Abstract
Studies of DNA methylation from fungi, plants, and animals indicate that gene body methylation is ancient and highly conserved in eukaryotic genomes, but its role has not been clearly defined. It has been postulated that regulation of alternative splicing of transcripts was an original function of DNA methylation, but a direct experimental test of the effect of methylation on alternative slicing at the whole genome level has never been performed. To do this, we developed a unique method to administer RNA interference (RNAi) in a high-throughput and noninvasive manner and then used it to knock down the expression of DNA methyl-transferase 3 (dnmt3), which is required for de novo DNA methylation. We chose the honey bee (Apis mellifera) for this test because it has recently emerged as an important model organism for studying the effects of DNA methylation on development and social behavior, and DNA methylation in honey bees is predominantly on gene bodies. Here we show that dnmt3 RNAi decreased global genomic methylation level as expected and in addition caused widespread and diverse changes in alternative splicing in fat tissue. Four different types of splicing events were affected by dnmt3 gene knockdown, and change in two types, exon skipping and intron retention, was directly related to decreased methylation. These results demonstrate that one function of gene body DNA methylation is to regulate alternative splicing.
Keywords: epigenetics, gene regulation, gene silencing, insect
One of the greatest discoveries of the genomic era is environmental regulation of gene expression; DNA is not just inherited, it is also environmentally responsive. DNA cytosine methylation is an epigenetic mechanism that mediates many environmental influences on gene expression, affecting diverse aspects of organismal function and disease (1), and is one of the best studied mechanisms (2). DNA methylation of CG sites in the promoters of genes in plants and some species of animals acts to repress transcription (3). By contrast, methylation of gene bodies occurs in many species of fungi, plants, and animals (4, 5), but its function has not been clearly elucidated.
We chose to address this issue with the honey bee (Apis mellifera) because it has emerged as an important model organism for studying the effects of DNA methylation on development and social behavior (6–9). Moreover, DNA methylation in honey bees is predominantly on gene bodies (7, 8, 10). There are striking differences in methylation between alternative castes that develop from a totipotent female egg—worker and queen (11)—that relate to differences in diet, especially components of royal jelly (12). RNAi silencing of DNA methyl-transferase 3 (dnmt3), which encodes an enzyme involved in de novo methylation (13), alters developmental fate; worker larvae treated with dnmt3 RNAi show an increased likelihood of developing into queens (11).
Recent studies in both invertebrates and mammals support the idea that methylation is correlated with alternative splicing (AS) of transcripts (7, 14). By comparing brain methylomes of queen and worker bees, a correlation was revealed between DNA methylation and AS (7, 8). An in vitro study of mammalian cells showed that DNA methylation inhibited the binding of transcription factor CCCTC binding factor (CTCF), which affected alternative splicing (14). Regulation of AS is a complicated process that also involves spliceosome assembly (7, 15), chromatin structure (16, 17), siRNA activity (18), and transcriptional elongation (19), in addition to the apparent action of methylation.
To explore the causal relationship of DNA methylation and AS in vivo, we used the honey bee to test for a direct link between DNA methylation and AS. We decreased DNA methylation by dnmt3 RNA interference (RNAi) and hypothesized that there would be effects on AS.
RNAi has previously been administered to honey bees and other insects for gene knockdown (KD) by injection (20, 21), but this method is time consuming and highly invasive. We developed a unique method to treat large numbers of insects quickly and noninvasively. We coupled small interfering RNA (siRNA) to perfluocarbon-nanoparticles (PFC-NPs) and nebulized the mixture for aerosol application. PFC-NPs have been used in a variety of biomedical applications, including delivery of RNAi to tumor cells (22, 23), but had never before been nebulized.
To test the hypothesis that DNA methylation regulates AS, we used our method to knock down dnmt3 in honey bee abdominal fat tissue. We chose this tissue because abdominal injections of RNAi have proven especially effective (20, 21), and there are extensive gene expression data from honey bee fat body to compare our results to. Insect fat tissue has analogous functions to both liver and adipose tissue in mammals because they are crucial for both carbohydrate metabolism and lipid storage. We used RNAseq and new AS quantification software (24) to evaluate our hypothesis.
Results and Discussion
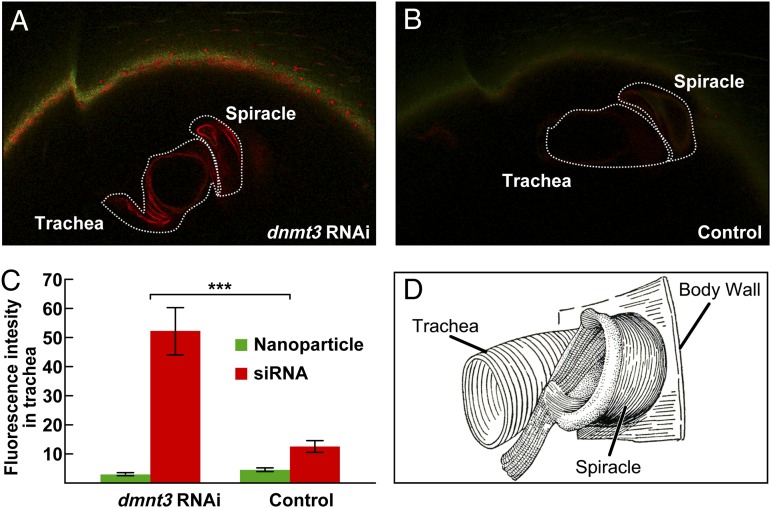
We first tested the efficacy of our method of administering RNAi. Image analysis indicates that the nebulized nanoparticle–siRNA complex penetrates the spiracles on the thorax and abdomen and travels through the tracheal respiratory system, which functions in insects to deliver oxygen directly to cells via ever-finer branches ending in tracheoles (25). This was shown by spraying caged bees (n = 17–18 bees/cage) with a mixture of PFC-NPs (labeled Alexa 488 in green) and dnmt3 siRNA (labeled Q-670 in red); control bees were sprayed with PFC-NPs (same label in green) and exogenous GFP siRNA (no label). Confocal microscopy revealed that trachea of dnmt3 siRNA-treated bees showed significantly higher levels of red fluorescence compared with nonlabeled control bees (Fig. 1). Using the same dnmt3 RNAi construct (Fig. S1) used previously for a prior honey bee RNAi injection study (11), aerosol application caused an ∼30% knockdown in fat tissue (n = 44 and 42 control and RNAi bees, respectively, across three biological replicates; Fig. 2). We obtained a similar KD by abdominal injection of the PFC-NP–siRNA complex (Fig. S2). By contrast to these results for the abdomen, no consistent KD was detected in the head, so brain analysis was not pursued. This result was unfortunate but not unexpected, given that the penetration points (spiracles) are located on the thorax and abdomen and not the head (25). These results demonstrate that it is possible to effectively exploit the insect tracheal system to deliver siRNA at least to abdominal cells via nebulizing spray in a rapid and noninvasive manner.
Fig. 1.
Noninvasive high-throughput method of delivering RNA interference in vivo. Spraying a nebulized mixture of nanoparticles (PFC-NP) and small interfering RNA (siRNA) on bees allows penetration through the insect tracheal respiratory system. (A) Treated: NP (labeled green with Alexa 488) and dnmt3 siRNA (labeled red with Q-670). (B) Control: NP (green) and GFP siRNA (no label). (C) Quantitative analysis of fluorescence reveals significantly higher levels of red signal in treated (n = 5) comparing to control bees (n = 6, P < 0.001, two-tailed t test), demonstrating penetration via the trachea. (D) Schematic of an inner view of trachea, spiracle atrium, and body wall. Diagram adapted from ref. 25.
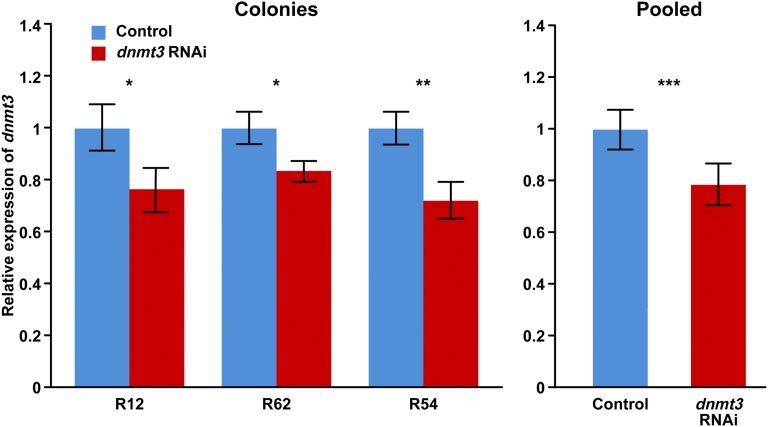
Fig. 2.
Effects of spraying a nebulized mixture of nanoparticles and small interfering RNA of dnmt3 on dnmt3 expression ± SEM in honey bee abdominal fat body. Results from three biological replicates (bees from unrelated colonies, n = 22 for colony R12; n = 31 for R62, and n = 33 for R54; two-tailed t tests). Analysis of pooled data: mixed-model ANOVA, df = 82, F = 15.52. *P < 0.05, **P < 0.01, ***P < 0.001.
dnmt3 siRNA-treated and control bees were used for genomewide analysis of gene expression, AS, and DNA methylation. We used the TrueSight program to analyze the RNA sequencing (RNA-seq) data and quantify AS. Detailed description of this application is in ref. 24. Across treated and control samples, RNA-seq yielded robust expression results for 13,008 genes, estimated to comprise 84.9% of the genes predicted from Assembly 4.5 and Official Gene Set 3.2 of the honey bee genome (http://hymenopteragenome.org/beebase/?q=gbrowse_amel). KD of dnmt3 caused strong effects on gene expression. A total of 2,613 (17.1%) genes showed significant differential expression due to RNAi treatment. Gene Ontology analysis of a subset of these differentially expressed genes (DEGs) that had orthologs in the Drosophila melanogaster genome (2,121) revealed that dnmt3 KD had particularly strong effects on RNA processing, intracellular transport, and protein catabolic processing (Table S1). Approximately 60.3% of the genes were found to be alternatively spliced, which is consistent with findings from Drosophila (26), and lower than in mammals (27, 28).
Comparing our results to previous gene expression studies of fat body, our DEG list also was enriched for genes related to honey bee behavioral maturation, which involves a shift from working in the hive when young to foraging when older (29). We compared our DEG list with DEG lists from fat tissue associated with (i) hive vs. forager bees; (ii) exposure to poor vs. rich diet (poor diet accelerates behavioral maturation); and (iii) exposure to queen pheromone (which delays behavioral maturation). There were significant overlaps between the dnmt3 KD DEG list and all three of these DEG lists [298, 368, and 363 genes, respectively; representation factor (RF) = 1.02, 1.37, and 1.37; P = 0.026, 2.36e−12, and 4.34e−12]. There also was a significant overlap of our dnmt3 KD DEG list and the DEG list from the caste determination study that used dnmt3-silenced larvae (11) (RF = 2.62, P = 5.77e−5). These results suggest that nutritional regulation of behavioral maturation involves differential methylation of genes in the honey bee fat body.
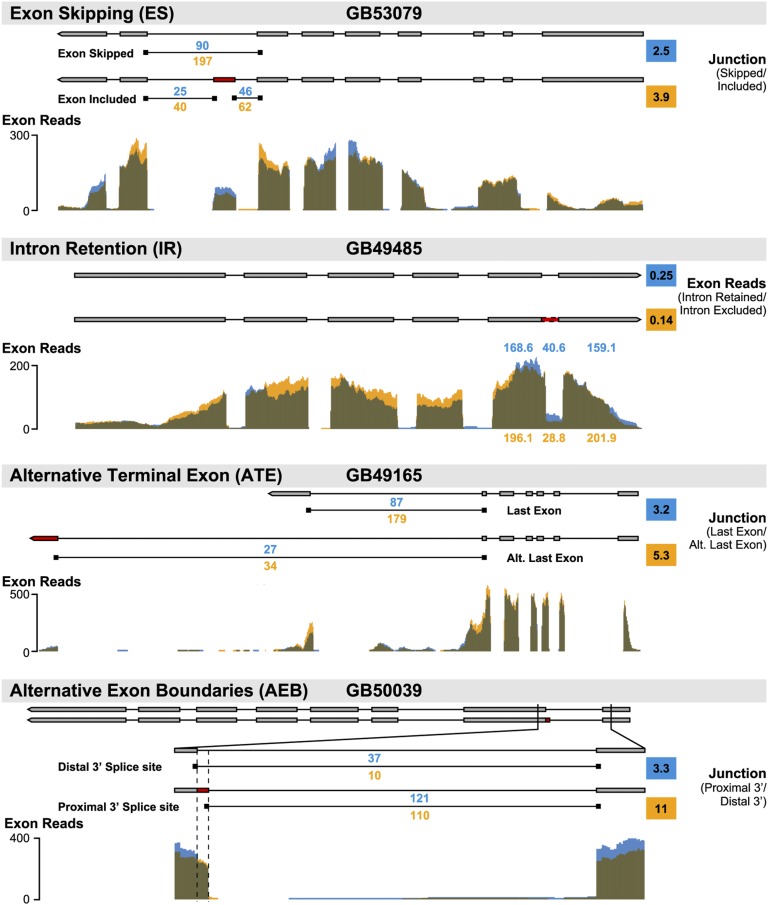
KD of dnmt3 caused strong effects on AS. A total of 524 genes showed significant differences in AS due to treatment [false discovery rate (FDR) < 0.1]. Moreover, there were significant differences for four different types of AS events: exon skipping (ES), alternative terminal exons (ATE), alternative exon boundaries (AEB), and intron retention (IR) (Table 1). This result revealed a broader effect of dnmt3 KD on AS than expected. Results with quantitative PCR (qPCR) for selected genes (Fig. 3; Fig. S3) confirmed the effects of dnmt3 KD on AS.
Table 1.
Effects of dnmt3 knockdown on DSGs, including types of ES, IR, ATE, and AEB on gene alternative splicing (AS)
| Types of AS event | No. of DSGs | FDR | Gene example | P value of qPCR validation |
| ES | 192 | 0.01 | GB53079 | 0.002 |
| IR | 27 | 0.1 | GB49485 | 0.03 |
| ATE | 141 | 0.01 | GB49165 | 0.003 |
| AEB | 225 | 0.01 | GB50039 | 0.004 |
Fig. 3.
Effects of dnmt3 knockdown on differentially spliced genes. (Upper) Gene model in both splicing isoforms with the number of RNAseq reads mapped to each splicing junction. (Lower) Exon coverage from RNAseq. Blue, control; orange, dnmt3 knockdown. Derivation of the AS ratios in the colored squares are in SI Materials and Methods.
To examine the possibility that dnmt3 KD exerted its effects by altering the expression of genes known to act as AS factors, we investigated the RNA-seq results for all known AS factors or AS factor-like genes in honey bees (by orthology to Drosophila). Of 72 AS factor or AS factor-like genes, 16 were differentially expressed (Table S2). There was no correlation between DNA methylation changes in gene bodies or promoter regions and the expression of these 16 genes (r = 0.25 and −0.12, P = 0.35 and 0.66, respectively). We cannot rule out the possibility that changes in expression of AS factor genes caused all of the above-reported change in AS, but we consider this unlikely, given the close association between the RNAi KD–induced changes in methylation and AS that we detected, as we report in the following paragraphs.
To address the issue of whether the effects of dnmt3 KD on AS were due to decreases in DNA methylation, dnmt3 siRNA-treated and control bees were used for genome-wide analysis of DNA methylation by bisulfite sequencing (BS-seq). Using pooled samples for a single analysis of treated and control bees, dnmt3 KD caused a 21% decrease in DNA methylation, similar to the level of KD of dnmt3 itself (∼30%). Sequencing coverage was comprehensive, covering 86.5% of all cytosine-phosphate-guanines (CpGs) with at least one read in the control and 80.1% in the dnmt3 KD sample (Fig. S4).
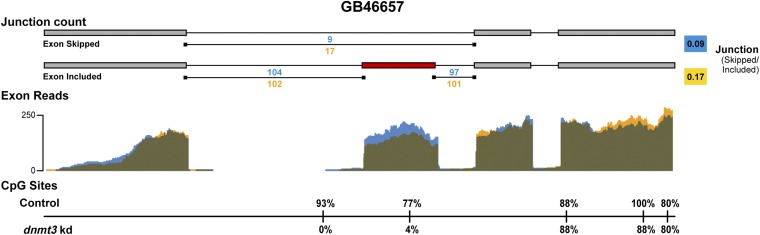
BS-seq results support the contention that the effects of dnmt3 KD on AS are at least partly due to decreases in DNA methylation. There was a significant relationship between dnmt3 KD, DNA methylation, and exon skipping: decreased DNA methylation (due to dnmt3 KD) was associated with increased ES (Table 2; P = 0.019, two-tailed Fisher’s exact test, n = 72 differential exon skipping events; Fig. 4). A galactokinase-like gene GB46657 is represented in Fig. 4 as an example of DNA methylation affecting ES. These genome-wide findings are consistent with results from honey bees for a single gene (8). A study in human cell lines (14) reported that DNA methylation prevented CTCF from binding to the alternative exon, leading to an increase in ES. The specific effect of DNA methylation on AS may vary in different genomic landscapes and in different species (30). Our results provide experimental evidence at the whole-genome level for a direct link between alternative splicing and DNA methylation in a dnmt3 KD background.
Table 2.
Effects of dnmt3 knockdown (KD) on DNA methylation of DESs and DIRs
| Splicing type | Direction | More methylated by dnmt3 KD | Less methylated by dnmt3 KD | P value |
| DES | More skipped | 13 | 35 | 0.019 |
| Less skipped | 14 | 10 | ||
| DIR | Less retained | 3 | 9 | 0.044 |
| More retained | 3 | 0 |
DES, differentially exon skipped gene; DIR, differentially intron retained gene.
Fig. 4.
Effects of dnmt3 knockdown on exon skipping and methylation in gene GB46657 (galactokinase-like). (Top and Middle) Same information as in Fig. 3. (Bottom) mCpG sites with vertical lines, together with percent differences between treatments.
BS-seq also revealed a significant association with decreased DNA methylation and less IR (P = 0.044, two-tailed Fisher’s exact test; Table 2). We did not test for a relationship between DNA methylation levels and AEB or ATE because no practical method has been developed yet: alternative exon boundaries are very short in cases of AEB, and ATE is often close to the regulatory regions. Other mechanisms in addition to DNA methylation are thus likely involved in the regulation of different forms of AS. Our results with BS-seq data demonstrate that DNA methylation of gene bodies affects alternative splicing on a genome-wide scale, specifically for ES and IR.
Alternative splicing of mRNA transcripts is an important mechanism for increasing the diversity and complexity of phenotypes that are generated from a genome (31) and can show strong species-specific differences (32, 33). AS has been shown to play fundamental roles in sex determination, development, behavior, and disease, and in worker honey bees, sterility, which is a hallmark of eusociality (1, 34, 35). Recently, DNA methylation has been detected in the genomes of other important social insects including several ants and termites (36, 37) and has been implicated in caste differentiation and behavioral maturation (8, 38). Further studies of DNA methylation and AS will help understand the mechanisms and evolution of phenotypic plasticity.
Materials and Methods
Bees.
Honey bees were a mix of European races typical of Illinois, predominantly Apis mellifera ligustica. They were maintained according to standard practices at the University of Illinois Bee Research Facility. We used worker bees derived from single-drone inseminated queens for RNA-seq analysis to reduce interindividual genetic variability and worker bees from a naturally mated queen colony for BS-seq analysis. Treated and control bees for each biological replicate were from the same colony.
RNA Interference Treatment.
We administered a nebulized aerosol mixture of PFC-NPs made in the Wickline laboratory and small siRNA for dnmt3 (Sigma). The control was siRNA for EGFP-S1 (IDT). siRNA oligo sequences are given in Table S3. The doses of PFC-NP and siRNA were 200 pM and 1 µM, respectively, in a total volume of 1,368 µL (mixture of PFC-NP, siRNA, and molecular graded water). A higher ratio of dnmt3 siRNA/NP (200 pM/2 µM) also was tested, but did not appear as efficacious. The reason may be that the number of siRNA molecules per nanoparticle was increased at a higher dose and the charge of the transfection complex changed, so the delivery of the transfection complex was not efficient. This treatment was administered to groups of 17–18 bees for 5 min. After treatment, the bees were placed in a standard Plexiglas cage and held in a dark incubator at 32 °C, 40% relative humidity, for 96 h. They were fed pollen paste [50% (wt/vol) honey/50% (wt/vol) pollen] and sugar syrup [50% sucrose (wt/vol) in water], which was replaced daily. Bees were collected 96 h after spray and frozen in liquid nitrogen, and then kept at −80 °C for analysis. Abdominal fat bodies were stored in RNAlater-ICE (catalog no. AM7030; Ambion by Life Technologies) overnight and then dissected and stored at −80 °C. For injection experiments, PFC-NP:siRNA (10 pM:10 nM) was administered in 400 nL of insect saline solution and administered to 1-d-old bees abdominally. Holding conditions were as for the sprayed bees.
Image Processing to Determine Mode of Action of Nanoparticle–RNAi Spray.
To determine whether the nanoparticle-RNAi spray acts by penetrating the insect tracheal respiratory system, 1-d-old bees (n = 17–18) were sprayed with both 1 µM siRNA of GFP control (no labeling) and 200 pM Alexa 488 fluorescently labeled nanoparticles (green) or with 1 µM Q-670 siRNA of dnmt3 (red) and nanoparticles (green) with the above dose. Abdominal spiracles and trachea were dissected in insect saline solution 24 h after spraying. Samples were dehydrated in solutions of 25%, 50%, 75%, 100%, and 100% methanol for 20 min, respectively, and maintained in 100% methyl salicylate until imaging. The images were captured by a Zeiss LSM 700 confocal microscope at 20×.
RNA-seq.
We selected individuals showing typical KD for RNA sequencing (6 treated and 6 control). Libraries were made with the TruSeq RNA sample preparation kit per the manufacturer’s instructions (Illumina). One library was made per individual and barcoded. The 12 libraries were pooled and quantitated by qPCR, and the pool was sequenced on two lanes for 100 cycles, in paired-end mode, on an Illumina HiSeq2000 machine (Illumina). We used the TruSeq SBS sequencing kit (version 3) and analyzed the results with Casava1.8 (pipeline 1.9). Library construction and RNA-seq sequencing were performed at the University of Illinois W.M. Keck Center for Comparative and Functional Genomics. The number of sequencing reads was ∼56–72 M/sample, and the average cDNA length was 220 bp. Reads from each sample were aligned onto the A. mellifera genome assembly v4.5 using TrueSight (24), allowing up to two mismatches for both exonic and junction spanning reads. These reads were mapped to 14,124 genes of 15,314 annotated genes in the Official Gene Set v3.2. TrueSight assigns a probability score to each of the predicted splice junctions for each gene, which indicates their reliability based on coding potentials and RNA-seq mapping quality. We used all splice junctions with TrueSight score greater than 0.5 (on a scale from 0 to 1). BEDTools (39) was used to calculate the read coverage of the nonredundant exon models based on TrueSight mapping results (in binary alignment/map format). Complete methods and further analyses are provided in SI Materials and Methods.
BS-seq Analysis.
Genomic DNA (1 μg/group) extracted from pooled abdominal fat body samples (18 bees per group) was used to generate BS-seq libraries using the premethylated adapter method based on a previously published protocol (40). The single-end libraries were sequenced on an Illumina HiSeq2000, following the manufacturer’s manual. Data processing, aligning Bisulfite-converted reads, and methylation assessments are described in ref. 40. Identical reads were collapsed into single reads. The methylation level was calculated as the ratio of methylated reads over all reads covering each CpG site, using a threshold of 20% methylation and read coverage ≥4 in either control or treated samples. We used this threshold because it resulted in a comparable number of methylated cytosine-phosphate-guanine (mCpG) sites (123,023) to previous studies (7). At this threshold there was no methylated cytosine signal from the mitochondrial genome. Dnmt3 KD caused an overall decrease in CpG methylation levels (an average of −0.045 per mCpG).
Supplementary Material
Acknowledgments
We thank C. Nye for bee colony assistance; A. Duque and D. Dalpiaz for technical assistance in the laboratory; A. Hernandez and the W. M. Keck Center for Comparative and Functional Genomics for library preparation and RNAseq; M. Akhavan for technical assistance; the UCLA BSCRC BioSequencing Core Facility for BS-Seq; Y. Liu for advice on DEG analysis; D. M. Muzny and C. L. Kovar and the BCM-HGSC genome sequencing and assembly teams for the improved Apis mellifera genome assembly; A. Bennett, K. Hoff, F. Camara, R. Guigo, T. Murphy, K. Pruitt, V. Soloveyev, and M. Stanke for preparing the Official Gene Set v3.2; and P. L. Jones, R. Maleszka, and members of the G.E.R. laboratory for comments that improved the manuscript. This project was supported by National Institutes of Health (NIH) Director’s Pioneer Award 1DP1OD006416 (to G.E.R.), National Science Foundation Grant 1054309 (to J.M.), NIH Grant 1R21HG006464 (to J.M.), NIH Grant R01 HL073646 (to S.A.W.), NIH Grant GM60398 (to S.E.J.), an American Association of University Women American postdoctoral fellowship (to H.L.-B.), and an American Heart Association Heartland Affiliate predoctoral training grant (to M.K.). S.F. is a Special Fellow of the Leukemia and Lymphoma Society. S.E.J. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the National Center for Biotechnology Information Sequence Read Archive database (accession no. SRP024289).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310735110/-/DCSupplemental.
References
- 1.Edwards TM, Myers JP. Environmental exposures and gene regulation in disease etiology. Environ Health Perspect. 2007;115(9):1264–1270. doi: 10.1289/ehp.9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suzuki MM, Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 3.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11(3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328(5980):916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 5.Jones PA. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, et al. Functional CpG methylation system in a social insect. Science. 2006;314(5799):645–647. doi: 10.1126/science.1135213. [DOI] [PubMed] [Google Scholar]
- 7.Foret S, et al. DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees. Proc Natl Acad Sci USA. 2012;109(13):4968–4973. doi: 10.1073/pnas.1202392109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyko F, et al. The honey bee epigenomes: Differential methylation of brain DNA in queens and workers. PLoS Biol. 2010;8(11):e1000506. doi: 10.1371/journal.pbio.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lyko F, Maleszka R. Insects as innovative models for functional studies of DNA methylation. Trends Genet. 2011;27(4):127–131. doi: 10.1016/j.tig.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Consortium HGS. Honeybee Genome Sequencing Consortium Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443(7114):931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319(5871):1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 12.Kamakura M. Royalactin induces queen differentiation in honeybees. Nature. 2011;473(7348):478–483. doi: 10.1038/nature10093. [DOI] [PubMed] [Google Scholar]
- 13.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 14.Shukla S, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479(7371):74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 16.Schor IE, Rascovan N, Pelisch F, Alló M, Kornblihtt AR. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci USA. 2009;106(11):4325–4330. doi: 10.1073/pnas.0810666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hnilicová J, et al. Histone deacetylase activity modulates alternative splicing. PLoS ONE. 2011;6(2):e16727. doi: 10.1371/journal.pone.0016727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13(9):793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 19.Selth LA, Sigurdsson S, Svejstrup JQ. Transcript elongation by RNA polymerase II. Annu Rev Biochem. 2010;79:271–293. doi: 10.1146/annurev.biochem.78.062807.091425. [DOI] [PubMed] [Google Scholar]
- 20.Nelson CM, Ihle KE, Fondrk MK, Page RE, Amdam GV. The gene vitellogenin has multiple coordinating effects on social organization. PLoS Biol. 2007;5(3):e62. doi: 10.1371/journal.pbio.0050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ament SA, et al. The transcription factor ultraspiracle influences honey bee social behavior and behavior-related gene expression. PLoS Genet. 2012;8(3):e1002596. doi: 10.1371/journal.pgen.1002596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaneda MM, Sasaki Y, Lanza GM, Milbrandt J, Wickline SA. Mechanisms of nucleotide trafficking during siRNA delivery to endothelial cells using perfluorocarbon nanoemulsions. Biomaterials. 2010;31(11):3079–3086. doi: 10.1016/j.biomaterials.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soman NR, et al. Molecularly targeted nanocarriers deliver the cytolytic peptide melittin specifically to tumor cells in mice, reducing tumor growth. J Clin Invest. 2009;119(9):2830–2842. doi: 10.1172/JCI38842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, et al. TrueSight: A new algorithm for splice junction detection using RNA-seq. Nucleic Acids Res. 2013;41(4):e51. doi: 10.1093/nar/gks1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snodgrass RE. Anatomy of the Honey Bee. New York: Comstock Publishing Associates; 1956. pp. 230–240. [Google Scholar]
- 26.Graveley BR, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471(7339):473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 28.Wang Z, Burge CB. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14(5):802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ament SA, et al. Mechanisms of stable lipid loss in a social insect. J Exp Biol. 2011;214(Pt 22):3808–3821. doi: 10.1242/jeb.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gelfman S, Cohen N, Yearim A, Ast G (2013) DNA-methylation effect on co-transcriptional splicing is dependent on GC-architecture of the exon-intron structure. Genome Res 23(5):789–799. [DOI] [PMC free article] [PubMed]
- 31.Marden JH. Quantitative and evolutionary biology of alternative splicing: How changing the mix of alternative transcripts affects phenotypic plasticity and reaction norms. Heredity (Edinb) 2008;100(2):111–120. doi: 10.1038/sj.hdy.6800904. [DOI] [PubMed] [Google Scholar]
- 32.Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338(6114):1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barbosa-Morais NL, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338(6114):1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- 34.Jarosch A, Stolle E, Crewe RM, Moritz RF. Alternative splicing of a single transcription factor drives selfish reproductive behavior in honeybee workers (Apis mellifera) Proc Natl Acad Sci USA. 2011;108(37):15282–15287. doi: 10.1073/pnas.1109343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salz HK. Sex determination in insects: A binary decision based on alternative splicing. Curr Opin Genet Dev. 2011;21(4):395–400. doi: 10.1016/j.gde.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonasio R, et al. Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr Biol. 2012;22(19):1755–1764. doi: 10.1016/j.cub.2012.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glastad KM, Hunt BG, Goodisman MA. Evidence of a conserved functional role for DNA methylation in termites. Insect Mol Biol. 2013;22(2):143–154. doi: 10.1111/imb.12010. [DOI] [PubMed] [Google Scholar]
- 38.Herb BR, et al. Reversible switching between epigenetic states in honeybee behavioral subcastes. Nat Neurosci. 2012;15(10):1371–1373. doi: 10.1038/nn.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng S, Rubbi L, Jacobsen SE, Pellegrini M. Determining DNA methylation profiles using sequencing. Methods Mol Biol. 2011;733:223–238. doi: 10.1007/978-1-61779-089-8_16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.