Significance
Exosomes are released from tumor cells at high levels, and multiple studies have determined that the secreted exosomes enter recipient cells and can affect their biologic and biochemical properties. In this study, the specific effects of the oncogenic herpesviruses, EBV and Kaposi sarcoma-associated virus, on the proteomes of B-cell exosomes were determined using global quantitative proteomics. The data indicate that the viruses greatly impact the protein content of exosomes with common and distinct changes induced by both viruses. It is likely that these alterations in exosome content modulate the tumor environment, potentially to enhance viral infection and promote tumorigenesis.
Keywords: microparticles, microvesicles, oncosomes
Abstract
The human gamma herpesviruses, Kaposi sarcoma-associated virus (KSHV) and EBV, are associated with multiple cancers. Recent evidence suggests that EBV and possibly other viruses can manipulate the tumor microenvironment through the secretion of specific viral and cellular components into exosomes, small endocytically derived vesicles that are released from cells. Exosomes produced by EBV-infected nasopharyngeal carcinoma cells contain high levels of the viral oncogene latent membrane protein 1 and viral microRNAs that activate critical signaling pathways in recipient cells. In this study, to determine the effects of EBV and KSHV on exosome content, quantitative proteomics techniques were performed on exosomes purified from 11 B-cell lines that are uninfected, infected with EBV or with KSHV, or infected with both viruses. Using mass spectrometry, 871 proteins were identified, of which ∼360 were unique to the viral exosomes. Analysis by 2D difference gel electrophoresis and spectral counting identified multiple significant changes compared with the uninfected control cells and between viral groups. These data predict that both EBV and KSHV exosomes likely modulate cell death and survival, ribosome function, protein synthesis, and mammalian target of rapamycin signaling. Distinct viral-specific effects on exosomes suggest that KSHV exosomes would affect cellular metabolism, whereas EBV exosomes would activate cellular signaling mediated through integrins, actin, IFN, and NFκB. The changes in exosome content identified in this study suggest ways that these oncogenic viruses modulate the tumor microenvironment and may provide diagnostic markers specific for EBV and KSHV associated malignancies.
Microvesicles are membrane-enclosed vesicles secreted from cells that participate in intracellular communication events through the transfer of biologically active proteins, lipids, and RNAs (1). Perhaps the best-studied class of microvesicles is exosomes which are 40- to 100-nm vesicles that originate from internal endosomal-derived membranes of multivesicular bodies (MVBs). Upon fusion of MVBs with the cell surface, exosomes are released into the extracellular space and can be taken up by neighboring cells, degraded, or enter connecting bodily fluids and travel to distal sites within the body. To date, exosomes have been found in almost every bodily fluid and increasingly are evaluated for their potential as diagnostic biomarkers. Exosomes are thought potentially to modulate many physiological processes including development, cell growth, immune regulation, angiogenesis, neuronal communication, cell migration, and invasion (1). Accumulating evidence supports the hypothesis that disruption of or alterations in normal exosome function may contribute to disease pathogenesis, and unique properties have been identified in exosomes released from malignant cells.
The human gamma herpesviruses, Kaposi sarcoma-associated virus (KSHV) and EBV, are considered the etiologic agents for several lymphoid malignancies (2–4). EBV is associated with Burkitt lymphoma, Hodgkin lymphoma, posttransplantation lymphoproliferative diseases, and AIDS-associated lymphomas, whereas KSHV is found in all primary effusion lymphomas (PELs) and multicentric Castleman’s disease. A subset of PELs contains both KSHV and EBV (5). Previous studies have shown that EBV affects exosome content and function, and virally modified exosomes can contain the viral oncoproteins latent membrane proteins 1 and 2 (LMP1 and LMP2) and virally encoded microRNAs (miRNAs) (6, 7). The transfer of LMP1-containing exosomes induces the activation of phosphoinositide 3-kinase (PI3K)/AKT and MAPK/ERK pathways in target cells (6). Additionally, exosomal transfer of EBV miRNAs has been shown to reduce target gene expression in recipient cells (8). Therefore, EBV-infected cells can modulate the cellular microenvironment through the transfer of virally modified exosomes. This transfer may be a significant mechanism through which herpesviruses maintain a latent and persistent infection within the host. Additionally, it is likely that tumorigenic herpesviruses such as EBV and KSHV modulate exosome content and function so that exosomes produced by the infected cells affect the tumor microenvironment and contribute to cancer progression.
Multiple studies have begun to define the protein, RNA, and lipid components of exosomes secreted from various cell types (9–11). In this study, exosomes were purified from EBV-infected, KSHV-infected, and dually infected B-cell lines and were analyzed using quantitative proteomics approaches to determine how EBV and KSHV infection alters B-cell exosome components. Mass spectrometry analysis of purified exosomes from 11 different B-cell lines revealed 871 proteins. Data from spectral counting determined that the B-cell exosome proteome from the virally infected cells was significantly different from the exosomes produced by the uninfected B-cell control exosomes, with 345 proteins unique to the virally infected cells and with significant changes specific to each virus. These data show that EBV and KSHV potently affect the host exosome pathway.
Results
Viral Expression and Exosome Purification.
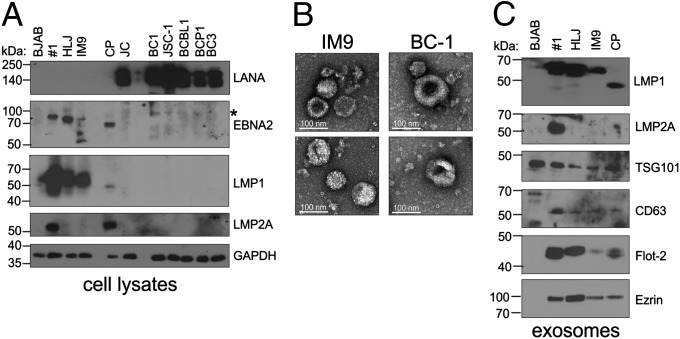
The type of latent infection in the EBV-infected cells was determined by evaluating expression of Epstein–Barr nuclear antigen 2 (EBNA2) and LMP1, considered indicative of Type III latency (12). EBNA2 was detected readily in #1, HLJ, and CP cell lines and at lower levels in IM9 cells but was not detected in the KSHV-infected or dually infected lines. A background band with a higher molecular weight was detected at varying levels in all cell lines tested (Fig. 1A, asterisk). LMP1 was detected in the four EBV+ cell lines (a smaller form was detected consistently in CP cells), whereas LMP2 was detected in #1 and CP cells. The dually infected KSHV+/EBV+ cell lines did not express these viral proteins. Dually infected PEL cell lines have been shown previously to lack expression of these EBV proteins and are considered representative of Type I latency, which frequently is found in Burkitt lymphoma (Fig. 1A). Latency-associated nuclear antigen (LANA) was detected in all KSHV cell lines (Fig. 1A). Exosomes were purified using a sucrose cushion from the conditioned media of various B-cell lines, either uninfected (BJAB cell line) or infected with EBV, KSHV, or EBV and KSHV. The purity of the exosome preparations was determined by electron microscopy (EM) and immunoblot analysis for exosome specific markers. EM images of the preparations did not detect any virions but did reveal many membrane-enclosed vesicles with sizes ranging from 40–100 nm that exhibited the typical cup-shaped morphology previously described for exosomes after negative staining (Fig. 1B). Multiple exosome-specific markers were detected by immunoblot analysis of exosome lysates (Fig. 1C). Interestingly, the levels of some exosome markers varied among the cell lines, suggesting that viral infection could alter exosome components. Ezrin, a protein previously shown to be up-regulated by LMP1, was significantly increased in EBV+/LMP1+ exosomes compared with the uninfected BJAB control cells (Fig. 1C) (13). Additionally, Flotillin 2, a major lipid raft resident protein, was increased in LMP1+ exosomes (14, 15). LMP1 traffics to and alters the components of lipid rafts for signal transduction (16, 17) and therefore may contribute to exosomal targeting of lipid raft proteins such as Flotillin 2 (18). LMP1 was detected by immunoblotting in exosomes isolated from all four EBV-infected B cells at levels approximately proportional to those found in whole-cell lysates (Fig. 1C). LMP2 was detected in exosomes from one of the LMP2+ cell lines (Fig. 1C). These data indicate that both EBV- and KSHV-infected B cells secrete microvesicles with biophysical and biochemical properties consistent with exosomes.
Fig. 1.
Characterization of exosomes secreted from B cells. (A) Whole-cell lysates from various B-cell lines that were uninfected (BJAB), EBV infected (#1, HLJ, IM9, CP), KSHV infected (JC, BCBL1, BCP1, BC3), or dually infected (BC1 and JSC-1) were separated by SDS/PAGE and analyzed by immunoblot analysis for the indicated proteins. GAPDH was used as a loading control. (B) Exosomes were purified from the conditioned media of the B cells and analyzed by electron microscopy. Representative images of exosomes produced from the IM9 and BC1 cell lines are shown. (C) Lysates from purified exosomes were separated by SDS/PAGE and analyzed by immunoblot for expression of indicated viral proteins and common exosome markers. Asterisks indicate a with a higher molecular weight detected using EBNA2 antibody.
Mass Spectrometric Analyses.
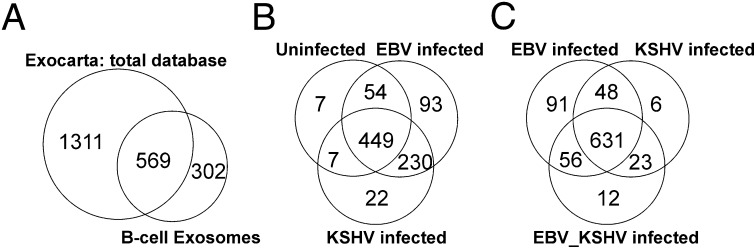
To determine the effect of viral infection on exosome content, purified exosome proteins were separated by strong anion-exchange and reverse-phase chromatography before mass spectrometry. Data analysis following multidimensional protein identification technology (19) uncovered a total of 871 specific proteins throughout all B-cell exosome lines (Dataset S1). Of these 871 proteins detected in all preparations of B-cell exosomes, 569 were present in the Exocarta database of published exosome proteomic results, including 21 of the 25 most frequently identified proteins (11). These data confirm that the preparations do indeed contain abundant levels of exosomes. The mass spectrometry analysis also uncovered 302 newly identified exosome components not present in Exocarta as indicated by Venn diagram (Fig. 2A and Dataset S1). Interestingly of the 302 proteins uniquely identified in this study, 164 had significantly (P ≤ 0.05) altered levels because of virus infection as compared with the uninfected control cell line (BJAB) (Dataset S1). Viral proteins were not identified in this unbiased mass spectrometry analysis, which is based on the 15 most intense peptide ion peaks for MS/MS during a mass spectrometry acquisition cycle. These findings indicate that proteins representing potential virion contamination of the exosome preparations were below the threshold of detection for MS/MS.
Fig. 2.
Venn diagrams of proteins identified in B-cell exosomes by mass spectrometry. (A) The total number of proteins identified throughout all B-cell exosome samples was compared with results from the entire Exocarta database of published exosome proteomics. (B) Exosome components identified from the EBV- or KSHV-infected cells were compared with those found in the uninfected BJAB control cell line. (C) Comparison of proteins specific to each infection type and those proteins common to infection types.
Presentation of the distinct exosome protein content according to the infection status of the B-cell lines indicated that of the 871 total proteins identified, 449 were common to uninfected BJAB B-cell exosomes and EBV- or KSHV-infected exosomes (Fig. 2B and Dataset S2). As anticipated, latent herpesvirus infection significantly altered exosome content; 230 proteins were identified in both EBV and KSHV exosomes that were not present in the uninfected exosomes, 93 proteins were specific to EBV-infected exosomes and 22 were specific to the KSHV exosomes (Fig. 2B). An additional 16 distinct proteins were present in the dually infected cell lines that were not detected in the uninfected control cells (Dataset S2). Many of the differences determined between exosome groups are likely to be specific to viral infection; however, some may be unique to the BJAB exosomes.
Comparison of the exosomes from the KSHV-, EBV-, and dually infected cell lines indicated that the virus-modified exosomes had 631 proteins affected in common (Fig. 2C). The EBV exosomes had multiple additional changes, with 91 uniquely identified components, whereas 41 changes were detected in the exosomes from KSHV or dually infected cells, including 23 proteins that were present in both the KSHV-infected and KSHV/EBV dually infected exosomes, six proteins specific for the KSHV exosomes, and 12 proteins found only in the dually infected exosomes (Fig. 2C and Dataset S2). These data further support the hypothesis that virus infection has major effects on exosome content and that these changes likely modulate their functional properties.
2D Gel Electrophoresis.
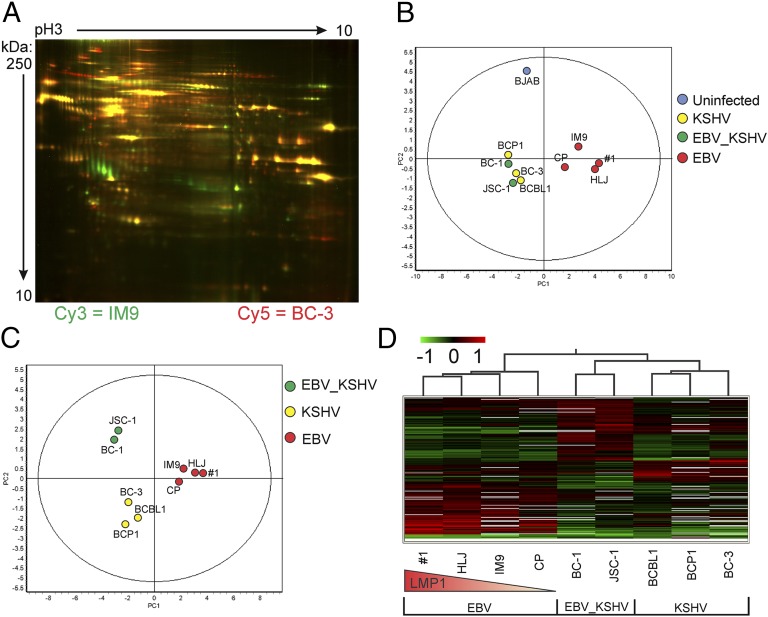
To confirm the potential viral-specific differences in exosome content, 2D difference gel electrophoresis (2D DIGE) was used (20). Changes in protein-expression levels in B-cell exosomes revealed by 2D-DIGE were analyzed using DeCyder software which identified 2,131 protein spots matched across all gels (for a representative gel, see Fig. 3A). A t-test analysis of LMP1+ versus LMP1− cell lines revealed 217 protein spots with significantly different expression (P < 0.05). When cell lines were grouped according to infection type, differential expression analysis (ANOVA) revealed 209 protein spots with significantly different expression (P < 0.05).
Fig. 3.
2D DIGE and Decyder analysis of B-cell exosome proteomes. Exosomal proteins were labeled with fluorescent dyes and separated by 2D DIGE in pH 3–10 immobilized gradients and SDS 12.5% polyacrylamide gels. (A) Representative gel shown (1 of 10) with IM9 exosome proteins labeled with Cy3 (green) and BC-3 exosome proteins labeled with Cy5 (red). PCA was performed on the 2D DIGE data and plotted based on (B) LMP1 expression or (C) viral groups. (D) Hierarchical cluster analysis of significantly (P ≤ 0.05, ANOVA) altered exosome components grouped by viral infection status. Red lines represent proteins whose expression levels were up-regulated relative to the group, green lines represent down-regulated proteins, and black lines represent unchanged proteins. The red triangle depicts relative LMP1 expression in the EBV cell lines.
These protein spot differences were examined further by principal component analysis (PCA) and hierarchical cluster analysis using the DeCyder software (Fig. 3). In the PCA of LMP1+ versus LMP1− data (Fig. 3B), principal component 1 (PC1), which contained 58.8% of the variance between samples, confirmed that the expression of EBV infection with LMP1 expression represented the dominant component affecting the grouping of B-cell line exosomes. The second principle component (PC2), with 15.7% of variance, was primarily between the uninfected BJAB control cells and all other (infected) cell lines. When the cell lines were grouped according to infection differences, PCA revealed that PC1, containing 56.9% of the variance, was found between EBV alone and other types of infection (Fig. 3C). PC2, with 19.4% of variance, was primarily between KSHV-infected and the dually infected cell lines. In contrast, EBV-infected B-cell lines showed little influence of PC2, as shown by their proximity to the PC2 axis. Similarly, hierarchical cluster analysis (Fig. 3D) grouped B-cell exosomes into three distinct branches (clusters) representing the type of infection (i.e., EBV, KSHV, or dual infection). EBV infection alone produced the most distinct cluster, with these B-cell lines branching off first from the KSHV- and dually infected lines. Interestingly, these exosomes also branched within the distinct EBV cluster based on LMP1 expression levels in the producing cell lines. KSHV- and dually infected lines also segregated as a second branching event (Fig. 3D).
Label-Free Spectral Count-Based Quantitative Proteomic Analysis.
To evaluate further the exosome proteome changes caused by virus infection, the grouped virally modified exosome proteins were analyzed by spectral counting with changes represented relative to the uninfected BJAB exosomes (21). Regardless of the infection types compared, ∼21% of the B-cell exosome proteome was altered. The levels of 187 of the 871 proteins identified in the dataset were altered significantly (P ≤ 0.05; Fisher’s exact t-test) in exosomes secreted from EBV-infected cells (Dataset S1, indicated in yellow). Similar numbers of changes also were observed with KSHV (188 of 871) or KSHV+EBV (207 of 871) infection as compared with uninfected BJAB exosomes (Dataset S1). Ezrin, which was not detected by immunoblotting in the BJAB exosomes, also was not detected by liquid chromatography tandem mass spectrometry (LC-MS/MS) in BJAB exosomes. However, it was detected in all the exosome preparations from the virally infected cell lines. Thus, ezrin is indicated with an infinite fold change with P values of 4.2 × 10−6 for the EBV exosomes and 0.015 for KSHV exosomes (Dataset S1).
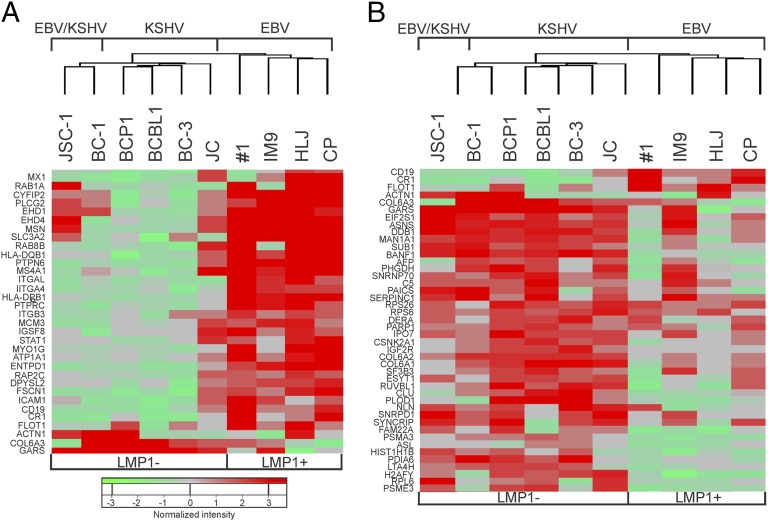
Unbiased hierarchical clustering analysis of the differentially expressed exosome components separated the samples into groups based on virus infection, confirming the 2D-DIGE analyses. The unique clustering pattern and variable levels of EBNA2 and LMP2 in the cell lines suggested that LMP1 was a major factor in the induction of specific changes in exosome content. Major differences in expression correlated highly with Type 3 latency and levels of LMP1 expression, with LMP1− exosomes isolated from EBV and KSHV-infected PELs clustering distinctly from those isolated from the EBV-infected, LMP1-expressing lymphoblastoid cell lines (LCLs) (Fig. 4 A and B). These data further indicate that EBV and KSHV have distinct effects on exosome content. In EBV-infected cells, Type III latency and LMP1 expression are major factors contributing to the changes in exosome content, whereas the exosomes produced by the cells dually infected with EBV and KSHV are closely linked to those from KSHV-infected cells. This result suggests that the highly restricted EBV infection within the dually infected PEL does not have a major effect beyond KSHV on exosome content. However, the two cell lines did group distinctly, and 12 proteins were detected only in exosomes from the dually infected cells, which also contained 54 proteins that were significantly altered compared with BJAB cells (Fig. 2C and Dataset S1).
Fig. 4.
Hierarchical clustering of B-cell exosome proteins. (A) Unsupervised hierarchical clustering of B-cell exosome proteins that show up-regulation in EBV-infected B-cell lines as determined by spectral count-based quantitative proteomic analysis. (B) Unsupervised hierarchical clustering of B-cell exosome proteins that show up-regulation in KSHV- or KSHV+EBV-infected cell lines that also were LMP1−.
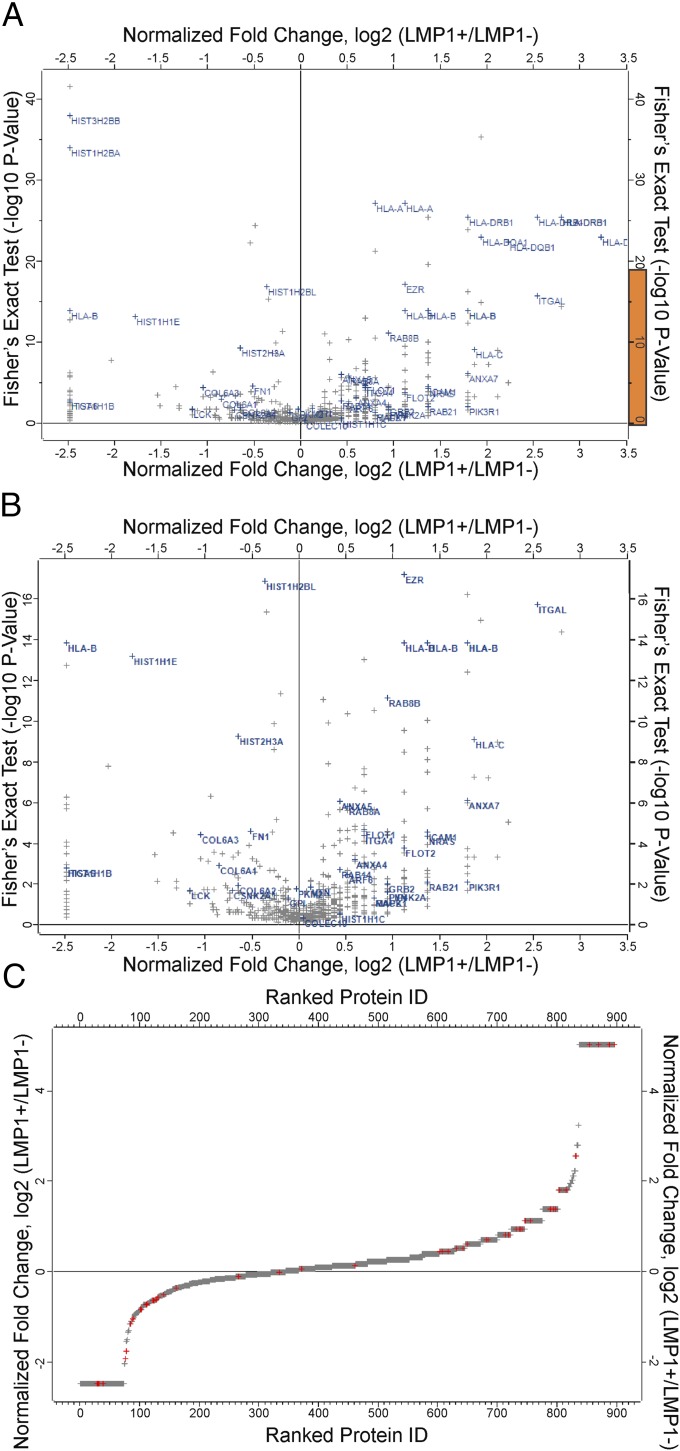
To enhance the statistical power of the analyses the quantitative mass spectrometry, data from each individual cell line were grouped into either LMP1+ or LMP1− datasets. In this analysis, the exosomes from the EBV/KSHV dually infected cells are grouped with the KSHV-infected cell exosomes because they lacked LMP1 expression. There were 358 significant changes (P ≤ 0.05, Fisher’s exact t test) identified between the groups with log twofold changes ranging from 5 to −2.5 (Fig. 5 and Dataset S3). This analysis reveals that the LMP1− exosomes had 30% of the amount of ezrin contained in LMP1+ exosomes (Fig. 5 A and B). There also were 60 proteins present in LMP1+ exosomes not found in LMP1− exosomes and were given an arbitrary fold change of 5 (log2) (Fig. 5C and Dataset S3). Interestingly, more of the significantly altered proteins were increased in LMP1+ exosomes than in LMP1− exosomes (Fig. 5C and Dataset S3). This difference may reflect the specific recruitment of protein complexes into exosomes and the potent effects of LMP1 on cellular protein expression.
Fig. 5.
Label-free spectral count-based quantitative proteomic analysis. (A) Volcano plot showing normalized differences in protein expression in LMP1+ and LMP1− B-cell lines infected with EBV, KSHV, or EBV+KSHV. Note that the ANOVA P value of 0.05 (−log10 = 1.3) is used to identify significant differences in protein expression. (B) Enlarged view of the region highlighted in orange in A. (C) Overall differences in protein expression in LMP1+ and LMP1− B-cell lines ranked by protein ID. (Insets) Number of down-regulated and up-regulated proteins that show a log2 twofold change in the LMP1+/LMP1− ratio. Points marked in red represent proteins highlighted in A and B. Infinity units were given arbitrary values of log2 5 and −2.5.
Viral-Specific Effects.
The cellular proteins that were specifically up-regulated in the EBV+ LMP1+ exosomes included multiple HLA class I and class II proteins (Fig. 5 A and B). LMP1 has been shown to up-regulate HLA proteins, and their enriched secretion into exosomes may modulate viral entry or immune recognition (22–24). Furthermore, the EBV/LMP1 exosomes contained multiple kinases including the TNF receptor-associated factor 2 and NF-κB–inducing kinase (TNIK) (25), the fgr kinase, which has been linked to EBV infection (26), and the protein 85 (p85) regulatory subunit of PI3K, a protein recently shown to be induced into lipid rafts by LMP1 expression (16). Additional proteins previously shown to be up-regulated by LMP1, including intercellular adhesion molecule 1 and ezrin, also were increased in the EBV exosomes (13, 27) (Figs. 1A and B and Dataset S3). Other exosome components potentially regulated by LMP1 include proteins involved with membrane and protein trafficking [annexins, Rab GTPases, and ADP-ribosylation factor 6 (ARF6)], binding (integrins), lipid rafts (Flotillin 1 and 2), and signaling [growth factor receptor-bound protein 2 (GRB2), NRAS, LYN, MAPK1, RAC2, and phosphatidylinositol-5-phosphate 4-kinase type-2 alpha (PIP4K2A)] (Fig. 5 A and B and Datasets S1 and S2). These findings support previous studies that have indicated functional effects of EBV exosomes on signaling and immune function (6, 28–30).
The exosome components from KSHV-infected PEL with P values <0.05 are indicated in yellow in Dataset S1 with fold increase in comparison with exosomes from uninfected BJAB cell. Although histones previously have been shown to be present in exosomes from different cell types (31), the exosomes from KSHV-infected PEL cells showed a preferential increase in many histone proteins including histones H1, H2A, H2B, H3, H4, and variants of each of these core histones as compared with BJAB cells (Dataset S1) or EBV LCLs (LMP1+) (Fig. 5 A and B). Additionally, several enzymes involved with glycolysis also were significantly up-regulated in PEL exosomes compared with BJAB exosomes. These enzymes included pyruvate kinase, enolase, phosphoglyceromutase, phosphoglycerate kinase, glyceraldehyde dehydrogenase, triose phosphate isomerase, fructose biphosphate aldolase, phosphoglucose isomerase, and lactate dehydrogenase. These data are concordant with other reports showing KSHV-infected cells have increased glycolysis and fatty acid synthesis (32–34). Therefore, it is conceivable that exosomal transfer of glycolytic enzymes could enhance glycolysis in recipient cells.
Factors involved in protein translation also were increased in exosomes from KSHV-infected cells as compared with uninfected BJAB exosomes. The 40S and 60S ribosomal subunits, several different tRNA synthetases, as well as several translation factors (e.g., Eukaryotic translation initiation factor 3) were increased in exosomes from PEL compared with BJAB cells. This increase may reflect the known effects of KSHV and some of its viral proteins including K1 and viral G protein-coupled receptor, which have been shown to modulate the cellular protein synthesis machinery through activation of the PI3K/mammalian target of rapamycin (mTOR) pathway (35–40). Of interest, up-regulation of PI3K signaling also is involved in activation of glycolysis by KSHV, as described above (32, 41). Moreover, additional factors involved in cell migration (integrin alpha 6, fibronectin, vitronectin, and collagen) and PI3K signaling (lymphocyte-specific protein tyrosine kinase) were found to be preferentially up-regulated in the exosomes from KSHV-infected cells (Fig. 5 A and B and Dataset S3).
Collectively, these data suggest that exosomes from KSHV-infected B cells preferentially incorporate proteins involved in metabolism, translation, migration, and chromatin modeling as compared with uninfected B cells. This exclusive loading may reflect alterations of these pathways within infected cells and suggests that the secreted exosomal proteins from KSHV-infected cells could participate in paracrine or autocrine mechanisms.
Predicted Viral-Specific Effects on Exosome Content and Function.
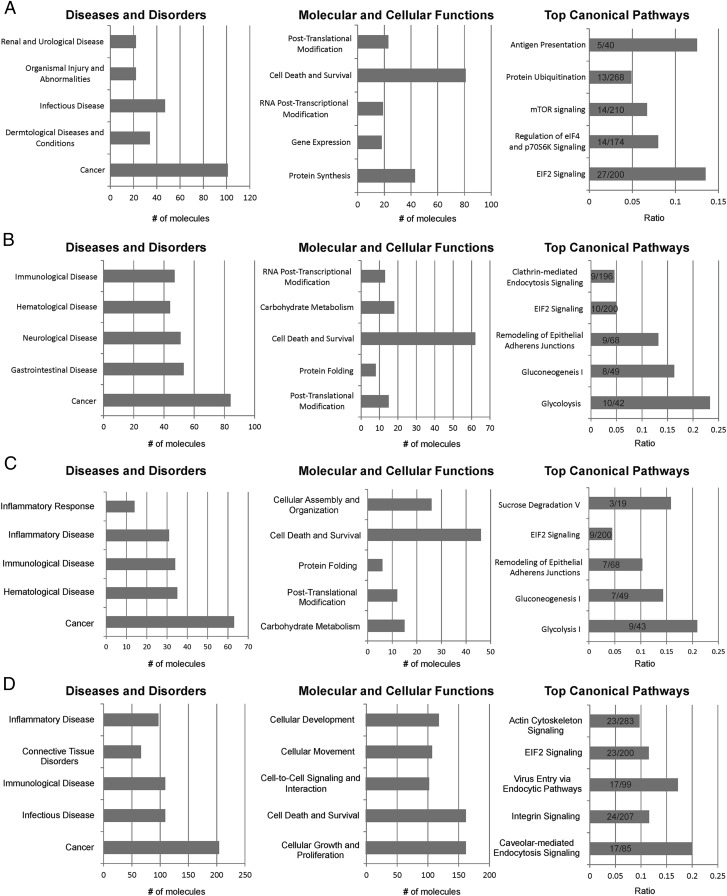
To gain potential insight into how modulation of exosome components may contribute to exosome function and potentially to disease, the proteins that were only identified in the viral exosome preparations or that had significantly altered expression in comparison with BJAB were analyzed with Ingenuity Pathway Analysis (IPA) software (Fig. 6). The gene sets include the 230 virally induced genes (Fig. 2B and Dataset S2) that are unique to the EBV and KSHV singly infected cell lines and that are not present in the viral-negative BJAB cell line (Fig. 6A), the 264 genes significantly altered (P ≤ 0.05; Dataset S1) by KSHV and KSHV/EBV (Fig. 6B), the 190 genes significantly altered (P ≤ 0.05; Dataset S1) by KSHV alone (Fig. 6C), and the 358 genes that are changed significantly (Dataset S3) when grouped by LMP1 expression (Fig. 6D). Strikingly, IPA indicated that for all four populations Cancer was the highest represented Diseases and Disorder category (Fig. 6), and Cell Death and Survival was the top Molecular and Cellular Function predicted to be affected. The EBV/LMP1 exosomes also were enriched for proteins that affect the Molecular and Cellular Functions of Cell Growth and Proliferation, Cellular Movement, and Cell-to-Cell Signaling. The Top Canonical Pathways had more variation among groups, although all were enriched for genes predicted to affect eukaryotic initiation factor 2 (EIF2) signaling. IPA also predicted KSHV effects on glycolysis, gluconeogenesis, remodeling of adherens junctions, and clathrin-mediated endocytosis. Confounding the interpretation of the IPA analysis is that glycolysis and glyconeogenesis are competing pathways. However, the two pathways contain many of the same enzymes, which are dimeric and can catalyze in both directions. The KSHV exosomes contained seven enzymes that participate in both pathways and additionally contained hexokinase, pyruvate kinase, and lactose dehydrogenase, which are specific to glycolysis. Enzymes specific for gluconeogenesis, such as glucose-6-phosphatase, fructose 1,6-biphosphatase, pyruvate carboxylase, and phosphoenolpyruvate carboxykinase, were not detected in the exosomes. The EBV/LMP1 exosomes were enriched for proteins in Canonical Pathways that likely affect exosome formation and uptake such as Caveolar-Mediated Endocytosis Signaling, Virus Entry via Endocytic Pathway, Integrin Signaling, and Actin Cytoskeleton (Fig. 6).
Fig. 6.
IPA of virally modified exosome components. IPA of (A) proteins only identified within EBV and KSHV exosomes compared to the BJAB control, (B) genes significantly altered by KSHV and KSHV/EBV relative to BJAB, (C) genes significantly altered by KSHV relative to BJAB, and (D) genes significantly changed when grouped by LMP1 expression. The graphs for the top Diseases and Disorders (Left) and Molecular and Cellular Functions (Center) categories show the total number of molecules identified by mass spectrometry that fall into the designated Ingenuity classification. The Top Canonical Pathways graphs (Right) represent the ratio of the number of molecules identified in this study to the total number of proteins in each Ingenuity pathway.
Discussion
Exosomes’ potential roles in disease pathogenesis are increasingly appreciated, and many studies are focusing on exosomes produced by malignant cells as potential biomarkers. The data presented here identify specific and global effects of the human oncogenic viruses, EBV and KSHV, on exosome content. The data indicate that exosome content is highly complex, with 871 proteins identified by LC-MS/MS, including more than 360 that were not detected in the BJAB uninfected control cell line and that potentially are unique to the viral exosomes. As the exosome proteome is further defined, it is likely that many more intriguing differences will be identified. For example, cluster of differentiation 63 (CD63) and LMP1 were readily detected by immunoblotting of exosome preparations, even though neither protein was identified by the mass spectrometry analysis. Based on the proteins identified in this study, KSHV and EBV exosomes are predicted to have multiple properties in common, with Cancer identified as the predicted Disease and Disorder, Cell Death and Survival as the commonly affected Cellular Function, and EIF2 Signaling as the commonly affected Canonical Pathway. These findings suggest that both viruses affect exosome content to modulate both cell death and protein synthesis. Additionally, the viruses had distinct predicted effects on molecular and cellular functions. Interestingly, many of the predicted modulated properties have been linked previously to the distinct viruses. KSHV recently has been shown to reprogram the host B-cell metabolism toward glycolysis, and the exosomes produced by the KSHV-infected cells are highly enriched in proteins in the Glycolysis Pathway. Importantly, it has been demonstrated experimentally that overexpression of pyruvate kinase, lactate dehydrogenase, or phosphoglucose isomerase increases glycolysis, and therefore exosomal transfer of these enzymes also could modulate metabolism in the recipient cells (42, 43). Intriguingly, the KSHV exosomes also include proteins involved in remodeling epithelial adherens junctions, suggesting that they may modulate cell anchorage or movement through these effects.
The 2D DIGE and the spectral counting and LC-MS/MS data grouped the KSHV/EBV dually infected exosomes closely with the KSHV exosomes and not with the EBV singly positive exosomes. These data indicate that the limited EBV expression in the PEL cell lines has a lesser effect on exosome content than the robust effects mediated in Type III latency with expression of LMP1. However, PCA revealed that the dually infected cells are distinct from the singly infected KSHV cells and the EBV-infected cells (Fig. 3C). This difference may reflect the effect of EBV functions that are expressed in the PEL lines, including Epstein–Barr nuclear antigen 1 and the BamHI-A region (BART) miRNAs, which are expressed at considerably higher levels in the PEL cells than in most EBV-transformed B-cell lines (in which BART miRNA expression is very low) (44).
The predicted Molecular Functions for the EBV+/LMP1+ exosomes are all cellular properties that have been shown to be affected by LMP1. In various in vitro models LMP1 can induce cellular growth and proliferation, modulate cell death and survival, activate cell-to-cell signaling, and induce cellular movement (3). The Canonical Pathways predicted to be affected by LMP1 expression in exosomes include Endocytosis, Cytoskeleton Signaling, and Integrin Signaling. The effects on endocytosis may be related to the enhanced binding and uptake described for LMP1 exosomes, and it is known that LMP1 can induce actin remodeling (6, 45). These data suggest that the functional properties of the exosomes may mimic many of the properties of LMP1-expressing cells. Through its effects on exosome content, LMP1 could influence these molecular functions or pathways in both autocrine and paracrine mechanisms.
One mechanism through which virally modified exosomes could affect these cellular functions is the transfer of biologically active signal transduction components. Importantly, when the significantly modified exosome components were analyzed within IPA, many pathways were predicted to be activated by LMP1+ exosomes (Table S1). These pathways included several already known to be regulated by LMP1 within infected cells, such as protein 53 (P53), JAK/STAT, NFκB, interferon regulatory factor 7, and MAPK (Table S1). Interestingly, TNF and cluster of differentiation 40 (CD40), cellular equivalents to LMP1, also were predicted to be activated by LMP1+ exosomes. Constitutive CD40 signaling phenocopies the transforming function of the EBV oncoprotein LMP1 in vitro (46, 47). Insulin receptor signaling, which activates the PI3K/AKT and rat sarcoma (Ras)/MAPK pathways, two pathways affected by LMP1 signaling (16), was predicted to be activated by LMP1+ exosomes. The identification of multiple signaling pathways that are predicted to be activated confirms previous studies that have shown that LMP1+ expression alters exosome content so that LMP1+ exosomes induce activation of ERK and AKT in recipient cells (6). Similarly, the pathways predicted to be activated specifically by the KSHV+ exosomes included MAPK1 and PI3K.
To look more closely at known LMP1-regulated pathways, the dataset was overlaid with the ERK/MAPK, PI3K, and JAK/STAT pathway within Ingenuity. As expected from the predicted activated pathways, many exosome components also were present in these pathways and were altered based on LMP1 expression. These included important upstream activators such as Ras, FYN, LYN, p85 subunit of PI3K, ERK1/2, STAT, mTOR, and GRB2 (Dataset S3). These data support the hypothesis that LMP1 modulates exosome content and that critical growth-inducing pathways may be induced in recipients’ cells with the exosomal transfer of LMP1 and other key cellular components (6).
The changes common to the exosome proteomes of both EBV and KSHV indicate that both viruses target specific cellular functions and that the altered exosomes likely modulate cell death and survival with additional predicted effects on ribosome function, protein synthesis, and mTOR signaling. Additionally, the viruses have individual specific effects, with KSHV exosomes predicted to modulate cellular metabolism and EBV exosomes predicted to activate signaling mediated through integrins, actin, IFN, and NFκB. It is clear that the virally infected cells can manipulate the cellular microenvironment via exosomes and that KSHV and EBV exosomes may have distinct effects on the recipient cells. This activity is supported by the unique proteomes of KSHV and EBV exosomes and previous studies that have shown that exosomes released from EBV-infected epithelial cells and lymphocytes can transfer their contents and modulate cellular functions (6, 8). It is likely that these effects contribute to viral persistence and pathogenesis.
Materials and Methods
Cells.
B-cell lines were cultured in Roswell Park Memorial Institute (RPMI) medium 1640 supplemented with 10% (vol/vol) FBS (Thermo), penicillin-streptomycin (Gibco), antibiotic-antimycotic (Gibco), 0.075% sodium bicarbonate, and 0.05 mmol/L 2-mercaptoethanol at 37 °C in 5% (vol/vol) C02. The cell lines include EBV-infected LCLs (#1, HLJ, IM9, and CP), KSHV-infected PELs (JC, BC3, BCP1, and BCBL1), and dually infected PELs (JSC-1 and BC1) (48, 49). The EBV-infected lines were established as spontaneous transformed lines from peripheral blood samples. HLJ, CP, and JC cells were kindly provided by Clio Rooney (Baylor College of Medicine, Houston, TX), and #1 cells were obtained from Richard O’Reilly (Memorial Sloan–Kettering Cancer Center, New York).
Exosome Purification.
Cells were grown to an approximate concentration of 0.5 × 106 cells/mL in 100 mL of RPMI medium that was supplemented with 10% (vol/vol) exosome-depleted serum, antibiotics, 0.1% β-mercaptoethanol, and sodium bicarbonate. The medium then was harvested and replaced with 150 mL of fresh RPMI medium, and the cells were grown for an additional 5 d. The conditioned medium was subjected to differential centrifugation of 480 × g for 5 min, 2,000 × g for 10 min, and 10,000 × g for 30 min and then was filtered (0.2-uM pore size) and stored at 4 °C for up to 1 wk. To purify exosomes, the medium was concentrated with a Centricon Plus-70 filter (Millipore) according to the manufacturer’s instructions, placed over a 30% (wt/vol) sucrose:deuterium oxide (D20) cushion (5 mL) in a SW32 polyallomer tube (Beckman Coulter), and subjected to ultracentrifugation at 100,000 × g for 1 h. After centrifugation, the sucrose cushion (∼4.75 mL) was removed with a needle syringe without disturbing the pellet on the bottom of the tube or the sucrose/medium interphase, and the medium was diluted with 30 mL of PBS. The exosomes then were pelleted at 100,000 × g for 1 h, dissolved in lysis buffer [9 M Urea, 2 M Thiourea, 2% (wt/vol) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate hydrate (CHAPS), 60 mM n-octyl-β-d-glucopyranocide, 30 mM Tris, pH8.5], and frozen at −80 °C for proteomic analysis. To obtain enough protein for mass spectrometry (100 μg), exosomes preps were pooled from individual harvests before digestion.
Protein Extraction and Digestion.
Proteins were prepared for digestion using the filter-assisted sample preparation method (50). Briefly, the sample was brought to 2% (wt/vol) SDS, 50 mM Tris⋅HCl (pH 7.6), 10 mM DTT and was heated at 95 °C for 10 min. Samples then were transferred to a 30,000 (30-k) Vivaspin molecular weight cut off (MWCO) device (Vivaproducts) and centrifuged at 13,000 × g for 10 min. The remaining sample was buffer exchanged with 6 M urea, 100 mM Tris⋅HCl (pH 7.6) and then alkylated with 55 mM iodoacetamide. Concentrations were measured using a Qubit fluorometer (Invitrogen). Trypsin was added at an enzyme-to-substrate ratio of 1:40, and the sample was incubated overnight on a heat block at 37 °C. The device was centrifuged, and the filtrate was collected.
Peptide Desalting and Fractionation.
Digested peptides were desalted using C18 stop-and-go extraction (STAGE) tips (51). Briefly, for each sample a C18 STAGE tip was activated with methanol, and then conditioned with 60% (vol/vol) acetonitrile, 0.5% acetic acid followed by 5% (vol/vol) acetonitrile, 0.5% acetic acid. Samples were loaded onto the tips and desalted with 0.5% acetic acid. Peptides were eluted with 60% acetonitrile, 0.5% acetic acid and vacuum centrifuged in a SpeedVac (Thermo Savant) for ∼1 h to a final volume of ∼5 μL.
Peptides were fractionated by strong anion exchange STAGE tip chromatography. Briefly, each sample was dissolved in Britton Robinson buffer, pH 10, and loaded on to the STAGE tip. Flow-through was collected using a C18 STAGE tip. Subsequent fractions were taken by eluting peptides with Britton–Robinson buffers at pH 8, 6, 5, 4, and 3.2 and capturing with C18 STAGE tips. Peptides were eluted from the C18 STAGE tip and dried as described above.
LC–MS/MS.
Each fraction was analyzed by LC-MS/MS. LC was performed on an Easy Nano LC II system (Thermo Scientific). Mobile phase A was 97.5% MilliQ water, 2% (vol/vol) acetonitrile, 0.5% acetic acid. Mobile phase B was 90% (vol/vol) acetonitrile, 9.5% MilliQ water, 0.5% acetic acid. The 120-min LC gradient ran from 0% B to 35% B over 90 min, with the remaining time used for sample loading and column regeneration. Samples were loaded to a trap column, 2 cm × 75 μm i.d. The analytical column was 13 cm × 75 μm i.d. fused silica with a pulled tip emitter. Both trap and analytical columns were packed with 3 μm C18 resin (Magic C18AQ; Michrom). The LC system was interfaced to a dual-pressure linear ion trap mass spectrometer (LTQ Velos; Thermo Fisher) via nano-electrospray ionization. An electrospray voltage of 1.8 kV was applied to a precolumn tee. The mass spectrometer was programmed to acquire, by data-dependent acquisition, tandem mass spectra from the top 15 ions in the full scan from 400–1400 m/z.
Mass Spectrometry Data Processing.
Mass spectra were processed, and peptide identification was performed using the Mascot search engine (ver. 2.3) (Matrix Science) found in Proteome Discoverer (ver. 1.3) (Thermo Scientific) against a composite database containing the human Uniprot database appended with EBV and KSHV entries plus the common Repository of Adventitious Proteins database (52). All searches were carried out with cysteine carbamidomethylation as static modification, along with methionine oxidation and protein N-terminal acetylation as dynamic modifications. Spectral count-based label-free quantitation was performed in Scaffold (ver. 3.0) (Proteome Software). Peptides were confidently identified using a peptide false-discovery-rate (FDR) of 0.01 and a protein FDR of 0.01 with at least two unique peptides. Protein quantitation was performed by grouping B-cell lines based on infection status or the expression status of LMP1. Statistically significant differences in protein expression were reported by a two-tailed ANOVA test with a P value cutoff of 0.05. Data processing, functional categorization, and Kyoto Encyclopedia of Genes and Genomes pathway analysis were performed in Perseus (ver. 1.2.0.17) (53).
IPA software was used for predicting viral specific effects on exosome content and function and for calculating enrichment of differential gene lists to IPA-curated Diseases and Disorder, Biological Functions, Canonical Signaling Pathways, and Upstream Regulators categories. Differential gene lists with fold changes were generated using Scaffold Q+S (Proteome Software).
2D DIGE.
Exosomes were purified from the conditioned medium of cells grown in serum-free RPMI medium for 24 h by differential centrifugation as previously described. Proteins isolated from the concentrated exosome pellets of two biological and technical replicates were cleaned by methanol/chloroform precipitation and dissolved in lysis buffer [9 M urea, 2 M Thiourea, 20 mM Tris⋅HCl (pH 8.5), 2% (wt/vol) CHAPS, and 60 mM n-Octyl-β-d-glucopyranoside] (54). Aliquots (15 μg) of exosome samples from biological replicates of each B-cell line were labeled with 200 pmol of either Cy3 or Cy5 fluorescent dye, alternating Cy3 and Cy5 between replicates. An internal control was prepared by pooling equal amounts of protein (7.5 µg) from all samples which then was labeled with 200 pmol of Cy2 for every 15 µg of protein. The labeling reaction was carried out on ice for 30 min, protected from light. To quench the reaction, 1 µL of 10-mM lysine was added, and then the reaction was incubated for an additional 10 min on ice in the dark. After labeling, corresponding samples were combined so that each gel contained an internal control (Cy2) and two samples from distinctly infected cell lines (Cy3 and Cy5). An equal volume of 2× sample buffer [9 M urea, 2 M Thiourea, 2% (wt/vol) CHAPS, 60 mM n-Octyl-β-d-glucopyranoside, 30 mg/mL DTT, 2% (vol/vol) IPG buffer (pH 3–10)] was added, and the mixture was placed on ice for 15 min. Rehydration buffer [9 M urea, 2 M Thiourea, 2% (wt/vol) CHAPS, 60 mM n-Octyl-β-d-glucopyranoside, 15 mg/mL DTT, 2% (vol/vol) IPG buffer (pH 3–10)] was added to a final volume of 450 µL.
The resulting mix was loaded onto an immobilized pH gradient (IPG) strip (24 cm, pI range 3–10), and the strip was allowed to rehydrate overnight at room temperature. Isoelectric focusing and the subsequent SDS/PAGE, using precast 12.5% gels (Jule Biotechnologies Inc.), were performed as originally described (55). After SDS/PAGE, gels were scanned using a Typhoon Trio Plus scanner (GE Healthcare) and analyzed using DeCyder 7.0 software (GE Healthcare). Enhanced statistical power (up to an n of 12) was achieved by including biological and technical replicates and by grouping the samples based on infection status or LMP1 expression.
EM.
Aliquots of purified exosome samples were absorbed for 5 min directly onto glow-charged thin carbon foils on 400-mesh copper grids without fixation and were stained with 2% (wt/vol) uranyl acetate dissolved in water. The grids were examined in an FEI Tecnai 12 electron microscope at 80 kV. Images were captured on an Orius CCD Camera (Gatan) using Digital Micrograph software. Images for publication were arranged and contrast optimized using CorelDrawX5.
Immunoblot Analysis.
Cells were harvested by centrifugation at 500 × g for 5 min, washed in PBS, repelleted, and lysed with radioimmunoprecipitation assay buffer [20 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 0.1% SDS, 0.1% deoxycholic acid] supplemented with 0.5 mM phenylmethylsulfonyl fluoride, protease, and phosphatase inhibitor mixtures (Sigma) and 1 mM sodium orthovanadate for 30 min on ice. Insoluble material was removed by centrifugation for 5 min at 16,100 × g, and equal amounts of soluble protein were mixed with 5X Laemmli sample buffer and subjected to immunoblot analysis as previously described (56). Equal protein loading was confirmed by Ponceau S (Sigma) staining of blots before blocking. Similarly, lysates from purified exosomes were mixed with Laemmli sample buffer before SDS/PAGE and immunoblot analysis. Primary antibodies used include LANA (Advanced Biotechnologies), EBNA2 (PE2), LMP1 (CS1-4; Dako), LMP2A (Thermo), GAPDH (Santa Cruz), tumor susceptibility gene 101 (TSG101) (Santa Cruz), CD63, Flotillin 2 (Cell Signaling), and Ezrin (Cell Signaling).
Supplementary Material
Acknowledgments
We thank Brian Balgley at Bioproximity for performing the mass spectrometry data acquisition. This study was funded by National Institutes of Health Grants CA32979 (to N.R.-T.), CA096500 (to B.D.), and CA019014 (to N.R.-T, B.D., and J.D.G.). D.G.M. was supported by Training Grant T32CA009156 and American Cancer Society Fellowship PF-11-158-01-MPC. P.R.H. was supported by Training Grant T32CA009156.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Commentary on page 12503.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303906110/-/DCSupplemental.
References
- 1.Meckes DG, Jr, Raab-Traub N. Microvesicles and viral infection. J Virol. 2011;85(24):12844–12854. doi: 10.1128/JVI.05853-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raab-Traub N. Novel mechanisms of EBV-induced oncogenesis. Curr Opin Virol. 2012;2(4):453–458. doi: 10.1016/j.coviro.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raab-Traub N. 2007. EBV-induced oncogenesis. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis, eds Arvin A, et al. (Cambridge, NY)
- 4.Damania B. DNA tumor viruses and human cancer. Trends Microbiol. 2007;15(1):38–44. doi: 10.1016/j.tim.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Cesarman E, Knowles DM. The role of Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) in lymphoproliferative diseases. Semin Cancer Biol. 1999;9(3):165–174. doi: 10.1006/scbi.1998.0118. [DOI] [PubMed] [Google Scholar]
- 6.Meckes DG, Jr, et al. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci USA. 2010;107(47):20370–20375. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikeda M, Longnecker R. Cholesterol is critical for Epstein-Barr virus latent membrane protein 2A trafficking and protein stability. Virology. 2007;360(2):461–468. doi: 10.1016/j.virol.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pegtel DM, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA. 2010;107(14):6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raimondo F, Morosi L, Chinello C, Magni F, Pitto M. Advances in membranous vesicle and exosome proteomics improving biological understanding and biomarker discovery. Proteomics. 2011;11(4):709–720. doi: 10.1002/pmic.201000422. [DOI] [PubMed] [Google Scholar]
- 10.Choi D-S, Kim D-K, Kim Y-K, Gho YS. 2013. Proteomics, transcriptomics, and lipidomics of exosomes and ectosomes. Proteomics 13(11–12):1554–1571.
- 11.Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: Database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40(Database issue) D1:D1241–D1244. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rickinson A, Kieff E. Epstein-Barr Virus and Its Replication. 5th Ed. Philadelphia: Lippincott Williams & Wilkins; 2007. [Google Scholar]
- 13.Endo K, et al. Phosphorylated ezrin is associated with EBV latent membrane protein 1 in nasopharyngeal carcinoma and induces cell migration. Oncogene. 2009;28(14):1725–1735. doi: 10.1038/onc.2009.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bickel PE, et al. Flotillin and epidermal surface antigen define a new family of caveolae-associated integral membrane proteins. J Biol Chem. 1997;272(21):13793–13802. doi: 10.1074/jbc.272.21.13793. [DOI] [PubMed] [Google Scholar]
- 15.Morrow IC, et al. Flotillin-1/reggie-2 traffics to surface raft domains via a novel golgi-independent pathway. Identification of a novel membrane targeting domain and a role for palmitoylation. J Biol Chem. 2002;277(50):48834–48841. doi: 10.1074/jbc.M209082200. [DOI] [PubMed] [Google Scholar]
- 16.Meckes DG, Jr, Menaker NF, Raab-Traub N. Epstein-Barr virus LMP1 modulates lipid raft microdomains and the vimentin cytoskeleton for signal transduction and transformation. J Virol. 2013;87(3):1301–1311. doi: 10.1128/JVI.02519-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ardila-Osorio H, et al. TRAF interactions with raft-like buoyant complexes, better than TRAF rates of degradation, differentiate signaling by CD40 and EBV latent membrane protein 1. Int J Cancer. 2005;113(2):267–275. doi: 10.1002/ijc.20503. [DOI] [PubMed] [Google Scholar]
- 18.de Gassart A, Géminard C, Février B, Raposo G, Vidal M. Lipid raft-associated protein sorting in exosomes. Blood. 2003;102(13):4336–4344. doi: 10.1182/blood-2003-03-0871. [DOI] [PubMed] [Google Scholar]
- 19.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 20.Minden J. DIGE: Past and future. Difference gel electrophoresis (DIGE) Methods Mol Biol. 2012;854:3–8. doi: 10.1007/978-1-61779-573-2_1. [DOI] [PubMed] [Google Scholar]
- 21.Neilson KA, et al. Less label, more free: Approaches in label-free quantitative mass spectrometry. Proteomics. 2011;11(4):535–553. doi: 10.1002/pmic.201000553. [DOI] [PubMed] [Google Scholar]
- 22.Rowe M, et al. Restoration of endogenous antigen processing in Burkitt’s lymphoma cells by Epstein-Barr virus latent membrane protein-1: Coordinate up-regulation of peptide transporters and HLA-class I antigen expression. Eur J Immunol. 1995;25(5):1374–1384. doi: 10.1002/eji.1830250536. [DOI] [PubMed] [Google Scholar]
- 23.Murray PG, Constandinou CM, Crocker J, Young LS, Ambinder RF. Analysis of major histocompatibility complex class I, TAP expression, and LMP2 epitope sequence in Epstein-Barr virus-positive Hodgkin’s disease. Blood. 1998;92(7):2477–2483. [PubMed] [Google Scholar]
- 24.Mullen MM, Haan KM, Longnecker R, Jardetzky TS. Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Mol Cell. 2002;9(2):375–385. doi: 10.1016/s1097-2765(02)00465-3. [DOI] [PubMed] [Google Scholar]
- 25.Shkoda A, et al. The germinal center kinase TNIK is required for canonical NF-κB and JNK signaling in B-cells by the EBV oncoprotein LMP1 and the CD40 receptor. PLoS Biol. 2012;10(8):e1001376. doi: 10.1371/journal.pbio.1001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein C, Busson P, Tursz T, Young LS, Raab-Traub N. Expression of the c-fgr related transcripts in Epstein-Barr virus-associated malignancies. Int J Cancer. 1988;42(1):29–35. doi: 10.1002/ijc.2910420107. [DOI] [PubMed] [Google Scholar]
- 27.Wang F, et al. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J Virol. 1990;64(5):2309–2318. doi: 10.1128/jvi.64.5.2309-2318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keryer-Bibens C, et al. Exosomes released by EBV-infected nasopharyngeal carcinoma cells convey the viral latent membrane protein 1 and the immunomodulatory protein galectin 9. BMC Cancer. 2006;6(1):283. doi: 10.1186/1471-2407-6-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klibi J, et al. Blood diffusion and Th1-suppressive effects of galectin-9-containing exosomes released by Epstein-Barr virus-infected nasopharyngeal carcinoma cells. Blood. 2009;113(9):1957–1966. doi: 10.1182/blood-2008-02-142596. [DOI] [PubMed] [Google Scholar]
- 30.Dukers DF, et al. Direct immunosuppressive effects of EBV-encoded latent membrane protein 1. J Immunol. 2000;165(2):663–670. doi: 10.4049/jimmunol.165.2.663. [DOI] [PubMed] [Google Scholar]
- 31.Simpson RJ, Jensen SS, Lim JWE. Proteomic profiling of exosomes: Current perspectives. Proteomics. 2008;8(19):4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 32.Bhatt AP, et al. Dysregulation of fatty acid synthesis and glycolysis in non-Hodgkin lymphoma. Proc Natl Acad Sci USA. 2012;109(29):11818–11823. doi: 10.1073/pnas.1205995109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delgado T, Sanchez EL, Camarda R, Lagunoff M. Global metabolic profiling of infection by an oncogenic virus: KSHV induces and requires lipogenesis for survival of latent infection. PLoS Pathog. 2012;8(8):e1002866. doi: 10.1371/journal.ppat.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delgado T, et al. Induction of the Warburg effect by Kaposi’s sarcoma herpesvirus is required for the maintenance of latently infected endothelial cells. Proc Natl Acad Sci USA. 2010;107(23):10696–10701. doi: 10.1073/pnas.1004882107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Damania B. Kaposi’s sarcoma-associated herpesvirus confers a survival advantage to endothelial cells. Cancer Res. 2008;68(12):4640–4648. doi: 10.1158/0008-5472.CAN-07-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L, Dittmer DP, Tomlinson CC, Fakhari FD, Damania B. Immortalization of primary endothelial cells by the K1 protein of Kaposi’s sarcoma-associated herpesvirus. Cancer Res. 2006;66(7):3658–3666. doi: 10.1158/0008-5472.CAN-05-3680. [DOI] [PubMed] [Google Scholar]
- 37.Sodhi A, et al. The TSC2/mTOR pathway drives endothelial cell transformation induced by the Kaposi’s sarcoma-associated herpesvirus G protein-coupled receptor. Cancer Cell. 2006;10(2):133–143. doi: 10.1016/j.ccr.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 38.Sin S-H, et al. Rapamycin is efficacious against primary effusion lymphoma (PEL) cell lines in vivo by inhibiting autocrine signaling. Blood. 2007;109(5):2165–2173. doi: 10.1182/blood-2006-06-028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhatt AP, et al. Dual inhibition of PI3K and mTOR inhibits autocrine and paracrine proliferative loops in PI3K/Akt/mTOR-addicted lymphomas. Blood. 2010;115(22):4455–4463. doi: 10.1182/blood-2009-10-251082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy D, Dittmer DP. Phosphatase and tensin homolog on chromosome 10 is phosphorylated in primary effusion lymphoma and Kaposi’s sarcoma. Am J Pathol. 2011;179(4):2108–2119. doi: 10.1016/j.ajpath.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 42.Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84(6):1014–1020. doi: 10.1016/j.ygeno.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 43.Kondoh H, et al. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65(1):177–185. [PubMed] [Google Scholar]
- 44.Cai X, et al. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2(3):e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dawson CW, Tramountanis G, Eliopoulos AG, Young LS. Epstein-Barr virus latent membrane protein 1 (LMP1) activates the phosphatidylinositol 3-kinase/Akt pathway to promote cell survival and induce actin filament remodeling. J Biol Chem. 2003;278(6):3694–3704. doi: 10.1074/jbc.M209840200. [DOI] [PubMed] [Google Scholar]
- 46.Hatzivassiliou EG, Kieff E, Mosialos G. Constitutive CD40 signaling phenocopies the transforming function of the Epstein-Barr virus oncoprotein LMP1 in vitro. Leuk Res. 2007;31(3):315–320. doi: 10.1016/j.leukres.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 47.Mosialos G, et al. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80(3):389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 48.Nador RG, Cesarman E, Knowles DM, Said JW. Herpes-like DNA sequences in a body-cavity-based lymphoma in an HIV-negative patient. N Engl J Med. 1995;333(14):943. doi: 10.1056/NEJM199510053331417. [DOI] [PubMed] [Google Scholar]
- 49.Renne R, et al. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2(3):342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 50.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 51.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75(3):663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 52.Cox J, et al. Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 53.Cox J, Mann M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu Rev Biochem. 2011;80(1):273–299. doi: 10.1146/annurev-biochem-061308-093216. [DOI] [PubMed] [Google Scholar]
- 54.Kim K-B, Lee J-S, Ko Y-G. The Isolation of Detergent-Resistant Lipid Rafts for Two-Dimensional Electrophoresis. 2008;424:413–422. doi: 10.1007/978-1-60327-064-9_32. [DOI] [PubMed] [Google Scholar]
- 55.Unlü M, Morgan ME, Minden JS. Difference gel electrophoresis: A single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18(11):2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- 56.Kung C-P, Meckes DG, Jr, Raab-Traub N. Epstein-Barr virus LMP1 activates EGFR, STAT3, and ERK through effects on PKCdelta. J Virol. 2011;85(9):4399–4408. doi: 10.1128/JVI.01703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.