Abstract
Auxin is a key phytohormone regulating central processes in plants. Although the mechanism by which auxin triggers changes in gene expression is well understood, little is known about the specific role of the individual members of the TIR1/AFB auxin receptors, Aux/IAA repressors, and ARF transcription factors and/or molecular pathways acting downstream leading to plant responses to the environment. We previously reported a role for AFB3 in coordinating primary and lateral root growth to nitrate availability. In this work, we used an integrated genomics, bioinformatics, and molecular genetics approach to dissect regulatory networks acting downstream of AFB3 that are activated by nitrate in roots. We found that the NAC4 transcription factor is a key regulatory element controlling a nitrate-responsive network, and that nac4 mutants have altered lateral root growth but normal primary root growth in response to nitrate. This finding suggests that AFB3 is able to activate two independent pathways to control root system architecture. Our systems approach has unraveled key components of the AFB3 regulatory network leading to changes in lateral root growth in response to nitrate.
Keywords: systems biology, nitrogen, Sungear
Auxin is a key controller of root growth processes. The auxin response is initiated by binding of the hormone to TIR1/AFB receptors. Auxin receptors are part of the SCFTIR1/AFB E3 ubiquitin ligase complex (1). Auxin binding to TIR1/AFB receptors triggers the recognition and degradation by polyubiquitination of the Aux/IAA repressors, releasing the inhibition of the ARF transcription factors that mediate auxin-responsive gene transcription (2–5). The Arabidopsis genome encodes 29 Aux/IAA and 23 ARF factors. Genetics studies have shown that these factors may have both unique and redundant functions on plant growth and development, which likely depend on the specific patterns of expression of these factors on tissues or cells (6). The previous work on auxin signaling has focused mainly on development, and little is known about the specific role of the auxin receptors, ARF, and Aux/IAA proteins on plant responses to environmental cues.
Nitrogen (N) is an essential macronutrient and a limiting factor for agricultural productivity owing to its enormous impact on plant growth and development. In agricultural soils, the main N source available for plants is nitrate. Given the importance of nitrate as a plant nutrient, several studies have focused on the effect of nitrate treatment on gene expression. Nitrate is able to regulate more than 1,000 genes in roots and shoots, including genes involved in its transport, reduction, and assimilation as well as genes involved in hormone transduction pathways and diverse transcription factors, kinases, and phosphatases, among other genes (7, 8). A significant proportion of these genes are able to respond to nitrate directly, indicating that nitrate is the signal controlling their gene expression (9). However, the exact mechanisms by which nitrate is sensed and triggers changes in mRNA levels are not well understood. Auxin has been identified as an important player in the root response to nitrate. Nitrate is able to control lateral root growth by controlling the provision of auxin to lateral roots by the NRT1.1 nitrate transporter (10). In the absence of nitrate, NRT1.1 favors basipetal transport of auxin, preventing accumulation of auxin in the lateral root tip; in the presence of nitrate, this transport is inhibited, leading to accumulation of auxin and subsequent growth of laterals (10).
Lateral root growth in response to organic N has been shown to depend on miR167 and its target, the auxin response factor ARF8 mRNA (11). This module acts specifically in the pericycle to control a connected network of genes, leading to induction of lateral root initiation and repression of lateral root elongation in response to N (11). We previously described another microRNA (miRNA)/target regulatory module consisting of miR393 and the auxin receptor AFB3 in root system architecture (RSA) control by nitrate (12). AFB3 is induced by nitrate and repressed by miR393. miR393 is induced by N metabolites produced by nitrate reduction and assimilation (12). This regulatory network module is an example of a motif commonly present in regulatory networks of mammals, bacteria, and yeast known as the incoherent type I feed-forward loop (12).
In this work, we used integrated genomics, systems biology, and molecular genetics approaches to identify molecular mechanisms downstream of the miR393/AFB3 module that lead to RSA modulation by nitrate. We found that AFB3 acts specifically in the context of the nitrate response regulating a connected gene network controlled by the NAC4 transcription factor. Phenotypical analysis of nac4 mutants further implicates NAC4 in the lateral root response to nitrate in a pathway that requires AUX/IAA signaling.
Results
Transcriptomic Analysis of the Nitrate Response in afb3-1 Mutant Plants Uncovers a Specific Role of AFB3 in the Nitrate Response of Arabidopsis Roots.
AFB3 has been identified as an important regulator of primary root and lateral root modulation by nitrate (12). AFB3 controls auxin-responsive gene transcription by promoting protein degradation of the Aux/IAA transcriptional repressors in the presence of auxin (2, 13, 14). Thus, loss of AFB3 function would lead to changes in transcript abundance of its direct and indirect targets. To identify downstream targets of AFB3 in the context of the nitrate response, and to find molecular factors involved in primary and/or lateral root responses that are dependent on AFB3 regulation, we analyzed the transcriptome of the afb3-1 mutant (1) in response to nitrate treatments and compared it with the transcriptome of WT plants. Plants were grown with ammonium succinate as the only N source for 14 d and were treated at the beginning of the light period on day 15 with 5 mM KNO3 or 5 mM KCl. Under these experimental conditions, AFB3 demonstrated a rapid and transient response to nitrate, with a peak of induction at 1 h after nitrate exposure, as described previously (12).
Because we anticipated that changes in transcript abundance in response to nitrate of downstream target genes would be delayed compared with AFB3, we treated plants with nitrate for 2 h for transcriptome analysis. Total RNA was isolated from roots and prepared for Affymetrix ATH1 GeneChip hybridization. Gene expression data were normalized using robust multiarray analysis (RMA) (15), and differential gene expression was determined using two-way ANOVA as described previously (16), considering the treatment (T) and genotype (G) as factors and controlling type I error using the false discovery rate (17). Our analysis identified 445 genes responding in our experiments, including 442 genes with a significant T factor, 38 genes with a significant G factor, and 39 genes with a significant TG interaction factor.
To simplify the analysis of our results and to provide an initial insight into how AFB3 regulates gene expression in response to nitrate, we analyzed our data using the Sungear tool (18) available at the VirtualPlant webpage (http://www.virtualplant.org) (19). Sungear allows for the visualization and analysis of multiple datasets to identify genes that are unique or are shared by different gene lists (18, 20). We used Sungear to generate a triangle representing the 445 genes with significant T, G, or TG factors (Fig. S1). Each vertex in the triangle represents a factor of the ANOVA model, and the circles inside the triangle (vessels) represent the number of genes controlled by the different factors, as indicated by the arrows around the vessels.
Using Sungear, we found that treatment was the sole significant factor for 390 genes (87.6% of the total genes regulated), indicating that the nitrate response of these genes was not altered by the afb3 mutation under our experimental conditions. Analysis of the genes in this group suggested that basic N metabolic functions are not affected in the afb3 mutant, given that several genes in the nitrate transport, reduction, and assimilation pathways responded similarly to the treatments in WT and afb3 mutant plants (e.g., nitrate reductase genes NIA1 and NIA2; nitrite reductase gene NiR; NADH-dependent glutamate synthase gene GLT1; glutamine synthetase genes GSR2, GLN2, and GLN1;4; and nitrate transporters NRT1.1, NRT2.1, and NRT3.1). This result suggests that modulation of RSA by AFB3 in response to nitrate does not depend on alterations of N transport or metabolism. Interestingly, we found no genes with G as the sole significant factor, indicating that under our experimental conditions, the effects of the afb3-1 mutation were evident only in the context of the nitrate response. These results are consistent with a specific role of AFB3 in the root nitrate response of Arabidopsis thaliana.
Network Analysis Identifies a Highly Connected Nitrate-Responsive Regulatory Module Controlled by AFB3.
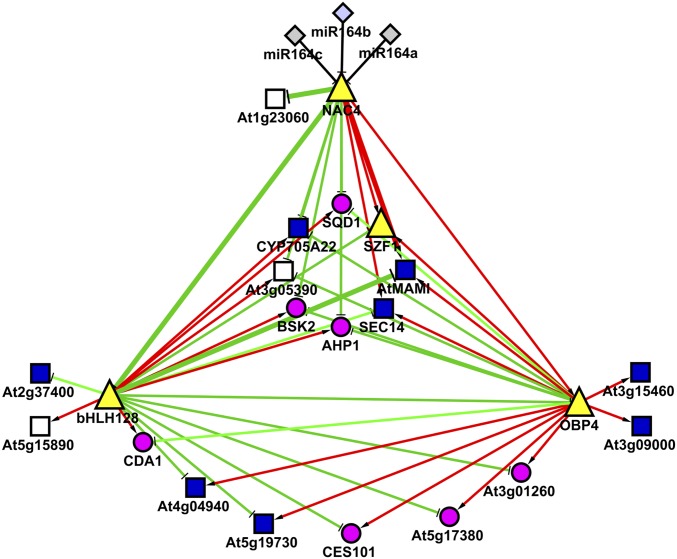
To identify targets of AFB3, we focused on the 39 genes that showed an altered response to nitrate in the afb3-1 mutant (i.e, significant TG factor). Altered expression of these genes could account for the altered root phenotype seen in the afb3-1 mutants in response to nitrate (12). To predict possible regulatory relationships between these differentially regulated genes, we generated a network view using the Gene Networks tool available at the VirtualPlant Web page. This integrative network bioinformatics approach has been used and validated previously (21, 22). Cytoscape software (23) was used to visualize the network, in which genes are represented as nodes and the edges linking these nodes represent the functional relationships between nodes. Twenty-eight genes with a significant TG factor have predicted regulatory interactions and were included in our analysis. The genes were grouped into two gene networks: a small network containing six genes, basic helix–loop–helix transcription factor bHLH64 (24), five target genes, and a miRNA (Fig. S2) and a larger network including 22 genes, NAM/ATAF/CUC transcription factor NAC4, basic helix–loop–helix transcription factor bHLH128 (24), and zinc finger transcription factors OBP4 and SZF1, along with their predicted target genes (Fig. 1). We focused our analysis in the larger network because it contained most of the TG genes. Interestingly, NAC4 is predicted to target all of the genes in the large network by direct binding to their promoters or indirectly by controlling the OBP4 or bHLH128 transcription factors (Fig. 1); thus, NAC4 might be an important component of a coordinated regulatory network controlling root nitrate response downstream of AFB3.
Fig. 1.
A connected network of regulatory factors and their potential targets is differentially regulated in afb3-1. The nodes represent genes (gray squares, miRNA; purple circles, enzyme- coding genes; blue squares, protein-coding genes; white squares, unknown protein-coding genes; yellow triangles, transcription factor-coding genes), and the edges represent miRNA/TARGET regulation or predicted regulatory interactions based on the occurrence of a transcription factor-binding site on the gene promoter. Green edges represent repression, and red edges represent induction based on correlation analysis of our Affymetrix data.
NAC4 Transcription Factor Acts Downstream of AFB3 to Control Root Nitrate Responses.
NAC4 is a member of a family of transcription factors present only in plants (25). Although NAC4 has no reported function, the closely related NAC1 and NAC2 factors are known to be involved in lateral root development in Arabidopsis (26, 27); thus, NAC4 represented an attractive candidate for mediating the effects of nitrate over RSA downstream of AFB3.
NAC4 is predicted to both positively or negatively regulate different genes in the network directly or indirectly by regulating expression of the bHLH128 and OBP4 transcription factors. bHLH128 and OBP4 target the remaining genes in the network, possibly accounting for most of the changes seen in our microarray experiment (Fig. 1). To validate our network predictions, we analyzed mRNA levels of NAC4, bHLH128, and OBP4 over time after nitrate treatment in WT plants and in the afb3-1 mutant using quantitative RT-PCR (qRT-PCR).
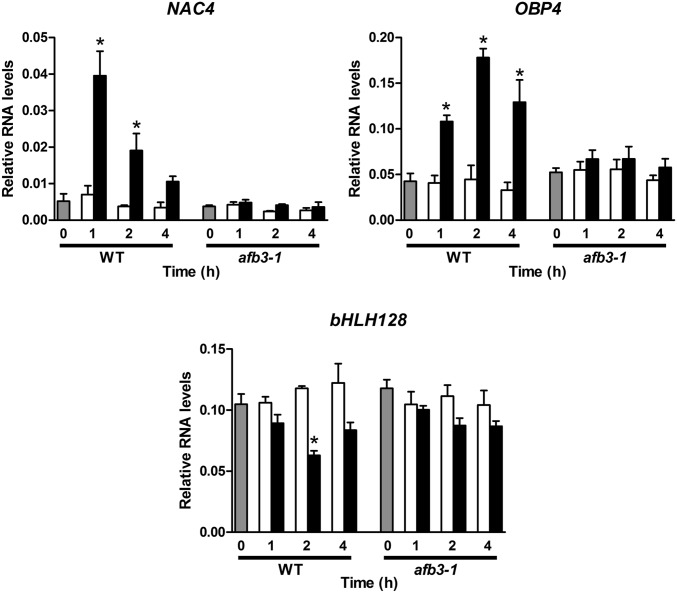
NAC4 exhibited an early peak of induction at 1 h after nitrate treatment in WT plants (Fig. 2). NAC4 response over time was similar to the transient response of AFB3 (12), as expected for an AFB3 target. No changes in RNA levels were detected in the afb3-1 mutant even after 4 h of nitrate treatment. This result clearly indicates that AFB3 function is required for nitrate regulation of NAC4.
Fig. 2.
Nitrate response of the NAC4, OBP4, and bHLH128 transcription factors is altered in the afb3-1 mutant. WT Ws and afb3-1 mutant plants were grown in ammonium succinate for 2 wk and then treated with 5 mM KNO3 or 5 mM KCl for 1, 2, and 4 h. RNA levels of the NAC4, OBP4, and bHLH128 transcription factors were measured using qRT-PCR. Values are mean ± SE of three biological replicates. Gray bars represent time 0 (before treatment), white bars represent KCl treatment, and black bars represent KNO3 treatment. Asterisks denote means that are statistically different between control and treatment (P < 0.05).
OBP4 and bHLH128 exhibited transient responses to nitrate similar to AFB3 and NAC4 but delayed, with a peak of regulation at 2 h after treatment in WT plants (Fig. 2). However, in NAC4, the nitrate response of these transcription factors was seriously compromised in the afb3-1 mutant (Fig. 2). Our qRT-PCR data are consistent with a model in which nitrate induction of NAC4 triggers changes in the levels of OBP4 and bHLH128.
We previously reported that AFB3 is regulated directly by nitrate, given that AFB3 is induced by nitrate in a nitrate reductase (NR)-null A thaliana mutant, nia1/nia2 (12). Considering that our network predicts that NAC4, OBP4, and bHLH128 are acting downstream of AFB3, we expected these transcription factors to also be regulated by nitrate directly as a signal. We analyzed the expression of NAC4, OBP4 and bHLH128 in nia1/nia2 plants after nitrate treatments using qRT-PCR. These transcription factors were still regulated by nitrate in the NR-null mutant, indicating that, as AFB3, they respond to nitrate and not to N metabolites generated by nitrate reduction or assimilation (Fig. S3A). Accordingly, nitrite or ammonium treatments had no effect on NAC4, OBP4, and bHLH128 mRNA levels (Fig. S3 B and C). NAC4, OBP4, and bHLH128 mRNA levels in the NR-null mutant differed from those described in the WT plant after 4 h of treatment (compare Fig. 2 and Fig. S3A), suggesting a complex regulation of these transcription factor levels by both nitrate and N metabolites other than nitrate. We found a similar regulation for AFB3, which is transcriptionally induced by nitrate and posttranscriptionally repressed by N metabolites produced by N reduction/assimilation.
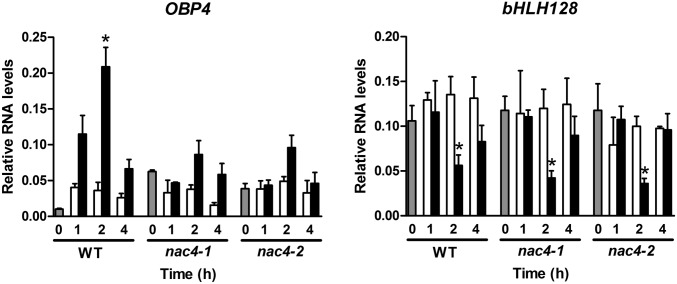
To validate OBP4 and/or bHLH128 as NAC4 targets in the context of the nitrate response, we analyzed two nac4 T-DNA insertion mutant lines obtained from the Arabidopsis Biological Resource Center, nac4-1 (SALK_040204) and nac4-2 (SALK_006735). We measured OBP4 and bHLH128 mRNA levels in WT plants and in the nac4-1 and nac4-2 mutants over time after nitrate treatment. bHLH128 response was not affected in the mutants, indicating that this gene is repressed by AFB3 in a NAC4-independent manner; however, OBP4 response was altered in nac4 mutants, indicating that this transcription factor acts downstream of NAC4 (Fig. 3).
Fig. 3.
Nitrate response of the OBP4 transcription factor is altered in nac4 mutants. WT Col-0, nac4-1, and nac4-2 mutant plants were grown in ammonium succinate for 2 wk and then treated with 5 mM KNO3 or 5 mM KCl for 1, 2, and 4 h. RNA levels of the OBP4 and the bHLH128 transcription factors were measured using qRT-PCR. Values are mean ± SE of three biological replicates. Gray bars represent time 0 (before treatment), white bars represent KCl treatment, and black bars represent KNO3 treatment. Asterisks denote means that are statistically different between the KCl and KNO3 treatments (P < 0.05).
NAC4 Regulates Lateral Root Responses to Nitrate Downstream of AFB3.
Based on our network analysis predicting NAC4 as an important regulator of the AFB3 regulatory network, we wished to determine whether this transcription factor is involved in primary and/or lateral root responses to nitrate as AFB3. To evaluate the impact of NAC4 in RSA modulation, we grew plants under the experimental conditions used to analyze the effect of AFB3 in RSA (12). Because in our previous work we used a medium containing 1× Murashige and Skoog (MS) salts without N, and because under some experimental conditions the salt concentration in this medium can inhibit root growth (28), we also performed the experiments using 0.5× and 0.2× MS salts (Fig. S4). We found no major differences between plants grown on 1× MS salt and those grown on 0.5× MS salt. However, plant growth was affected in 0.2× MS salt (Fig. S4 A and B), likely owing to the limiting nutrient concentration in this medium under our experimental conditions.
We measured primary root length and lateral root density, two parameters affected by the afb3 mutation (12), after KNO3 or KCl treatment in WT Col-0 and nac4 mutants. Given the previously reported effects of nitrate on lateral root elongation (29–32), we also measured lateral root length after KNO3 or KCl treatment. We found no significant differences between the KCl and KNO3 conditions in WT or nac4 plants (Fig. S4 E and F); thus, nitrate treatment had no effect on visible lateral root length under our experimental conditions. The inhibitory effect of nitrate on primary root elongation was not affected by the nac4 mutation (Fig. 4A), indicating that this response occurs through an AFB3-dependent signaling pathway that does not involve NAC4.
Fig. 4.
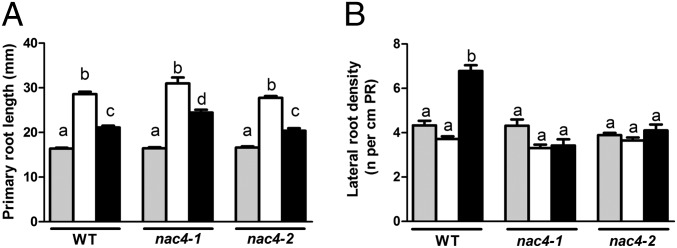
NAC4 is involved in lateral root development, but not in primary root development, in response to nitrate treatment. (A) Primary root lengths of Col-0 WT plants and nac4-1 and nac4-2 mutant plants were measured using ImageJ at day 0 (gray bars) and after 3 d of treatment with 5 mM KCl (white bars) or 5 mM KNO3 (black bars). (B) The number of lateral roots (initiating and emerging) of Col-0 WT and nac4-1 and nac4-2 mutant plants at day 0 (gray bars), treated for 3 d with 5 mM KCl (white bars) or 5 mM KNO3 (black bars), was counted using DIC optics. Values are mean ± SE of three biological replicates (n = 15). Different letters represent means that are statistically different (P < 0.05).
We have previously shown that repression of root growth by nitrate correlates with an induction of AFB3 in root tips (12). Our analysis of AFB3 regulation in the meristematic zone and in the elongation zone of the primary root revealed that after 2 h of nitrate treatment, AFB3 was induced in the meristematic zone but not in the elongation zone (Fig. S5). However, our analysis of the nitrate response of NAC4 and OBP4 in meristematic and elongation zones found that these factors did not respond to the treatments (Fig. S5), in accordance with our results showing that nac4-1 has no primary root phenotype in response to nitrate. However, the effect of nitrate over lateral root density was altered in the nac mutants (Fig. 4B).
We evaluated the density of initiating and emerging lateral roots using differential interference contrast (DIC) microscopy as described previously (12). We found that in WT plants, nitrate treatment increased the density of initiating and emerging lateral roots, but this response was altered in the nac4 mutants (Fig. S6), suggesting a specific role for NAC4 in controlling lateral root initiation and emergence in response to nitrate. This root phenotype is similar to the afb3-1 phenotype that we described previously (12), further supporting NAC4’s function downstream of the AFB3 auxin receptor.
Nitrate treatment regulates AFB3 expression in pericycle cells in accordance with AFB3’s role in mediating changes in lateral root growth in response to nitrate (12). Considering that NAC4 and OBP4 are regulated by AFB3, we expected to find these factors spatially coregulated with AFB3. We analyzed the expression of AFB3, NAC4, and OBP4 in different cell types of the root, using GFP-expressing lines and fluorescence-activated cell sorting after 2 h of nitrate treatment. AFB3 was induced in pericycle cells after 2 h of nitrate treatment (Fig. S7). This result complements our previous findings using a pAFB3:GUS line (12). We found a similar nitrate induction in pericycle cells for NAC4 and OBP4 (Fig. S7), suggesting that AFB3-NAC4-OBP4 might represent a regulatory module that acts specifically in the pericycle to control lateral root growth in response to changes in nitrate availability.
Nitrate Regulation of NAC4 Depends on AUX/IAA Signaling Function.
Nitrate-specific induction of AFB3 in roots might control a specific combination of Aux/IAA and ARF factors that control NAC4 induction and lateral root growth. In Arabidopsis, lateral root development depends on multiple Aux/IAA-ARF modules that act in sequential steps to generate new lateral roots. Lateral root founder cell specification occurs in a zone between the meristem and elongation zone called the basal meristem and depends on IAA28 and ARF proteins that might include ARF5, ARF6, ARF7, ARF8, and ARF19 (33-35). The IAA14(SLR)-ARF7-ARF19 module operates in the zone above the basal meristem and regulates the coordinated nuclear migration and posterior asymmetric cell division of the founder cells for lateral root initiation (36–38). IAA12(BDL)-ARF5(MP) also activates lateral root formation, acting after the IAA14(SLR)-ARF7-ARF19 module (39).
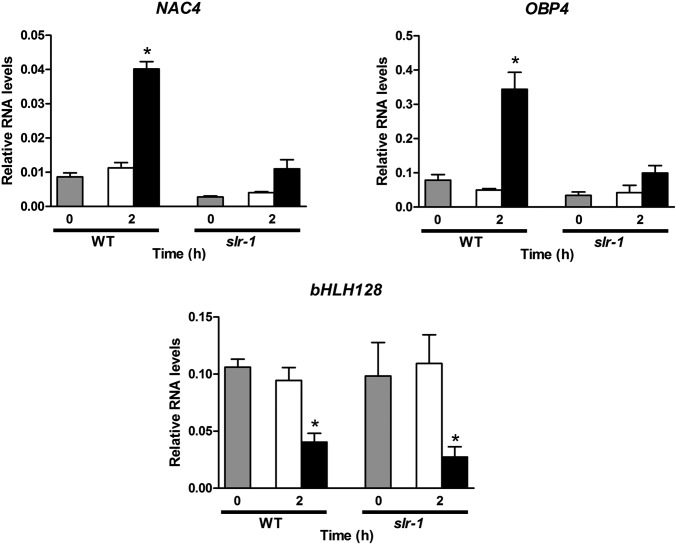
To determine whether these IAA-ARF modules participate upstream of NAC4, we analyzed the NAC4 response to nitrate in plants with altered function of these factors. We found that only the IAA14/SLR gain-of-function mutation altered NAC4 response to nitrate (Fig. 5 and Fig. S8). Accordingly, IAA14 is expressed in the xylem pole pericycle cells (38). This suggests that normal AUX/IAA-mediated auxin signaling in pericycle cells is required for induction of NAC4 by nitrate. We found that the OBP4 response was also altered in the slr-1 mutant, supporting its role as an NAC4 target acting in the AFB3-NAC4 pathway in the pericycle (Fig. 5). As expected, the nitrate response of bHLH128, which is not a NAC4 target, was not altered in this mutant (Fig. 5).
Fig. 5.
Nitrate response of NAC4 and its target OBP4 is altered in IAA14/SLR gain-of-function mutant slr-1. WT Col-0 and slr-1 mutant plants were grown in ammonium succinate for 2 wk and then treated with 5 mM KNO3 or 5 mM KCl for 2 h. RNA levels of the NAC4, OBP, and bHLH128 transcription factors were measured using qRT-PCR. Values are mean ± SE of three biological replicates. Gray bars represent time 0 (before treatment), white bars represent KCl treatment, and black bars represent KNO3 treatment. Asterisks represents means that are statistically significantly different between control and treatment (P < 0.05).
In summary, our work has identified important components of an AFB3-dependent network that controls lateral root growth in response to changes in nitrate availability (Fig. S9). These factors are specifically regulated in pericycle cells and act downstream of AUX/IAAs for lateral root development.
Discussion
Auxin controls key aspects of plant growth and development and is specifically important for the control of root growth and development (5, 40, 41). These changes in root morphology are caused mainly by changes in gene expression triggered by auxin (41). Although the mechanism by which auxin is able to increase or decrease mRNA levels of auxin-responsive genes has been characterized, the specific combinations of auxin receptors, Aux/IAAs, ARFs, and underlying regulatory networks controlling auxin dependent processes remain incompletely understood. In this work, we used an integrated approach including genomics, bioinformatics, and molecular genetics to identify the molecular networks acting downstream of the miR393/AFB3 nitrate-responsive regulatory module in RSA modulation in response to nitrate.
To determine which nitrate-responsive genes were affected by the afb3-1 mutation, leading to the changes in RSA response observed in this mutant (12), we performed a transcriptomic analysis of WT and afb3-1 nitrate-treated and control plants. Most of the genes responded to nitrate independent of AFB3, indicating that the afb3-1 mutation affects only a small proportion of root genes regulated by nitrate. Genes with a T-only model included those involved in nitrate transport, reduction, and assimilation, suggesting that the RSA response to nitrate is related to a signaling effect, not to a metabolic effect of nitrate. The absence of genes with a G-only model indicates that there are no genes whose basal expression depends on AFB3, and that under our experimental conditions gene expression alterations are detectable only in a nitrate-dependent context. This observation is in agreement with a previous phenotypic analysis of afb3-1 showing that this mutant has no visible root phenotype when grown under standard conditions (1). This is likely related to functional redundancy between the TIR1/AFB auxin receptors in plant development. Consistently, a RSA phenotype was evident under our experimental conditions only when afb3-1, and not the other auxin receptor mutants, was treated with 5 mM KNO3 (12), supporting an AFB3-specific role in root nitrate response.
Given our focus on identifying nitrate regulatory networks affected in the afb3-1 mutant that might explain the altered lateral and/or primary root response to nitrate, we centered our analysis on genes exhibiting an altered nitrate response in this mutant. Network analysis of the TG genes showed that the genes formed two networks composed of transcription factors and their putative targets. The larger network, containing most of the altered genes, is predicted to have a main regulator, NAC4. This transcription factor is part of the plant-specific NAM/ATAF/CUC family of transcription factors but has no reported function on plant development. Our network predicted that NAC4 could regulate the genes included in its network either directly or by regulating the OBP4 or bHLH128 transcription factors. We found that NAC4, OBP4, and bHLH128 are regulated directly by nitrate as a signal. This was expected based on our previous finding that AFB3 is also induced directly by nitrate (12). Continuous exposure to nitrate in the nia1/nia2 mutant caused altered expression of NAC4, OBP4, and bHLH128 compared with their response over time in WT plants. This suggests that N metabolites other than nitrate modulate the expression of these transcripts after their initial nitrate response, integrating signals from both external and internal N availability. This is similar to the regulation of AFB3 by miR393, in which N metabolites induce this miRNA to decrease AFB3 levels (12).
We found that NAC4 was necessary for the nitrate response of OBP4 gene, but not for the response of the bHLH128 transcription factor. Considering we verified that AFB3 controls bHLH128 nitrate response, a possible explanation for this finding is that AFB3 controls bHLH128 directly through an AuxIAA/ARF mediated pathway independent of NAC4. Consistently, bHLH128 has two ARF-binding sites in its promoter according to the Arabidopsis Gene Regulatory Information Server (AGRIS) (http://arabidopsis.med.ohio-state.edu).
We found that NAC4 acts downstream of AFB3 to mediate the lateral root response to nitrate. NAC1 and NAC2 also have been implicated in lateral root responses to auxin (26, 27). Thus, NAC4 also works in an auxin-related pathway by regulating lateral root growth, but in the context of the nitrate response. As we reported previously, NAC4 is necessary for the nitrate response of OBP4. Similar to NAC4, OBP4 does not have a reported function in plant development. Other members of the OBP family—the OBP1, OBP2, and OBP3 proteins—are able to interact with the OBF4 protein (also known as TGA4), and have been identified as salicylic acid- and auxin-responsive (42). Lateral root initiation is known to be controlled by auxin through activation of the asymmetrical division of precursor xylem-pole pericycle cells (43). Thus, the nitrate-AFB3-NAC4-OBP4 pathway might transduce the auxin signal into activation of lateral root initiation by controlling cell cycle progression in the pericycle. Auxin control of the cell cycle involving changes in TIR1 auxin receptor levels in the pericycle has been reported in lateral root initiation in response to low phosphate (44). Furthermore, OBP1 is involved in cell cycle regulation (45). Our mechanism does not preclude other possible mechanisms contributing to the developmental effects of NAC4, such as control of other hormonal signaling pathways. We have found that components of the cytokinin and brassinosteroid pathways have an altered nitrate response in the afb3-1 mutant, and these hormones are known to modulate RSA in Arabidopsis (46–51).
IAA14/SLR is considered a master regulator of lateral root initiation, controlling the initial pericycle cell divisions that give birth to new roots (36, 52). We found that a IAA14/SLR gain-of-function mutation altered NAC4’s response to nitrate in Arabidopsis roots. IAA14 is known to interact with the ARF factors ARF7 and ARF19 to regulate lateral root initiation (39, 52); however, we found no altered NAC4 response in arf7-1, arf19-1, or arf7-1/arf19-1 mutants. This may suggest that in the context of the nitrate response, IAA14 is able to interact with a different set of ARF factors to control NAC4 expression. Alternatively, the accumulation of IAA14 protein caused by the lack of repression by the auxin receptors might lead to an unspecific binding to ARF factors that normally would not interact with IAA14. In either case, we can conclude that normal AUX/IAA-dependent signaling, possibly IAA14 itself, is required for the proper regulation of NAC4 and its target OBP4.
AFB3 is also able to repress primary root growth in response to nitrate. We found that NAC4 was not involved in this response, with the nac4 primary root demonstrating a normal nitrate response. Consistently, we did not find regulation of NAC4 or OBP4 in root tips, although AFB3 is regulated in the meristematic zone. Given that IAA14 is expressed primarily in xylem pole pericycle cells (53), the lack of Aux/IAA expression in root tips might explain the absent nitrate response of NAC4 and OBP4 even when AFB3 is induced in this root zone. Thus, AFB3 regulation of different regulatory networks in root tips and pericycle might independently control the growth of lateral and primary roots, finely modulating RSA in response to external and internal N availability.
Methods
Plant Material and Growth Conditions.
WT A thaliana ecotypes Ws and Col-0 were used in these experiments. The afb3-1 mutant (1), arf7-1, arf7-1/arf19-1 double mutant (54), arf6-2, arf8-3 (55), and slr-1 gain-of-function mutant (36) were kindly donated by Dr. Mark Estelle, Indiana University, Bloomington, IN. The iaa28-1 (35) mutant was kindly donated by Dr. Bonnie Bartel, Rice University, Houston. arf19-1 (CS24617) (54), iaa12-1(CS25213) (56), mp-S319 (SALK_021319) (57), nac4-1 (SALK_040204), and nac4-2 (SALK_006735) were obtained from the Arabidopsis Biological Resource Center. GFP lines that mark the epidermis (pWER:GFP), cortex (pAT1G09750:GFP), endodermis (pSCR:GFP), and stele (pWOL:GFP) were kindly donated by Dr. Phil Benfey, Duke University, Durham, NC. The GFP line that marks the pericycle (E3754) was obtained from http://enhancertraps.bio.upenn.edu. The nia1/nia2 mutant line (58) was kindly donated by Dr. Nigel Crawford, University of California at San Diego, La Jolla, CA.
Plants were grown in hydroponic cultures using modified MS basal salt medium without N (M531; Phytotechnology Laboratories), supplemented with 0.5 mM ammonium succinate and 0.1% sucrose. The plants were grown for 14 d under long-day conditions at 22 °C in Percival incubators. The plants were treated at the onset of the light cycle of the 15th day as indicated in the figure legends. For primary and lateral root length measurements, plant images were acquired using an Epson Perfection V700 scanner, and roots were measured using the ImageJ program (http://rsbweb.nih.gov/ij/). Lateral roots were counted using DIC optics on a Nikon Eclipse 80i microscope.
Gene Expression Using Affymetrix ATH1 Microarrays and Network Data Analysis.
Biotinylated cRNA was synthesized from 5 µg of total RNA from Arabidopsis roots using the Affymetrix IVT Kit according to the manufacturer’s instructions. cRNA was used to hybridize ATH1 GeneChip expression microarrays. Affymetrix data were normalized in R (http://www.r-project.org/) using RMA (15). Normalized data were subjected to two-way ANOVA, with a false discovery rate of 5%. For the ANOVA, we used a model considering the expression of a given gene Y as Yi = β0 + β1T + β2G + β3TG + ε, where β0 is the global mean; β1, β2, and β3 are the effects of T, G, and the TG interaction; and the variable ε is the unexplained variance. The data were analyzed with bioinformatics tools available at the VirtualPlant Web site (http://www.virtualplant.org).
Genes containing significant T, G, or TG factors were analyzed using the Sungear tool, and a molecular network for genes with a significant TG factor was created using the Gene Networks tool. The network includes miRNA–RNA interactions from miRBase (http://www.mirbase.org) and the protein/DNA regulatory interactions from AGRIS (http://arabidopsis.med.ohio-state.edu). To improve the regulatory predictions, the protein–DNA interactions were filtered to include only transcription factor/target pairs with significantly correlated (P ≤ 0.05) expression values of ≥0.7 or ≤−0.7 in our data. Network modeling was performed using Cytoscape software (23).
Supplementary Material
Acknowledgments
This work was funded by the International Early Career Scientist program from the Howard Hughes Medical Institute (Award 55007421), Fondo de Desarrollo de Areas Prioritarias Center for Genome Regulation (Grant 1509007), Millennium Nucleus Center for Plant Functional Genomics (Grant P10-062-F), Fondo Nacional de Desarrollo Científico y Tecnológico (Grant 1100698), and Agence Nationale de la Recherche (ANR)-Comisión Nacional de Investigación Científica y Tecnológica Program (Grant ANR-07). E.A.V. is supported by Proyecto de Inserción en la Academia (PSD74) and Fondo de Desarrollo Científico y Tecnológico (Grant 11121225).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. N.M.C. is a guest editor invited by the Editorial Board.
Data deposition: Microarray data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE35544).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310937110/-/DCSupplemental.
References
- 1.Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9(1):109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 2.Maraschin FdosS, Memelink J, Offringa R. Auxin-induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J. 2009;59(1):100–109. doi: 10.1111/j.1365-313X.2009.03854.x. [DOI] [PubMed] [Google Scholar]
- 3.Dharmasiri S, Estelle M. The role of regulated protein degradation in auxin response. Plant Mol Biol. 2002;49(3-4):401–409. [PubMed] [Google Scholar]
- 4.Chapman EJ, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet. 2009;43(1):265–285. doi: 10.1146/annurev-genet-102108-134148. [DOI] [PubMed] [Google Scholar]
- 5.Vanneste S, Friml J. Auxin: A trigger for change in plant development. Cell. 2009;136(6):1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Rademacher EH, et al. A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J. 2011;68(4):597–606. doi: 10.1111/j.1365-313X.2011.04710.x. [DOI] [PubMed] [Google Scholar]
- 7.Vidal EA, Gutiérrez RA. A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr Opin Plant Biol. 2008;11(5):521–529. doi: 10.1016/j.pbi.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Krouk G, Crawford NM, Coruzzi GM, Tsay Y-F. Nitrate signaling: Adaptation to fluctuating environments. Curr Opin Plant Biol. 2010;13(3):266–273. doi: 10.1016/j.pbi.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Wang R, et al. Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 2004;136(1):2512–2522. doi: 10.1104/pp.104.044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krouk G, et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell. 2010;18(6):927–937. doi: 10.1016/j.devcel.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 11.Gifford ML, Dean A, Gutiérrez RA, Coruzzi GM, Birnbaum KD. Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA. 2008;105(2):803–808. doi: 10.1073/pnas.0709559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vidal EA, et al. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107(9):4477–4482. doi: 10.1073/pnas.0909571107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446(7136):640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 14.Mockaitis K, Estelle M. Auxin receptors and plant development: A new signaling paradigm. Annu Rev Cell Dev Biol. 2008;24(1):55–80. doi: 10.1146/annurev.cellbio.23.090506.123214. [DOI] [PubMed] [Google Scholar]
- 15.Irizarry RA, et al. Summaries of Affymetrix GeneChip probe-level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krouk G, et al. A systems approach uncovers restrictions for signal interactions regulating genome-wide responses to nutritional cues in Arabidopsis. PLOS Comput Biol. 2009;5(3):e1000326. doi: 10.1371/journal.pcbi.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc, B. 1995;57:289–300. [Google Scholar]
- 18.Poultney CS, et al. Sungear: Interactive visualization and functional analysis of genomic datasets. Bioinformatics. 2007;23(2):259–261. doi: 10.1093/bioinformatics/btl496. [DOI] [PubMed] [Google Scholar]
- 19.Katari MS, et al. VirtualPlant: A software platform to support systems biology research. Plant Physiol. 2010;152(2):500–515. doi: 10.1104/pp.109.147025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutiérrez RA, et al. Insights into the genomic nitrate response using genetics and the Sungear Software System. J Exp Bot. 2007;58(9):2359–2367. doi: 10.1093/jxb/erm079. [DOI] [PubMed] [Google Scholar]
- 21.Gutiérrez RA, et al. Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA. 2008;105(12):4939–4944. doi: 10.1073/pnas.0800211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nero D, Krouk G, Tranchina D, Coruzzi GM. A system biology approach highlights a hormonal enhancer effect on regulation of genes in a nitrate responsive “biomodule”. BMC Syst Biol. 2009;3(1):59. doi: 10.1186/1752-0509-3-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15(8):1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005;10(2):79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 26.He XJ, et al. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2005;44(6):903–916. doi: 10.1111/j.1365-313X.2005.02575.x. [DOI] [PubMed] [Google Scholar]
- 27.Xie Q, Frugis G, Colgan D, Chua N-H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000;14(23):3024–3036. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dubrovsky JG, Soukup A, Napsucialy-Mendivil S, Jeknić Z, Ivanchenko MG. The lateral root initiation index: An integrative measure of primordium formation. Ann Bot (Lond) 2009;103(5):807–817. doi: 10.1093/aob/mcn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Jennings A, Barlow PW, Forde BG. Dual pathways for regulation of root branching by nitrate. Proc Natl Acad Sci USA. 1999;96(11):6529–6534. doi: 10.1073/pnas.96.11.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linkohr BI, Williamson LC, Fitter AH, Leyser HMO. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J. 2002;29(6):751–760. doi: 10.1046/j.1365-313x.2002.01251.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Forde BG. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science. 1998;279(5349):407–409. doi: 10.1126/science.279.5349.407. [DOI] [PubMed] [Google Scholar]
- 32.Remans T, et al. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc Natl Acad Sci USA. 2006;103(50):19206–19211. doi: 10.1073/pnas.0605275103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Smet I, et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development. 2007;134(4):681–690. doi: 10.1242/dev.02753. [DOI] [PubMed] [Google Scholar]
- 34.De Rybel B, et al. A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr Biol. 2010;20(19):1697–1706. doi: 10.1016/j.cub.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 35.Rogg LE, Lasswell J, Bartel B. A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell. 2001;13(3):465–480. doi: 10.1105/tpc.13.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29(2):153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- 37.Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19(1):118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanneste S, et al. Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell. 2005;17(11):3035–3050. doi: 10.1105/tpc.105.035493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Smet I, et al. Bimodular auxin response controls organogenesis in Arabidopsis. Proc Natl Acad Sci USA. 2010;107(6):2705–2710. doi: 10.1073/pnas.0915001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woodward AW, Bartel B. Auxin: Regulation, action, and interaction. Ann Bot (Lond) 2005;95(5):707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benjamins R, Scheres B. Auxin: The looping star in plant development. Annu Rev Plant Biol. 2008;59(1):443–465. doi: 10.1146/annurev.arplant.58.032806.103805. [DOI] [PubMed] [Google Scholar]
- 42.Kang H-G, Singh KB. Characterization of salicylic acid-responsive, Arabidopsis Dof domain proteins: Overexpression of OBP3 leads to growth defects. Plant J. 2000;21(4):329–339. doi: 10.1046/j.1365-313x.2000.00678.x. [DOI] [PubMed] [Google Scholar]
- 43.Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997;124(1):33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- 44.Pérez-Torres CA, et al. Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell. 2008;20(12):3258–3272. doi: 10.1105/tpc.108.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skirycz A, et al. The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. Plant J. 2008;56(5):779–792. doi: 10.1111/j.1365-313X.2008.03641.x. [DOI] [PubMed] [Google Scholar]
- 46.Kuderová A, et al. Effects of conditional IPT-dependent cytokinin overproduction on root architecture of Arabidopsis seedlings. Plant Cell Physiol. 2008;49(4):570–582. doi: 10.1093/pcp/pcn029. [DOI] [PubMed] [Google Scholar]
- 47.Laplaze L, et al. Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell. 2007;19(12):3889–3900. doi: 10.1105/tpc.107.055863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, Mo X, Shou H, Wu P. Cytokinin-mediated cell cycling arrest of pericycle founder cells in lateral root initiation of Arabidopsis. Plant Cell Physiol. 2006;47(8):1112–1123. doi: 10.1093/pcp/pcj082. [DOI] [PubMed] [Google Scholar]
- 49.Hutchison CE, et al. The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell. 2006;18(11):3073–3087. doi: 10.1105/tpc.106.045674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müssig C, Shin G-H, Altmann T. Brassinosteroids promote root growth in Arabidopsis. Plant Physiol. 2003;133(3):1261–1271. doi: 10.1104/pp.103.028662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bao F, et al. Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol. 2004;134(4):1624–1631. doi: 10.1104/pp.103.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fukaki H, Taniguchi N, Tasaka M. PICKLE is required for SOLITARY-ROOT/IAA14-mediated repression of ARF7 and ARF19 activity during Arabidopsis lateral root initiation. Plant J. 2006;48(3):380–389. doi: 10.1111/j.1365-313X.2006.02882.x. [DOI] [PubMed] [Google Scholar]
- 53.Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M. Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J. 2005;44(3):382–395. doi: 10.1111/j.1365-313X.2005.02537.x. [DOI] [PubMed] [Google Scholar]
- 54.Okushima Y, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17(2):444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nagpal P, et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development. 2005;132(18):4107–4118. doi: 10.1242/dev.01955. [DOI] [PubMed] [Google Scholar]
- 56.Overvoorde PJ, et al. Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell. 2005;17(12):3282–3300. doi: 10.1105/tpc.105.036723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cole M, et al. DORNROSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development. 2009;136(10):1643–1651. doi: 10.1242/dev.032177. [DOI] [PubMed] [Google Scholar]
- 58.Wang R, Xing X, Crawford N. Nitrite acts as a transcriptome signal at micromolar concentrations in Arabidopsis roots. Plant Physiol. 2007;145(4):1735–1745. doi: 10.1104/pp.107.108944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.