Abstract
Primary ciliary dyskinesia (PCD) is an autosomal recessive disease caused by defective cilia motility. The identified PCD genes account for about half of PCD incidences and the underlying mechanisms remain poorly understood. We demonstrate that Reptin/Ruvbl2, a protein known to be involved in epigenetic and transcriptional regulation, is essential for cilia motility in zebrafish. We further show that Reptin directly interacts with the PCD protein Lrrc6/Seahorse and this interaction is critical for the in vivo function of Lrrc6/Seahorse in zebrafish. Moreover, whereas the expression levels of multiple dynein arm components remain unchanged or become elevated, the density of axonemal dynein arms is reduced in reptinhi2394 mutants. Furthermore, Reptin is highly enriched in the cytosol and colocalizes with Lrrc6/Seahorse. Combined, these results suggest that the Reptin-Lrrc6/Seahorse complex is involved in dynein arm formation. We also show that although the DNA damage response is induced in reptinhi2394 mutants, it remains unchanged in cilia biogenesis mutants and lrrc6/seahorse mutants, suggesting that increased DNA damage response is not intrinsic to ciliary defects and that in vertebrate development, Reptin functions in multiple processes, both cilia specific and cilia independent.
Keywords: ciliopathy, chromatin remodeling complex, dynein arm assembly factor
Although the sensory function of the cilium has garnered significant attention only in the past decade, defective cilia motility was recognized nearly four decades ago as the cause of primary (genetic) ciliary dyskinesia (PCD) (1). PCD is a group of rare human genetic diseases characterized by recurrent infections of the respiratory system, male infertility, and frequently laterality defects, all of which are tightly linked to cilia motility abnormalities. Up to now, 19 PCD genes have been identified (2–20). However, combined they account for about 50% of all PCD cases, suggesting the existence of multiple additional causative genes. Despite such significant advances, our understanding of how cilia motility is regulated is limited and the list of genes involved in cilia motility remains incomplete.
In motile cilia or flagella, dynein arms are large protein complexes powering cilia motility. Not surprisingly, multiple PCD genes encode dynein arm components, including DNAL1, DNAI1, DNAI2, DNAH5, and DNAH11 (2–6). In addition, it is understood that dynein arm subunits are preassembled in the cytosol, transported into cilia/flagellum, and docked onto the axoneme, although the underlying mechanisms are poorly understood (21). In recent years, three PCD genes, i.e., DNAAF1/LRRC50 (7, 8), DNAAF2/KTU (9), and DNAAF3/PF22 (10) have been shown to be involved in the assembly of dynein arm subunits, highlighting the relevance and importance of this process in cilia motility and PCD.
Very recently, mutations in human LRRC6/SEAHORSE were linked to PCD (18). Similar to the three known dynein arm assembly factors, mutations in LRRC6/SEAHORSE lead to the absence of both the outer and the inner dynein arms, suggesting that this might be an additional dynein arm assembly factor (18). Strikingly, lrrc6/seahorse mutants were previously identified in a genetic screen in zebrafish together with reptin mutants and the two mutants share nearly identical abnormalities: ventral body curvature and kidney cysts (22, 23).
Reptin (or Reptin52/Ruvbl2/Ino80J/Tip48) contains an AAA-family ATPase domain resembling the bacterial RuvB, a DNA helicase (for a review, see ref. 24). It regulates transcription through multiple chromatin remodeling complexes (25–28). It is also found in protein complexes involved in telomere maintenance, small nucleolar RNA assembly and DNA damage responses (29–32). Interestingly, the transcription of reptin is up-regulated during flagellum regeneration in Chlamydomonas (33). However, the role of Reptin in cilia biology and vertebrate development was rarely studied before this study.
In this study, we demonstrate that Reptin is essential for cilia motility in zebrafish. We further show that Reptin directly interacts with Lrrc6/Seahorse and that this interaction is important for the in vivo function of Lrrc6/Seahorse, indicating that reptin functions together with lrrc6/seahorse in a process essential for cilia motility and is a candidate gene for PCD. We further provide evidence that although the expression levels of multiple dynein arm components are not decreased, the number of dynein arms on the axoneme is reduced in reptin mutants. Moreover, Reptin is highly enriched in the cytosol and colocalizes with Lrrc6/Seahorse. Combined, these results suggest that the Reptin-Lrrc6/Seahorse complex functions in dynein arm assembly. Additionally, we investigated the level of the DNA damage response (DDR) because of its recent association with ciliopathies (34). Our results show that although DDR is induced in reptin mutants, it remains unchanged in both lrrc6/seahorse and the cilia biogenesis mutant ift172, suggesting that DDR induction is not intrinsic to defective cilia and that the Reptin-Lrrc6/Seahorse interaction is not involved in Reptin’s role in DDR.
Results
Inactivation of reptin Leads to Cilia-Associated Phenotypes.
hi2394 was identified in a large scale insertional mutagenesis screen conducted in zebrafish (22, 23). The homozygous mutants showed ventral body curvature and kidney cyst (Fig. 1A), whereas heterozygous carriers are viable with no obvious phenotypes. Cross-sections verified large cysts in the glomerular–tubular region, as well as grossly dilated pronephric duct, which was confirmed by whole-mount immunostaining using a renal epithelial marker (Fig. S1 A and B). Notably, this specific combination of phenotypes was also detected in a collection of cilia mutants, including mutants of intraflagellar transport (IFT) genes (23, 35), which encode IFT particle components essential for cilia biogenesis, suggesting that hi2394 may interfere with cilia biogenesis or function.
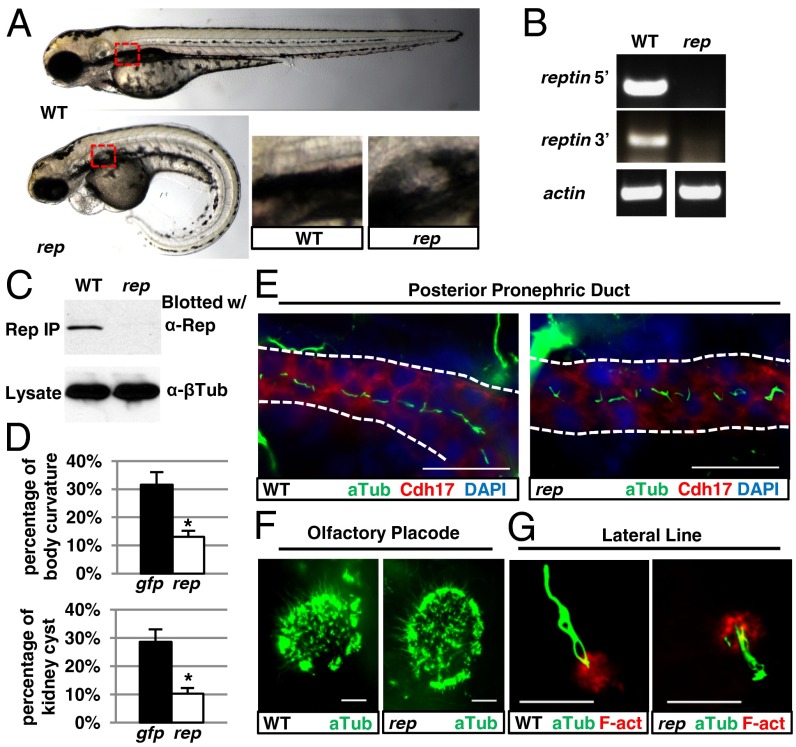
Fig. 1.
Phenotypes of reptinhi2394 mutants. (A) Reptinhi2394 mutant (rep) at 3 d postfertilization (dpf) showing kidney cyst (red box, magnified in Lower Right) and body curvature, compared with wild-type (WT) fish. (B) RT-PCR using lysates of 2-dpf reptinhi2394 (rep) and WT siblings. Two pairs of primers, one to the 5′ side of the proviral insertion and one to the 3′ side, were used. (C) The greatly reduced Reptin protein in 4-dpf reptinhi2394 mutant (rep). (Upper) Samples precipitated and blotted with anti-Reptin. (Lower) Lysates blotted with anti–β-tubulin as a loading control. (D) Microinjection of reptin-eGFP mRNA (rep) reduced the percentages of body curvature (Upper) and kidney cysts (Lower) in embryos from reptinhi2394+/− crosses, with eGFP mRNA (gfp) microinjection as a negative control. Data are represented as mean + SD, from three replicates. *P < 0.05. (E–G) Immunostaining showing cilia (in green) in reptinhi2394 mutants (rep) in the posterior pronephric duct (E) (stained with anti-Cdh17, a marker for kidney epithelial cells, in red and lined with dotted lines), the olfactory placode (F), and the lateral line organ (G) [F-actin (F-act) stained with rhodamine phalloidin in red]. (Scale bar, 20 µm in E and F and 10 µm in G.)
Inverse PCR revealed that the proviral insertion hi2394 was located in the first intron of reptin. To determine the impact of the insertion on reptin expression, we performed RT-PCR. Although in wild-type embryos, RT-PCR led to bands of predicted sizes, no transcript was detected in hi2394 mutants (Fig. 1B). The absence of reptin mRNA in hi2394 mutants was further supported by in situ hybridization using antisense reptin probe. Strong staining is detected in wild-type embryos at multiple stages, whereas the signal is lost in hi2394 mutants (Fig. S2C). Consistently, whereas in lysate of wild-type fish immunoprecipitation (IP) followed by Western Blot using anti-Reptin detected a clear band at around 50 kDa, the predicted size of Reptin, this signal was greatly reduced in hi2394 mutants (Fig. 1C). Taken together, these findings suggest that reptin expression is dramatically reduced, if not completely abolished, in hi2394 mutants.
Although the expression of reptin is greatly diminished in hi2394 mutants, it remains possible that hi2394 disrupts additional genes that are responsible for the mutant phenotypes. We therefore designed a morpholino oligo (MO) against the translational initiation site of reptin. Results showed that wild-type embryos injected with 400 pg reptin MO displayed similar phenotypes as hi2394 mutants, including ventral body curvature and kidney cysts (Fig. S1C). Further, we introduced a 5-nucleotide mismatch into a reptin cDNA clone so that the encoded mRNA was no longer a target of reptin MO. Coinjection of the mismatched mRNA rescued the morphant (Fig. S1D). Finally, we tested whether expression of reptin could rescue hi2394 mutants. Specifically, we injected 150 pg reptin-egfp mRNA or as a control, 150 pg myc-egfp mRNA into embryos from crosses of heterozygous carriers of hi2394. From three repeated experiments, an average of 13.0% body curvature and 10.3% kidney cyst phenotypes were observed in the reptin-egfp–injected group, compared with an average of 31.5% body curvature and 28.6% kidney cyst phenotypes in the control group (Fig. 1D). Combined, the above results demonstrate that hi2394 is a loss-of-function allele of reptin.
Reptin Transcripts Are Enriched in Tissues with Motile Cilia.
Reptin’s function in vertebrate development, aside from its potential role in heart growth (36), has not been examined. To understand the role of reptin in development, we examined the expression pattern of reptin through multiple developmental stages via in situ hybridization. Reptin RNA was present as early as one- to two-cell stage in wild-type embryos (Fig. S2A). Because in zebrafish, zygotic transcription does not commence until around the 500–1,000-cell stage, this result indicates that reptin transcripts are deposited maternally. Up to the bud stage, reptin was expressed almost ubiquitously (Fig. S2A). Later, although still widely distributed, reptin transcripts became more concentrated in highly ciliated tissues, including the neural tube, the lateral line organ, and the pronephric duct (Fig. S2A). In contrast, the control reptin sense mRNA probe failed to detect any signal, verifying the specificity of the antisense probe (Fig. S2B).
Reptin Is Essential for Cilia Motility.
Because reptinhi2394 mutants manifested similar phenotypes as cilia-specific mutants and reptin transcripts are enriched in ciliated tissues, we examined whether reptin is involved in cilia-mediated pathways by testing potential genetic interactions between reptin and the cilia biogenesis gene ift172.
We first inspected reptinhi2394−/−; ift172hi2211−/− double mutants and found ventral body curvature and cystic kidney phenotypes (Fig. S3A). The cyst size and onset time in double mutants is comparable with that in each single mutant. The body curvature phenotype in double mutants resembles that in reptinhi2394 single mutants, in which it tends to be more severe than in ift172hi2211 single mutants. Furthermore, combining suboptimal doses of morpholino oligos against ift172 and reptin significantly increased the percentage of phenotypic embryos compared with the same dose of morpholino against either of the two genes (Fig. S3B). Combined, these results suggest that reptin genetically interacts with ift172.
To investigate the precise function of reptin in cilia biology, we first examined cilia morphology in reptinhi2394 mutants by immunostaining with the cilia marker antiacetylated tubulin. No difference in cilia formation was detected in single-ciliated cells (SCCs) in the kidney duct, the olfactory placode, and the lateral line in reptinhi2394 mutants (Fig. 1 E–G). In multiciliated cells (MCCs) of the pronephric duct, abundant cilia were detected in reptinhi2394 mutants, albeit in a disorganized fashion in the grossly dilated lumen of the pronephric duct compared with wild-type embryos (Fig. S3C).
Because abundant cilia form in reptinhi2394 mutants, we hypothesized that reptin may be involved in cilia motility. To test this hypothesis, we performed differential interference contrast (DIC) videomicroscopy to examine cilia motility at 3 dpf. Strikingly, in all reptinhi2394 mutants examined, cilia in the pronephric duct were completely paralyzed and orientated in random directions; whereas in wild-type siblings, cilia were tightly bundled and beat in a coordinated and cyclic fashion (Fig. 2A and Movie S1). Because the lumen of the kidney duct was grossly enlarged in reptinhi2394 mutants, to rule out the possibility that the observed cilia motility defect was secondary to the grossly enlarged lumen size, we performed the same assay in the olfactory placode, in which cilia were freely exposed to the environment and not confined by the organ structure. Consistently, motile cilia on the border of the placode were entirely paralyzed in reptinhi2394 mutants. By contrast, cilia in wild-type siblings beat vigorously (Movie S2). Combined, these results suggest that Reptin is essential for cilia motility.
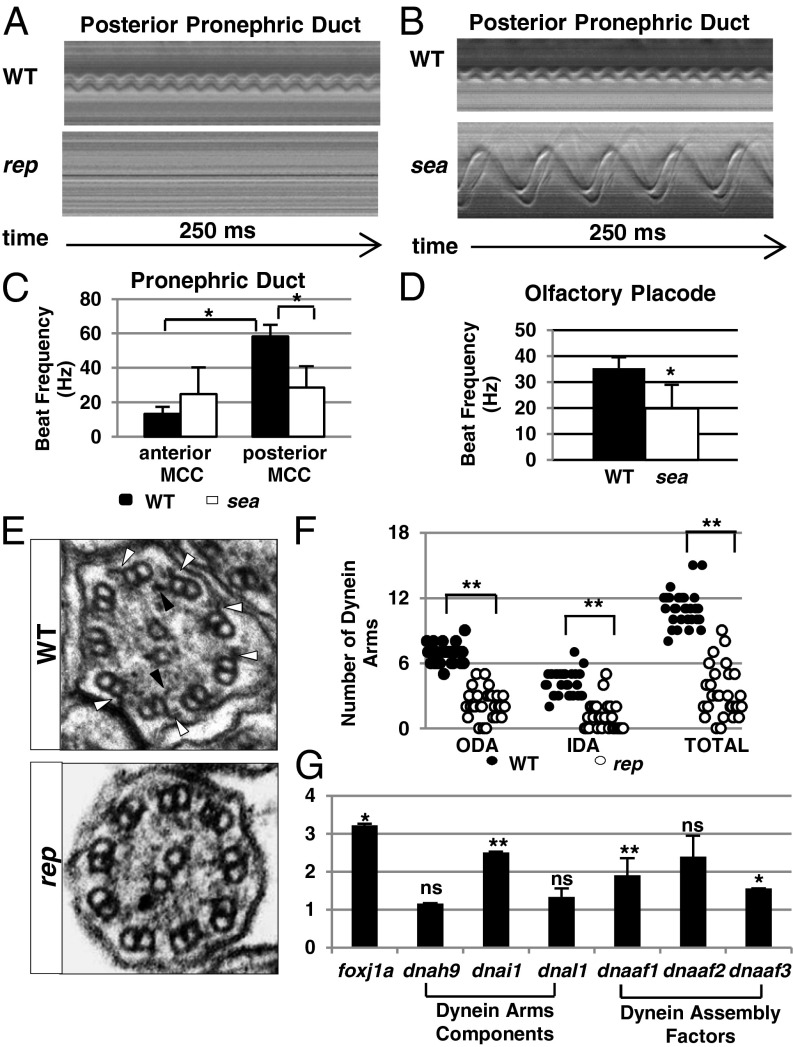
Fig. 2.
Reptin is essential for cilia motility. (A and B) Kymographs showing the rhythmic beating of a multicilia bundle in the posterior pronephric duct in a wild-type (WT) embryo and paralyzed cilia in a reptinhi2394 mutant (rep), or a slower beating cilium in a seahorsehi3308 mutant (sea) at 3 dpf. (C) Cilia beating frequency in wild-type (black bar) and seahorsehi3308 (white bar) pronephros at 3 dpf. Data are represented as mean + SD, from eight replicates. *P < 0.05. (D) Cilia beating frequency in the olfactory placode of wild-type (black bar, WT) and seahorsehi3308 mutant (white bar, sea) embryos at 3 dpf. Data are represented as mean + SD, from five replicates. *P < 0.05. (E) Electron micrographs of cross-sections of cilia in WT and reptinhi2394 mutants (rep) at 5 dpf. Outer and inner dynein arms are pointed out by white and black arrowheads, respectively. (F) Number of outer and inner dynein arms in WT and reptinhi2394 mutants (rep) per section at 5 dpf. Data are represented as mean + SD. **P < 0.01. (G) Expression levels of multiple factors involved in dynein arm formation as shown by quantitative PCR (qPCR). Y axis shows the fold changes in reptinhi2394 mutants. Data are represented as mean + SD, from three replicates. *P < 0.05, **P < 0.01. NS, nonsignificant.
Reptin Is a Binding Partner of Lrrc6/Seahorse.
Unexpectedly, Reptin was identified twice as the only hit in a yeast two-hybrid screen we performed aimed to search for binding partners of Seahorse (also known as leucine-rich repeat-containing 6, Lrrc6), encoded by another zebrafish cystic kidney gene identified in the same genetic screen (22, 23). This interaction was verified by co-IP experiments using lysates of zebrafish embryos overexpressing Reptin tagged with the Flag epitope. Anti-Flag IP was able to pull down endogenous Lrrc6/Seahorse (Fig. 3A). In contrast, no Lrrc6/Seahorse was precipitated by anti-Flag when Flag-tagged eGFP was overexpressed.
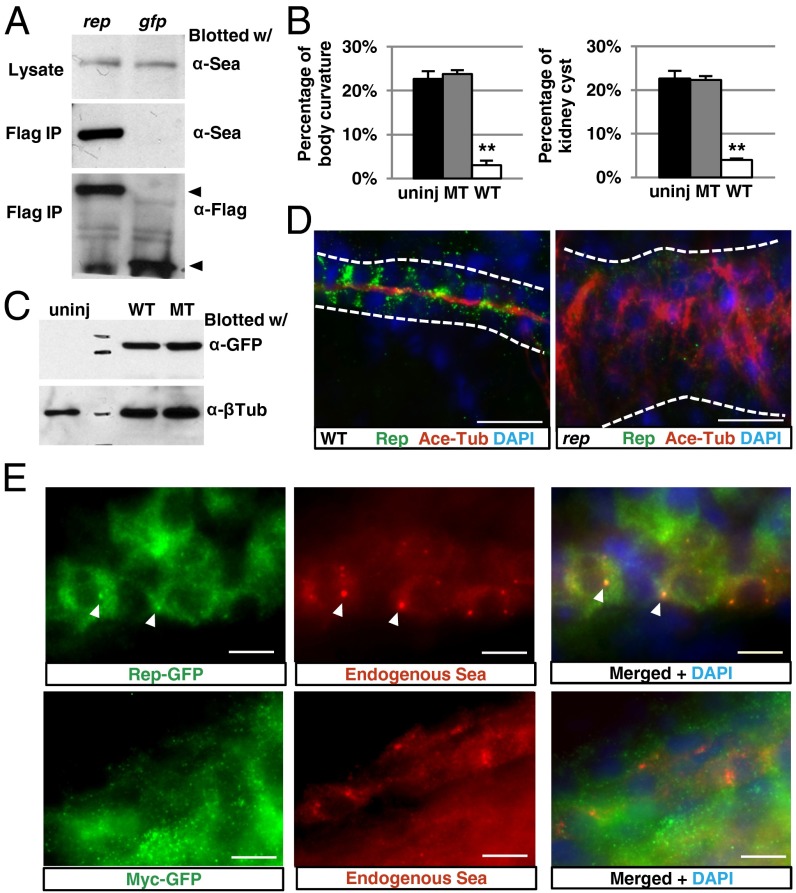
Fig. 3.
Reptin binds to and colocalizes with Lrrc6/Seahorse. (A) Anti-Flag IP brings down endogenous Lrrc6/Seahorse in lysates of 2-dpf embryos overexpressing Flag-tagged Reptin (rep), but not in embryos overexpressing Flag-tagged eGFP (gfp). Bottom arrow points to Reptin-Flag (Left) and Gfp-Flag (Right). To avoid interference from the signal of IgG light chain, the blot was cut at the bottom before exposure. (B) The percentage of body curvature (Left) and kidney cyst (Right) in embryos from seahorsehi3308+/− crosses uninjected (uninj), injected with wild-type seahorse mRNA (WT), and mutant seahorse mRNA encoding full-length Seahorse with the amino acids 392–396 alanine substitution (MT). Data are represented as mean + SD, from three replicates, **P < 0.01. (C) Western blot on whole embryo lysate of embryos from seahorsehi3308+/− crosses unjected (uninj), injected with GFP-tagged wild-type seahorse mRNA (WT), or GFP-tagged mutant seahorse mRNA as in B (MT). (D) Immunostaining of the pronephric duct (white line) in 4-dpf wild type (WT) and reptinhi2394 mutants (rep). Cilia are labeled with antiacetylated tubulin (Ace-Tub) in red, and Reptin is labeled with anti-Reptin (Rep) in green. (Scale bar, 20 µm.) (E) Immunostaining of the pronephric duct in 1-dpf embryo showing overexpressed Reptin-eGFP (Upper, green) and endogenous Seahorse (Sea, red). Overexpressed Myc-eGFP is shown (Lower) as a negative control. Arrowheads point to puncta positive for both Reptin eGFP and Seahorse. (Scale bar, 10 µm.)
Because reptin and lrrc6/seahorse encode two proteins that physically interact, we compared the phenotypes of reptinhi2394 and seahorsehi3308 double mutants with those in each single mutant. Results showed that the cyst size and onset time in double mutants was comparable with that in each single mutant. In addition, double mutants displayed similar body curvature phenotype as in reptin single mutants (Fig. S4A). We additionally performed a morpholino knockdown-based synergy assay between reptin and lrrc6/seahorse. Coinjection of suboptimal doses of seahorse MO and reptin MO leads to significantly increased percentage of embryos showing kidney cyst and body curvature compared with coinjection of the same doses of reptin or seahorse MO combined with a control MO (Fig. S4B). Combined, these results suggest that reptin genetically interacts with lrrc6/seahorse to modulate common processes underlying the two phenotypes.
The Lrrc6/Seahorse Reptin Binding Domain Is Essential for the in Vivo Function of Lrrc6/Seahorse.
We further investigated whether the physical interaction between Lrrc6/Seahorse and Reptin is functionally significant. Through a serial deletion analysis using the yeast two-hybrid system, we first narrowed down the Reptin-interacting domain in Lrrc6/Seahorse to a C-terminal 60-aa region (amino acids 381–440) (Fig. S5B). No functional domain could be identified in this region (Fig. S5B). However, sequence alignment revealed that two stretches of amino acids within this region, amino acids 392–396 and 435–439, were conserved (Fig. S5C). We separately substituted the two conserved epitopes in the minimum binding domain with alanines and tested the ability of the mutated proteins to interact with Reptin using the yeast two-hybrid system. Interestingly, the mutated protein with the amino acid 392–396 substitution could no longer interact with Reptin, whereas the one with the amino acid 435–439 substitution still could, suggesting that amino acid 392–396 in Lrrc6/Seahorse is essential for its interaction with Reptin (Fig. S5B). Using this knowledge, we performed rescue experiments to test the importance of the Lrrc6/Seahorse–Reptin interaction for the in vivo function of Lrrc6/Seahorse. Whereas injection of 4 pg wild-type full-length lrrc6/seahorse mRNA into embryos from crosses between heterozygous carriers of seahorsehi3308 reduced phenotypic embryos (22.7% in the control group) to 3.6% and 3.0% for cystic kidney and body curvature, respectively, the full-length lrrc6/seahorse mRNA with the amino acid 392–396 substitution failed to do so, with 22.3% of the injected embryos displaying kidney cysts and 23.8% body curvature (Fig. 3B). Western blot verified that both proteins were expressed at similar levels (Fig. 3C). Combined, these results suggest that the Lrrc6/Seahorse–Reptin interaction is essential for the in vivo function of Lrrc6/Seahorse.
Previously it was shown that two different alleles of seahorse seafa20r and seatg238a in zebrafish display a variable cilia motility defect, although we failed to detect such a defect in the pronephric duct of seahorsehi3308 mutants using standard DIC videomicroscopy techniques (37, 38). In light of the Lrrc6/Seahorse–Reptin interaction and the severe cilia motility defect in reptinhi2394 mutants, we reanalyzed cilia motility in seahorsehi3308 mutants using an improved ultraspeed DIC camera with an acquisition speed of 2,000 frames per second. Results showed that in the pronephric duct of wild-type embryos, cilia beat at 13.3 Hz in the anterior region, slower than the 58.3 Hz observed in the posterior region. In seahorsehi3308 mutants, whereas cilia motility was 50.8% reduced in the posterior region, beat rate became variable in the anterior region (Fig. 2 B and C and Movies S3 and S4). Further analysis in the olfactory placode revealed variable beating speed in seahorsehi3308 mutants as well (Fig. 2D and Movie S5), ruling out duct dilation as the cause of motility variation in seahorsehi3308 mutants. Together, these results support an important role for the Reptin–Lrrc6/Seahorse protein complex in cilia motility.
Reptin Is Enriched in Cytoplasmic Puncta.
Previously we demonstrated that Lrrc6/Seahorse is enriched in cytoplasmic puncta (37). Reptin was previously shown to be localized to both the cytosol and the nucleus in cultured mammalian cells (39). Prompted by the physical interaction between Reptin and Lrrc6/Seahorse, we analyzed the subcellular localization pattern of Reptin in the zebrafish pronephric duct by immunostaining. Results show that Reptin is localized in cytoplasmic puncta with no obvious enrichment on the cilium (Fig. 3D). The specificity of the anti-Reptin antibody is confirmed by the lack of Reptin signal in reptinhi2394 mutants (Fig. 3D).
We then examined whether Reptin and Lrrc6/Seahorse colocalize. As both the anti-Reptin and the anti-Seahorse antibodies were derived from rabbits, we compared localization patterns of overexpressed Reptin-eGFP and the endogenous Lrrc6/Seahorse in the zebrafish kidney duct. Although more diffusive than the endogenous Reptin, anti-GFP showed enrichment of Reptin-eGFP in cytoplasmic punta and more importantly, these punta colocalize with endogenous Lrrc6/Seahorse (Fig. 3E). In contrast, no puncta were detected with anti-GFP when Myc-eGFP was overexpressed, validating the specificity of this signal (Fig. 3E).
Reptin Is Involved in Dynein Arm Formation.
To search for the mechanism of the observed cilia motility defect in reptinhi2394 mutants, we performed transmission electron microscopy using 5-dpf embryos. In cross-sections, we frequently, but not always, observed missing dynein arms in reptinhi2394 mutant cilia (Fig. 2E). Due to imperfect sample preparation, dynein arms are not always visible on every microtubule doublet even in cross-sections of wild-type cilia. We therefore counted dynein arms in 28 cilia cross-sections in three reptinhi2394 mutants and 30 cilia cross-sections in three wild-type embryos at 5 dpf. Statistical analysis suggests that the number of both inner dynein arms (IDAs) and outer dynein arms (ODAs) decreased significantly in reptinhi2394 mutants cilia compared with wild-type cilia (Fig. 2F).
To directly test the role of Reptin in transcriptional regulation of cilia motility genes, we performed real-time PCR using reptinhi2394 mutants and wild-type siblings at 2 dpf. Surprisingly, the expression levels of dynein arm components are either not changed or up-regulated (Fig. 2G). Strikingly, the expression levels of all of the three known dynein arm assembly factors were increased, two of them significantly (Fig. 2G). Moreover, the level of foxj1 was increased threefold (Fig. 2G). Combined, these results suggest that the reduced dyneim arm defect is not caused by decreased expression of these genes. Interestingly, a previous study shows that the expression of cilia motility gene is up-regulated in response to injury and epithelial stretch (40), suggesting that there exists a compensatory transcriptional regulation of this group of genes.
Reptinhi2394 Mutants Show Additional Cilia-Independent Phenotypes, Including Increased DDR.
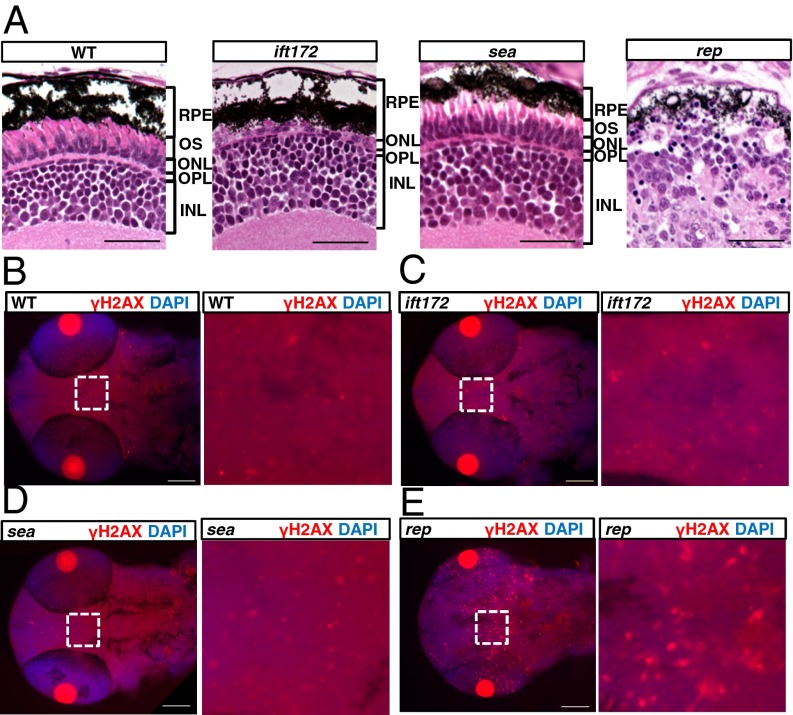
Compared with classic cilia mutants, such as cilia biogenesis mutants ift172hi2211, which show body curvature and kidney cysts (23), reptinhi2394 mutants display additional phenotypes. More than 80% of the mutants died at around 5 dpf with severe necrosis throughout the body, whereas most IFT mutants could survive to 7 dpf. In addition, we observed that the size of the eye in reptinhi2394 mutants was considerably reduced compared with either wild type or ift172hi2211 mutants. Histological sections of the eye showed that in ift172hi2211 mutants, whereas the outer segment, a form of specialized cilia of the photoreceptor layer, was greatly diminished, the other layers were relatively well maintained (Fig. 4A), consistent with previous reports of other IFT mutants (41, 42). Moreover, the structure of the eye in seahorsehi3308 mutants seems normal (Fig. 4A). In contrast, in reptinhi2394 mutants, the layered structure of the eye is grossly disrupted (Fig. 4A).
Fig. 4.
reptinhi2394 mutants show cilia-independent phenotypes. (A) Sections of the eye at 5 dpf. RPE, retinal pigment epithelium; OS, outer segment; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer. (Scale bar, 20 µm.) (B–E) The head and eye region at 2-dpf mutants stained with an anti-γH2AX antibody in red. The boxed regions (Left) of B–E are magnified on corresponding Right. The pair of bright red circles in each image is the pair of lenses. (Scale bar, 100 µm.) WT, wild type; ift172, ift172hi2211 mutant; sea, seahorsehi3308 mutant; rep, reptinhi2394 mutant.
In addition, because recently it was suggested that in some ciliopathies DDR was activated as shown by an increased level of phosphorylated H2AX (34), we performed the same immunostaining assay using antiphosphorylated H2AX. Results showed that whereas the signal in ift172hi2211 and seahorsehi3308 mutants is comparable to wild-type siblings, it is greatly increased in the brain of reptinhi2394 mutants (Fig. 4 B–E). Taken together, these results suggest that reptinhi2394 mutants have additional cilia-independent defects.
Discussion
In this study, we demonstrate that the known epigenetic factor Reptin is required for cilia motility. Biochemically, Reptin is a DNA-stimulated ATPase critical for transcriptional control, either as a key component in chromatin remodeling complexes, such as TIP60 and INO80 (25, 28) or by interacting with transcription factors, including β-catenin (43) and c-Myc (44). However, little is known about the precise function of Reptin in vertebrate development, except that a zebrafish reptin mutant showed hyperplastic heart, possibly via abnormal activation of the β-catenin pathway (36). Here we report that a loss-of-function allele of reptin develops characteristic phenotypes of ciliary mutants, including ventral body curvature and kidney cysts. We further show that reptin genetically interacts with ift172, a gene essential for cilia biogenesis and maintenance. However, in reptin mutants, abundant cilia are able to form in all of the organs we examined. Because wild-type transcripts of reptin are deposited into mutant eggs by heterozygous mothers, it is possible that a cilia biogenesis requirement is masked by the maternal contribution. However, in IFT mutants, whereas maternal contribution of the gene product is able to support cilia biogenesis in single ciliated cells in the kidney duct during early development, it is insufficient for cilia formation later in development, e.g., cilia in MCCs of the kidney duct and the lateral line organ (35), in contrast to reptin mutants, suggesting that Reptin is dispensable for cilia formation per se. In the zebrafish pronephros, cilia are motile. Strikingly, in the dilated pronephric duct of reptin mutants, cilia become completely paralyzed. To rule out the possibility that the observed cilia motility defect was secondary to ductal dilation, we further analyzed cilia motility in the zebrafish nose, which opens to the outer environment, and observed similar cilia motility defects in reptin mutants, suggesting that Reptin is essential for cilia motility.
We further show that Reptin directly interact with Lrrc6/Seahorse, a protein very recently linked to PCD (18, 45). Consistent with this physical interaction, mutants of reptin and lrrc6/seahorse were identified in the same genetic screen in zebrafish and both show similar body curvature and kidney cyst formation (22, 23). Moreover, we found that amino acid substitutions disrupting the Reptin–Lrrc6/Seahorse interaction also interfere with the ability of Lrrc6/Seahorse to rescue cilia-related phenotypes in seahorsehi3308 mutants. Together, these results suggest that Reptin and Lrrc6/Seahorse form a functional protein complex essential for cilia motility. Interestingly, previous yeast two-hybrid assay and affinity capture-mass spectrometry (46–48) identified Dpcd, encoded by a gene disrupted in a mouse PCD model (49), as a Reptin binding protein, providing additional support for a multiprotein complex in cilia motility control and REPTIN as a candidate gene for PCD.
To search for potential mechanisms of the newly identified Reptin–Lrrc6/Seahorse complex in cilia motility, we examined the ultrastructure of cilia in reptinhi2394 mutants. Results show that the numbers of both ODAs and IDAs are significantly reduced in the mutants. However, despite the known role of Reptin in transcriptional regulation, the expression levels of multiple dynein arm components and dynein arm assembly factors are not decreased in reptinhi2394 mutants, ruling out transcriptional regulation of these factors as a major mechanism for reduced dynein arm presence in reptinhi2394 mutants. Interestingly, Reptin was found to interact with PIH1D1 (47, 50), which contains a PIH (protein interacting with Hsp90) domain implicated in the preassembly of dynein arms (51), raising the possibility that the Reptin–Lrrc6/Seahorse complex regulates cilia motility via dynein arm assembly.
Zebrafish has been used as a model organism for studying cystic kidney diseases. A significant difference between mammalian kidney and the zebrafish pronephros is that, whereas cilia in mammalian kidney are immotile primary cilia of the “9+0” configuration, cilia in the zebrafish kidney are of the “9+2” configuration with a central pair of microtubules and are motile. Multiple previous studies suggest that cilia motility defect alone may be sufficient to cause cyst formation in zebrafish (16, 52–54). Our results that Reptin seems to affect cilia motility specifically and that mutants develop kidney cysts provide further support for this model. However, it is still possible that Reptin has additional function in cilia-mediated signaling. In agreement with this hypothesis, the cilia motility defect in seahorsehi3308 mutants is much milder than reptinhi2394 mutants and yet the severity of the cystic kidney phenotype is comparable between the two mutants.
As an increasing number of human diseases are being attributed to ciliary defects, a pressing question is why different ciliopathies show overlapping and yet distinct constellations of symptoms. Recent results suggest that multiple ciliopathies also show defective DDR (34). We therefore compared the level of DDR between wild type, the classic cilia biogenesis mutants ift172hi2211, seahorsehi3308, and reptinhi2394. Results show that whereas the level of DDR is comparable among IFT mutants, seahorsehi3308 mutants and wild-type siblings, the level of DDR is significantly increased in reptinhi2394 mutants. Interestingly, as an integral component of the Tip60 complex, an activator of the DDR pathway, Reptin is recruited to DNA damage sites and required for the histone acetyltransferase activity of the Tip60 complex on histone 4 (29). Combined, our results suggest that ciliary defect itself does not lead to DDR activation. Instead, some cilia-related genes, such as reptin, have multiple parallel functions, both cilia specific and cilia independent. Thus, one potential mechanism for the heterogeneous symptoms observed in ciliopathies is the diverse additional functions of individual ciliopathy genes, highlighting the importance of analyzing each ciliopathy gene in detail and resisting the temptation to attribute all of the symptoms to ciliary defects.
Methods
All materials and methods used in this study are detailed in SI Methods. This includes a detailed description of zebrafish husbandry, histological analysis, molecular cloning, microinjection, immunohistochemistry, RT-PCR, whole-mount in situ hybridization, protein extraction, immunoprecipitation, Western blotting, high-speed videomicroscopy of cilia, electron microscopy, and yeast two-hybrid, quantitative PCR, and statistical analysis. All antibodies and primers used in this study are also listed in SI Methods.
Supplementary Material
Acknowledgments
We thank M. Brueckner, M. Khokha, and S. Weatherbee for critical reading of this manuscript; N. Semanchik for maintenance of our fish facility; S. Mentone for histology and electron microscopy assistance; W. Niu, G. Wang, and P. G. Gallagher for advice on quantitative PCR; J. Amatruda for anti–phospho-H2AX; and S. Roy and I. Drummond for plasmids. This work was supported by National Institutes of Health (NIH) Grant R01 DK092808, Research Grant RSG-10-247-01-DDC from the American Cancer Society (to Z.S.), the Animal Core supported by NIH 1P30DK090744-01, and the China Scholarship Council–Yale World Scholars Program (to L.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300968110/-/DCSupplemental.
References
- 1.Afzelius BA. A human syndrome caused by immotile cilia. Science. 1976;193(4250):317–319. doi: 10.1126/science.1084576. [DOI] [PubMed] [Google Scholar]
- 2.Pennarun G, et al. Loss-of-function mutations in a human gene related to Chlamydomonas reinhardtii dynein IC78 result in primary ciliary dyskinesia. Am J Hum Genet. 1999;65(6):1508–1519. doi: 10.1086/302683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartoloni L, et al. Mutations in the DNAH11 (axonemal heavy chain dynein type 11) gene cause one form of situs inversus totalis and most likely primary ciliary dyskinesia. Proc Natl Acad Sci USA. 2002;99(16):10282–10286. doi: 10.1073/pnas.152337699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olbrich H, et al. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left-right asymmetry. Nat Genet. 2002;30(2):143–144. doi: 10.1038/ng817. [DOI] [PubMed] [Google Scholar]
- 5.Loges NT, et al. DNAI2 mutations cause primary ciliary dyskinesia with defects in the outer dynein arm. Am J Hum Genet. 2008;83(5):547–558. doi: 10.1016/j.ajhg.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mazor M, et al. Primary ciliary dyskinesia caused by homozygous mutation in DNAL1, encoding dynein light chain 1. Am J Hum Genet. 2011;88(5):599–607. doi: 10.1016/j.ajhg.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duquesnoy P, et al. Loss-of-function mutations in the human ortholog of Chlamydomonas reinhardtii ODA7 disrupt dynein arm assembly and cause primary ciliary dyskinesia. Am J Hum Genet. 2009;85(6):890–896. doi: 10.1016/j.ajhg.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loges NT, et al. Deletions and point mutations of LRRC50 cause primary ciliary dyskinesia due to dynein arm defects. Am J Hum Genet. 2009;85(6):883–889. doi: 10.1016/j.ajhg.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Omran H, et al. Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature. 2008;456(7222):611–616. doi: 10.1038/nature07471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchison HM, et al. 2012. Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia. Nat Genet 44(4): 381–389, S1–S2.
- 11.Duriez B, et al. A common variant in combination with a nonsense mutation in a member of the thioredoxin family causes primary ciliary dyskinesia. Proc Natl Acad Sci USA. 2007;104(9):3336–3341. doi: 10.1073/pnas.0611405104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castleman VH, et al. Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am J Hum Genet. 2009;84(2):197–209. doi: 10.1016/j.ajhg.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Budny B, et al. A novel X-linked recessive mental retardation syndrome comprising macrocephaly and ciliary dysfunction is allelic to oral-facial-digital type I syndrome. Hum Genet. 2006;120(2):171–178. doi: 10.1007/s00439-006-0210-5. [DOI] [PubMed] [Google Scholar]
- 14.Merveille AC, et al. CCDC39 is required for assembly of inner dynein arms and the dynein regulatory complex and for normal ciliary motility in humans and dogs. Nat Genet. 2011;43(1):72–78. doi: 10.1038/ng.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becker-Heck A, et al. The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation. Nat Genet. 2011;43(1):79–84. doi: 10.1038/ng.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panizzi JR, et al. CCDC103 mutations cause primary ciliary dyskinesia by disrupting assembly of ciliary dynein arms. Nat Genet. 2012;44(6):714–719. doi: 10.1038/ng.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horani A, et al. Whole-exome capture and sequencing identifies HEATR2 mutation as a cause of primary ciliary dyskinesia. Am J Hum Genet. 2012;91(4):685–693. doi: 10.1016/j.ajhg.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kott E, et al. Loss-of-function mutations in LRRC6, a gene essential for proper axonemal assembly of inner and outer dynein arms, cause primary ciliary dyskinesia. Am J Hum Genet. 2012;91(5):958–964. doi: 10.1016/j.ajhg.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olbrich H, et al. UK10K Consortium Recessive HYDIN mutations cause primary ciliary dyskinesia without randomization of left-right body asymmetry. Am J Hum Genet. 2012;91(4):672–684. doi: 10.1016/j.ajhg.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krawczyński MR, Dmeńska H, Witt M. Apparent X-linked primary ciliary dyskinesia associated with retinitis pigmentosa and a hearing loss. J Appl Genet. 2004;45(1):107–110. [PubMed] [Google Scholar]
- 21.Fowkes ME, Mitchell DR. The role of preassembled cytoplasmic complexes in assembly of flagellar dynein subunits. Mol Biol Cell. 1998;9(9):2337–2347. doi: 10.1091/mbc.9.9.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amsterdam A, et al. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci USA. 2004;101(35):12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Z, et al. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 2004;131(16):4085–4093. doi: 10.1242/dev.01240. [DOI] [PubMed] [Google Scholar]
- 24.West SC. Processing of recombination intermediates by the RuvABC proteins. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 25.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406(6795):541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 26.Gstaiger M, et al. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science. 2003;302(5648):1208–1212. doi: 10.1126/science.1088401. [DOI] [PubMed] [Google Scholar]
- 27.Wu WH, et al. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat Struct Mol Biol. 2005;12(12):1064–1071. doi: 10.1038/nsmb1023. [DOI] [PubMed] [Google Scholar]
- 28.Jin J, et al. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J Biol Chem. 2005;280(50):41207–41212. doi: 10.1074/jbc.M509128200. [DOI] [PubMed] [Google Scholar]
- 29.Jha S, Shibata E, Dutta A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol. 2008;28(8):2690–2700. doi: 10.1128/MCB.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King TH, Decatur WA, Bertrand E, Maxwell ES, Fournier MJ. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol Cell Biol. 2001;21(22):7731–7746. doi: 10.1128/MCB.21.22.7731-7746.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watkins NJ, et al. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol Cell. 2004;16(5):789–798. doi: 10.1016/j.molcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 32.Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132(6):945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stolc V, Samanta MP, Tongprasit W, Marshall WF. Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomonas reinhardtii identifies orthologs of ciliary disease genes. Proc Natl Acad Sci USA. 2005;102(10):3703–3707. doi: 10.1073/pnas.0408358102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaki M, et al. Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell. 2012;150(3):533–548. doi: 10.1016/j.cell.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao Y, Park A, Sun Z. Intraflagellar transport proteins are essential for cilia formation and for planar cell polarity. J Am Soc Nephrol. 2010;21(8):1326–1333. doi: 10.1681/ASN.2009091001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rottbauer W, et al. Reptin and pontin antagonistically regulate heart growth in zebrafish embryos. Cell. 2002;111(5):661–672. doi: 10.1016/s0092-8674(02)01112-1. [DOI] [PubMed] [Google Scholar]
- 37.Kishimoto N, Cao Y, Park A, Sun Z. Cystic kidney gene seahorse regulates cilia-mediated processes and Wnt pathways. Dev Cell. 2008;14(6):954–961. doi: 10.1016/j.devcel.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 38.Serluca FC, et al. Mutations in zebrafish leucine-rich repeat-containing six-like affect cilia motility and result in pronephric cysts, but have variable effects on left-right patterning. Development. 2009;136(10):1621–1631. doi: 10.1242/dev.020735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim JH, et al. Roles of sumoylation of a reptin chromatin-remodelling complex in cancer metastasis. Nat Cell Biol. 2006;8(6):631–639. doi: 10.1038/ncb1415. [DOI] [PubMed] [Google Scholar]
- 40.Hellman NE, et al. The zebrafish foxj1a transcription factor regulates cilia function in response to injury and epithelial stretch. Proc Natl Acad Sci USA. 2010;107(43):18499–18504. doi: 10.1073/pnas.1005998107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sukumaran S, Perkins BD. Early defects in photoreceptor outer segment morphogenesis in zebrafish ift57, ift88 and ift172 intraflagellar transport mutants. Vision Res. 2009;49(4):479–489. doi: 10.1016/j.visres.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Sun Z. Qilin is essential for cilia assembly and normal kidney development in zebrafish. PLoS ONE. 2011;6(11):e27365. doi: 10.1371/journal.pone.0027365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bauer A, et al. Pontin52 and reptin52 function as antagonistic regulators of beta-catenin signalling activity. EMBO J. 2000;19(22):6121–6130. doi: 10.1093/emboj/19.22.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Etard C, Gradl D, Kunz M, Eilers M, Wedlich D. Pontin and Reptin regulate cell proliferation in early Xenopus embryos in collaboration with c-Myc and Miz-1. Mech Dev. 2005;122(4):545–556. doi: 10.1016/j.mod.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 45.Horani A, et al. LRRC6 mutation causes primary ciliary dyskinesia with dynein arm defects. PLoS ONE. 2013;8(3):e59436. doi: 10.1371/journal.pone.0059436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ewing RM, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeronimo C, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27(2):262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437(7062):1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 49.Zariwala M, et al. Investigation of the possible role of a novel gene, DPCD, in primary ciliary dyskinesia. Am J Respir Cell Mol Biol. 2004;30(4):428–434. doi: 10.1165/rcmb.2003-0338RC. [DOI] [PubMed] [Google Scholar]
- 50.McKeegan KS, Debieux CM, Boulon S, Bertrand E, Watkins NJ. A dynamic scaffold of pre-snoRNP factors facilitates human box C/D snoRNP assembly. Mol Cell Biol. 2007;27(19):6782–6793. doi: 10.1128/MCB.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamoto R, Hirono M, Kamiya R. Discrete PIH proteins function in the cytoplasmic preassembly of different subsets of axonemal dyneins. J Cell Biol. 2010;190(1):65–71. doi: 10.1083/jcb.201002081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu X, Ng CP, Habacher H, Roy S. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat Genet. 2008;40(12):1445–1453. doi: 10.1038/ng.263. [DOI] [PubMed] [Google Scholar]
- 53.Sullivan-Brown J, et al. Zebrafish mutations affecting cilia motility share similar cystic phenotypes and suggest a mechanism of cyst formation that differs from pkd2 morphants. Dev Biol. 2008;314(2):261–275. doi: 10.1016/j.ydbio.2007.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Rooijen E, et al. LRRC50, a conserved ciliary protein implicated in polycystic kidney disease. J Am Soc Nephrol. 2008;19(6):1128–1138. doi: 10.1681/ASN.2007080917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.