Significance
In eukaryotes, the Atg1 kinase complex controls both autophagy induction and the recruitment of other autophagy proteins to the phagophore assembly site (PAS); however, it remains unclear how the Atg1 kinase complex itself is targeted to the PAS or regulated. In this study, we showed that the Atg17-Atg31-Atg29 complex displayed an elongated S-shaped structure, and the interaction of this complex with Atg11 was essential for recruiting the intact Atg1 kinase complex. The phosphorylation of Atg29 is the “switch” controlling the interaction, as it is required for both binding to Atg11 and the PAS targeting of the Atg17-Atg31-Atg29 complex.
Keywords: lysosome, organelle biogenesis, vacuole, yeast
Abstract
Macroautophagy (hereafter autophagy) functions in the nonselective clearance of cytoplasm. This process participates in many aspects of cell physiology, and is conserved in all eukaryotes. Autophagy begins with the organization of the phagophore assembly site (PAS), where most of the AuTophaGy-related (Atg) proteins are at least transiently localized. Autophagy occurs at a basal level and can be induced by various types of stress; the process must be tightly regulated because insufficient or excessive autophagy can be deleterious. A complex composed of Atg17-Atg31-Atg29 is vital for PAS organization and autophagy induction, implying a significant role in autophagy regulation. In this study, we demonstrate that Atg29 is a phosphorylated protein and that this modification is critical to its function; alanine substitution at the phosphorylation sites blocks its interaction with the scaffold protein Atg11 and its ability to facilitate assembly of the PAS. Atg29 has the characteristics of an intrinsically disordered protein, suggesting that it undergoes dynamic conformational changes on interaction with a binding partner(s). Finally, single-particle electron microscopy analysis of the Atg17-Atg31-Atg29 complex reveals an elongated structure with Atg29 located at the opposing ends.
Autophagy is the major lysosome/vacuole-dependent cellular degradative pathway. During autophagy, cytoplasmic constituents including proteins, lipids, and even entire organelles are surrounded by the phagophore, the initial sequestering compartment. The phagophore then expands to form double-membrane vesicles, termed autophagosomes. The completed autophagosomes fuse with lysosomes/vacuoles allowing access of the cargo to the degradative enzymes within this organelle; the resulting breakdown products are released back into the cytosol as building blocks or catabolic substrates (1–3). Autophagy is not only critical for survival during nutrient deprivation but is also involved in various human pathophysiologies, including cancer and neurodegeneration (4).
On autophagy induction, AuTophaGy-related (Atg) proteins accumulate at the phagophore assembly site (PAS) and initiate autophagosome formation. Among the 36 known Atg proteins, Atg1 is the only kinase, and it plays a particularly important role in autophagy induction by controlling the movement of other Atg proteins including Atg9 and Atg23 (5) and in the proper organization of the PAS (6, 7). Atg1 interacts with several proteins, including direct binding to Atg13 (which interacts with Atg17) and Atg11; the kinase activity of Atg1 is regulated in part by its binding to, and/or interaction with, some of these components (8, 9).
Atg17 constitutively forms a stable protein complex with Atg29 and Atg31 in both growing and nitrogen starvation conditions (10), and Atg31 directly interacts with Atg17 and Atg29 to bridge these two proteins (11). When autophagy is initiated, the Atg17-Atg31-Atg29 complex is first targeted to the PAS and recruits other Atg proteins, including Atg1 and Atg13, highlighting the significance of the ternary complex (12, 13). Along these lines, a single deletion of the ATG17, ATG29, or ATG31 genes results in a dramatic decrease in autophagy activity (14–16).
In this study, we examined the role of posttranslational modification of Atg29. We found that Atg29 is a phosphoprotein and that phosphorylation on the C-terminal domain is critical for autophagy activity; the N terminus of Atg29 contains the functional domain, whereas the C terminus plays a regulatory role. We continued and extended our study to include a structural and functional analysis of the Atg17-Atg31-Atg29 complex. Single-particle electron microscopy (EM) reveals that the recombinant Atg17-Atg31-Atg29 complex is present as an elongated S-shaped dimerized structure, with Atg17 forming the backbone. We further demonstrate that Atg29 has the characteristics of an intrinsically disordered protein (IDP), suggesting that the C-terminal half is flexible and capable of altering its conformation on binding to one or more interacting proteins. Finally, we determined that Atg11 is necessary and sufficient to recruit this complex to the PAS and that phosphorylation of Atg29 is required for its interaction with Atg11 and proper PAS localization.
Results
Atg29 Is a Phosphoprotein.
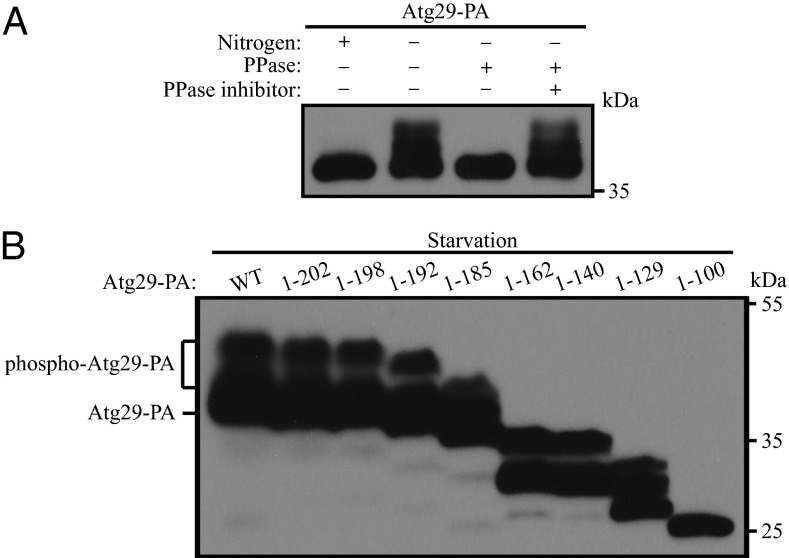
The activity of Atg1 kinase is regulated in part by various binding partners (8, 17). For example, Atg13 is critical for Atg1 kinase activity, and Atg13 function is regulated through phosphorylation (18–20). Similarly, a stable ternary complex composed of Atg17, Atg29, and Atg31 also interacts with the Atg1 kinase complex (13, 14); whereas the function of the Atg17-Atg31-Atg29 complex is not known, the absence of any of these proteins results in a substantial decrease in autophagy activity, suggesting that, similar to Atg13, they affect Atg1 kinase activity. Atg31 is a phosphoprotein (10), but the functional significance of its phosphorylation has not been demonstrated. To gain further insight into the mechanism of regulation of the Atg1 kinase complex, we decided to examine the phosphorylation status of its binding partner, Atg29. Following a shift from nutrient-rich to nitrogen starvation conditions, we noted that protein A (PA)-tagged Atg29 migrated as multiple bands (Fig. 1A). The highest molecular mass corresponded to an apparent increase of ∼9–10 kDa. To determine whether the reduced migration corresponded to phosphorylation, we performed a phosphatase treatment. Cells expressing Atg29-PA were shifted to starvation conditions, and lysates were generated. Treatment of cell lysates with λ protein phosphatase resulted in a collapse of the multiple bands to a single, lower mass band, whereas phosphatase treatment combined with phosphatase inhibitor resulted in a migration pattern that appeared similar to lysates without treatment (Fig. 1A). These data suggest that Atg29 is a phosphorylated protein and that phosphorylation, as assessed by SDS/PAGE, occurs when autophagy is induced.
Fig. 1.
Atg29 is a phosphoprotein. (A) Cells containing a chromosomal protein A-tagged Atg29 (HCY129) were grown in YPD and shifted to SD-N for 2 h. λ phosphatase and phosphatase inhibitor were added to the cell lysate as indicated. (B) Different forms of plasmid-encoded Atg29-PA with the indicated truncations were transformed into atg29∆ cells (HCY109). Cells were grown in SMD and shifted to SD-N for 2 h. Cell lysates were separated by SDS/PAGE and analyzed by Western blot.
To identify phosphorylation sites in Atg29 that affect autophagy activity, we generated several truncated forms of Atg29 and examined their migration pattern. All of the Atg29 constructs displayed multiple bands except for a construct that contained the N-terminal half of the protein, Atg29[1-100] (Fig. 1B). This finding suggested that most of the phosphorylated sites of Atg29, or at least those that contribute to the molecular mass shift detected by SDS/PAGE, are in the C terminus (amino acids 101–213), but not the N terminus (amino acids 1–100).
N-Terminal Domain of Atg29 Is Functional.
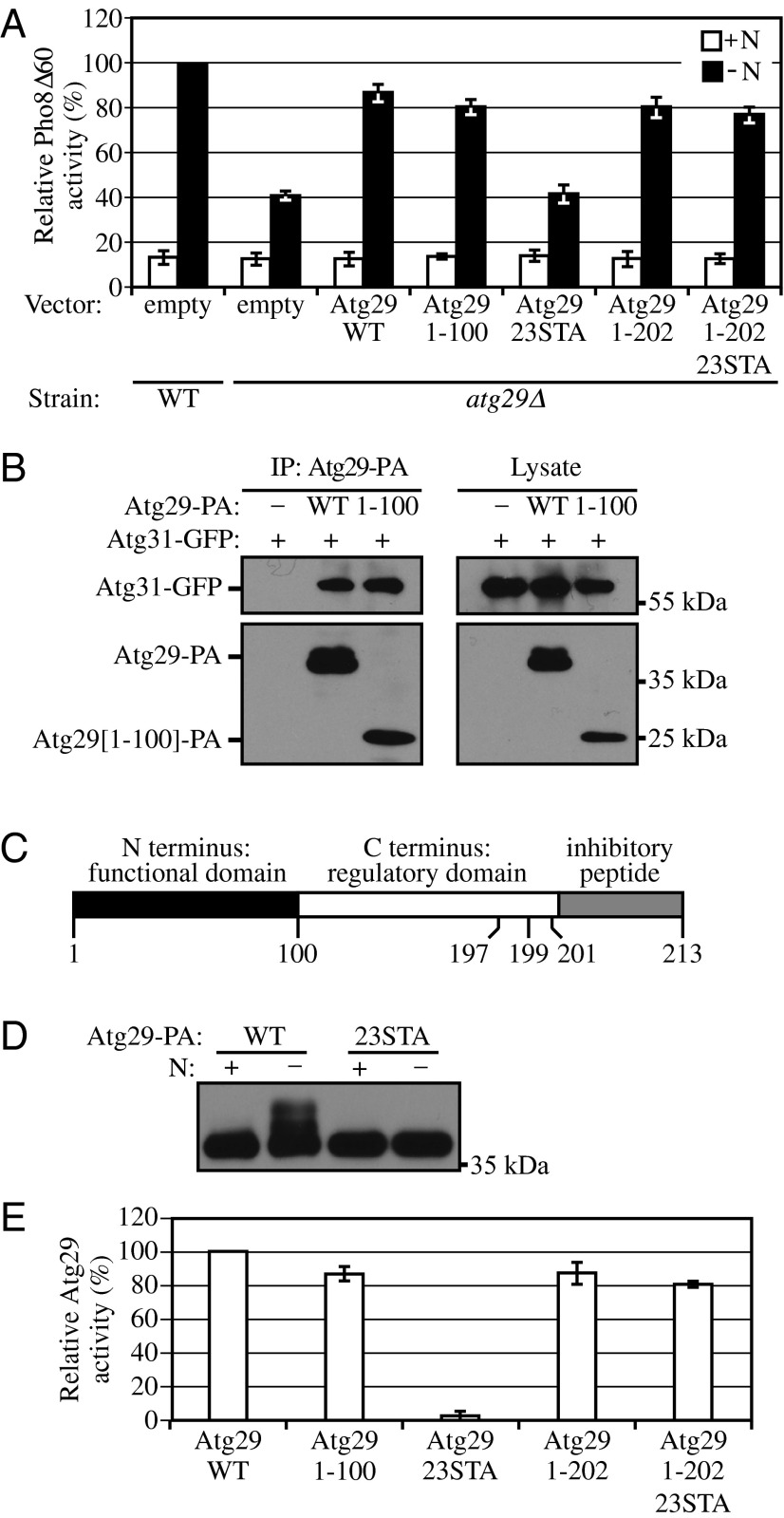
Considering that the two halves of Atg29 displayed different migration/phosphorylation characteristics, we decided to separately examine the two parts of the protein for their ability to complement the autophagy defect of the atg29∆ strain. The C terminus of Atg29 expressed by itself was unstable, whereas the N-terminal half of the protein displayed essentially normal stability (Fig. 1B). Therefore, we transformed the atg29∆ strain with a plasmid encoding either WT Atg29 or Atg29[1-100] or with the empty vector and monitored autophagy activity using the Pho8∆60 assay (21). In brief, Pho8∆60 is a truncated version of vacuolar alkaline phosphatase that lacks the N-terminal transmembrane domain. As a result of this deletion, Pho8∆60 cannot enter the endoplasmic reticulum and remains in the cytosol, where is serves as a nonselective cargo for autophagy. Pho8∆60-dependent alkaline phosphatase activity thus provides a quantitative readout for autophagy activity. The atg29∆ strain (atg29∆ with empty vector) displayed a significant block in autophagy activity under starvation conditions (Fig. 2A), in agreement with previous data (15), whereas transformation with the plasmid expressing WT Atg29 or the N-terminal half of the protein, Atg29[1-100], restored the autophagy activity to ∼80% of that seen in WT cells transformed with empty vector.
Fig. 2.
Atg29 contains distinct functional, regulatory, and inhibitory domains. (A) Pho8Δ60 WT (TN124) cells were transformed with empty vector; Pho8Δ60 atg29∆ (YIY36) cells were transformed with empty vector or a plasmid containing WT or different mutant forms of Atg29-PA as indicated. Cells were cultured in SMD to midlog phase (+N), and shifted to SD-N for 4 h. The Pho8Δ60 assay was performed as described in Materials and Methods. Error bars correspond to the SD and were obtained from three independent repeats. (B) Empty vector, plasmids encoding Atg29-PA, or Atg29[1-100]-PA was transformed into atg29∆ cells expressing Atg31-GFP (KDM1233). Cells were cultured in SMD to midlog phase, and cell lysates were prepared and incubated with IgG-Sepharose for affinity isolation as described in Materials and Methods. (C) A schematic diagram of the Atg29 protein. (D) The plasmid pAtg29-PA or pAtg29[23STA] was transformed into atg29∆ cells (HCY109). Cells were cultured in SMD, shifted to SD-N for 2 h, and analyzed by immunoblot. (E) Relative activities of different Atg29 mutants based on the results in A. The conversion from percent Pho8∆60 to relative Atg29 activity was carried out as described in the text.
Thus, Atg29[1-100] largely suppressed the autophagy defect of the atg29∆ mutant. Atg29 is associated with Atg17 and Atg31, through direct interaction with the latter (11). To further investigate the functionality of the N-terminal half of Atg29, we asked whether Atg29[1-100] retained the ability to associate with Atg31. Accordingly, we transformed the atg29∆ strain with plasmids encoding Atg29-PA and Atg31-GFP and carried out protein A affinity isolation with IgG-Sepharose. Both PA-tagged WT Atg29 and Atg29[1-100] coprecipitated Atg31-GFP (Fig. 2B). Therefore, we conclude that the N terminus of Atg29 constitutes a functional domain (Fig. 2C).
C Terminus of Atg29 Plays a Regulatory Role.
Because the N-terminal domain of Atg29 is able to complement the atg29∆ strain with regard to autophagy activity, we hypothesized that the C-terminal domain containing multiple phosphorylation sites plays a regulatory role. The C terminus of Atg29 contains 23 serine or threonine residues. Individual mutations of serine or threonine to alanine had no significant effect on Atg29 function, and deletions of individual genes encoding kinases did not completely eliminate phosphorylation. Because the number of possible permutations of kinase deletions is quite large, and the number of potential combinations of mutations in Atg29 phosphorylation sites makes systematic mutagenesis analysis impractical, we mutated all of these sites to alanine to generate Atg29[23STA]. This mutant migrated as a single lower mass band during SDS/PAGE when extracts were prepared from cells in either growing or nitrogen-starvation conditions (Fig. 2D). We then carried out a Pho8∆60 assay to assess the functionality of this construct. Atg29[23STA] was completely unable to complement the autophagy defect of the atg29∆ strain (Fig. 2A). The atg29∆ strain displays ∼40% of the autophagy activity of the WT strain. Therefore, for ease of comparison with the mutant constructs, we set the Pho8Δ60 activity of the atg29∆ strain expressing WT Atg29 to 100% and that of the atg29∆ strain expressing empty vector to 0 and normalized the results with the other forms of Atg29 as shown in Fig. 2E. The result with the Atg29[23STA] mutant indicated that the phosphorylation of Atg29 at its C terminus is crucial for activity. Furthermore, considering that the N-terminal half of Atg29 is able to complement the null strain for autophagy activity, these data suggest that the C terminus, or some portion of it, constitutes a negative regulatory domain (Fig. 2C).
During the course of mapping the phosphorylation sites in Atg29, we generated a series of C-terminal truncations (Fig. 1B). We began to systematically combine these truncations with the Atg29[23STA] construct to define a domain that no longer inhibited the function of the N terminus. The C-terminal 11 amino acids of Atg29 (residues 203–213) lack any potential phosphorylation sites, and Atg29[1-202] retained ∼90% of the Pho8∆60 activity of the WT protein (Fig. 2 A and E). When the truncation of the extreme C-terminal 11 amino acids was combined with the nonphosphorylable mutant, which by itself lacked autophagy activity, the double mutant construct Atg29[1-202/23STA] displayed ∼80% of the Pho8∆60 activity of the WT (Fig. 2 A and E). Therefore, we conclude that the last 11 amino acids play a negative role in Atg29 function, indicating that the extreme C terminus of Atg29 acts as an inhibitory peptide (Fig. 2C). The observation that the truncation of the extreme C terminus restores function to Atg29[23STA] suggests that at least one function of phosphorylation is to alter the conformation of Atg29 such that the inhibitory peptide is displaced, presumably altering its interaction with another component of the Atg1 kinase complex.
Residues Adjacent to the Inhibitory Peptide Are Important in Atg29 Function.
Of the 23 serine and threonine residues in the C-terminal half of Atg29, the last three serines (S197, S199, and S201) are very close to the inhibitory peptide. Therefore, we proposed that these residues would have a particularly important role in Atg29 function. Accordingly, we mutated S197, S199, and S201 to alanine to generate Atg29[3SA]. We also generated the complementary construct by mutating the first 20 serines and threonines to alanine to generate Atg29[20STA] in which the last three potential phosphorylation sites were left unchanged. In contrast to Atg29[23STA], which displayed essentially no Pho8∆60 activity, Atg29[20STA] retained ∼65% of the activity of WT Atg29, which was presumably due to phosphorylation of one or more of the last three serines (Fig. 3 A and B). Along these lines, mutation of only the last three serine residues had an even stronger effect, as Atg29[3SA] only exhibited ∼45% of the Pho8∆60 activity of WT (Fig. 3 A and B). These results suggest that the phosphorylation of the last three serines plays a particularly important role for Atg29 function.
Fig. 3.
Phosphorylation of serine residues adjacent to the inhibitory peptide is important for Atg29 function. (A) Pho8Δ60 WT (TN124) cells were transformed with empty vector; Pho8Δ60 atg29∆ (YIY36) cells were transformed with empty vector or a plasmid containing WT or different mutant forms of Atg29-PA as indicated. Cells were cultured in SMD (+N) to midlog phase and shifted to SD-N for 4 h. Protein extracts were analyzed using the Pho8Δ60 assay. Error bars correspond to the SD and were obtained from three independent repeats. (B and C) Relative activities of different Atg29 mutants based on the results in A.
To extend our analysis of the serines at positions 197, 199, and 201 of Atg29, we made phosphomimetic mutants, Atg29[3SD], or Atg29[3SE] by substituting them with either aspartic acid or glutamic acid, and examined the effect with regard to autophagy activity. Both Atg29[3SD] and Atg29[3SE] retained ∼80% of WT activity (Fig. 3 A and C), indicating that the phosphomimetic mutants functioned similar to WT Atg29. Even in the context of the Atg29 mutant where the remainder of the C-terminal sites could not be phosphorylated (i.e., Atg29[20STA]), the phosphomimetic mutants (Atg29[20STA/3SD] or Atg29[(20STA/3SE]) retained ∼80% of WT Atg29 activity (Fig. 3 A and C). Together, these data suggest that phosphorylation of multiple residues of the Atg29 C terminus results in a mass action effect that alters the conformation of the inhibitory peptide with respect to some other protein, possibly another component of the Atg1 kinase complex.
Atg29 Is an Intrinsically Disordered Protein.
The results of the above analysis suggest that Atg29 undergoes significant conformational changes on phosphorylation. This behavior is not unusual for a large group of functional proteins that are either entirely disordered or contain long disordered domains under physiological conditions. These polypeptides are referred to as intrinsically disordered proteins (IDPs) (22–25). Because IDPs can clearly be discriminated from the group of globular proteins that gain a fixed rigid 3D structure in solution, we examined whether or not Atg29 is an IDP.
The presence of long disordered regions makes IDPs very sensitive to proteolytic cleavage, and disordered regions correspond to missing electron density in protein structures (23). The ribbon model of Atg29 that was obtained as part of the crystal structure of the Atg17-Atg31-Atg29 complex (26) lacks the C-terminal half of the protein, due to sensitivity to proteolytic cleavage (Fig. S1A). Furthermore, from the 79 residues that were used in the attempted crystallization, residues 41–50 are missing, and residues 51–79 could not be assigned to specific amino acids. These results indicate that Atg29 is a solvent-accessible polypeptide with a high degree of mobility that leads to noncoherent X-ray scattering, making flexible atoms unobservable or hard to distinguish.
Unfoldedness of IDPs is encoded in their amino acid sequence (22–24). We carried out a disorder prediction for Atg29 (Fig. 4A) using IUPred, an algorithm that evaluates the tendency for intrinsic disorder based on the observation that globular proteins have an ordered structure that is stabilized by a large number of interresidue interactions, whereas intrinsically disordered proteins possess an amino acid composition that does not allow these interactions (27). IDPs contain random coil domains of at least 40 consecutive amino acid residues (23). The IUPred analysis predicts that only the first half of Atg29, up to Ser96, is structured, whereas the remaining part of Atg29 (>100 amino acids including the entire C terminus) is disordered when the protein is present alone in solution. However, in the presence of a structured binding partner, this domain may undergo a disorder-to-order transition. The ANCHOR predictor (28) identifies small hydrophobic domains (typically present as α-helical linear motifs) present within disordered regions; these domains are too small to favor intramolecular interactions (i.e., aggregation), but facilitate intermolecular binding, whereas the disordered regions themselves are generally low in binding capacity. Analysis using this algorithm revealed a high probability for disordered binding regions in the C-terminal regulatory domain (Fig. 4A). In this regard it is noteworthy that the extreme C terminus of Atg29 including the inhibitory peptide was identified as the longest disordered binding region and exhibited the highest binding tendency, in agreement with its proposed regulatory role involving interaction with another component of the Atg1 kinase complex.
Fig. 4.
Atg29 is an intrinsically disordered protein. (A) Tendency for intrinsic disorder and probability of an Atg29 residue being in a disordered binding region, as predicted by IUPred and ANCHOR, respectively. Gray areas, generated by ANCHOR, identify the disordered binding segments in Atg29 that are likely to be stabilized through binding to a globular protein partner. The Met residue at the Atg29 N terminus was not included in the calculations for this plot and the plots in B and Fig. S1B. (B) Compositional profiling of Atg29. The gray and black bars show the fractional difference [calculated as (CX − CPDB25)/CPDB25] in amino acid composition of Atg29 and IDPs from the DisProt database, respectively, relative to a reference set of proteins in PDB (31) that is biased toward the composition of proteins responsive to crystallization. CX is the content of amino acids in Atg29 or IDPs in DisProt and CPDB25 is the corresponding value for proteins in the PDB protein set. A negative/positive fractional difference indicates depletion/enrichment in the corresponding amino acid. Amino acids are arranged on the x axis from the most rigid to the most flexible according to the Vihinen’s flexibility scale (48). (C) Migration of Atg29 fusion proteins following SDS/PAGE. (Left) Atg29 fused to the MOCR solubilization tag was visualized by immunoblotting the E. coli cell lysate with antiserum to polyHis. (Right) Atg29 fused to protein A was detected by immunoblotting the yeast cell extract with antiserum that detects PA.
A very typical characteristic of IDPs is their special position in the charge-hydrophobicity (CH) plot given by a combination of their low mean hydrophobicity <H> and high mean net charge <R> (22, 23). Atg29 occupies the same area in the CH plot as known IDPs such as Sic1 (29) (Fig. S1B). Because disorder propensity is embedded directly in the amino acid sequence, it follows that IDPs in the DisProt database (30) are depleted in rigid, order-promoting residues and enriched in flexible, disorder-promoting residues (24). Compositional profiler (31) shows that Atg29 has an amino acid composition similar to IDPs in the DisProt database (Fig. 4B).
Another feature that Atg29 shares with IDPs is its anomalous migration during SDS/PAGE (Fig. 4C). The molecular mass of Atg29 calculated from its amino acid sequence is 24.7 kDa. With the 16.7-kDa MOCR [monomeric mutant of the Ocr protein (0.3 gene product) of bacteriophage T7] purification tag (Materials and Methods), the expressed and purified Atg29 migrated as an ∼48-kDa protein, and with the 14.5-kDa PA tag, the molecular mass of the Atg29-PA fusion protein in yeast was ∼46 kDa. Untagged Atg29 purified from Escherichia coli appeared to be ∼31 kDa (Fig. 5A). Thus, Atg29 migrated at a molecular mass ∼6.8 kDa larger than predicted. This type of aberrant migration typically results from poor interaction of the protein with SDS due to a higher content of charged amino acid residues compared with globular proteins (32). Taken together, these data suggest that Atg29 is an IDP.
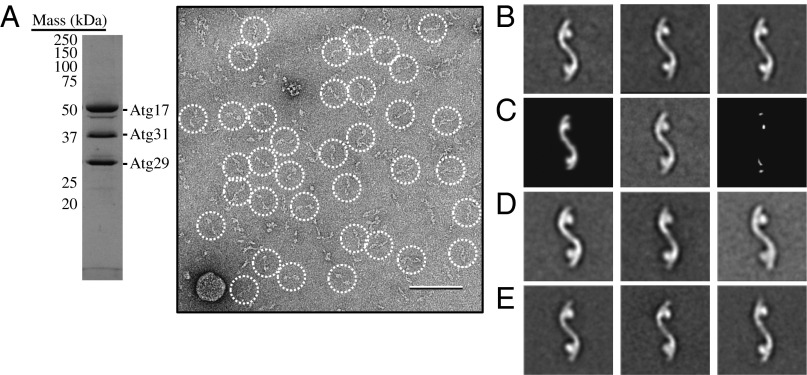
Fig. 5.
The Atg17-Atg31-Atg29 complex adopts an elongated S-shaped structure. (A) Coomassie blue–stained SDS/PAGE analysis of gel filtration-purified recombinant Atg17-Atg31-Atg29 (Left). A raw image taken from a negatively stained recombinant Atg17-Atg31-Atg29 specimen (Right). Particles are circled. (Scale bar, 100 nm.) (B) Representative class averages obtained from reference-free classification of 6,788 negatively stained Atg17-Atg31-Atg29 particles into 50 classes. Each of the three classes contains between 110 and 220 particles. The side length of each panel is 52 nm. (C) Difference mapping between the high-resolution crystal structure projection (Left) and the negative-stained EM average of the full-length components from S. cerevisiae (Center). The difference image (Right) indicates that the Atg29 C terminus is located in the globular domains. (D) Representative class averages obtained from classification of 4,671 negatively stained Atg29[3SD]-containing particles into 50 classes. Each of the three classes contains between 100 and 190 particles. (E) Representative class averages obtained from classification of 6,588 negatively stained Atg29[20STA/3SD]-containing particles into 50 classes. Each of the three classes contains between 150 and 170 particles.
Single-Particle EM Analysis of the Atg17-Atg31-Atg29 Complex.
Atg29 exists in a stable protein complex that also contains Atg17 and Atg31. To better understand the structure and function of Atg29, we carried out negative stain single-particle EM analysis. Coomassie blue–stained SDS/PAGE analysis confirmed that the ternary complex purified using the previously published procedure contained stoichiometric quantities of Atg17, Atg31, and Atg29 (Fig. 5A) (10). Next, negative-stained specimens were prepared by adsorbing purified Atg17-Atg31-Atg29 to glow discharged carbon-coated EM grids and staining with uranyl formate. Initial images obtained for these specimens revealed that the purified complex was relatively homogeneous and had an elongated shape (Fig. 5A).
To gain further insights into the structural features of the Atg17-Atg31-Atg29 complex, we selected a total of 6,788 particles from the raw images and subjected these particle images to reference-free alignment and classification. The gallery of average images revealed that Atg17-Atg31-Atg29 contained a backbone that had an overall shape resembling a stretched out letter S, with two circular domains attached to opposite ends (Fig. 5B). These averages also showed that Atg17-Atg31-Atg29 is dimeric, with the twofold symmetry axis located at the center of the complex. The observed stoichiometry is in agreement with yeast two-hybrid studies, which showed that Atg17 self-associates (14), and with recent analytical ultracentrifugation experiments, which showed that Atg17 forms a 2:2:2 complex with Atg29 and Atg31 (10). The overall architecture of this complex resembles the recently determined crystal structure of the Lachancea thermotolerans Atg17-Atg31-Atg29 core complex (26), suggesting that the Saccharomyces cerevisiae complex has a similar subunit organization as the L. thermotolerans complex. In other words, S. cerevisiae Atg17 forms the backbone, whereas Atg31 and Atg29 constitute the terminal globular domains.
The crystallized complex, however, lacks more than 100 C-terminal residues of Atg29, the domain that we determined was subject to regulatory phosphorylation and that contains the C-terminal inhibitory peptide, whereas the S. cerevisiae complex, we analyzed contains full-length components. To localize the Atg29 C-terminal domain, a difference mapping approach was implemented. This approach involves generating a series of 2D projections corresponding to different views of the high-resolution model and then experimentally determining the projection that correlates best with a representative 2D average obtained for the S. cerevisiae complex. Next, a difference image was calculated by subtracting the identified 2D projection from the experimental 2D average. Strong difference densities adjacent to each of the two globular domains were observed, suggesting that the C-terminal domains of Atg29 may be projecting away from these domains (Fig. 5C). The two weaker densities were also located on opposite ends of the complex, but these could be attributed to flattening of the particle by the negative staining procedure, leading to a slight extension to its overall length (Fig. 5C).
The recombinant Atg17-Atg31-Atg29 complex that we structurally characterized is nonphosphorylated and presumably in the non–autophagy-inducing state. To examine the autophagy-inducing state of Atg17-Atg31-Atg29, we reconstituted complexes containing the phosphomimetic mutants Atg29[3SD] or Atg29[20STA/3SD] and subjected the purified complexes to negative stain 2D EM analysis. Two galleries of class averages were obtained from the classification of 4,671 Atg29[3SD]-containing and 6,558 Atg29[20STA/3SD]-containing particles, respectively (Fig. 5 D and E). These averages indicated that the phosphomimetic mutations did not significantly alter the overall structure of the ternary complex. This finding is in agreement with the earlier assignment of the C-terminal regulatory domain of Atg29 to the periphery of the Atg17-Atg31-Atg29 complex (Fig. 5C). Although the Atg29 regulatory domain is not directly engaged in assembling and/or stabilizing this complex, it localizes to a highly accessible region suitable for engaging in interactions with other Atg proteins and/or components of the Atg1 signaling machinery.
In Vivo Reconstitution and Analysis of the Atg17-Atg31-Atg29 Complex.
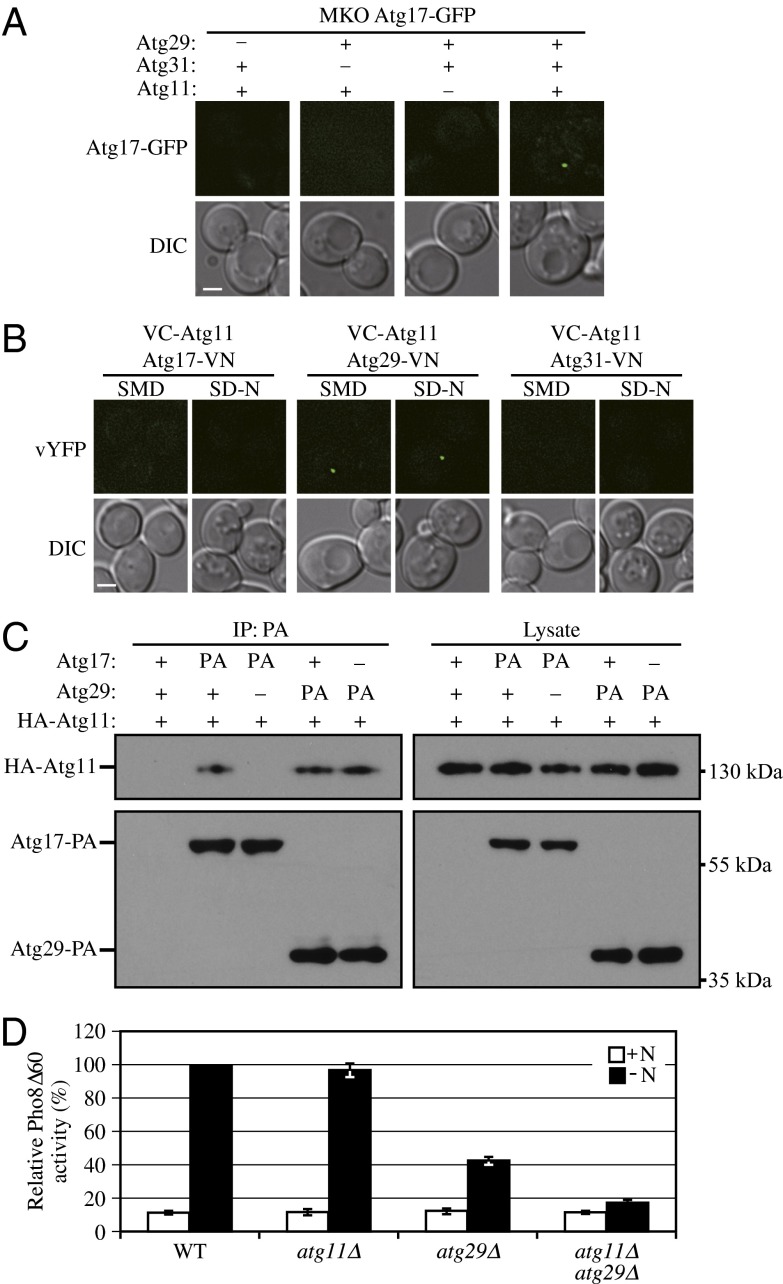
The Atg17-Atg31-Atg29 complex is considered to be one of the first sets of proteins that assemble at the PAS, and it plays a role in the recruitment of other Atg proteins. However, the complexity of the interaction network of Atg proteins has made it difficult to completely dissect the process of PAS organization. Therefore, we tried to reconstitute assembly of the ternary complex in vivo in our multiple KO (MKO) strain (33) and used Atg17-GFP as the reporter to observe the PAS localization of the complex. In the MKO strain, the ATG genes whose products are involved in autophagosome formation have been deleted; this allows us to eliminate contributions from other Atg proteins and determine the minimum components needed for the assembly and correct localization of the complex. When we expressed both Atg29 and Atg31 together with Atg17-GFP, we were unable to observe puncta corresponding to Atg17-GFP (Fig. 6A). Similarly, strains expressing Atg29 or Atg31 along with Atg11 did not display Atg17-GFP puncta. In contrast, if Atg11 was coexpressed with all three proteins, the Atg17-GFP puncta were now detected (Fig. 6A). Thus, in the absence of all other known Atg proteins, Atg11 is necessary and sufficient to recruit the Atg17-Atg31-Atg29 complex to the PAS.
Fig. 6.
Atg11 is important for PAS recruitment of the Atg17-Atg31-Atg29 complex. (A) An empty vector or a plasmid expressing Atg29-PA was coexpressed with empty vector or a plasmid encoding HA-Atg11 under the control of the CUP1 promoter (pCuHA-Atg11) in MKO ATG17-GFP (HCY107) or MKO ATG17-GFP atg31∆ (HCY108) cells as indicated. Cells were cultured in SMD to early log phase. (B) VC-ATG11 ATG17-VN (KDM1551), VC-ATG11 ATG29-VN (KDM1552), and VC-ATG11 ATG31-VN (KDM1553) cells were cultured in YPD and shifted to SD-N for 2 h. All of the cell samples in A and B were observed by fluorescence microscopy. The images are representative pictures from single Z-sections. DIC, differential interference contrast. (Scale bar, 2 μm.) (C) The plasmid pCuHA-Atg11(416) was transformed into WT (SEY6210), ATG17-PA (KDM1234), ATG17-PA atg29Δ (KDM1235), ATG29-PA (HCY129), or ATG29-PA atg17Δ (HCY125) cells. Cells were cultured in SMD, and cell lysates were prepared and incubated with IgG-Sepharose for affinity isolation as described in Materials and Methods. The eluted proteins were separated by SDS/PAGE and detected with monoclonal antibody that recognizes HA or PA. (D) Pho8Δ60 WT (TN124), atg11Δ (KDM1406), atg29Δ (YIY36), or atg11Δ atg29Δ (KDM1407) cells were cultured in YPD (+N) and shifted to SD-N for 4 h. The Pho8Δ60 assay was performed. Error bars correspond to the SD and were obtained from three independent repeats.
Previous studies from our laboratory suggested that Atg17 interacts with Atg11 (34); however, neither ATG29 nor ATG31 had been identified at that time. Therefore, we decided to reexamine which component of the Atg17-Atg31-Atg29 complex was the direct binding partner of Atg11. To carry out this analysis, we took advantage of the bimolecular fluorescence complementation assay (35). In this system, the Venus YFP is split into two fragments, corresponding to the N terminus (VN) and C terminus (VC). We fused VC to Atg11 and VN to Atg17, Atg29, or Atg31 by integrating the corresponding constructs at the ATG11, ATG17, ATG29, or ATG31 loci on the yeast genome. Fluorescence from these chimeras can only be observed when the two proteins interact and bring the two fluorophore fragments into close proximity. Bright dots were observed in cells expressing VC-Atg11 and Atg29-VN, but no fluorescent signal was observed in cells expressing either VC-Atg11 and Atg17-VN or VC-Atg11 and Atg31-VN (Fig. 6B). To confirm the interaction between Atg29 and Atg11, we carried out protein A affinity isolation with IgG-Sepharose. PA-tagged Atg17 and Atg29 were both able to coprecipitate HA-Atg11. When Atg17 was absent, Atg29 could still pull down Atg11; however, when Atg29 was absent, Atg17 no longer interacted with Atg11 (Fig. 6C). These results indicate that Atg29, but not Atg17, directly interacts with Atg11 to facilitate the association of Atg11 with the complex and to achieve its PAS localization.
Atg11 is a scaffold protein required for selective autophagy that appears to be dispensable for nonselective autophagy, as deletion of the ATG11 gene has no obvious effect on autophagy activity (9). However, some lines of evidence also suggest that Atg11 might have a role in nonselective autophagy. For example, a small number of autophagic bodies (the single-membrane vesicles that result from the fusion of autophagosomes with the vacuole) are detected in the vacuole in atg17Δ cells (7), consistent with the fact that autophagy is not completely blocked when Atg17 is absent. However, no autophagic bodies are detected in atg11Δ atg17Δ cells, suggesting that Atg11 contributes to autophagic body (and hence autophagosome) formation in the absence of Atg17. Atg11 is proposed to function as the initial component localizing to the PAS in vegetative conditions, facilitating the formation of the starvation-specific PAS on the induction of autophagy (7). In the present study, we found that Atg11 interacted with Atg29 and was required for the PAS targeting of the Atg17-Atg31-Atg29 complex in MKO cells. Therefore, we extended our analysis of the role of Atg11 in nonselective autophagy with the Pho8∆60 assay. In atg11Δ cells, autophagy activity was normal in agreement with previous studies, whereas in atg29Δ cells, autophagy activity was ∼40–45% the level of the WT. In contrast, when both ATG11 and ATG29 were deleted, autophagy was almost completely blocked (Fig. 6D). Therefore, Atg11 appears to contribute to nonselective autophagy when Atg29 is absent.
Phosphorylation of Atg29 Is Required for Its Binding to Atg11 and the PAS Targeting of the Atg17-Atg31-Atg29 Complex.
Because we found that the phosphorylation of Atg29 is important for autophagy activity, we decided to examine the role of Atg29 phosphorylation in the PAS targeting of the Atg17-Atg31-Atg29 complex. We coexpressed Atg11, Atg31, and either WT or mutant Atg29 in the MKO strain and again used Atg17-GFP as the reporter. With WT Atg29, ∼22% or ∼33% of the cells contained Atg17-GFP puncta in growing or nitrogen starvation conditions, respectively (Fig. 7 A and B). The phosphomimetic mutants Atg29[3SD] and Atg29[3SE] were also able to support Atg17-GFP puncta formation at essentially WT levels (Fig. 7 A and B). In contrast, no Atg17-GFP puncta were observed with the nonphosphorylable Atg29[23STA] mutant in either growing or nitrogen starvation conditions (Fig. 7 A and B). These results suggest that the phosphorylation of Atg29 is required for the PAS recruitment of the Atg17-Atg31-Atg29 complex.
Fig. 7.
Phosphorylation of Atg29 is required for binding to Atg11. (A) Plasmids encoding Atg29, Atg29[3SD], Atg29[3SE], or Atg29[23STA] were coexpressed with HA-Atg11 under the control of the CUP1 promoter (pCuHA-Atg11) in MKO ATG17-GFP (HCY107) cells. Cells were cultured in SMD and shifted to SD-N for 2 h, and cell samples were observed by fluorescence microscopy. The images are representative pictures from single Z-sections. DIC, differential interference contrast. (Scale bar, 2 μm.) (B) Quantification of Atg17-GFP dots. Twelve Z-section images were projected and the percentage of cells containing an Atg17-GFP puncta was determined. The SD was calculated from three independent experiments. (C) A plasmid encoding Atg29 or Atg29[23STA] was coexpressed with pCuHA-Atg11 in MKO ATG17-GFP (HCY107) cells. Cells were cultured in SMD, and cell lysates were prepared and incubated with IgG-Sepharose for affinity isolation.
Because Atg29[23STA] could not promote PAS targeting of the Atg17-Atg31-Atg29 complex, we suspected that the phosphorylation of Atg29 was required for its association with the complex or for interaction with Atg11. Therefore, we used WT Atg29-PA or Atg29[23STA]-PA to pull down Atg17-GFP and HA-Atg11 in MKO cells. Atg17-GFP was coprecipitated by both WT Atg29-PA and Atg29[23STA]-PA, in agreement with the observation that the Atg17-Atg31-Atg29 complex is present in vegetative conditions (i.e., when we do not detect phosphorylation of the C terminus) (10); however, only WT Atg29-PA was able to coprecipitate HA-Atg11 (Fig. 7C). Therefore, the phosphorylation of Atg29 is dispensable for its association with Atg31 and Atg17 but is required for interaction with Atg11 and the PAS targeting of the Atg17-Atg31-Atg29 complex.
Atg17-Atg31-Atg29 Complex and Atg11 Cooperate to Recruit the Intact Atg1 Kinase Complex.
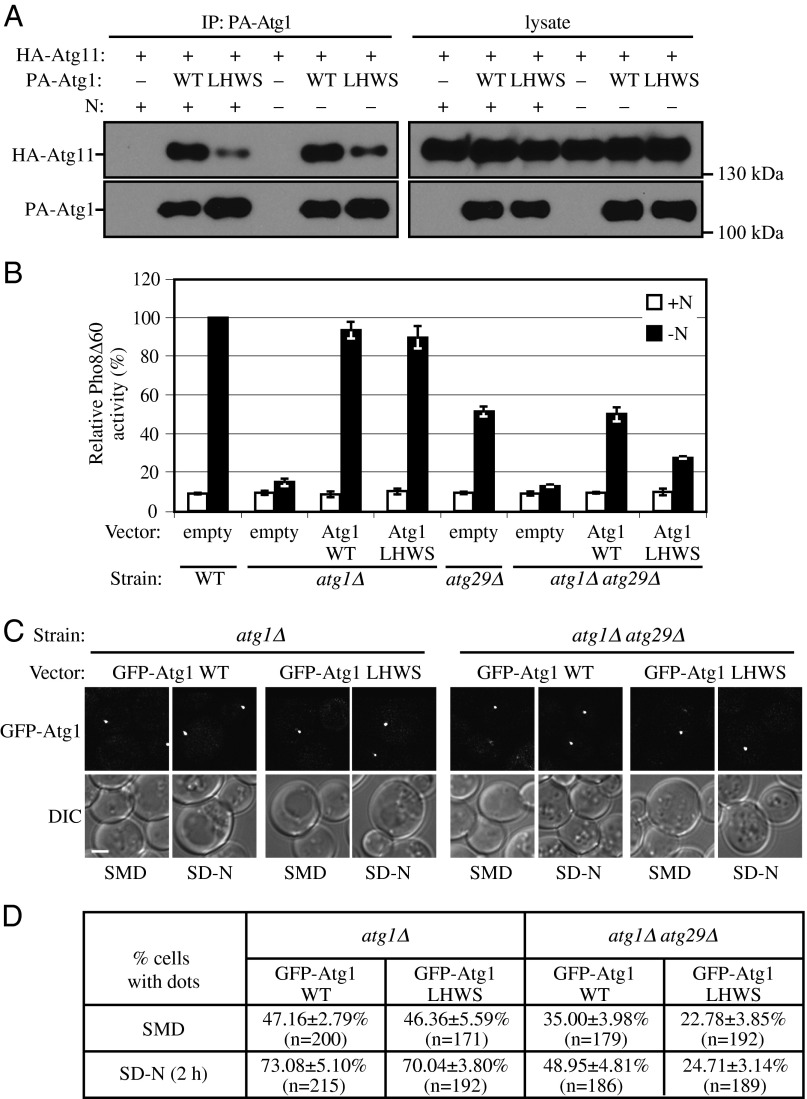
Previous work showed that Atg11 directly interacts with Atg1, although the physiological role of this interaction is unknown (7, 9). In the absence of Atg17 or Atg29, Atg11 permits the cells to retain residual autophagy activity (7). Therefore, we hypothesized that the intact Atg1 complex consists of (at least) six subunits, which form an interacting structure that controls autophagy induction. To test our hypothesis and gain further insight into the physiological role of the Atg1-Atg11 interaction, we combined random mutagenesis and yeast two-hybrid assays and identified an Atg1 mutant, Atg1L825H,W826S [hereafter Atg1(LHWS)]. Atg1(LHWS) showed a significant decrease in its interaction with Atg11, as detected by protein A affinity isolation (Fig. 8A). Importantly, Atg1(LHWS) had normal Atg1 function other than decreased interaction with Atg11, as the plasmid-driven Atg1(LHWS) was able to rescue the defect of autophagy activity essentially as well as WT Atg1 (Fig. 8B).
Fig. 8.
Atg29 and Atg11 have redundant roles in the recruitment of Atg1 to the PAS. (A) The plasmids pCuPA(416) or pCuPA-Atg1(416) and pCuHA-Atg11(414) were transformed into atg1∆ atg11∆ cells (KDM1270). Cells were cultured in SMD, and cell lysates were prepared and incubated with IgG-Sepharose for affinity isolation as described in Materials and Methods. The eluted proteins were separated by SDS/PAGE and detected with monoclonal antibody that recognizes HA or PA. (B) The plasmid pRS414, pAtg1(414), or pAtg1(LHWS)(414) was transformed into either Pho8Δ60 WT (WLY176), atg1Δ (KDM1408), atg29Δ (KDM1409), or atg11∆ atg29Δ (KDM1410) cells. Cells were cultured in SMD (+N) and shifted to SD-N (−N) for 4 h. The Pho8Δ60 assay was performed as described in Materials and Methods. Error bars correspond to the SD and were obtained from three independent repeats. (C) The plasmid pGFP-Atg1(416) or pGFP-Atg1(LHWS)(416) was transformed into either atg1∆ (WHY1) or atg1∆ atg29∆ (TKY12) cells. Cells were cultured in SMD and shifted to SD-N for 2 h, and cell samples were observed by fluorescence microscopy. The images are representative pictures from single Z-sections. DIC, differential interference contrast. (Scale bar, 2 μm.) (D) Quantification of C. Twelve Z-section images were projected, and the percentage of cells that contained GFP-Atg1 dots was determined. SD was calculated from three independent experiments.
Atg1 protein kinase is the key subunit of the Atg1 complex, and we showed that Atg11 is necessary and sufficient to recruit the Atg17-Atg31-Atg29 complex to the PAS (Fig. 6A). We hypothesized that the Atg17-Atg31-Atg29 complex and Atg11 together form a scaffold for the recruitment of Atg1 to the PAS. To test this possibility and to preclude interference from endogenous WT Atg1, we transformed plasmids expressing chimeras of GFP fused to either WT Atg1 or Atg1(LHWS) into atg1∆ cells. WT GFP-Atg1 and GFP-Atg1(LHWS) displayed similar levels of puncta formation in both growing and starvation conditions (Fig. 8 C and D). Loss of the Atg1-Atg11 interaction resulting from the presence of GFP-Atg1(LHWS) may not have had an obvious effect on autophagy activity (Fig. 8B) or puncta formation (Fig. 8 C and D) because of compensation from the Atg11-Atg29 interaction, which was able to retain Atg11 within the Atg1 complex. To test this hypothesis, we next deleted the ATG29 gene. In atg1∆ atg29∆ cells, WT GFP-Atg1 showed reduced puncta formation compared with that in atg1∆ cells in both growing and starvation conditions (Fig. 8 C and D), which suggested that Atg29 is important for the PAS recruitment of Atg1. GFP-Atg1(LHWS) showed a further decrease in puncta formation in the double deletion background (Fig. 8 C and D), which implied that the Atg1-Atg11 interaction is essential for Atg1 recruitment when Atg29 is absent (or when Atg11 loses its interaction with Atg29 due to the absence of Atg29 phosphorylation).
Finally, we asked whether the change in puncta formation corresponded to a change in autophagy activity. Although both WT Atg1 and Atg1(LHWS) expressed from plasmids were able to rescue the defect of autophagy activity in cells with only an ATG1 deletion (Fig. 8B), when we also deleted the ATG29 gene, only plasmid-driven WT Atg1, but not Atg1(LHWS), rescued the autophagy defect (Fig. 8B). Therefore, the Atg1-Atg11 interaction is also essential for autophagy activity when Atg29 is absent or cannot be phosphorylated.
Discussion
Autophagy is crucial for cellular metabolism, but either excessive or insufficient autophagy can be deleterious. Therefore, the activity of this process has to be tightly controlled. The Atg1 kinase complex, which includes Atg1, Atg13, Atg17, Atg29, and Atg31, is a key regulator of autophagy induction and inhibition, and the activity of these proteins is modulated in part by posttranslational modifications. For example, Atg1, Atg13, and Atg31 are all phosphoproteins, and the functions of at least Atg1 and Atg13 are regulated by phosphorylation (10, 19, 20, 36). Here, we show that Atg29 is also a phosphoprotein, and we determine the physiological role of its phosphorylation.
Based on our results, we suggest that in nutrient-rich conditions, when autophagy is at a basal level, the activity contributed by Atg29 is blocked by an inhibitory peptide located at its extreme C terminus. On autophagy induction, Atg29 is phosphorylated, and the modification of the last three serine residues in particular promotes a conformational change and release of the inhibition, thus allowing the full activity of the N-terminal functional domain.
The phosphorylation of Atg29 affected its ability to interact with Atg11. All these results are consistent with our protein analysis that Atg29 belongs to a new class of very flexible polypeptides called intrinsically disordered proteins. IDPs have a number of functional advantages over the function of structured proteins. For example, they have larger interacting surfaces compared with their ordered partners, exhibit increased speed of interactions and high specificity coupled with low affinity, carry recognition segments that can fold after binding to a native partner or cofactor, have accessible posttranslational modification sites, and possess conformational flexibility that allows for structural adaptation through interaction with several partners; they can also be involved in very distinct functional modes, for example, acting as both inhibitors and activators of their partner enzyme(s) (22, 32, 37, 38).
The lines of evidence that Atg29 is an IDP come mainly from the characteristics that originate in the protein’s amino acid sequence and that have been shown by numerous studies (22–25, 29) to be distinct for IDPs relative to ordered (globular) proteins. We were not able to apply a full set of experimental techniques that would provide comprehensive characterization of this protein, because the purified Atg29 could not be obtained in the monomeric form; the protein exhibited a very significant propensity for aggregation under different experimental conditions. This tendency to aggregate further supported our finding that Atg29 is disordered, because this type of proteins exists as an ensemble of conformations without a single minimum in the energy landscape model, and as a result is always more prone to intermolecular interactions or aggregation in the absence of a natural binding partner in solution (25). Given the enormous versatility of IDPs, we conclude that the presence of at least Atg29 as one component representing this class of proteins in the constitutively formed regulatory Atg1 kinase complex is consistent with the requirement for prompt regulation of autophagy induction in response to changing intracellular conditions.
Our single-particle EM analysis of the Atg17-Atg31-Atg29 complex revealed an S-shaped architecture that is consistent with the crystal structure of the recently published L. thermotolerans complex (26), although we note that the crystal structure does not include substantial portions of Atg29 or Atg31, including the entire C-terminal half of Atg29 that we show in the present study is subject to phosphorylation. Atg29 is located at the ends of the dimeric scaffold that is formed primarily by the rigid structure of Atg17. Considering this location, it is not surprising that the introduction of phosphomimetic mutations to substitute for the last three serine residues in Atg29 does not change the overall structure of the complex at this level of resolution. We have not yet succeeded in purifying the complex from yeast under autophagy-inducing conditions, so we cannot rule out the possibility that the phosphorylated protein will display a more obvious conformational change. At any rate, we found that the phosphorylation state of Atg29 affected its ability to interact with Atg11.
Atg11 is a selective autophagy scaffold protein, binding to a variety of cargo receptors to mediate different types of selective autophagy (39–41). Atg11 directly interacts with Atg1 and is involved in the activation of Atg1 kinase activity (8, 9). In the absence of Atg17 or Atg29, Atg11 permits the cells to retain residual autophagy activity, even though the deletion of the ATG11 gene by itself does not have an obvious effect on autophagy activity (7, 9). The hierarchical study of the Atg proteins along with other work suggests that the Atg17-Atg31-Atg29 complex comprises the first set of proteins localized to the PAS (12, 13). However, in the absence of other Atg proteins (as in our MKO strain), this complex is diffuse in the cytosol rather than accumulating at the PAS; under these conditions, Atg11 is necessary and sufficient for PAS recruitment.
Atg11 directly interacts with Atg1, although the physiological role of this interaction is unknown (9). With regard to Atg1 kinase activity, the Atg17-Atg31-Atg29 complex can interact with Atg1 via the binding of Atg13 to Atg17 or through the binding of Atg11 to Atg29. This finding may explain the residual autophagy activity seen in atg17Δ or atg29Δ cells, and the complete loss of autophagy activity in atg11Δ atg17Δ or atg11Δ atg29Δ cells. Atg1 and Atg13 are the core subunits within the complex; therefore, both nonselective and selective autophagy are blocked when ATG1 or ATG13 are deleted. Atg11 and the Atg17-Atg31-Atg29 complex have partly redundant roles as scaffolds for PAS organization (7); under starvation conditions, the Atg17-Atg31-Atg29 complex plays a major role, whereas Atg11 is critical for selective autophagy, and for facilitating the transition to nonselective autophagy.
In summary, the function and organization of the PAS, and the regulation of the Atg1 kinase complex are critical issues in the autophagy field. A continued structure–function analysis of the Atg17-Atg31-Atg29 complex will help us better understand the molecular mechanism of phagophore initiation. In particular, further structural studies that include additional components of the Atg1 kinase complex combined with molecular genetic data should provide tremendous insight into autophagy regulation.
Materials and Methods
Strains, Media, and Growth Conditions.
Yeast strains used in this paper are listed in Table S1. Yeast cells were grown in rich [YPD; 1% (wt/vol) yeast extract, 2% (wt/vol) peptone, and 2% (wt/vol) glucose] or synthetic minimal [SMD; 0.67% yeast nitrogen base, 2% (wt/vol) glucose, and auxotrophic amino acids and vitamins as needed] media. Autophagy was induced by shifting the cells to nitrogen starvation medium [SD-N; 0.17% yeast nitrogen base without ammonium sulfate or amino acids, and 2% (wt/vol) glucose].
Plasmids.
pCuHAAtg11(416), pCuHAAtg11(414), and pCuPA-Atg1(416) have been reported previously (7, 39). For constructing WT or truncation forms of the ATG29-PA plasmid, the full-length or corresponding truncation forms of the ATG29 gene with its endogenous promoter were amplified by PCR and ligated into pNopPA(314), which was described elsewhere (42), after removing the NOP1 promoter. The DNA fragments of ATG29-PA[23STA] and ATG29-PA[20STA] were synthesized by GenScript and ligated into pNopPA(314). The plasmids pAtg29[3SA]-PA, pAtg29[3SD]-PA, and pAtg29[3SE]-PA were generated by site-directed mutagenesis from pAtg29-PA. pAtg29[20STA/3SD]-PA and pAtg29[20STA/3SE]-PA were generated from pAtg29[23STA]. For pAtg29[1-202/23STA], the DNA fragment lacking the last 33 nucleotides was amplified by PCR and ligated into pNopPA(314). To construct pAtg1(414), the ORF along with 1,763 bp upstream and 566 bp downstream of the ATG1 gene was amplified from the yeast genome and inserted in the BamHI site of pRS414. pGFP-Atg1(316) was a kind gift from Yoshinori Ohsumi (Tokyo Institute Technology, Yokohama, Japan). pAtg1(LHWS)(414) was generated by site-directed mutagenesis from pAtg1(414), pGFP-Atg1(LHWS)(316) was from pGFP-Atg1(316), and pCuPA-Atg1(LHWS)(416) was from pCuPA-Atg1(416).
Fluorescence Microscopy.
For fluorescence microscopy, yeast cells were grown to OD600 ∼ 0.6 in YPD or SMD and shifted to SD-N for autophagy induction. Samples were then examined by microscopy (Olympus; or Delta Vision, Applied Precision) using a 100× objective, and pictures were captured with a CCD camera (CoolSnap HQ; Photometrics). For each microscopy picture, 12 Z-section images were captured with a 0.3-μm distance between two neighboring sections.
Expression and Purification of MOCR-Atg29.
The ATG29 gene was cloned using ligation-independent cloning technology (43) into the multiple cloning site of the pMOCR expression vector (44). The recombinant fusion protein MOCR-Atg29 was expressed for 3 h at 28 °C in BL21-AI E. coli cells after induction with 0.2 mM isopropyl β-d-1-thiogalactopyranoside and 0.2% arabinose. The protein was purified in the first step using the His tag. The soluble fraction of the bacterial lysate was batch bound to Ni-nitrilotriacetic acid agarose (Qiagen) for 3 h. Resin was washed with 20 mM imidazole in a buffer containing 50 mM sodium phosphate, pH 8, 300 mM NaCl, and 10% (wt/vol) betaine. The protein was eluted with 250 mM imidazole in the same buffer and dialyzed overnight in 50 mM sodium phosphate, pH 8, 10 mM NaCl, 1 mM DTT, and 5% (wt/vol) betaine. The second purification step was performed by gel filtration in dialysis buffer using a Superdex 200 column and the ÄKTApurifierTM FPLC system (GE Healthcare). The protein eluted in the void peak as a soluble aggregate.
Purification of Recombinant Atg17-Atg31-Atg29 Complexes and Variants.
N-terminal GST–tagged or N-terminal hexahistidine-maltose binding protein (MBP)–tagged Atg17 was coexpressed with N-terminal hexahistidine-tagged Atg31 and untagged Atg29 in the same E. coli expression host. The recombinant complex was purified using glutathione-agarose (for GST-tagged Atg17) or nickel Sepharose (for His-MBP–tagged Atg17) followed by PreScission or TEV protease cleavage to remove the affinity tag. The cleaved complex was further purified by gel filtration chromatography using a Superose 6 column.
Single-Particle EM.
Negatively stained specimens were prepared as previously described (45). Raw images were recorded at a nominal magnification of 49,000× on a 4K × 4K Eagle camera (FEI) with a Tecnai Spirit transmission electron microscope operated at an accelerating voltage of 120 kV. Images for 2D analysis were collected using the same instrument with operating parameters and at a defocus value of −1.2 μm. For 2D analysis, particles were interactively selected using Boxer (46). The selected particles were windowed into 112 × 112-pixel images, rotationally and translationally aligned, and subjected to 10 cycles of multireference alignment using SPIDER (47). Each round of alignment was followed by K-means classification specifying 50 classes.
Additional Assays.
The Pho8Δ60 assay and immunoprecipitation were performed as described previously (39).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant GM053396 (to D.J.K.); a University of Michigan Rackham predoctoral fellowship (to K.M.); a Natural Sciences and Engineering Research Council of Canada Discovery grant, a Canadian Institutes of Health Research Operating Grant, a Michael Smith Foundation for Health Research Career Investigator award, and a Canadian Institutes of Health Research New Investigator award (to C.K.Y.); and a Natural Sciences and Engineering Research Council of Canada postgraduate scholarship award (to L.H.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300064110/-/DCSupplemental.
References
- 1.Xie Z, Klionsky DJ. Autophagosome formation: Core machinery and adaptations. Nat Cell Biol. 2007;9(10):1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 2.Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol. 2012;22(1):R29–R34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6(1):79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 6.Abeliovich H, Zhang C, Dunn WA, Jr, Shokat KM, Klionsky DJ. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol Biol Cell. 2003;14(2):477–490. doi: 10.1091/mbc.E02-07-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19(2):668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamada Y, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150(6):1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J, et al. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001;153(2):381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kabeya Y, et al. Characterization of the Atg17-Atg29-Atg31 complex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2009;389(4):612–615. doi: 10.1016/j.bbrc.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 11.Cao Y, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. A multiple ATG gene knockout strain for yeast two-hybrid analysis. Autophagy. 2009;5(5):699–705. doi: 10.4161/auto.5.5.8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12(2):209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 13.Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19(5):2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheong H, et al. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell. 2005;16(7):3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawamata T, et al. Characterization of a novel autophagy-specific gene, ATG29. Biochem Biophys Res Commun. 2005;338(4):1884–1889. doi: 10.1016/j.bbrc.2005.10.163. [DOI] [PubMed] [Google Scholar]
- 16.Kabeya Y, Kawamata T, Suzuki K, Ohsumi Y. Cis1/Atg31 is required for autophagosome formation in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2007;356(2):405–410. doi: 10.1016/j.bbrc.2007.02.150. [DOI] [PubMed] [Google Scholar]
- 17.Nair U, Klionsky DJ. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J Biol Chem. 2005;280(51):41785–41788. doi: 10.1074/jbc.R500016200. [DOI] [PubMed] [Google Scholar]
- 18.Abeliovich H, Dunn WA, Jr, Kim J, Klionsky DJ. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol. 2000;151(5):1025–1034. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamada Y, et al. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30(4):1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott SV, et al. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000;275(33):25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 21.Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;210(1):126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- 22.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41(3):415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 23.Uversky VN. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002;11(4):739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunker AK, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19(1):26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 25.Turoverov KK, Kuznetsova IM, Uversky VN. The protein kingdom extended: Ordered and intrinsically disordered proteins, their folding, supramolecular complex formation, and aggregation. Prog Biophys Mol Biol. 2010;102(2-3):73–84. doi: 10.1016/j.pbiomolbio.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151(7):1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dosztányi Z, Csizmok V, Tompa P, Simon I. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21(16):3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 28.Dosztányi Z, Mészáros B, Simon I. ANCHOR: Web server for predicting protein binding regions in disordered proteins. Bioinformatics. 2009;25(20):2745–2746. doi: 10.1093/bioinformatics/btp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brocca S, et al. Order propensity of an intrinsically disordered protein, the cyclin-dependent-kinase inhibitor Sic1. Proteins. 2009;76(3):731–746. doi: 10.1002/prot.22385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sickmeier M, et al. DisProt: the database of disordered proteins. Nucleic Acids Res. 2007;35(Database issue):D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vacic V, Uversky VN, Dunker AK, Lonardi S. Composition Profiler: A tool for discovery and visualization of amino acid composition differences. BMC Bioinformatics. 2007;8:211. doi: 10.1186/1471-2105-8-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27(10):527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 33.Cao Y, Cheong H, Song H, Klionsky DJ. In vivo reconstitution of autophagy in Saccharomyces cerevisiae. J Cell Biol. 2008;182(4):703–713. doi: 10.1083/jcb.200801035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16(4):1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sung MK, Huh WK. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast. 2007;24(9):767–775. doi: 10.1002/yea.1504. [DOI] [PubMed] [Google Scholar]
- 36.Yeh YY, Wrasman K, Herman PK. Autophosphorylation within the Atg1 activation loop is required for both kinase activity and the induction of autophagy in Saccharomyces cerevisiae. Genetics. 2010;185(3):871–882. doi: 10.1534/genetics.110.116566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579(15):3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 38.Uversky VN. A decade and a half of protein intrinsic disorder: Biology still waits for physics. Protein Sci. 2013;22(6):693–724. doi: 10.1002/pro.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17(1):98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shintani T, Huang W-P, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3(6):825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mijaljica D, et al. Receptor protein complexes are in control of autophagy. Autophagy. 2012;8(11):1701–1705. doi: 10.4161/auto.21332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He C, et al. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J Cell Biol. 2006;175(6):925–935. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stols L, et al. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25(1):8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 44.DelProposto J, Majmudar CY, Smith JL, Brown WC. Mocr: A novel fusion tag for enhancing solubility that is compatible with structural biology applications. Protein Expr Purif. 2009;63(1):40–49. doi: 10.1016/j.pep.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification: Powerful tools in modern electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128(1):82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 47.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 48.Vihinen M. Relationship of protein flexibility to thermostability. Protein Eng. 1987;1(6):477–480. doi: 10.1093/protein/1.6.477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.