Abstract
Viral infections are common causes of fever without an apparent source in young children. Despite absence of bacterial infection, many febrile children are treated with antibiotics. Virus and bacteria interact with different pattern recognition receptors in circulating blood leukocytes, triggering specific host transcriptional programs mediating immune response. Therefore, unique transcriptional signatures may be defined that discriminate viral from bacterial causes of fever without an apparent source. Gene expression microarray analyses were conducted on blood samples from 30 febrile children positive for adenovirus, human herpesvirus 6, or enterovirus infection or with acute bacterial infection and 22 afebrile controls. Blood leukocyte transcriptional profiles clearly distinguished virus-positive febrile children from both virus-negative afebrile controls and afebrile children with the same viruses present in the febrile children. Virus-specific gene expression profiles could be defined. The IFN signaling pathway was uniquely activated in febrile children with viral infection, whereas the integrin signaling pathway was uniquely activated in children with bacterial infection. Transcriptional profiles classified febrile children with viral or bacterial infection with better accuracy than white blood cell count in the blood. Similarly accurate classification was shown with data from an independent study using different microarray platforms. Our results support the paradigm of using host response to define the etiology of childhood infections. This approach could be an important supplement to highly sensitive tests that detect the presence of a possible pathogen but do not address its pathogenic role in the patient being evaluated.
Analysis of the transcriptional profile of the host may provide an indirect approach to pathogen detection and disease assessment that can supplement direct approaches, such as culture or nucleic acid amplification (1). This analysis may be especially useful when a pathogen is not detected or the pathogenic role of a detected microbial agent is in question. Circulating blood leukocytes react to pathogens by recognizing pathogen-specific molecular patterns through pattern recognition receptors leading to up- or down-regulation of the expression of host genes associated with immune functions (2, 3), with differential activation of host transcriptional programs with different pathogens (4). Thus, host blood transcriptional profiles and representative biomarkers may be powerful tools for categorizing infection (5).
In previous studies, host transcriptional analysis has been applied successfully to distinguish acute influenza A infection from specific bacterial infections (6) and Kawasaki disease from adenovirus infections (7), characterize acute invasive Staphylococcus aureus infections (8), distinguish active tuberculosis from other infectious and inflammatory diseases (9), and distinguish septicemic melioidosis from sepsis from other causes (10). In addition, specific blood transcriptional signatures were defined in adult volunteers challenged with specific respiratory viruses (11). Analysis of host transcriptional profiles has also been applied to the diagnosis of inflammatory and hematological diseases (12–15), including the identification of potential biomarkers for specific disease entities (5).
In recent studies, we have been able to confirm infection with specific viruses in most children with fever without a source (FWS) (16). We hypothesized that these children would have distinctive blood leukocyte transcriptional profiles associated with specific viral infection and that it would also be possible to use blood transcriptional profiles to distinguish children with viral infection from children with bacterial infection. We also hypothesized that symptomatic and asymptomatic children infected with the same virus would have distinctively different leukocyte transcriptional profiles. To test these hypotheses, we analyzed the blood leukocyte transcriptional profiles of febrile children with systemic DNA and RNA viral infections compared with those profiles of afebrile children positive for the same viruses, febrile children with acute bacterial infection, and virus-negative afebrile children.
Results
Transcriptional Profiles of Virus-Positive Febrile Children and Febrile Children with Acute Bacterial Infection Differed from Profiles of Afebrile Virus-Positive and -Negative Children.
We analyzed microarray data covering the whole human transcriptome from Illumina Human HT12 BeadChips consisting of 47,300 probes hybridized with RNA samples extracted from whole-blood specimens from 30 febrile children [8 children positive for human herpesvirus 6 (HHV-6), 8 children positive for adenovirus, 6 children positive for enterovirus, and 8 children with acute bacterial infection]. The same microarray assay was performed on blood samples from 35 afebrile children (2 children positive for HHV-6, 3 children positive for adenovirus, 8 children positive for rhinovirus, and 22 virus-negative control children).
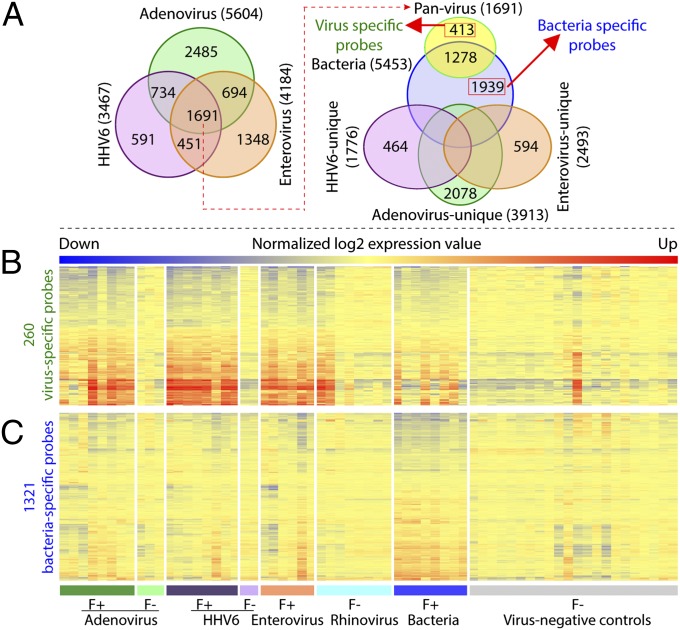
By using a strategy based on intersecting various probe sets as described previously (17), we identified sets of probes that were significantly up- or down-regulated for each of the virus-positive groups and the febrile acute bacterial infection group compared with afebrile virus-negative control children (Fig. 1A). Using this method, we identified 260 probes with significant up- or down-regulation specifically in virus-positive febrile children and 1,321 probes with significant up- or down-regulation specifically in children with febrile acute bacterial infection. Analysis of 260 viral probes revealed substantial overlap in gene expression profiles for febrile children who were positive for adenovirus, HHV-6, or enterovirus infection, all of which were very different from the profiles of most afebrile children (Fig. 1B). Profiles of virus-positive and -negative afebrile children were indistinguishable. Analysis using 1,321 bacterial probes showed similar patterns of gene expression for most of the children with fever and acute bacterial infection that differed from those patterns of the other groups, with a few exceptions (Fig. 1C).
Fig. 1.
Identification of virus- and bacteria-specific probes for distinguishing virus-positive febrile children and febrile children with acute bacterial infection from virus-negative afebrile children. (A) Venn diagram showing identification of virus- and bacteria-specific probes. Sets of probes differentially expressed in febrile children positive for one or more of three viruses compared with virus-negative afebrile control children were intersected, and 1,691 pan-virus probes were identified and intersected with the sets of probes that were differentially expressed in children with febrile acute bacterial infection compared with virus-negative afebrile control children. From this analysis, 413 virus- and 1,939 bacteria-specific probes were identified, and probes specific for individual viruses were also identified. (B and C) Heatmaps showing gene expression based on (B) 260 virus- and (C) 1,321 bacteria-specific probes in children with febrile and afebrile viral and bacterial infections and afebrile virus-negative control children. These probes were selected in the same manner as 413 virus- and 1,939 bacteria-specific probes described above, except that, for this selection, we excluded 2 of 22 virus-negative controls, because they had expression profiles very similar to those profiles with viral/bacterial infections. We also excluded any probes that were not annotated in GenBank Build36 (National Center for Biotechnology Information). F+, febrile, F−, afebrile. Each row represents a gene with expression value that is normalized to the mean of the afebrile virus-negative control group, and each column represents one individual. Red represents up-regulation, and blue represents down-regulation.
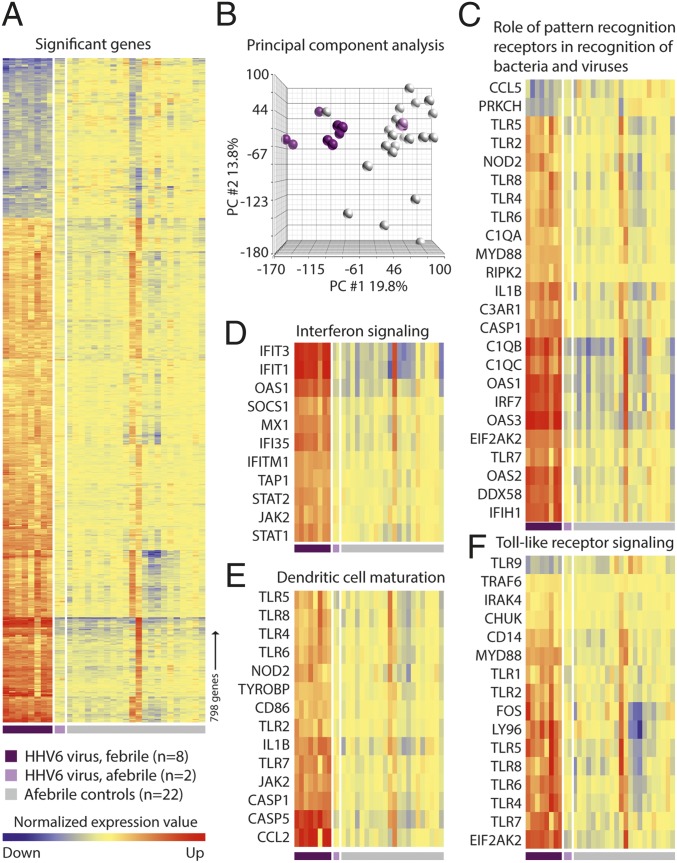
The extensive differences in gene expression profiles between febrile and afebrile children were further analyzed for each virus and acute bacterial infection by principal component analysis and analysis of genes grouped into Ingenuity canonical pathways (www.ingenuity.com). Results for HHV-6 are shown in Fig. 2, and results for adenovirus, enterovirus, and acute bacterial infection are available in Figs. S1, S2, and S3. Pathways with the most significant transcriptional changes for children with each of the three viral infections and acute bacterial infection are shown in Fig. S4.
Fig. 2.
Blood leukocyte transcriptional profiles of febrile and afebrile HHV-6–positive children compared with those profiles of afebrile virus-negative children. Microarray analysis was conducted on RNA extracted from blood samples of 10 children positive for HHV-6 (8 febrile and 2 afebrile children) and 22 afebrile virus-negative children. (A) Hierarchical clustering of all probe sets with a statistically significant more than twofold difference between HHV-6–positive febrile and afebrile virus-negative control children (FDR at 5%). (B) Principal component analysis of differentially expressed genes, with each oval representing one child. (C–F) Hierarchical clustering of differentially expressed genes in A according to their expression intensity in four Ingenuity canonical pathways of particular interest, which were among the most strongly activated pathways in HHV-6–positive febrile children. Each row represents a gene with expression value that is normalized to the mean of the afebrile virus-negative control group. Gene names are listed to the left. Each column represents one individual. Red represents up-regulation, and blue represents down-regulation.
HHV-6.
Comparison of individual probes between HHV-6–positive febrile children and virus-negative afebrile children yielded 3,467 probes with significant transcriptional changes, including 798 probes with twofold or greater changes (606 up- and 192 down-regulated) (Fig. 2A). Principal component analysis of the transcriptional profiles revealed clear differences between the febrile and afebrile HHV-6–positive children (Fig. 2B). The sample from the one child in the virus-negative afebrile control group with a transcriptional pattern in the heatmaps (shown in Fig. 1 B and C) that was similar to the patterns of those children with febrile infection was classified with febrile infections in the principal component analysis. The most up-regulated gene was interferon alpha-inducible protein 27 (IFI27)/interferon-stimulated gene 12A (ISG12A), which mediates IFN-induced apoptosis by the release of cytochrome C from the mitochondria and activation of BAX and caspases (18). Analysis of transcriptional pathways showed a number of pathways with up-regulation of many component genes (Fig. 2 C–F).
Adenovirus.
The gene expression profile of adenovirus-positive febrile children (Fig. S1) showed major overlap with the profile of HHV-6–positive febrile children. Statistical comparison of transcriptional profiles between adenovirus-positive febrile children and virus-negative afebrile children showed 5,604 probes with significant transcriptional changes, including 847 probes with a twofold or greater increase (576) or decrease (271) in expression level. Principal component analysis revealed the clear differences between the febrile and afebrile adenovirus-positive children. As was also true for HHV-6, the most up-regulated gene was IFI27/ISG12A. The pathways with the most significant transcriptional changes were also similar to those pathways for HHV-6 infection (Fig. S4).
Enterovirus.
Comparison of transcriptional profiles of enterovirus-positive febrile children and virus-negative afebrile control children yielded 4,184 probes with significant changes (Fig. S2). The magnitude of these transcriptional changes was generally less than the magnitude for adenovirus and HHV-6, and therefore, false discovery rate (FDR) was set at 20% to maximize the possibility of detecting either up- or down-regulated genes. This procedure yielded 678 probes with twofold or greater transcriptional change (559 up- and 119 down-regulated). As in HHV-6 and adenovirus infections, IFI27/ISG12A was the most up-regulated gene. The pathways with the most significant transcriptional changes in febrile enterovirus-positive children were similar to those pathways for HHV-6– and adenovirus-positive children (Fig. S4).
Bacteria.
Statistical comparison of microarray data between febrile children with acute bacterial infection and virus-negative afebrile control children yielded 1,234 probes with twofold or greater change with either up- (850) or down-regulation (384) (Fig. S3). We did not detect probes with significant differential expression between Gram-positive and -negative bacterial infections. The most up-regulated gene was Annexin A3 (14.6-fold), which regulates calcium-dependent neutrophil secretion (19), suggesting a defense mechanism against bacteria.
Transcriptional Pathways Were Differentially Activated in Febrile Children with Viral and Bacterial Infections.
A number of Ingenuity canonical pathways had significant transcriptional changes in febrile children positive for one of three viruses (HHV-6, adenovirus, and enterovirus) or with acute bacterial infection. Pathways that were activated in each of four infection groups were role of pattern recognition receptors in recognition of bacteria and viruses, TREM1 signaling, and toll-like receptor signaling. A notable number of genes from the natural killer cell signaling pathway was down-regulated in each of four infection groups. The IFN signaling pathway and the activation of IFN regulatory factors by cytosolic pattern recognition receptors pathway were more activated in febrile virus-positive children compared with febrile children with acute bacterial infection. In contrast, genes in the integrin signaling pathway were activated only in bacterial infection. Transcriptional changes in each of these pathways are displayed in Fig. S4.
Unique Sets of Genes Were Associated with Specific Viral and Bacterial Infections in Febrile Children.
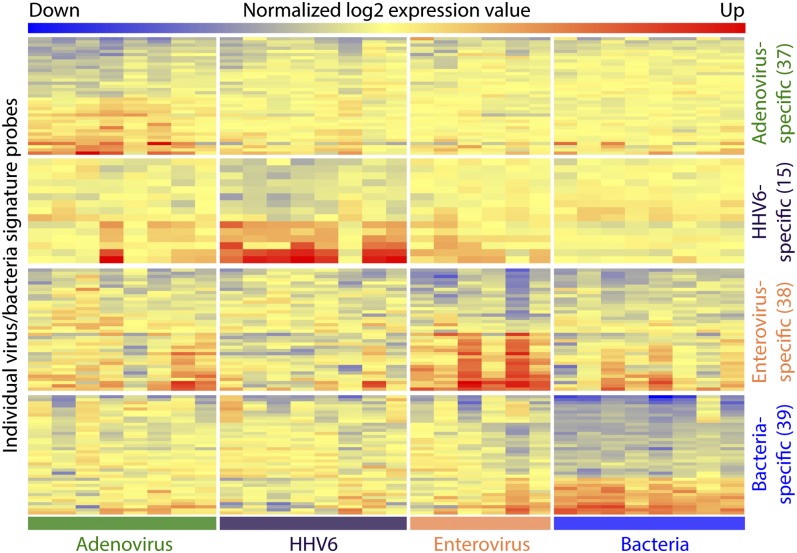
Although signaling pathways tended to be similarly activated among the different viruses tested in this study, there were significant variations in the expression level of many individual genes. We identified 2,078 probes with significant transcriptional changes uniquely present in adenovirus-positive febrile children, 464 probes uniquely present in HHV-6–positive febrile children, 594 probes uniquely present in enterovirus-positive febrile children, and 1,939 probes uniquely present in febrile children with acute bacterial infection (Fig. 1A). Using the shrunken centroid algorithm (20), we identified the most informative subsets of these specific gene probes for each of the individual viruses and acute bacterial infection (Table S1). These virus-specific transcriptional profiles and the profile specific for acute bacterial infection are shown in Fig. 3.
Fig. 3.
Probes specific for individual viruses and bacteria. Virus- and bacteria-specific probes were subjected to the shrunken centroid algorithm individually for each of four pathogen groups to find the minimum number of probes with the greatest ability to differentiate among pathogen groups. Each row represents a probe, and each column displays probes for one febrile child positive for the indicated virus or with acute bacterial infection.
Classifier Probes Were Identified to Distinguish Viral and Bacterial Infections in Febrile Children with Validation on Independent Datasets.
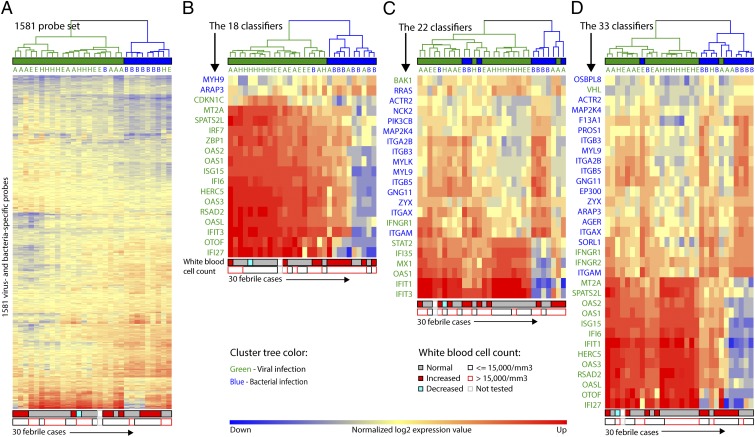
Several of our findings related to pathways and individual genes suggested that transcriptional profiles unique to either viral or bacterial infection could be characterized to assist in making this clinically important discrimination. We compared individual gene- and pathway-based approaches for selecting probes. For the gene-based approach, we used a master set consisting of 1,581 (260 viral- and 1,321 bacterial-specific) probes described above, and a limited subset of 18 of 1,581 was selected using the shrunken centroid algorithm as the most efficient classifiers. For the pathway-based approach, we used the shrunken centroid algorithm to select 22 probes from the IFN signaling pathway (selectively activated in virus-positive febrile children) and the integrin signaling pathway (selectively activated in febrile children with acute bacterial infection). The hybrid approach used 33 probes selected using the shrunken centroid algorithm from the master set and the IFN signaling and integrin signaling pathways. We selected nine key classifiers from three sets of classifiers described above and validated them by quantitative RT-PCR (RT-qPCR). High correlation in expression level was found for all nine classifier genes between RT-qPCR and microarray results (Fig. S5 and Table S2). Classification of cases was carried out using unsupervised hierarchical clustering and the K-nearest neighbor algorithm. The true class of each case was based on virus-specific PCRs and bacterial cultures as previously described (16). The classifications derived from the use of probes selected by each approach are shown in Fig. 4, the signal intensity of the probes is shown in Fig. S5, and classification performance of each set is summarized in Table 1. Correct classification based on hierarchical clustering ranged from 77% to 90% and from 83% to 90% based on the K-nearest neighbor method.
Fig. 4.
A limited number of classifier probes discriminate febrile children positive for viruses from febrile children with acute bacterial infections: (A) 1,581 gene-based classifiers, (B) 18 gene-based classifiers, (C) 22 pathway-based classifiers, and (D) 33 classifiers selected from gene- and pathway-based classifier sets. Patients are shown as columns, and probes are shown as rows. Gene symbols are shown in blue for bacterial infection-specific genes and green for viral infection-specific genes. Expression values presented in the heatmap were normalized to the mean of the afebrile virus-negative control cases. Hierarchical clustering was used to classify patients into two groups, with the majority of cases classified as either viral (green tree branch) or bacterial (blue tree branch). Classification as predicted using the K-nearest neighbor algorithm is shown as a bar above each heatmap, with green showing classification as viral and blue showing classification as bacterial. True class was determined by virus-specific PCR and bacterial cultures, and it is designated by green (viral) or blue (bacterial) letters: A, adenovirus; B, bacteria; E, enterovirus; H, HHV-6. Classification based on patients’ white blood cell count is shown beneath each heatmap. The upper strip shows classification based on age-specific normal values, and the lower strip shows classification based on a cutoff of 15,000 cells/mm3.
Table 1.
Prediction accuracy of four sets of classifier probes
| Dataset | Microarray | N* | 1,581 virus- and bacteria-specific probes without selection† | 18 classifiers from 1,581 virus- and bacteria-specific probes† | 22 classifiers from Ingenuity IFN and Integrin pathway genes‡ | 33 classifiers selected using a combined gene- and pathway-based approach†‡ |
| Present study | Human-HT12 | 30 | 27 (90%)§ | 26 (87%) | 27 (90%) | 25 (83%) |
| Ramilo et al. (6) | U133Plus2 | 22 | 21 (95%) | 21 (95%) | 19 (86%) | 18 (82%) |
| Ramilo et al. (6) | Human-WG6 | 24 | 23 (96%) | 22 (92%) | 18 (75%) | 21 (88%) |
| Ramilo et al. (6) | U133A | 91 | 86 (95%) | 82 (90%) | 81 (89%) | 85 (93%) |
| Ramilo et al. (6) and present study | All three sets | 137 | 130 (95%) | 123 (90%) | 121 (88%) | 124 (91%) |
We used independent datasets from the literature (6) to test the clinical validity and robustness of our classifier probes for distinguishing viral and bacterial infection. The validation data included three different cohorts analyzed using three different microarray platforms, each of which differed from our platform. We achieved 95% accuracy in distinguishing viral and bacterial infection using 1,581 probes and 88–91% accuracy in using the other three sets of probes (Table 1 and Fig. S6).
Transcriptional Profile Was Superior to Traditional White Blood Cell Count for Discriminating Bacterial from Viral Infection.
Previous studies indicated that white blood cell count is an imperfect tool for distinguishing between viral and bacterial infection, a distinction often used to determine whether to treat the patient with antibiotics (21, 22). We compared classification based on transcriptional profiles with classification based on white blood cell count using a cutoff of 15,000/mm3 as recommended by the American Academy of Pediatrics in their guideline for the management of febrile young children 0–36 mo of age (23). We also analyzed a different set of cutoffs based on age-specific normal values for white blood cell count used by the clinical laboratory at St. Louis Children's Hospital (Materials and Methods). The classifier gene probes were more accurate for distinguishing bacterial and viral infections than either of the white blood cell criteria (Fig. 4).
To explore the association between gene expression level and total white blood cell count and counts of specific leukocyte types, we performed a Pearson correlation test for 4,716 probes that were significantly different from virus-negative controls in 30 febrile children. Overall, the total white blood cell count was not associated with gene expression level, but expression of several clusters of genes was significantly associated with neutrophil, lymphocyte, or monocyte counts, suggesting that those clusters of genes might be activated in specific types of cells (Fig. S7). Of note, only a few of the classifier probes selected by the various approaches described above fell into neutrophil or lymphocyte clusters with borderline significance (P = 0.043–0.049).
Discussion
Our study on groups of young children presenting with FWS addressed three questions: (i) could we use host transcriptional profiles to distinguish symptomatic from asymptomatic viral infection, (ii) could we characterize virus-specific transcriptional profiles for two important DNA viruses and one RNA virus that cause systemic infection in young children, and (iii) could we discover viral- and bacterial-specific transcriptional profiles that distinguish between infections caused by these large groups of pathogens.
We detected transcriptional changes in multiple genes in multiple pathways in febrile children who were infected with DNA viruses, an RNA virus, or bacteria, with substantial overlap in the specific activated pathways and genes. Interestingly, the transcriptional profile of febrile children positive for HHV-6 or adenovirus was dramatically different from the profile of afebrile children positive for the same viruses, which was indistinguishable from the profile of virus-negative afebrile children. This finding has potential practical importance, because the application of sensitive molecular viral detection tests to clinical medicine may detect asymptomatic as well as symptomatic infection (16) and thus, has created a need to determine the clinical significance of the detection of viral nucleic acid in an individual patient. In an effort to detect virus-specific transcriptional profiles, we analyzed host response at the level of up- and down-regulation of individual genes and functional gene pathways. Despite very substantial overlap in transcriptional profiles in febrile children positive for any of the three viruses and with acute bacterial infection, we were able to detect differences in up- and down-regulation of individual genes. Additional studies are needed to validate these virus-specific profiles. In contrast, because of overlap in patterns of pathway activation, pathway analysis was not useful for distinguishing among febrile children positive for each of the three viruses that were represented in the study population.
Other studies have attempted to use host transcriptional signatures to distinguish viral from bacterial infection (6, 11). In the present study, we used several approaches to developing a panel of probes that could make this important distinction. First, we developed a panel of individual gene probes selected simply on the strength of their statistical association with type of infection. Second, we drew selectively on genes from two pathways that were differentially activated: the IFN signaling pathway, activated in febrile virus-positive children, and the integrin signaling pathway, activated in children with acute bacterial infection. Third, we used a hybrid approach, in which we selected genes from each of the gene- and pathway-based approaches. Overall, the best classification was achieved with the large set of gene-based probes (1,581) selected on the basis of statistical association with type of infection. However, each of the three shrunken approaches that used between 18 and 33 probes functioned almost as well.
Because we did not have a sufficient number of subjects in our study to have both a training set and a validation set, we validated the performance of our probe sets using three previously published microarray datasets (6). This approach was complicated by the difficulty of comparing across different microarray platforms. Despite this limitation, our probes were still able to achieve classifications that were 88–95% concordant with the classifications of the validation sets. It is also important to recognize that the etiology of infection in subjects in the validation sets was not as rigorously defined as in our study, and it is possible that some of the subjects in those studies may have been misclassified.
Our study has shown that host blood transcriptional signatures associated with broad categories of infectious etiology can be defined in young children with FWS and are of superior predictive value than current white blood cell count-based criteria in discriminating febrile children with viral from bacterial infection. Presently, the use of gene expression microarray as a practice for bedside decision making would not be practical. However, individual predictive biomarkers may be of more clinical value. Studies to evaluate candidate biomarkers derived from our dataset are underway.
Our study has a number of limitations. Most importantly, the number of children positive only for each virus under study was limited, reflecting the difficulty of recruiting children into experimental studies and also, the fact that a large number of febrile children in our original study was infected with more than one virus, thus disqualifying the children from the present study. Also, samples were obtained at different times and were not always from the same time period with respect to the onset of the illness. However, the children were all highly febrile at the time that the sample was obtained, and therefore, we are confident that most children were in the acute phase of their infection. Another concern is that cases and controls were not matched according to demographic characteristics, especially race. However, our statistical analysis did not reveal race as an important confounding variable. It is possible that some study subjects were infected with other agents for which we did not test. Lastly, the RNA that was analyzed was extracted from whole-blood samples, thus representing a pool of a variety of leukocyte populations. It is possible that differences among study groups might reflect differences in the distribution of leukocytes in the peripheral blood. However, our statistical analysis of possible correlations between gene expression and leukocyte composition of the sample did not support this concern. It is also possible that differences among groups would be sharpened by extracting RNA from specific leukocyte populations. Despite these considerations, the identity of many virus-specific classifier genes was similar to a previous study on respiratory viral infections (11). Finally, as is true for many gene expression microarray studies, our study was challenged by dimensionality, because the number of parameters measured vastly exceeded the number of study subjects (15). We used statistical corrections for multiple comparisons. Nevertheless, additional studies to replicate our findings are required.
Studies on host blood transcriptional profiles can be considered as a paradigm shift, providing clues about infectious pathogens through interrogation of host gene expression patterns (1). Host transcriptional analysis may prove to be a useful test method, supplementing sensitive pathogen-based nucleic acid amplification assays and also providing clues about etiology when no pathogens are confirmed from the direct detection of microbial pathogens.
Materials and Methods
Subjects.
Subjects were drawn from a study of children between 2 and 36 mo of age with FWS (Table S3) and afebrile children having ambulatory surgery who were recruited at St. Louis Children’s Hospital as described previously (16). The febrile and afebrile groups were similar with respect to age, sex, and season of recruitment, but they differed with respect to race, with more African-American children in the febrile group (57% vs. 13%). Patients were enrolled according to Institutional Review Board-approved protocol. Informed consent was obtained from parents or guardians of all patients. The study was approved by the Washington University Human Research Protection Office. Each subject was tested for multiple viruses in blood and nasopharyngeal samples using panels of virus-specific PCR assays as described (16). Subjects were selected for the study of gene expression profiles if they were positive for only a single virus in one or both samples. The viruses included were adenovirus, HHV-6, enterovirus, and rhinovirus. Also included were subjects who had a definite bacterial infection (bacteremia, urinary tract infection, skin and soft tissue infection, or bone or joint infection) as well as a group of subjects selected from the afebrile control group whose samples were also negative for all viruses tested (16).
Specimens.
In addition to whole-blood and nasopharyngeal samples for virus-specific PCR and high-throughput sequencing, a blood sample was collected in a Tempus Blood RNA Tube (Applied Biosystems) and stored at −80 °C for subsequent gene expression analysis.
RNA Preparation.
Total RNA was isolated from whole blood collected in Tempus Blood RNA Tubes (Applied Biosystems) according to the manufacturer’s instructions. RNA quality was determined by electropherogram (showing 28S, 18S, and 5S bands) and RNA integrity number (RIN; generally, a >7 RIN indicates good quality RNA) using an Agilent 2100 Bioanalyzer (Agilent). All but three of the RNA preparations had RIN scores ≥7.0. Total RNA concentration was obtained from an absorbance ratio at 260 and 280 nm using a NanoDrop ND-100 spectrometry instrument (NanoDrop Inc.).
Gene Expression Microarrays.
The microarray assays were carried out at the Genome Technology Access Center in Washington University in St. Louis (https://gtac.wustl.edu). Briefly, RNA transcripts were amplified by T7 linear amplification with the Illumina 3′IVT Direct Hybridization Assay Kit (Illumina Inc.), and biotin-labeled cRNA targets were hybridized to the Illumina Human-HT12 v4 Expression BeadChips (>47,000 probes), which were scanned on an Illumina BeadArray Reader. Scanned images were quantitated by Illumina Beadscan software (version 3). Quantitated data were imported into Illumina GenomeStudio software (version 2011.1) to generate expression profiles and make data quality assessment. These data have been deposited into Gene Expression Omnibus database at the National Center for Biotechnology Information (accession no. GSE40396).
Microarray Data Analysis.
Expression profiles generated in the GenomeStudio were imported into Partek Genome Suite (version 6.6; Partek Inc.), and data quality was assessed across individual samples using principal component analysis and hierarchical clustering analysis that could identify any specific sample clusters that were associated with nonexperimental factors (such as chip effect, age, sex, and array quality control metrics); 5 of 70 samples that had the lowest number of detectable probes were identified as outliers and eliminated without additional analysis. A total of ∼26,300 probes that had been detected (detection P value < 0.01) in at least 1 of 70 samples was kept in downstream statistical analysis and quantile-normalized for differential expression analysis.
Differential Expression Analysis.
ANOVA was performed in Partek Genomics Suite in order to derive genes with differential expression in viral and bacterial infection and afebrile controls, and to account for variance from hybridization date and individual chips. P values from the ANOVA were corrected for FDR q value. With the exception of symptomatic enterovirus infection as stated in Results, all analysis was conducted with P value < 0.05 and FDR at 5%. Partek was also used to generate all heatmaps and principal component analysis plots.
Pathway Analysis.
Pathways that were most activated for each virus and bacterial infection were identified from the Ingenuity pathway analysis (Ingenuity Systems) library of canonical pathways. The significance of the association between the dataset and the canonical pathway was assessed in two ways: (i) the ratio of the number of up- and down-regulated probes from the dataset included in the pathway divided by the total number of probes that included in the canonical pathway and (ii) statistical evaluation using Fisher exact test of the probability that the association between the genes in the dataset and the canonical pathway is explained by chance alone.
Identification of Classifier Genes, Class Prediction, and Unsupervised Hierarchical Clustering.
We used the K-nearest neighbor classification algorithm embedded in the Prediction Analysis of Microarrays tool to identify classifier genes presenting the highest capability to discriminate the two classes of bacterial and viral infection (20). With 10 nearest neighbors and 10-fold cross-validation, Prediction Analysis of Microarrays calculates misclassification error rate in the training set of data for each of two classes according to varying thresholds (a unique statistical parameter). A threshold is chosen when the misclassification error is minimized for both classes to define a subset of probes from the entire training dataset designated as classifier probes. These classifier probes were used for class prediction on all testing datasets. We also used hierarchical clustering with the complete linkage algorithm to evaluate the accuracy of classification.
RT-qPCR Validation Assays.
Primers and probes were purchased from Life Technologies (Applied Biosystems), and master mix was from Quanta Biosciences (Gaithersburg, MD) for RT-qPCR assays. The assays were carried out in triplicate on an ABI 7500 real-time PCR instrument following the manufacturer’s protocols. All assays had >80% PCR efficiency and <15% coefficient of variance in triplicate reactions.
Correlation Between Gene Expression Profile and White Blood Cell Counts and Differentials.
The Pearson test was used to find significant correlations of differentially expressed genes with white blood cell and differential counts in 30 febrile cases. The differential expression was defined with P < 0.05 and fold change >1.5 in comparisons between febrile groups and healthy controls. The original P value of the correlation coefficient was adjusted by multiple test correction, and the adjusted P value was set at 0.05 for significance. Age-specific normal values for white blood cell count used by the clinical laboratory at St. Louis Children's Hospital were <1 wk: 5.0–30.0 K/cu mm; 1 wk to 1 mo: 5.0–20.0 K/cu mm; 1 mo to 2 y: 6.0–17.5 K/cu mm; 2–6 y: 5.0–15.5 K/cu mm; 6–12 y: 4.5–13.5 K/cu mm; and >12 y: 3.8–9.8 K/cu mm.
Supplementary Material
Acknowledgments
We thank Maria Cannella for technical assistance, Avraham Smason for recruiting subjects for this study, and Dennis Dietzen, PhD, for providing the normal white blood cell counts in use at St. Louis Children's Hospital. We thank the Genome Technology Access Center at Washington University School of Medicine for help with genomic analysis. The Genome Technology Access Center is partially supported by National Cancer Institute Cancer Center Support Grant P30 CA91842 to the Siteman Cancer Center and Institute for Clinical and Translational Sciences/Clinical and Translational Sciences Award Grant UL1RR024992 from the National Center for Research Resources, a component of the National Institutes of Health, and the National Institutes of Health Roadmap for Medical Research. Funding for this study was provided by National Institute of Allergy and Infectious Diseases Grant 1UAH2AI083266-01, National Institutes of Health–National Center for Research Resources, Washington University Institute for Clinical and Translational Sciences Grant UL1 RR024992, and National Institute of Child Health and Human Development Training Grant T32HD049338.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in the paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE40396).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302968110/-/DCSupplemental.
References
- 1.Ramilo O, Mejías A. Shifting the paradigm: Host gene signatures for diagnosis of infectious diseases. Cell Host Microbe. 2009;6(3):199–200. doi: 10.1016/j.chom.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 3.Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. Pattern recognition receptors and the innate immune response to viral infection. Viruses. 2011;3(6):920–940. doi: 10.3390/v3060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paschos K, Allday MJ. Epigenetic reprogramming of host genes in viral and microbial pathogenesis. Trends Microbiol. 2010;18(10):439–447. doi: 10.1016/j.tim.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stojanov S, et al. Periodic fever, aphthous stomatitis, pharyngitis, and adenitis (PFAPA) is a disorder of innate immunity and Th1 activation responsive to IL-1 blockade. Proc Natl Acad Sci USA. 2011;108(17):7148–7153. doi: 10.1073/pnas.1103681108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramilo O, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109(5):2066–2077. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Popper SJ, et al. Gene transcript abundance profiles distinguish Kawasaki disease from adenovirus infection. J Infect Dis. 2009;200(4):657–666. doi: 10.1086/603538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ardura MI, et al. Enhanced monocyte response and decreased central memory T cells in children with invasive Staphylococcus aureus infections. PLoS One. 2009;4(5):e5446. doi: 10.1371/journal.pone.0005446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berry MP, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466(7309):973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pankla R, et al. Genomic transcriptional profiling identifies a candidate blood biomarker signature for the diagnosis of septicemic melioidosis. Genome Biol. 2009;10(11):R127. doi: 10.1186/gb-2009-10-11-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaas AK, et al. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe. 2009;6(3):207–217. doi: 10.1016/j.chom.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allantaz F, et al. Blood leukocyte microarrays to diagnose systemic onset juvenile idiopathic arthritis and follow the response to IL-1 blockade. J Exp Med. 2007;204(9):2131–2144. doi: 10.1084/jem.20070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aarøe J, et al. Gene expression profiling of peripheral blood cells for early detection of breast cancer. Breast Cancer Res. 2010;12(1):R7. doi: 10.1186/bcr2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 15.Chaussabel D, et al. Blood transcriptional fingerprints to assess the immune status of human subjects. In: Marincola FM, editor. Immunologic Signatures of Rejection. New York: Springer; 2011. pp. 105–125. [Google Scholar]
- 16.Colvin JM, et al. Detection of viruses in young children with fever without an apparent source. Pediatrics. 2012;130(6):e1455–e1462. doi: 10.1542/peds.2012-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaussabel D, et al. Analysis of significance patterns identifies ubiquitous and disease-specific gene-expression signatures in patient peripheral blood leukocytes. Ann N Y Acad Sci. 2005;1062:146–154. doi: 10.1196/annals.1358.017. [DOI] [PubMed] [Google Scholar]
- 18.Rosebeck S, Leaman DW. Mitochondrial localization and pro-apoptotic effects of the interferon-inducible protein ISG12a. Apoptosis. 2008;13(4):562–572. doi: 10.1007/s10495-008-0190-0. [DOI] [PubMed] [Google Scholar]
- 19.Rosales JL, Ernst JD. Calcium-dependent neutrophil secretion: Characterization and regulation by annexins. J Immunol. 1997;159(12):6195–6202. [PubMed] [Google Scholar]
- 20.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA. 2002;99(10):6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudinsky SL, et al. Serious bacterial infections in febrile infants in the post-pneumococcal conjugate vaccine era. Acad Emerg Med. 2009;16(7):585–590. doi: 10.1111/j.1553-2712.2009.00444.x. [DOI] [PubMed] [Google Scholar]
- 22.Herz AM, et al. Changing epidemiology of outpatient bacteremia in 3- to 36-month-old children after the introduction of the heptavalent-conjugated pneumococcal vaccine. Pediatr Infect Dis J. 2006;25(4):293–300. doi: 10.1097/01.inf.0000207485.39112.bf. [DOI] [PubMed] [Google Scholar]
- 23.Baraff LJ, et al. Practice guideline for the management of infants and children 0 to 36 months of age with fever without source. Ann Emerg Med. 1993;22(7):1198–1210. doi: 10.1016/s0196-0644(05)80991-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.