Abstract
Photoreceptors are biologically important for sensing changes in the color and intensity of ambient light and, for photosynthetic organisms, processing this light information to optimize food production through photosynthesis. Cyanobacteria are an evolutionarily and ecologically important prokaryotic group of oxygenic photosynthesizers that contain cyanobacteriochrome (CBCR) photoreceptors, whose family members sense nearly the entire visible spectrum of light colors. Some cyanobacteria contain 12 to 15 different CBCRs, and many family members contain multiple light-sensing domains. However, the complex interactions that must be occurring within and between these photoreceptors remain unexplored. Here we describe the regulation and photobiology of a unique CBCR called IflA (influenced by far-red light), demonstrating that a second CBCR called RcaE strongly regulates IflA abundance and that IflA uses two distinct photosensory domains to respond to four different light colors: blue, green, red, and far-red. The absorption of red or far-red light by one domain affects the conformation of the other domain, and the rate of relaxation of one of these domains is influenced by the conformation of the other. Deletion of iflA results in delayed growth at low cell density, suggesting that IflA accelerates growth under this condition, apparently by sensing the ratio of red to far-red light in the environment. The types of complex photobiological interactions described here, both between unrelated CBCR family members and within photosensory domains of a single CBCR, may be advantageous for species using these photoreceptors in aquatic environments, where light color ratios are influenced by many biotic and abiotic factors.
Keywords: chromatic acclimation, chromatic adaptation, phytochrome, signal transduction, tetrapyrrole
Members of the phytochrome superfamily of photoreceptors are broadly distributed in photosynthetic and nonphotosynthetic eukaryotes and bacteria, where they control an enormous range of physiological responses (1–4). Plant phytochromes, which possess covalently bound bilin chromophores and photoreversibly respond to red and far-red light, control many aspects of morphogenesis and development (5). Cyanobacteria often contain large numbers of phytochrome family proteins, with as many as 15 different members in a single species. A subgroup of the phytochrome superfamily within cyanobacteria is the cyanobacteriochromes (CBCRs). These proteins are the most structurally diverse subgroup of this superfamily and respond to the greatest number of different light colors. CBCRs contain from one to six GAF [cGMP phosphodiesterase/adenylate cyclase/formate hydrogen lyase activator (FhlA)] domains. Most, but not all, of these GAF domains are photosensory and contain a cysteine through which a photoconvertible phycocyanobilin (PCB) or phycoviolobilin chromophore is covalently attached (2, 6–8). The cellular roles of CBCRs are diverse and include the regulation of gene expression, cAMP levels, phototaxis, and blue-light–dependent growth (4). The most studied CBCR response, type III chromatic acclimation (CA3) (9, 10), controls the expression of genes in response to red and green light in the filamentous cyanobacterial species Fremyella diplosiphon and acts through the CBCR RcaE (11, 12). RcaE controls the activity of RcaC, an OmpR/PhoB-class transcription factor that binds a direct-repeat DNA sequence called the L box upstream of CA3-regulated genes (13, 14).
Although the structural and functional complexity of CBCRs has been intensively studied (6, 7, 15, 16), nothing is understood about how their levels are regulated, how they interact with each other, or even how the information from separate light-sensing GAF domains is integrated within a single CBCR. We show that the cellular levels of IflA (influenced by far-red light), a previously undescribed, four-color-sensing CBCR that appears to accelerate growth at low cell densities by sensing the ratio of red to far-red light within the ambient light, are strongly regulated by the two-color-sensing CBCR RcaE. This example of hierarchical control of the abundance of one CBCR by another is unique within the prokaryotic phytochrome family and establishes the existence of interactions between different photoreceptors within cyanobacteria. We also analyze the effects of the two IflA photosensory domains on each other after the absorption of four different colors of light, providing unique insights into how multiple GAF photosensory domains interact within a single CBCR. These studies suggest that complex interactions between and within photoreceptors often may be advantageous in aquatic environments, where light color ratios and irradiance levels vary greatly at different depths.
Results
iflA (Fig. 1A) was identified in F. diplosiphon. It encoded a protein with a predicted molecular mass of 85 kDa. IflA contained three GAF domains as well as a C-terminal region of unknown structure (Fig. S1A). The GAF domains were similar to those present in cyanobacterial phytochromes, such as the Cph2s (17) and other CBCRs (Fig. S1B). Canonical phytochrome chromophore attachment site cysteines were present in the GAF1 domain at position 141 (C141) and in GAF3 at C539, but not in GAF2, suggesting it did not attach a chromophore (Fig. S1B). In addition, a second chromophore attachment cysteine found in CBCR GAF domains with blue-green photoreversibility was located in GAF3 at C511 (6–8, 16, 18–23).
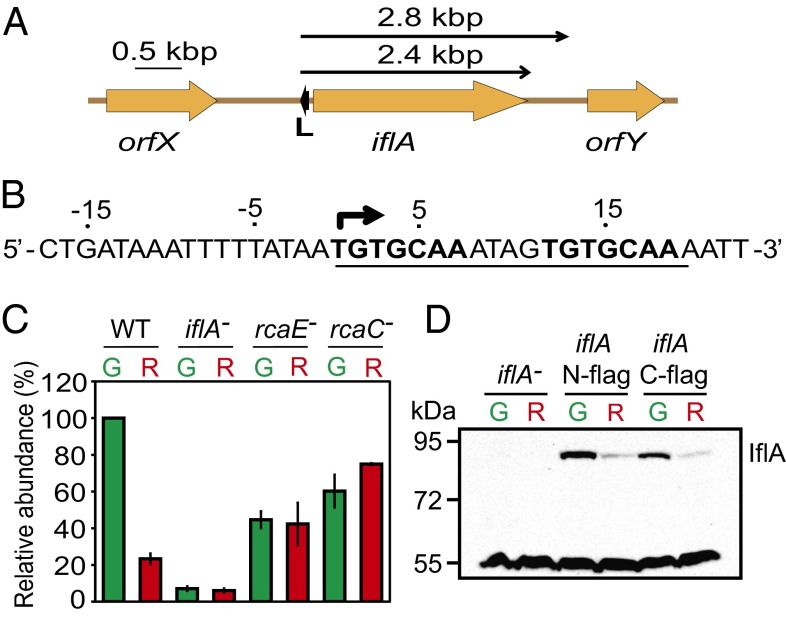
Fig. 1.
The Rca system regulates iflA mRNA and IflA protein abundance. (A) Genomic location of iflA and L box (small black arrow). Upper arrows, transcript locations/sizes. (B) iflA L box sequence, underlined; direct repeat, bold; transcription start site, bent arrow. (C) Histogram showing mean iflA transcript levels for strains and light colors indicated (G, green light; R, red light), normalized using the 16S ribosomal values for each lane. Error bars represent SEM. (D) Representative immunoblot showing IflA abundance during growth in the light colors indicated in C.
DNA sequence matching the L box was identified upstream of iflA (Fig. 1B), suggesting that the expression of this gene might be regulated by the Rca system. The iflA transcription start site was mapped (Fig. S2A) and shown to be within the L box (Fig. 1B), indicating that iflA expression could be repressed by RcaC binding and blocking iflA transcription initiation in red light. RNA blot analysis demonstrated that in wild-type cells, iflA transcripts were five times higher during growth in green light than red light (Fig. 1C). These transcripts, which were 2.4 and 2.8 kbp and apparently include only iflA (Fig. 1A), were absent in an iflA deletion mutant (Fig. S2B). In an rcaE null mutant, iflA RNA accumulated to intermediate levels and light regulation was abolished, demonstrating that iflA expression is controlled by the CBCR RcaE (Fig. 1C). CA3 regulation was also lost in an rcaC mutant because of increased iflA transcript abundance during growth in red light, supporting the proposal that the Rca system controls iflA expression via repression during growth in red light. The regulation of iflA by RcaE was reflected at the protein level, as IflA was six to seven times more abundant in green light than red light (Fig. 1D).
To further establish the role of IflA, we examined the spectral properties of various forms of IflA expressed in an Escherichia coli strain synthesizing PCB. These produced a wide range of brightly colored cells (Fig. 2A), and zinc-dependent fluorescence analysis of these purified proteins revealed that covalent bilin attachment to IflA required C141, C511, and C539 (Fig. 2B), with C511 and C539 likely doubly linked to the GAF3 bilin as demonstrated for blue-green CBCR GAF domains (6, 7, 16, 18–20, 24). The IflA GAF1 domain alone was converted to the red-absorbing (Pr) form by far-red or blue light and to a mix of the Pr and the far-red-absorbing (Pfr) form by red or green light (Fig. 2C). (The absorbance maxima of all IflA forms are provided in Table S1.) In contrast, the GAF3 domain was photoconverted by blue and green light between predominantly the green-absorbing (Pg) and blue-absorbing (Pb) forms, respectively, whereas red and far-red light did not photoconvert this domain (Fig. 2D). Attempts to express full-length IflA were unsuccessful, but the first 608 residues of IflA, containing the three GAF domains and lacking the C-terminal 142 residues (hereafter called IflA608), absorbed in the blue, green, red, and far-red regions (Fig. 2E).
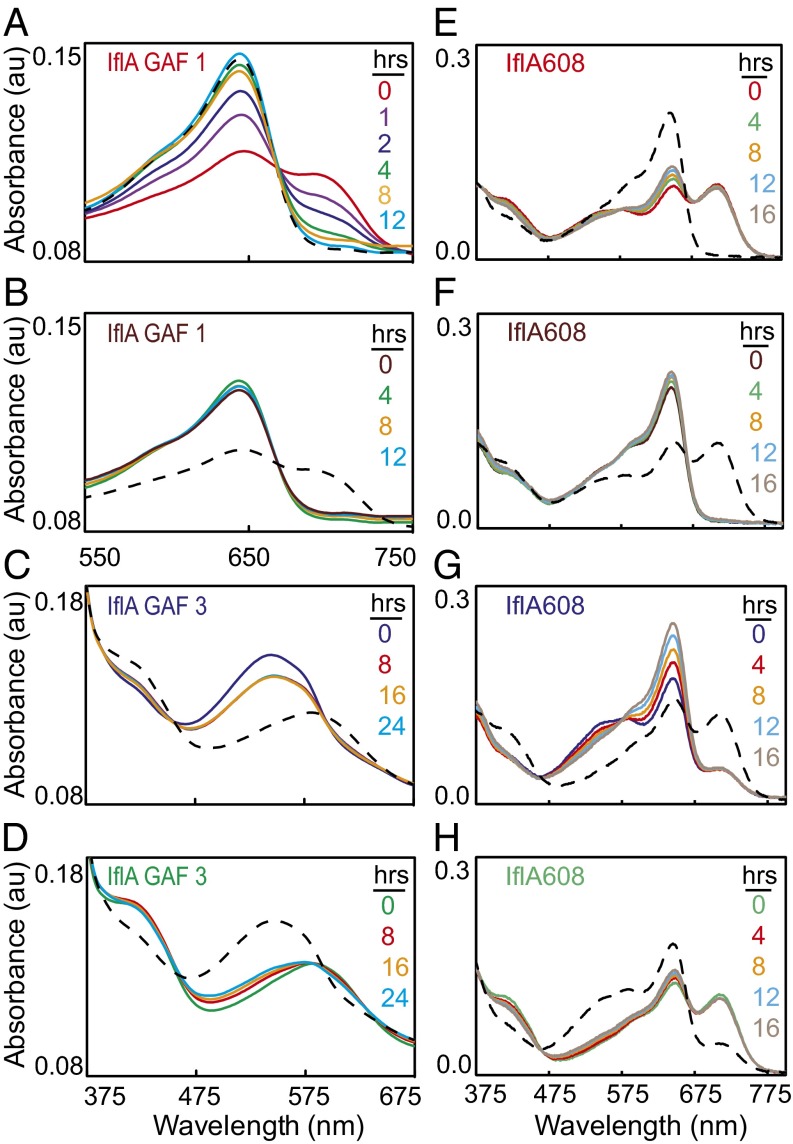
Fig. 2.
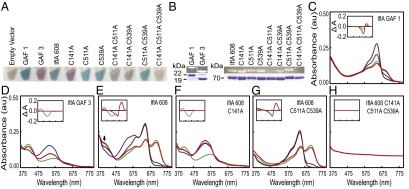
IflA is a four-color-sensing CBCR. (A) PCB-producing E. coli cell pellets containing wild-type and cysteine mutant forms of IflA608, GAF1, and GAF3. (B) Zn2+ blots (Upper) and stained bands (Lower) of wild-type and cysteine mutants of purified IflA608, GAF1, and GAF3. Upper and lower image sets were each spliced together from nonadjacent lanes of the same blot or gel. (C–H) Absorption spectra of purified wild-type and select cysteine mutants of IflA608, GAF1, and GAF3. Line colors indicate the blue, green, red, and far-red light treatment provided to that sample before scanning. Spectral distributions of the light sources are provided in Fig. S2C. Difference spectra for green minus blue (green lines) and red minus far red (red lines) treatments are shown (Insets).
Purified IflA608 protein was eluted from a size-exclusion column as an apparent dimer (Fig. S1C). Red and green light both shifted IflA608 to a relatively higher Pfr:Pr and Pb:Pg ratio, whereas far-red and blue light changed it to a relatively higher Pr:Pfr and Pg:Pb ratio. Several interesting features were apparent for IflA608 that were not observed for the individual GAF domains. First, both red and green light drove IflA608 to a much higher Pfr:Pr ratio than seen for GAF1 alone, suggesting that domain interactions within IflA608 modify the behavior of GAF1 in the context of the longer protein. The double cysteine to alanine mutant IflA608-C511A/C539A was also converted to a higher Pfr:Pr ratio by red and green light than was GAF1 alone (Fig. 2 G vs. C), confirming the results mentioned earlier and demonstrating that the higher ratio did not require a GAF3 chromophore or photoactivity. Second, GAF3 behavior was also changed within the context of IflA608, as it was not differentially responsive to red versus far-red light as an individual domain (Fig. 2D) but, within IflA608, was red/far-red photoreversible in the blue region (peak at 407 nm; Fig. 2E, arrow). The red and far-red light effects on GAF3 were clearly operating through the chromophore within GAF1, as in an IflA608-C141A mutant, this red/far-red responsiveness was absent, whereas blue/green light responsiveness was retained (Fig. 2F). Results very similar to those obtained with the IflA608-C511A/C539A mutant were recorded for the IflA608-C511A and IflA-C539A mutants (Fig. S3 A and B). IflA608-C141A/C511A/C539A, which contained replacements of all three cysteines required for bilin attachment in both GAF1 and GAF3, was unresponsive to all four light colors (Fig. 2H), and forms with a single substitution in both of these domains yielded similar results (Fig. S3 C and D). Collectively, these data demonstrate that IflA is a four-color-sensing CBCR in which domain interactions alter the photobiology of the light-sensing GAF domains within this protein, relative to the GAF domains by themselves, which is a previously undescribed capability of CBCRs.
We examined dark reversion rates of the IflA photosensory GAF domains individually and within the IflA608 wild-type and mutants after blue, green, red, and far-red light treatments. GAF1 alone nearly completely reverted to Pr within 4 h after photoconversion to Pfr (Fig. 3A), whereas Pr was stable for 12 h (Fig. 3B). Thus, the thermodynamically stable state of GAF1 is Pr. For GAF3, there was little dark reversion from Pg to Pb over the course of 16 h (Fig. 3C) and a similarly minor amount of reversion of Pb to Pg during the same time (Fig. 3D), demonstrating that Pb and Pg are nearly equivalent thermodynamically stable states for GAF3. Overall, these results suggest that GAF1 must absorb red light to form Pfr, whereas GAF3 is equally likely to exist as either Pg or Pb. Within IflA608, however, after red light treatment, GAF1 failed to strongly revert to Pr in the dark, and the small increase in Pr over the course of 16 h was not accompanied by a corresponding decrease in Pfr (Fig. 3E), as measured for GAF1 alone (Fig. 3A). However, this protein could be completely photoconverted to Pr at the end of the dark treatment (Fig. 3E). The Pb that was formed by red light was also relatively stable but was eliminated by far-red light, as seen previously (Fig. 2E). Initial far-red light treatment also photoconverted IflA608 to Pr, and no reversion to Pfr was measured after 16 h of darkness (Fig. 3F). Again, a small increase in Pr was measured during this dark treatment.
Fig. 3.
GAF1 dark reversion is influenced by other IflA608 domains and the Pg:Pb ratio of GAF3. Twelve-hour dark reversion analysis of IflA GAF1 after (A) red or (B) far-red light exposure, 24-h dark reversion analysis of IflA GAF3 after (C) blue or (D) green light exposure, and 16-h dark reversion analysis of IflA608 after red (E), far-red (F), blue (G), or green (H) light exposure. For all, line color denotes length of time in the dark after the initial light treatment (insets), and black dashed lines are spectra after irradiation with opposite light color (red/far-red or blue/green) postdark treatment.
Blue light treatment of IflA608 initially increased the Pg:Pb and Pr:Pfr ratio, and during the dark period, Pg decreased, whereas Pr increased significantly (Fig. 3G). Interestingly, both of these changes occurred without reciprocal changes in Pb or Pfr levels. The relatively high Pb:Pg and Pfr:Pr ratios driven by green light were predominantly stable for the 16 h dark treatment (Fig. 3H). IflA608-C141A was unresponsive to either red or far-red light and showed no dark reversion (Fig. S4 A and B). However, it was strongly photoconverted by blue and green light and only weakly dark reverted (Fig. S4 C and D), similar to the GAF3 domain alone (Fig. 3 C and D). For the IflA608-C511A/C539A mutant treated with red or far-red light, the results (Fig. S4 E and F) were nearly identical to those obtained for IflA608 (Fig. 3 E and F), except that virtually no increase in Pr occurred during the dark treatment, and this difference was particularly apparent after red light treatment. Blue-light–treated IflA608-C511A/C539A also strongly converted to Pr (Fig. S4G), but unlike IflA608 after the same treatment (Fig. 3G), there was no large additional increase in Pr during the dark treatment. A similar trend for IflA608-C511A/C539A compared with IflA608 was measured during the dark treatment after green light exposure (Fig. S4H).
All of these proteins remained functional after these treatments, because each was efficiently reversible at the end of its dark treatment period. Overall, these dark reversion data indicate that the rapid and complete reversion of GAF1 to Pr is highly constrained within the IflA608 protein, but that this reversion is enhanced by the presence of chromophorylated GAF3, and particularly by the formation of Pg in GAF3, whereas the dark reversion of GAF3 from either Pb or Pg appears to be generally equivalent for this domain by itself or in the context of IflA608. The influence of a single chromophore-binding domain on the dark reversion kinetics of another is unique among phytochrome-class photoreceptors and is certain to influence the activity of this photoreceptor in environments with changing ratios of the four light colors tested here.
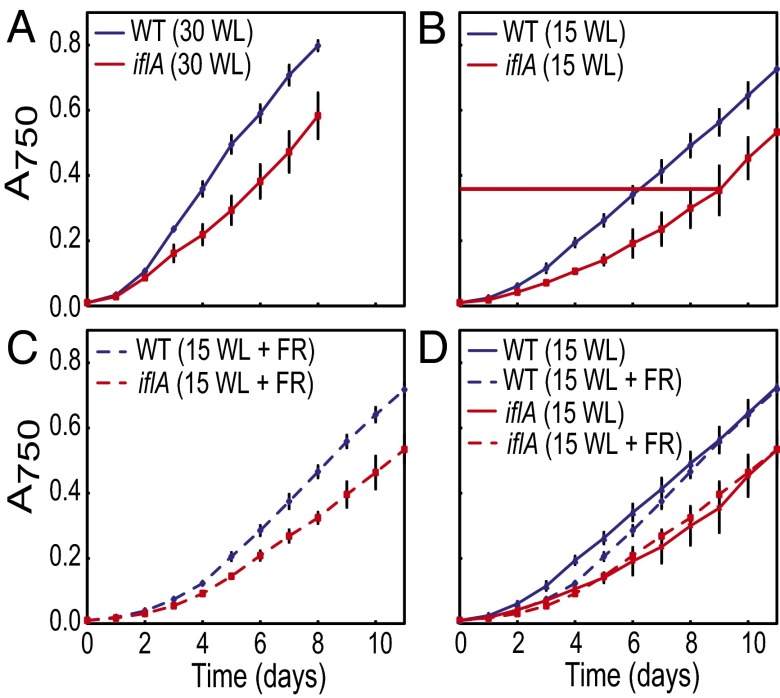
Because the cellular function of IflA could not be established by its predicted domain structure (Fig. S1A) or genome context (Fig. 1A), we created an iflA deletion mutant and examined it for irregularities in its physiological responses. The mutant possessed normal light-harvesting pigments, chlorophyll content, and CA3 capacity, as assayed by pigment accumulation profiles (Fig. S5). However, in white light, the growth of wild-type cells was more rapid than iflA cells for the first 5 d [mwt (wild-type growth curve slope), 0.116 (R2, 0.99); miflA (iflA mutant growth curve slope), 0.070 (R2, 0.99)] (Fig. 4A). After the sixth day, when iflA cells reached an absorbance at 750 nm (A750) ∼0.4, growth was equivalent for both strains [mwt, 0.104 (R2, 0.99); miflA, 0.101 (R2, 0.99)]. We tested whether this was a light-intensity effect by decreasing the irradiance from 30 to 15 μmol m−2⋅s−1. Although the growth of both wild-type and iflA cells slowed, the same patterns emerged (Fig. 4B). For the first 8 d, wild-type cells grew more rapidly (mwt, 0.073; R2, 0.99) than iflA cells (miflA, 0.045; R2, 0.98), but growth was equivalent [mwt, 0.082 (R2, 0.99); miflA, 0.089 (R2, 0.99)] after the iflA cells reached an A750∼ 0.4. The slower growth of the iflA mutant at low cell densities was not because of the higher irradiance level, as 15 μmol m−2⋅s−1-grown cells with an A750 of 0.2–0.4 grew slowly (Fig. 4B) yet received equivalent or less light than 30 μmol m−2⋅s−1-grown cells at an A750 of 0.4 (Fig. S6A). Media composition and age also did not cause this lag, as fresh media had no effect on growth (Fig. S7A).
Fig. 4.
Supplemental far-red light affects wild-type, but not iflA mutant, growth. Wild-type and iflA cultures grown in (A) 30- or (B) 15-µmol photons m−2⋅s−1 white light, or (C) 15-µmol photons m−2⋅s−1 white light plus 5-µmol photons m−2⋅s−1 of supplemental far-red light. (D) Comparison of growth curves for wild-type and iflA mutant grown in 15-µmol photons m−2⋅s−1 white light, with or without 5-µmol photons m−2⋅s−1 of supplemental far-red light. Horizontal red line in B indicates the A750 at which iflA cells shift from slower to wild-type growth. Error bars represent SEM.
Finally, we tested the effects of the four light colors that are best absorbed by IflA on the growth of wild-type and iflA cells. No differences in growth were measured in blue, green, or red light (Fig. S7 B–D). Because F. diplosiphon grows poorly in far-red light, white light with or without far-red light supplementation was used to test the effect of far-red light. Wild-type cells grown in 15 μmol m−2⋅s−1 of white plus far-red light had a longer lag in their initial growth phase than cells grown in white light only, and this difference was lost by the mid- to late-exponential growth phase (Fig. 4 C and D). However, this difference was not observed for iflA cells (Fig. 4 C and D). It also did not result from the development of suppressor mutations, as the cells used in this experiment, which were grown to an A750 of 0.9, were reused in the same experiment, with identical results. These data suggest that IflA increases growth in response to sensing an elevated red/far-red light ratio in the environment. Thus, we conclude that for iflA mutant cells growing in white light, the absence of IflA eliminates the ability of the cells to increase the rate of growth at low cell densities, when the red/far-red light ratio is elevated (Fig. S6B and Table S2).
Discussion
Our studies establish that the abundance of a newly discovered, multi-GAF-domain CBCR called IflA is regulated by green and red light through the action of another CBCR called RcaE (Fig. 1, C and D). This regulation occurs through the repression of iflA expression in red light, leading to a six to seven times higher level of IflA protein in green light. We determined that IflA accelerates cell growth, apparently by primarily sensing the red/far-red ratio of ambient light. We also discovered that in addition to the red/far-red photoreversibility of GAF1 and blue/green photoreversibility of GAF3, the behavior of these two domains in the context of IflA608 is complex.
Although genetic evidence exists for cross-regulation of the expression of plant phytochrome family members (25–28), the regulation of one prokaryotic phytochrome superfamily member by another has not been previously described. The RcaE-mediated increase in IflA abundance as the green to red light ratio increases (Fig. 1D) may have dramatic physiological effects. During Arabidopsis seedling development, phytochrome family members phyA and phyB activate mutually antagonistic responses until the degradation of phyA allows phyB to dominate growth regulation (29). F. diplosiphon is found in aquatic environments, where the decrease in irradiance with depth may lead to a reduction in the activation of IflA deeper in the water column. Because the Rca system senses the red/green light ratio, it may be used as a proxy for depth sensing. Water absorbs red light more efficiently than it does green light, increasing the green/red ratio with depth (30). Thus, RcaE may increase IflA abundance with depth to offset the decrease in the number of photoactivated IflA photoreceptors. The depth to which this increase might be effective can be calculated: If IflA is six times more abundant in green than red light (Fig. 1D), this would offset an 83% decrease in red light irradiance. Transmittance of red (650 nm) light through surface water ranges from 47% to 70% m−1 (30), so an 83% decrease in red light would typically occur between 2 and 4 m, making this a potentially useful physiological response in an aquatic environment.
The presence of IflA led to more rapid growth that could be countered by decreasing the red/far-red light ratio (Fig. 4D), suggesting that IflA exerts its effect in the Pfr form. However, the influence of IflA on growth was no longer measurable once the cells grew beyond an A750 of 0.4. Because the red/far-red ratio decreases with increasing culture density, this may be the density at which this ratio is no longer adequate to maintain a sufficient amount of the Pfr form of IflA (Table S2, asterisk). Because Pr is the thermodynamically stable state of GAF1 when it is part of IflA608 and is exposed to blue light (Fig. 3G), the absence of sufficient red light to maintain the Pfr form should lead to its reversion to the Pr form if blue light is also present. IflA-mediated change in the growth of F. diplosiphon cells in response to changes in the red/far-red light ratio is reminiscent of the phytochrome-mediated shade avoidance response of many angiosperms, in which a decrease in the red/far-red light ratio reduces the inhibition of stem elongation (31). IflA may be responsible for accelerating growth when the absorption of red light by nearby organisms is reduced, as gauged by an increase in the red/far-red light ratio.
The photoconversion characteristics of GAF1 and GAF3 are modified by other regions of the protein, as red light drove GAF1 to a higher Pfr:Pr ratio when it was part of IflA608 than when it was a single domain (Fig. 3 E vs. A), and GAF3 was converted to Pb by red light in IflA608 (Fig. 2E), which did not occur with either isolated GAF3 protein (Fig. 2D) or IflA608-C141A, in which GAF1 lacks a chromophore (Fig. 2F). In addition, the relatively rapid and complete dark reversion of the GAF1 domain to Pr after far-red light treatment was significantly reduced when it was part of IflA608 (Fig. 3 A vs. E). Interestingly, the reduction in Pr formation was mitigated when GAF3 was in the Pg form (Fig. 3G), although it is not yet clear how these two events are linked. Light-mediated interactions between GAF domains of a CBCR family member have not been previously shown.
The dark reversion characteristics of the two photosensory GAF domains within IflA608 were unusual because during the dark treatment, Pr accumulated without a corresponding decrease in Pfr (Fig. 3 E–G) and a decrease in Pg was not accompanied by an increase in Pb (Fig. 3G). The increase in Pr depended on GAF3 being chromophorylated (Fig. S4 E, F, and H). One possible explanation for this is that GAF3 in the Pg form subtly influences the shift of GAF1 to Pr. Another possibility is that the GAF3 chromophore is being deprotonated and converting to Pr (32), although this is unlikely, as in the IflA608-C141A mutant, conversion of GAF3 to Pg by blue light did not lead to any accumulation of Pr during the dark reversion period (Fig. S4C). It is also possible that an increase of one isomer without a concomitant decrease of the other isomer during dark reversion could result from the accumulation of a colorless intermediate(s). These surprising results highlight the fact that the interactions between domains of multichromic CBCRs can result in unique forms of photochemistry that cannot be generated in phytochrome family members with a single chromophore domain.
It is unclear why cyanobacteria possess so many phytochrome superfamily members with multiple photosensory GAF domains when photoreceptors in fungi, other eubacteria, and plants do not (4, 7, 24). Our lack of understanding is partly because the interactions between such GAF domains have not been described until now. Although such interactions must certainly be used by cyanobacteria to deal with many different environmental situations, unraveling the interactions between the photosensory domains and understanding the roles of these interactions in the natural history of these organisms will be a challenge. For IflA, it is not clear why GAF1 subtly influences the Pb:Pg ratio of GAF3 (Fig. 2E) or why blue light increases the ratio of Pr:Pfr in the dark (Fig. 3G). Generally, however, it seems reasonable that the evolution of multichromic phytochrome family members has allowed phytochrome-class photoreceptors to be more effectively used in aquatic environments, where cyanobacteria are predominantly found.
On land, the red/far-red ratio of sunlight remains relatively constant during much of the day, allowing plants to use changes in this ratio to trigger, for example, shade-avoidance responses (31). However, in water, where the red/far-red ratio increases with depth, the use of this information could be problematic, as it would shift a plant-type phytochrome further into the Pfr form, thereby modifying the signal output (30). However, the ratio of the other light colors of the visible spectrum also changes with depth, including the blue/green ratio, which increases with depth. We propose that multichromic phytochrome family members allow more robust sensing in such environments by simultaneously sampling multiple light color ratios, which may provide information not only about depth but also about competing photosynthesizers, the time of day (which strongly affects the relative amount of light of each wavelength in the visible spectrum), and other biotic and abiotic parameters. Because multichromic CBCRs are also found in nonaquatic cyanobacteria, it is likely that they are useful in additional settings, and as not all phytochrome-class photoreceptors in cyanobacteria contain multiple photosensory domains, there must be situations in which sensing only a single light color ratio is adequate. However, the abundance and diversity of photoreceptors with multiple light-sensing domains, along with the results presented here, strongly suggest that cyanobacteria are highly evolved to sense and respond to complex light color environments.
Materials and Methods
Strains and Growth Conditions.
The wild-type strain of F. diplosiphon UTEX 481 (also called Tolypothrix sp. PCC 7601) used was the shortened filament mutant strain SF33 (33). The rcaE and rcaC null mutant strains used were previously described (11–14, 34). Cultures were grown in BG-11 media from an A750 of 0.01, as described (35), in light irradiances of 15-, 20-, or 30-µmol photons m−2⋅s−1. Custom-built light-emitting diode (LED) panels provided red light (www.digikey.com, 160-1415-2-ND), green light (www.digikey.com, 754-1099-2-ND), blue light (Philips, LED-60L/2WB-1), and far-red light (Optotech, L-D-735-H), and Solux 4700 Kelvin halogen lamps (Eiko Ltd., Q50MR16/CG/47/36) provided white light that closely matched the natural visible spectrum. The spectral distribution and emission maxima of all LEDs are provided in Fig. S2C, and for the white light source they are found in Fig. S6B (black line). At least three independent growth experiments were conducted for each cell type and light condition. Media replacement experiments were performed once by harvesting cells growing in 15-µmol photons m−2⋅s−1 white light by centrifugation at 5,000 × g for 5 min and resuspending in either the original or fresh BG-11 media. A750 values were monitored before and after the media replacement.
Construction of the iflA Null Mutants, Expression Plasmids, and Transformations.
To create iflA null mutant strains, two iflA fragments were PCR amplified from F. diplosiphon genomic DNA. Primers 5′tgcGAATTCC CTCACCAACG TCGAAGGGCT3′ (EcoRI site bolded) and 5′cgaCTGCAGT CAGTCACTGC TGTCATGTAT TTCTACG3′ (PstI site bolded) were used to amplify the iflA N-terminal region. Primers 5′gatCTGCAGC AGCAAATCCA ACAAACACAA GCACA3′ (PstI site bolded) and 5′ggtCCATGGT TACATCCGCA GACTCGACAG CC3′ (NcoI site bolded) were used to amplify the iflA C-terminal region. Each iflA fragment was then cut with restriction enzymes that cleaved within the primer sequences and ligated into pJCF276 cut with EcoRI and NcoI. The sequence of the clean deletion construct of iflA was confirmed by sequencing. F. diplosiphon was transformed by triparental mating, and three iflA null mutant strains were selected as previously described (36, 37). All parent E. coli strains used for triparental mating were DH5α MCR except 803, which carried the RP4 conjugative plasmid (38). To create iflA strains containing a FLAG-tag at either the N or C terminus, iflA fragments were PCR amplified from F. diplosiphon genomic DNA. Primers 5′cttgtcatcg tcatccttat aatccaTGTA TTTCTACGTA GGTATAATGT ATAAGTACG3′ (Fusion overlap is lowercase) and 5′tcaTCCGGAG CGTTACCTTT CTCGAAGACA TATCCG3′ (BspEI site bolded) were used to amplify the iflA N-terminal region with a FLAG tag. Primers 5′atggattata aggatgacga tgacaagACA GCAGTGACTG AGTACTCACA GGAA3′ (Fusion overlap is lowercase) and 5′gtaCCATGGC GTCTCAATTA CTAATCACTT GTGTGTTC3′ (NcoI site bolded) were used to amplify the iflA C-terminal region with a FLAG-tag in the N terminus. Primers 5′ttacttgtca tcgtcatcct tataatccat ATTTTCATGC TGATTTACAC TTTTCAT3′ (Fusion overlap is lowercase) and 5′tcaTCCGGAG TTGATGTCAA CAGCCAATAC AAAT3′ (BspEI site bolded) were used to amplify the iflA N-terminal region with a FLAG-tag in the C terminus. Primers 5′atggattataa ggatgacgat gacaagtaaG ATTATTTGAT TCAGTTAACC CACATC3′ (Fusion overlap is lowercase) and 5′gtaCCATGGC GTCTCAATTA CTAATCACTT GTGTGTTC3′ (NcoI site bolded) were used to amplify the iflA C-terminal region with a FLAG tag. Fusion PCR was performed, and both iflA N-FLAG and iflA C-FLAG fragments were then cut with restriction enzymes that cleaved within the primer sequences and ligated into pJCF276 cut with BspEI and NcoI. The sequence of each iflA construct was confirmed by sequencing and was transformed into F. diplosiphon iflA mutant cells, and three isolates of each were selected. For the expression of various regions of IflA in E. coli, iflA fragments were generated by PCR amplification of F. diplosiphon genomic DNA and cloned into the pETDuet vector (Novagen) after digestion with BamHI and SacI. Plasmids and cloning primers used in this study are provided in Table S3. All junctions and PCR amplification products were confirmed by sequencing. These plasmids encode 6×-histidine-tagged IflA608, GAF1, and GAF3 with and without the C141A and/or C511A and/or C539A mutation or mutations and were transformed into E. coli BL21(DE3) cells containing pPcyA (39).
Expression, Purification, and Detection of his- or FLAG-Tagged Regions of IflA.
E. coli BL21(DE3) colonies containing pPcyA (39) and the pETDuet constructs listed in Table S3 were selected on Luria-Bertani medium plates containing 30 µg⋅mL−1 chloramphenicol (Cm) and 50 µg⋅mL−1 ampicillin (Ap). A 25-mL overnight starter culture grown with shaking at 37 °C was added to 1 L LB containing 30 µg⋅mL−1 Cm and 50 µg⋅mL−1 Ap and grown with shaking at 16 °C for 6 h. Addition of 0.1 mM isopropyl-β-d-thiogalactoside induced expression of heme oxygenase 1 (ho1)/phycocyanobilin ferredoxin-dependent oxidoreductase (pcyA) and 6×-histidine-tagged IflA variants. Cells were grown overnight with shaking at 16 °C and then harvested by centrifugation at 15,000 × g for 20 min at 4 °C. IflA proteins from E. coli were then isolated as previously described (12, 34). Size-exclusion chromatography was performed using a Superdex 200 10/300 GL (GE Healthcare) column run at 0.5 mL⋅min−1 with 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4⋅2H2O, and 2 mM KH2PO4, using a 50-µL sample loop. The column was calibrated with the molecular mass standards thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.35 kDa) (BioRad). Elution of IflA608 holoprotein and the standards were monitored at 280 nm. Total cellular proteins were isolated from F. diplosiphon cells (34), and Western analyses using the anti-FLAG antibody (Sigma-Aldrich) and goat anti-rabbit IgG-HRP antibody (Santa Cruz Biotechnology) were performed according to the manufacturer’s instructions. IflA proteins were detected using SuperSignal West Femto chemiluminescent substrate (Thermo Scientific), following the manufacturer’s instructions, and the images were viewed using the BioRad Chemidoc MP and quantified using Image Lab 4.1 software (BioRad).
Zinc-Induced Fluorescence Analysis.
Isolated proteins from E. coli were separated by SDS-PAGE. Gels were soaked in 10 mM Zn2+ acetate for 30 min in the dark and then examined for fluorescence, as previously described (40). Gels were then stained with Coomassie brilliant blue R250 to visualize proteins.
Spectral Measurements.
A Beckman DU640B spectrophotometer was used to measure the A750 of F. diplosiphon cultures. A Synergy Mx plate reader (BioTek Instruments) was used to measure the absorbance spectra, difference spectra, and dark reversion spectra of each form of IflA protein. The plate reader was checked for the effect of the measuring beam on the photochemistry of the samples, and no effect was found. The light sources used for each of these experiments were provided using LEDs with narrow spectral bandwidths for blue (λmax, 458 nm), green (λmax, 521 nm), red (λmax, 640 nm), and far-red (λmax, 740 nm) light. The spectral distributions of these light sources are provided in Fig. S2C. Light irradiances of each LED source were measured using a Jaz spectrometer (Ocean Optics) and a LI-250 photometer (LI-COR). For the absorbance measurements, the IflA proteins were irradiated for 5 min with 20-µmol photons m−2⋅s−1 of the appropriate light wavelength before each spectral measurement. For dark reversion measurements, IflA proteins were irradiated with 20-µmol photons m−2⋅s−1 light for 5 min, and then absorbance was measured every 2 h for 24 h while samples were kept at 21 °C in darkness. After 24 h of dark reversion measurements, samples were irradiated with the light color opposing their initial light treatment (red/far-red or blue/green), and absorbance was measured. Irradiance and wavelength distribution measurements of light passing through a culture tube were recorded using a Jaz spectrometer (Ocean Optics), with the probe attached to the outside wall of the culture tube positioned 180° away from the wall receiving the light and shielded from other light sources. Each scan shown in the manuscript is representative of at least three biologically independent replicates that were conducted for each analysis.
RNA Blot Analysis.
RNA blot analysis was performed as previously described (35). At least three independent experiments were carried out for each cell type and condition. The probe for iflA was made by PCR amplification using primers 5′CAGCAAATGT ACCAACAGGT ACAAG3′ and 5′CTGTCTGTTT TCAGCAGCTT CCGCA3′. Blots were imaged and quantified as previously described (41).
Primer Extension.
The transcription start site of iflA was determined using a primer extension system (Promega) with primer 5′CGCCTGTCCC TGCTGCGAAT T3′. The primer extension reactions were performed and sequenced as previously described (41).
Supplementary Material
Acknowledgments
We thank Nathan Rockwell, J. Clark Lagarias, and the members of the D.M.K. laboratory for their thoughtful discussions and comments on IflA and the manuscript. This research was entirely supported by National Science Foundation Grant MCB-1029414 (to D.M.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession nos. KF316936 (iflA), KF316937 (orfX), and KF316938 (orfY)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303371110/-/DCSupplemental.
References
- 1.Purschwitz J, Müller S, Kastner C, Fischer R. Seeing the rainbow: Light sensing in fungi. Curr Opin Microbiol. 2006;9(6):566–571. doi: 10.1016/j.mib.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 2.Rockwell NC, Lagarias JC. A brief history of phytochromes. ChemPhysChem. 2010;11(6):1172–1180. doi: 10.1002/cphc.200900894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vierstra RD, Zhang JR. Phytochrome signaling: Solving the Gordian knot with microbial relatives. Trends Plant Sci. 2011;16(8):417–426. doi: 10.1016/j.tplants.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 4.Auldridge ME, Forest KT. Bacterial phytochromes: More than meets the light. Crit Rev Biochem Mol Biol. 2011;46(1):67–88. doi: 10.3109/10409238.2010.546389. [DOI] [PubMed] [Google Scholar]
- 5.Franklin KA, Quail PH. Phytochrome functions in Arabidopsis development. J Exp Bot. 2010;61(1):11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikeuchi M, Ishizuka T. Cyanobacteriochromes: A new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem Photobiol Sci. 2008;7(10):1159–1167. doi: 10.1039/b802660m. [DOI] [PubMed] [Google Scholar]
- 7.Rockwell NC, Martin SS, Feoktistova K, Lagarias JC. Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proc Natl Acad Sci USA. 2011;108(29):11854–11859. doi: 10.1073/pnas.1107844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rockwell NC, Martin SS, Gulevich AG, Lagarias JC. Phycoviolobilin formation and spectral tuning in the DXCF cyanobacteriochrome subfamily. Biochemistry. 2012;51(7):1449–1463. doi: 10.1021/bi201783j. [DOI] [PubMed] [Google Scholar]
- 9.Tandeau de Marsac N. Occurrence and nature of chromatic adaptation in cyanobacteria. J Bacteriol. 1977;130(1):82–91. doi: 10.1128/jb.130.1.82-91.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutu A, Kehoe DM. Emerging perspectives on the mechanisms, regulation, and distribution of light color acclimation in cyanobacteria. Mol Plant. 2012;5(1):1–13. doi: 10.1093/mp/ssr054. [DOI] [PubMed] [Google Scholar]
- 11.Kehoe DM, Grossman AR. Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science. 1996;273(5280):1409–1412. doi: 10.1126/science.273.5280.1409. [DOI] [PubMed] [Google Scholar]
- 12.Terauchi K, Montgomery BL, Grossman AR, Lagarias JC, Kehoe DM. RcaE is a complementary chromatic adaptation photoreceptor required for green and red light responsiveness. Mol Microbiol. 2004;51(2):567–577. doi: 10.1046/j.1365-2958.2003.03853.x. [DOI] [PubMed] [Google Scholar]
- 13.Li L, Alvey RM, Bezy RP, Kehoe DM. Inverse transcriptional activities during complementary chromatic adaptation are controlled by the response regulator RcaC binding to red and green light-responsive promoters. Mol Microbiol. 2008;68(2):286–297. doi: 10.1111/j.1365-2958.2008.06151.x. [DOI] [PubMed] [Google Scholar]
- 14.Chiang GG, Schaefer MR, Grossman AR. Complementation of a red-light-indifferent cyanobacterial mutant. Proc Natl Acad Sci USA. 1992;89(20):9415–9419. doi: 10.1073/pnas.89.20.9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma Q, et al. A rising tide of blue-absorbing biliprotein photoreceptors: Characterization of seven such bilin-binding GAF domains in Nostoc sp. PCC7120. FEBS J. 2012;279(21):4095–4108. doi: 10.1111/febs.12003. [DOI] [PubMed] [Google Scholar]
- 16.Ulijasz AT, et al. Cyanochromes are blue/green light photoreversible photoreceptors defined by a stable double cysteine linkage to a phycoviolobilin-type chromophore. J Biol Chem. 2009;284(43):29757–29772. doi: 10.1074/jbc.M109.038513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshihara S, Katayama M, Geng X, Ikeuchi M. Cyanobacterial phytochrome-like PixJ1 holoprotein shows novel reversible photoconversion between blue- and green-absorbing forms. Plant Cell Physiol. 2004;45(12):1729–1737. doi: 10.1093/pcp/pch214. [DOI] [PubMed] [Google Scholar]
- 19.Rockwell NC, et al. A second conserved GAF domain cysteine is required for the blue/green photoreversibility of cyanobacteriochrome Tlr0924 from Thermosynechococcus elongatus. Biochemistry. 2008;47(27):7304–7316. doi: 10.1021/bi800088t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishizuka T, et al. The cyanobacteriochrome, TePixJ, isomerizes its own chromophore by converting phycocyanobilin to phycoviolobilin. Biochemistry. 2011;50(6):953–961. doi: 10.1021/bi101626t. [DOI] [PubMed] [Google Scholar]
- 21.Yoshihara S, et al. Reconstitution of blue-green reversible photoconversion of a cyanobacterial photoreceptor, PixJ1, in phycocyanobilin-producing Escherichia coli. Biochemistry. 2006;45(11):3775–3784. doi: 10.1021/bi051983l. [DOI] [PubMed] [Google Scholar]
- 22.Ishizuka T, Narikawa R, Kohchi T, Katayama M, Ikeuchi M. Cyanobacteriochrome TePixJ of Thermosynechococcus elongatus harbors phycoviolobilin as a chromophore. Plant Cell Physiol. 2007;48(9):1385–1390. doi: 10.1093/pcp/pcm106. [DOI] [PubMed] [Google Scholar]
- 23.Song JY, et al. Near-UV cyanobacteriochrome signaling system elicits negative phototaxis in the cyanobacterium Synechocystis sp. PCC 6803. Proc Natl Acad Sci USA. 2011;108(26):10780–10785. doi: 10.1073/pnas.1104242108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Savakis P, et al. Light-induced alteration of c-di-GMP level controls motility of Synechocystis sp. PCC 6803. Mol Microbiol. 2012;85(2):239–251. doi: 10.1111/j.1365-2958.2012.08106.x. [DOI] [PubMed] [Google Scholar]
- 25.Clack T, Mathews S, Sharrock RA. The phytochrome apoprotein family in Arabidopsis is encoded by five genes: The sequences and expression of PHYD and PHYE. Plant Mol Biol. 1994;25(3):413–427. doi: 10.1007/BF00043870. [DOI] [PubMed] [Google Scholar]
- 26.Somers DE, Quail PH. Phytochrome-mediated light regulation of PHYA- and PHYB-GUS transgenes in Arabidopsis thaliana seedlings. Plant Physiol. 1995;107(2):523–534. doi: 10.1104/pp.107.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tepperman JM, et al. Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J. 2004;38(5):725–739. doi: 10.1111/j.1365-313X.2004.02084.x. [DOI] [PubMed] [Google Scholar]
- 28.Hirschfeld M, Tepperman JM, Clack T, Quail PH, Sharrock RA. Coordination of phytochrome levels in phyB mutants of Arabidopsis as revealed by apoprotein-specific monoclonal antibodies. Genetics. 1998;149(2):523–535. doi: 10.1093/genetics/149.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quail PH, et al. Phytochromes: Photosensory perception and signal transduction. Science. 1995;268(5211):675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- 30.Jerlov NG. Elsevier Oceanography Series. Amsterdam: Elsevier Scientific; 1976. Marine Optics; pp. 127–150. [Google Scholar]
- 31.Kutschera U, Briggs WR. From Charles Darwin’s botanical country-house studies to modern plant biology. Plant Biol (Stuttg) 2009;11(6):785–795. doi: 10.1111/j.1438-8677.2009.00243.x. [DOI] [PubMed] [Google Scholar]
- 32.Hirose Y, et al. Green/red cyanobacteriochromes regulate complementary chromatic acclimation via a protochromic photocycle. Proc Natl Acad Sci USA. 2013;110(13):4974–4979. doi: 10.1073/pnas.1302909110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cobley JG, et al. Construction of shuttle plasmids which can be efficiently mobilized from Escherichia coli into the chromatically adapting cyanobacterium, Fremyella diplosiphon. Plasmid. 1993;30(2):90–105. doi: 10.1006/plas.1993.1037. [DOI] [PubMed] [Google Scholar]
- 34.Li L, Kehoe DM. In vivo analysis of the roles of conserved aspartate and histidine residues within a complex response regulator. Mol Microbiol. 2005;55(5):1538–1552. doi: 10.1111/j.1365-2958.2005.04491.x. [DOI] [PubMed] [Google Scholar]
- 35.Seib LO, Kehoe DM. A turquoise mutant genetically separates expression of genes encoding phycoerythrin and its associated linker peptides. J Bacteriol. 2002;184(4):962–970. doi: 10.1128/jb.184.4.962-970.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cobley JG, et al. CpeR is an activator required for expression of the phycoerythrin operon (cpeBA) in the cyanobacterium Fremyella diplosiphon and is encoded in the phycoerythrin linker-polypeptide operon (cpeCDESTR) Mol Microbiol. 2002;44(6):1517–1531. doi: 10.1046/j.1365-2958.2002.02966.x. [DOI] [PubMed] [Google Scholar]
- 37.Noubir S, et al. Co-ordinated expression of phycobiliprotein operons in the chromatically adapting cyanobacterium Calothrix PCC 7601: A role for RcaD and RcaG. Mol Microbiol. 2002;43(3):749–762. doi: 10.1046/j.1365-2958.2002.02783.x. [DOI] [PubMed] [Google Scholar]
- 38.Kuhlemeier CJ, et al. A host-vector system for gene cloning in the cyanobacterium Anacystis nidulans R2. Plasmid. 1983;10(2):156–163. doi: 10.1016/0147-619x(83)90068-9. [DOI] [PubMed] [Google Scholar]
- 39.Biswas A, et al. Biosynthesis of cyanobacterial phycobiliproteins in Escherichia coli: Chromophorylation efficiency and specificity of all bilin lyases from Synechococcus sp. strain PCC 7002. Appl Environ Microbiol. 2010;76(9):2729–2739. doi: 10.1128/AEM.03100-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berkelman TR, Lagarias JC. Visualization of bilin-linked peptides and proteins in polyacrylamide gels. Anal Biochem. 1986;156(1):194–201. doi: 10.1016/0003-2697(86)90173-9. [DOI] [PubMed] [Google Scholar]
- 41.Gutu A, Alvey RM, Bashour S, Zingg D, Kehoe DM. Sulfate-driven elemental sparing is regulated at the transcriptional and posttranscriptional levels in a filamentous cyanobacterium. J Bacteriol. 2011;193(6):1449–1460. doi: 10.1128/JB.00885-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.