Scholar]
- 7.Riou J.F., Guittat,L., Mailliet,P., Laoui,A., Renou,E., Petitgenet,O., Megnin-Chanet,F., Hélène,C. and Mergny,J.L. (2002) Cell senescence and telomere shortening induced by a new series of specific G-quadruplex DNA ligands. Proc. Natl Acad. Sci. USA, 99, 2672–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomez D., Mergny,J.L. and Riou,J.F. (2002) Detection of telomerase inhibitors based on G-quadruplex ligands by a modified telomeric repeat amplification protocol assay. Cancer Res., 62, 3365–3368. [PubMed] [Google Scholar]
- 9.Shin-ya K., Wierzba,K., Matsuo,K., Ohtani,T., Yamada,Y., Furihata,K., Hayakawa,Y. and Seto,H. (2001) Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus. J. Am. Chem. Soc., 123, 1262–1263. [DOI] [PubMed] [Google Scholar]
- 10.Gowan S.M., Harrison,J.R., Patterson,L., Valenti,M., Read,M.A., Neidle,S. and Kelland,L.R. (2002) A G-quadruplex-interactive potent small-molecule inhibitor of telomerase exhibiting in vitro and in vivo antitumor activity. Mol. Pharmacol., 61, 1154–1162. [DOI] [PubMed] [Google Scholar]
- 11.Mergny J.L. and Maurizot,J.C. (2001) Fluorescence resonance energy transfer as a probe for G-quartet formation by a telomeric repeat. Chembiochem. Eur. J. Chem. Biol., 2, 124–132. [DOI] [PubMed] [Google Scholar]
- 12.Mergny J.L., Phan,A.T. and Lacroix,L. (1998) Following G-quartet formation by UV-spectroscopy. FEBS Lett., 435, 74–78. [DOI] [PubMed] [Google Scholar]
- 13.Gomez D., Aouali,N., Londono-Vallejo,A., Lacroix,L., Megnin-Chanet,F., Lemarteleur,T., Douarre,C., Shin-ya,K., Mailliet,P., Trentesaux,C. et al. (2003) Resistance to the short-term antiproliferative activity of the G-quadruplex ligand 12459 is associated with telomerase overexpression and telomere capping alteration. J. Biol. Chem., 278, 50554–50562. [DOI] [PubMed] [Google Scholar]
- 14.Yi X., Shay,J.W. and Wright,W.E. (2001) Quantitation of telomerase components and hTERT mRNA splicing patterns in immortal human cells. Nucleic Acids Res., 29, 4818–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wick M., Zubov,D. and Hagen,G. (1999) Genomic organization and promoter characterization of the gene encoding the human telomerase reverse transcriptase (hTERT). Gene, 232, 97–106. [DOI] [PubMed] [Google Scholar]
- 16.Kim M.Y., Vankayalapati,H., Shin-Ya,K., Wierzba,K. and Hurley,L.H. (2002) Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular g-quadruplex. J. Am. Chem. Soc., 124, 2098–2099. [DOI] [PubMed] [Google Scholar]
- 17.Leem S.H., Londono-Vallejo,J.A., Kim,J.H., Bui,H., Tubacher,E., Solomon,G., Park,J.E., Horikawa,I., Kouprina,N., Barrett,J.C. et al. (2002) The human telomerase gene: complete genomic sequence and analysis of tandem repeat polymorphisms in intronic regions. Oncogene, 21, 769–777. [DOI] [PubMed] [Google Scholar]
- 18.Phan A.T. and Mergny,J.L. (2002) Human telomeric DNA: G-quadruplex, i-motif and Watson–Crick double helix. Nucleic Acids Res., 30, 4618–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim M.Y., Gleason-Guzman,M., Izbicka,E., Nishioka,D. and Hurley,L.H. (2003) the different biological effects of telomestatin and TMPyP4 can be attributed to their selectivity for interaction with intramolecular or intermolecular g-quadrplex structures. Cancer Res., 63, 3247–3256. [PubMed] [Google Scholar]
- 20.Rezler E.M., Bearss,D.J. and Hurley,L.H. (2003) Telomere inhibition and telomere disruption as processes for drug targeting. Annu. Rev. Pharmacol. Toxicol., 43, 359–379. [DOI] [PubMed] [Google Scholar]
- 21.Siddiqui-Jain A., Grand,C.L., Bearss,D.J. and Hurley,L.H. (2002) Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA, 99, 11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darnell J.C., Jensen,K.B., Jin,P., Brown,V., Warren,S.T. and Darnell,R.B. (2001) Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell, 107, 489–499. [DOI] [PubMed] [Google Scholar]
- 23.Bonnal S., Schaeffer,C., Créancier,L., Clamens,S., Moine,H., Prats,A.C. and Vagner,S. (2003) A single IRES containing a G-quartet RNA structure drives FGF-2 gene expression at four alternative translation initiation codons. J. Biol. Chem., 278, 39330–39336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sirand-Pugnet P., Durosay,P., Brody,E. and Marie,J. (1995) An intronic (A/U)GGG repeat enhances the splicing of an alternative intron of the chicken β-tropomyosin pre-mRNA. Nucleic Acids Res., 23, 3501–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cogan J.D., Prince,M.A., Lekhakula,S., Bundey,S., Futrakul,A., McCarthy,E.M. and Phillips,J.A.,3rd (1997) A novel mechanism of aberrant pre-mRNA splicing in humans. Hum. Mol. Genet., 6, 909–912. [DOI] [PubMed] [Google Scholar]
- 26.Fukuda H., Katahira,M., Tsuchiya,N., Enokizono,Y., Sugimura,T., Nagao,M. and Nakagama,H. (2002) Unfolding of quadruplex structure in the G-rich strand of the minisatellite repeat by the binding protein UP1. Proc. Natl Acad. Sci. USA, 99, 12685–12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dallaire F., Dupuis,S., Fiset,S. and Chabot,B. (2000) Heterogeneous nuclear ribonucleoprotein A1 and UP1 protect mammalian telomeric repeats and modulate telomere replication in vitro. J. Biol. Chem., 275, 14509–14516. [DOI] [PubMed] [Google Scholar]
- 28.Cocco M.J., Hanakahi,L.A., Huber,M.D. and Maizels,N. (2003) Specific interactions of distamycin with G-quadruplex DNA. Nucleic Acids Res., 31, 2944–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weisman-Shomer P., Cohen,E., Hershco,I., Khateb,S., Wolfovitz-Barchad,O., Hurley,L. and Fry,M. (2003) The cationic porphyrin TMPyP4 destabilizes the tetraplex form of the fragile X syndrome expanded sequence d(CGG)n. Nucleic Acids Res., 31, 3963–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]