Abstract
Nitrous oxide (N2O) is a powerful greenhouse gas implicated in climate change. The dominant source of atmospheric N2O is incomplete biological dentrification, and the enzymes responsible for the release of N2O are NO reductases. It was recently reported that ambient emissions of N2O from the Great Boiling Spring in the United States Great Basin are high, and attributed to incomplete denitrification by Thermus thermophilus and related bacterial species [Hedlund BP, et al. (2011) Geobiology 9(6)471–480]. In the present work, we have isolated and characterized the NO reductase (NOR) from T. thermophilus. The enzyme is a member of the cNOR family of enzymes and belongs to a phylogenetic clade that is distinct from previously examined cNORs. Like other characterized cNORs, the T. thermophilus cNOR consists of two subunits, NorB and NorC, and contains a one heme c, one Ca2+, a low-spin heme b, and an active site consisting of a high-spin heme b and FeB. The roles of conserved residues within the cNOR family were investigated by site-directed mutagenesis. The most important and unexpected result is that the glutamic acid ligand to FeB is not essential for function. The E211A mutant retains 68% of wild-type activity. Mutagenesis data and the pattern of conserved residues suggest that there is probably not a single pathway for proton delivery from the periplasm to the active site that is shared by all cNORs, and that there may be multiple pathways within the T. thermophilus cNOR.
Keywords: proton pathway, thermophilic
Nitric oxide (NO) is important as a signaling molecule in many eukaryotes (animals, plants, and fungi), serves as an immune defense agent in many animals (1), and is also an obligate intermediate of microbial denitrification. NO reduction by prokaryotes is an important environmental process, in part because incomplete denitrification produces nitrous oxide (N2O) as a final product, which is directly released from cells without being converted into N2. The unrestrained use of nitrogen fertilizers has led to the release of large amounts of N2O to the atmosphere as a result of this process. N2O is a more potent greenhouse gas than CO2 (2) and contributes to the depletion of the ozone layer (3). In the present work, the enzyme that is at least partially responsible for the N2O ambient emissions from the Great Boiling Spring (and presumably other hot springs) has been isolated and characterized (4).
In denitrification, NO reduction is carried out by NO reductase (NOR) members of the heme-copper oxidoreductase (HCO) superfamily, which also includes oxygen reductases (quinol and cytochrome oxidases). There are two closely related heme-copper NOR families: the cNOR and qNOR families. These are integral membrane enzymes that catalyze the two-electron reduction of NO to N2O:
The cNORs are composed of two subunits, NorC (∼17 kDa) and NorB (∼56 kDa). NorC is a monoheme c-type cytochrome that possesses an N-terminal transmembrane helix that anchors the heme domain to the periplasmic face of the cytoplasmic membrane. Its function is to relay electrons from the periplasmic donor proteins (cytochromes and cupredoxins) to the catalytic subunit, NorB (5, 6). NorB is structurally related to the catalytic subunit (subunit I) of the respiratory HCOs (5, 7, 8). NorB contains 12 transmembrane helices and binds three metal centers: a low-spin heme b and a binuclear active site composed of a high-spin heme b3, and a nonheme iron (FeB). FeB is coordinated by three histidine residues that are conserved in all typical HCOs and a glutamate residue that is conserved in all cNORs and qNORs (5). The qNOR family, which receives electrons from quinols, is very similar to the cNOR family, with the exception that NorB and NorC are fused into one subunit and the cytochrome c binding site is missing (9). Unlike the oxygen reductases, the cNORs are not electrogenic and are unable to pump protons (10, 11). The protons required for chemistry are delivered from the periplasmic side of the membrane and not from the cytoplasmic side, as in the case of the oxygen reductases.
Enzymes belonging to the cNOR family have been isolated from five denitrifying bacteria: Pseudomonas stutzeri (12, 13), Paracoccus denitrificans (14, 15), Halomonas halodenitrificans (16, 17), Roseobacter denitrificans (18, 19), Pseudomonas nautica (20), and Pseudomonas aeruginosa, for which an X-ray structure has been reported (5, 21). All of these strains are in the Proteobacteria phylum. Mutagenesis studies have been done (6, 22), but before the report of the X-ray structure of cNOR from P. aeruginosa (5).
Members of the Thermus order are thermophilic bacteria of great biotechnological interest as a source of thermostable enzymes (23). The most characterized strains belonging to the Thermus genus are strict aerobes; however, a few isolates have been shown to be facultative anaerobes, growing by reducing nitrate to nitrite (nitrate reduction) or by reducing nitrate and nitrite to either N2O (incomplete denitrification) or N2 (complete denitrification) (24). Denitrifying Thermus thermophilus strains are a contributor of the N2O released from thermal systems (24), and in this work the operon encoding the enzyme responsible for this has been cloned from the denitrifying strain T. thermophilus PRQ25 (25) and has been expressed in both T. thermophilus and Escherichia coli.
Results
cNOR Expression and Purification.
The operon encoding the cNOR from T. thermophilus (TtcNOR) was cloned and expressed homologously in T. thermophilus. The enzyme was purified by affinity chromatography using a His-tag placed on the C terminus of the NorB subunit. However, because the yield obtained was very low (0.04 mg from 1 L of culture), heterologous expression in E. coli was attempted. Expression of cNOR in E. coli required the presence in the host strain of a plasmid containing the genes required for the assembly of cytochromes c, pEC86 (26), and another plasmid carrying tRNA genes needed for reading rare codons (pRARE) (27). This process was successful, and the yield of TtcNOR using the E. coli expression system was about 20-times greater than in T. thermophilus. SDS/PAGE (Fig. S1B) shows the two bands corresponding to NorB and NorC, and heme-staining shows that the NorC subunit contains heme c. The reduced-minus-oxidized spectrum has diagnostic peaks for both the heme c (553 nm) and low-spin heme b (560 nm) components of TtcNOR. Without the pEC86 plasmid in the E. coli host strain, the heme c component of TtcNOR is absent (Fig. S1A) and the relative intensity of the NorC subunit in the SDS/PAGE gel is low (Fig. S1B), suggesting that the NorC subunit is unstable if the heme c is not properly assembled into the protein.
TtcNOR is active with different electron donors (Table 1). NOR activity could not be measured at 75 °C, the optimum growth temperature for T. thermophilus (28), because of limitations of the NO-sensing electrode. Therefore, assays were performed at 42 °C, which is below the optimum growth temperature. The turnover number (kcat) obtained at 42 °C for the TtcNOR is considerably smaller than those reported for other cNORs (5, 6, 13, 16, 18, 20). The highest activity obtained with the TtcNOR was about 9 electrons (e−)/min (at 42 °C) with the electron donor PMS (phenazine methosulfate) (Table 1), which presumably donates electrons directly to the active site in the NorB subunit without involving the heme c in the NorC subunit (5). The turnover number of TtcNOR in the presence of 2.5 mM TMPD (N,N,N′,N′-tetramethyl-p-phenylenediamine), which donates electrons to the heme c component of the enzyme, was about 5.5 e−/min (at 42 °C) (Table 1). The recombinant form of the soluble cytochrome c552 of T. thermophilus, was expressed in E. coli, purified as previously described (29), and tested as an electron donor to TtcNOR. In the presence of 30 μM cytochrome c552 (plus 0.5 mM TMPD to maintain the cytochrome c552 reduced) TtcNOR had a turnover number of about 2.5 electrons per minute (at 42 °C) (Table 1). In the absence of cytochrome c552 no activity is observed at this low concentration of TMPD. Unless indicated, all assays of TtcNOR were performed with TMPD (2.5 mM) as the electron donor. The TtcNOR exhibited no detectable oxygen reductase activity.
Table 1.
NOR activity of the TtcNOR with different electron donors
| Electron donors | kcat (e−/min) | n | % Activity |
| TMPD 2.5 mM | 5.5 ± 0.5 | 15 | 100 |
| PMS 10 μM | 9 ± 0.7 | 5 | 163 |
| cyt c552 30 μM + TMPD 0.5 mM | 2.5 | 1 | 46 |
Data are expressed as average ± SD of n independent experiments.
Regardless of the electron donor, the enzyme activity was completely inhibited by the addition of 100 μM KCN (Fig. S2), demonstrating that the observed NO consumption is enzyme-dependent and not an artifact of the activity assays. The cNOR obtained by heterologous expression in E. coli has the same turnover number (6 ± 0.8 e−/min at 42 °C with 2.5 mM TMPD as electron donor) as the enzyme expressed homologously in T. thermophilus (Table 1).
Sequence Analysis.
A phylogenetic tree of the cNOR family (Fig. S3), shows that the TtcNOR belongs to a different clade from the previously characterized cNORs (5, 6, 13, 16, 18). The TtNorC is ∼75 aa longer than the homologs from Proteobacteria and contains an N-terminal sequence with two predicted transmembrane helices instead of one as found in the cNORs from Proteobacteria (5). Homologs from other Thermus species and from Aquificae also have two predicted transmembrane helices at the N terminus of the NorC subunit. In addition, the TtcNOR operon contains an additional (third) ORF following the norB gene, which encodes a hypothetical protein containing 85 amino acids and three putative transmembrane helices. This ORF was included in the constructs used for expression of cNOR from T. thermophilus, but the putative third subunit was not detected in the isolated cNOR.
Table S1 lists the most conserved residues in cNORs (over 150 sequences) and their possible functions, inferred in part from the structure of the P. aeruginosa cNOR (PacNOR) (5). Table S1 also includes residues in the NorC subunit that have been postulated to be involved in the proton delivery from the periplasm to the active site (5). These residues are not conserved within the cNORs.
Site-Directed Mutagenesis of the Calcium Ligands.
The crystal structure of PacNOR shows a single Ca2+ that is buried within the protein at the interface between NorB and NorC and ligated by two amino acid side chains, E135 in NorB and Y73 in NorC, in addition to being bound to one propionate each in heme b and heme b3 (Fig. 1) (5). Whereas Y73NorC is totally conserved in cNORs (corresponding to Y137NorC in TtcNOR) (Table S1), E135 (E134 in TtcNOR) is replaced by an uncharged or basic amino acid in about 20% of the cNOR sequences (Table S1). In addition, a second glutamate that is near the Ca+2 but not ligated to it, is also present in >90% of the cNORs (E138 in PacNOR; E137 in TtcNOR), replaced only by aspartate in the remaining sequences.
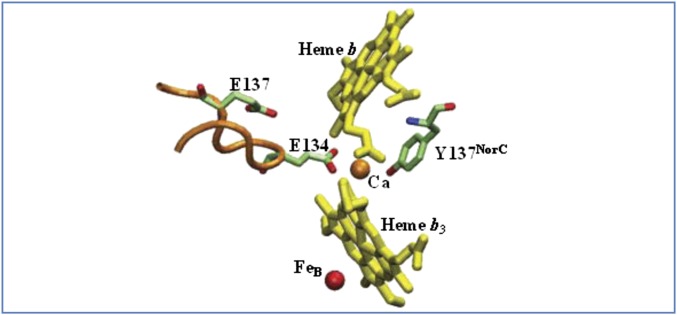
Fig. 1.
Calcium binding site at the interface of NorB and NorC. Ca2+ is directly ligated to E134 in NorB and to Y137NorC in NorC, along with propionates from heme b and from heme b3. E137 is important to maintain the conformation of the protein loop containing E134 and connecting transmembrane helices III and IV of NorB. The residue numbers are for the cNOR from T. thermophilus, and the structure is drawn from the coordinates of the cNOR from P. aeruginosa PDB ID code 3O0R (4).
The Ca2+ should be structurally important because it bridges both NorB and NorC as well as the propionates of hemes b and b3, which was confirmed by site-directed mutagenesis (Table 2). The E134Q, E137Q, and Y137FNorC mutants each exhibit less than 25% of the wild-type activity. The E134D and E137D mutants of TtcNOR were apparently not assembled or not stable because no protein was observed in either case. Both the size and charge of the two glutamates are important to maintain the structural integrity of the protein. Although E137 is not directly involved in the coordination of Ca2+, it may be important to maintain the conformation of the loop stabilizing the coordination of E134 to Ca2+ (Fig. 1), as previously suggested (30).
Table 2.
NOR activity of TtcNOR mutants and the wild-type enzyme
| Site involved | % Conserved | Ttc NOR | kcat e−/min | n | % Activity |
| Wild-type | WT | 5.5 ± 0.5 | 15 | 100 | |
| Sites involved In Ca binding | |||||
| E135 | >80 | E134D | — | — | — |
| E134Q | 1.0 ± 0.1 | 2 | 19 | ||
| E138 | >90 | E137D | — | — | — |
| E137Q | 1.32 ± 0.15 | 2 | 24 | ||
| Y73NorC | 100 | Y137FC | 0.90 ± 0.05 | 5 | 17 |
| Sites near the active site | |||||
| E215 | >95 | E215D | 1.70 ± 0.11 | 3 | 31 |
| E215Q | 3.1 ± 0.21 | 9 | 56 | ||
| E280 | >90 | E280D | 1.70 ± 0.20 | 3 | 31 |
| E280Q | 1.43 ± 0.32 | 8 | 26 | ||
| E280M | 0 | 4 | 0 | ||
| E211 | 100 | E211D | 1.87 ± 0.15 | 7 | 34 |
| E211Q | 1.88 ± 0.20 | 7 | 34 | ||
| E211A | 3.74 ± 0.35 | 8 | 68 | ||
| H259 | 100 | H259N | 0 | 6 | 0 |
| Sites in putative proton pathways | |||||
| D198 | >85 | D198E | 8.30 ± 1.0 | 5 | 151 |
| D198N | 7.50 ± 0.55 | 4 | 137 | ||
| D198L | —– | — | — | ||
| T66NorC | >80 | T130VC | 0.77 ± 0.16 | 3 | 14 |
| E77NorC | 35 | D141NC | – | — | — |
| D141LC | 0.99 ± 0.18 | 4 | 18 | ||
| N80NorC | <10 | R144MC | 3.30 ± 0.22 | 4 | 60 |
| R84NorC | <10 | D148LC | 1.11 ± 0.17 | 3 | 20 |
| E145NorC | >50 | D209EC | 1.92 ± 0.25 | 5 | 35 |
| D209NC | 1.65 ± 0.23 | 3 | 30 | ||
| D209LC | 0 | 3 | 0 | ||
| H339 | 100 | H339F | 0 | 5 | 0 |
| N335 | 100 | N335L | 0.82 ± 0.11 | 4 | 15 |
| S277 | 100 | S277L | 2.47 ± 0.26 | 4 | 45 |
| T330 | 100 | T330L | 2.36 ± 0.30 | 6 | 43 |
| T330L/S277L | 0 | 9 | 0 | ||
Data are expressed as the average of ± SD of n independent experiments. All mutants had normal UV-visble spectra with the exception of H259N (Fig. S4).
Site-Directed Mutagenesis of Glutamic Acids Near the Active Site.
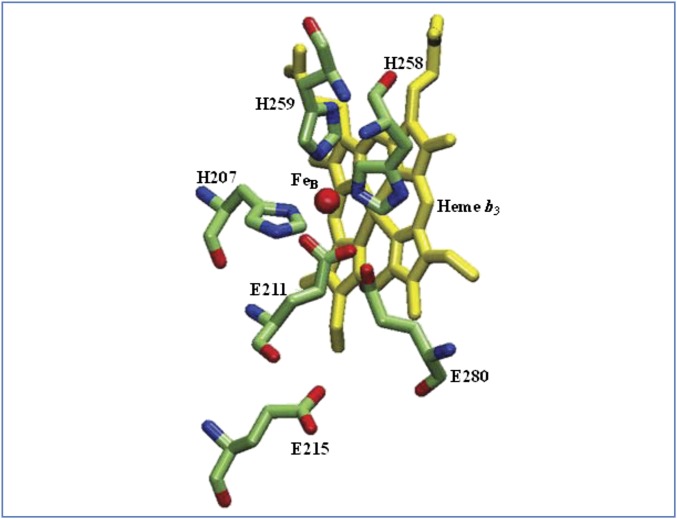
In addition to E134 and E137 (TtcNOR), there are three additional glutamates that are conserved to varying degrees in cNORs: E211 (100% conserved), E215 (>95% conserved), and E280 (>90% conserved). According to the PacNOR crystal structure, E211 is a ligand to FeB and, along with the three totally conserved histidines (H207, H258, and H259) form a trigonal-bipyramidal coordination structure (Fig. 2). It has been proposed that the E211 carboxylate in cNOR could also function as the shuttle for protons from E280 to the bound-NO (5, 30). E211 was mutated to D, Q, and A. Both E211D and E211Q mutants have 34% of the wild-type activity and, remarkably, E211A has 68% of the wild-type NO reductase activity (Table 2). Hence, the glutamate ligand to FeB is not absolutely essential for function in this enzyme. In contrast, when one of the histidine ligands to FeB, H259 (Fig. 2 and Table S1), is mutated to an asparagine, the purified H259N mutant has no detectable activity (Table 2).
Fig. 2.
The binding site of the nonheme iron (FeB) in cNOR. FeB is coordinated by E211, H207, H258, and H259. Also shown are E280 and E215. These residue numbers are the same for cNORs from T. thermophilus and P. aeruginosa, and the structure is drawn from the coordinates of the cNOR from P. aeruginosa PDB ID code 3O0R (4).
Metal analysis of two different preparations indicates that the iron of E211A (4 ± 0.32 irons per molecule of protein) is identical to that of the wild-type (4 ± 0.25 irons per molecule of protein), and no zinc or copper was detected. Furthermore, the UV-visible spectra of the wild-type and E211A mutant cNORs are similar [e.g., peak positions and peak ratios of the Soret (418 nm) and α (550 nm) bands]. In comparison, the spectrum of the fully reduced H259N mutant cNOR has a significantly higher A418/A550 ratio (Fig. S4), suggesting a significant perturbation of the structure of this mutant.
E280 is hydrogen bonded to E211 (Fig. 2) (5). E280D and E280Q mutations show a significant decrease (to about 30%) in NOR activity, and E280M is totally inactive (Table 2). E215 is near E211 but the two glutamates are not within hydrogen bonding distance (Fig. 2). The E215 carboxylate group could contribute to the electronegative environment of the binuclear center of cNOR and be a factor in determining the low midpoint potential of heme b3 iron (60 mV), which is lower than those of heme b (345 mV) and heme c (310 mV) in cNOR (31). The E215D and E215Q mutations retain 31% and 56% of the wild-type activity, respectively (Table 2). These results are similar to previous reports of mutating the equivalent residue (E202) in P. denitrificans (6, 22).
Site-Directed Mutagenesis of Putative Proton Entry Channels.
The protons required for chemistry in the NORs are delivered from the periplasmic side of the membrane. Three possible pathways have been proposed from the bulk water in the periplasm to the region of the heme b3 propionates, and several pathways have been suggested for proton transfer from the heme propionates to the NO bound at the active site.
Bulk water to propionates.
Two hydrophilic channels connecting the periplasm to the propionates of heme b3 were observed in the PacNOR crystal structure (5), and a third channel has been proposed based on molecular dynamics (32). Channel 1 starts from the E57NorC (PacNOR numbering) at the periplasmic surface and includes D198 and E135 (Ca2+ ligand), and ends at a propionate-A of heme b3. However, E57NorC is only about 75% conserved among the cNOR family (Fig. S5), and is a glutamine TtcNOR (Q121 in TtNorC). D198 (same number in TtNorB) is 85% conserved within the cNORs. The D198E, D198N, and D198L mutations were made in TtNorB. D198E and D198N mutants had no deleterious effect in the enzyme activity but D198L was not assembled (Fig. 3 and Table 2).
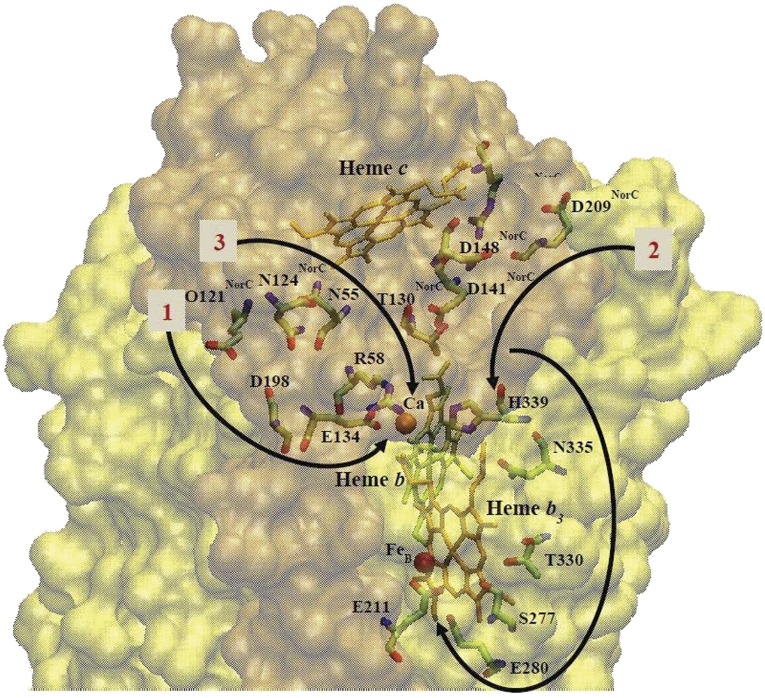
Fig. 3.
Residues putatively involved in facilitating proton transfer from the periplasm to the active site (4, 32). The residue numbers are for the cNOR from T. thermophilus, and the structure is drawn from the coordinates of the cNOR from P. aeruginosa PDB ID code 3O0R (4). The surfaces of NorC and NorB subunits are shown in orange and yellow, respectively. Arrows illustrate the proposed proton pathways.
Channel 2, observed in the PacNOR structure (5), includes polar residues in the NorC subunit: E145NorC; E77NorC, and T66NorC. These residues correspond to D209NorC, D141NorC, and T130NorC in in TtcNOR (Fig. 3). The putative entrance of channel 2, E145NorC (D209NorC in TtcNOR), is only about 50% conserved and is not an acidic residue in many cNOR sequences (Table S1). T130NorC (TtcNOR) is about 80% conserved in cNORs (often replaced by a serine). E77NorC (D141NorC in TtcNOR) is either a glutamate or an aspartate in the corresponding position in all other cNORs (Fig. S5 and Table S1). In addition, N80NorC and R84NorC are in the same region of the PacNOR channel 2, although they are both very poorly conserved (<10%); these correspond to R144NorC and D148NorC, respectively, in TtcNOR (Fig. 3).
Substitutions for D209NorC, the putative channel entrance, did reduce NOR activity: D209ENorC (30%), D209NNorC (30%), and D209LNorC (20%) (Table 2). Mutations T130VNorC, D141LNorC, and D148LNorC similarly exhibited less than 20% activity, whereas D141NNorC was not assembled. R144MNorC retained about 60% of the wild-type activity (Table 2). Aside from D141NNorC, which is not assembled, all of the mutations have SDS/PAGE patterns and UV/vis spectra that are similar to the wild-type enzyme. This pattern of phenotypes suggests that the selected residues are important for the full function of the cNOR. It is noted that these residues are near the heme c component of the NorC subunit, for example D141NorC (4.86 Å) and R144NorC (2.35 Å).
Channel 3 was proposed based on computational studies (32) and involves N54, N60NorC, E135 (Ca2+ ligand), E138, and R57 in PacNOR; these correspond to N55, N124NorC, E134, E137, and R58 in TtcNOR. There are internal water molecules interacting with E135, E138, and R57 (PacNOR), which connect to the bulk water when N54 and N60NorC (PacNOR) move away from their observed positions in the crystal structure (5) and assume an “open” conformation (32). N54 and N60NorC (PacNOR) are 76% and 55% conserved, respectively, within cNORs (Table S1). E138 (PacNOR) is always either E or D in other cNORs, whereas R57 (PacNOR) is either R or K. The only mutations testing channel 3 were those in the two glutamates, E134 and E137 in TtcNOR, which are also implicated in Ca2+ binding (Table 2), where it is clear that they are each important for both stability and for function.
Propionates-to-active site.
Four different pathways have been suggested for protons to get to the metal-bound NO and bound to the heme b3 Fe or FeB (32). Two pathways involve only internal water molecules, one involves the H259 ligand to FeB (which is an essential residue), and the fourth channel involves several amino acid residues that, along with internal water molecules, could facilitate proton transfer from propionate-D of heme b3 to E211, which is ligated to FeB: T330→ S277→ E280 → E211. Propionate-D of heme b3 is hydrogen-bonded to N335 and H339, which could also be part of this pathway.
S277, T330, N335, and H339, are totally conserved in all cNORs, and E280 is found in >90% of cNORs (Fig. 3 and Table S1). These residues are present in the TtcNOR and have the same residue numbers as in the PacNOR. In the NorB subunit, N335 and H339 are each hydrogen-bonded to propionate-D of the active-site heme b3 and are fully conserved in all cNORs. The N335L and H339F mutants were assembled but exhibited less than 15% NOR activity. The S277L and T330L mutations each retained about half of the wild-type activity, whereas the double-mutant was inactive (Table 2). The S227L/T330L double-mutant was also obtained in very low yield, suggesting that this mutant is unstable. As previously discussed, mutations of E280 substantially reduced the activity, E280Q (26%), E280D (31%), and E280M (0%), whereas E211A is 68% active (Table 2).
Discussion
The cNOR from T. thermophilus belongs to a different clade within the cNOR family than the proteobacterial cNORs previously characterized (Fig. S3) (5, 12–19, 21). The recombinant TtcNOR has the same properties whether it is isolated from an E. coli or from a T. thermophilus expression system. The purified enzyme has two subunits, NorB and NorC, and has the same heme and metal content as the proteobacterial cNORs: one heme c in the NorC subunit, and two hemes b plus a nonheme FeB in the NorB subunit. As with the proteobacterial cNORs, the TtcNOR is active with the artificial electron donors TMPD and PMS, but is also active with purified T. thermophilus cytochrome c552 (33), which is likely to be a natural substrate. The kcat, measured at 42 °C is low, about 0.15 e−/s, compared with the cNOR from P. aeruginosa, which has been reported to have a kcat of 7 e−/s with PMS (5, 6). The activity of the T. thermophilus enzyme could not be measured at the optimum growth temperature of the organism (75 °C) because of the limitations of the NO-sensing electrode.
The enzyme activity is the same for initial NO concentrations between 10 μM and 30 μM, demonstrating that the KM is less than 10 μM and also that there is no substrate inhibition by NO within this concentration range. Although an accurate measure of the KM for NO was not attempted, the steady-state rate clearly dropped during assays as the NO concentration went from 10 μM to 1 μM. Hence, the KM for NO at 42 °C is in the low micromolar range. This range is much higher than the KM reported for the cNOR from P. stutzeri (1–2 nM) (13).
The pattern of conserved residues shared within the proteobacterial cNORs is maintained in the TtcNOR. Site-directed mutagenesis was used to address several questions about the roles of specific residues.
Ca2+ Binding.
Y137NorC (100% conserved), E134 (80% conserved), and E137 (90% conserved) are important for the stability of TtcNOR, probably by stabilizing the binding of Ca2+. Both Y137NorC and E137 are direct ligands to Ca2+, whereas E137 probably stabilizes the protein conformation near the bound Ca2+ (30). It is interesting that the nonligating E137 is always an acidic residue (E or D), whereas a significant number of cNORs have nonacidic residues at the corresponding location of the ligating E134. This finding suggests that there are alternative ways to stabilize the Ca2+ at this site (or to stabilize the protein without Ca2+).
There is a single Ca2+ in the same location in the homologous qNORs (9) and also in the C-family oxygen reductases (cbb3 oxygen reductases) (8). Site-directed mutagenesis experiments have also concluded that the pair of glutamic acids corresponding to E134 and E137 in TtcNOR is required for structural stability of both the qNORs (9) and the cbb3 oxygen reductases (34). The A and B family of oxygen reductases do not have Ca2+ at the equivalent position but, rather, have a pair of arginine residues that appear to have a similar function (35–38).
FeB Binding.
E211, H207, H258, and H259 are ligands to the nonheme FeB in the cNOR active site and each of these residues is totally conserved within cNORs. The NO reductases are distinguished from the O2 reductases in the heme-copper oxidoreductase superfamily by the presence of FeB in the active site, replacing the CuB present in all of the oxygen reductases. In the A- and B-family oxygen reductases, the position corresponding to E211 is always a tyrosine, which is covalently cross-linked to the equivalent of H207 (TtcNOR). In the cbb3 oxygen reductases (C family), the histidine-tyrosine cross-linked pair of amino acids is also present in the active site but the tyrosine originates from a different position in the sequence (39, 40). Remarkably and unexpectedly, E211 can be replaced in the TtcNOR with substantial retention of activity (e.g., E211A is 68% active). It is clearly not essential to have a glutamate, an acidic residue, or even a potential metal-ligating amino acid, at this site.
The equivalent to E211 has been mutated to an alanine in the cNOR from P. denitrificans, with very different results showing a virtual elimination of catalytic function (6, 22, 41). In addition, computational (42, 43) as well as modeling studies (44) on NOR activity have emphasized the importance of the negative charge provided by E211 to NOR activity. One explanation for the relatively high activity of the E211A mutant is that the carboxylate group, at least in the TtcNOR, can be replaced by a hydroxyl (or water) ligand. Further work will be needed to test this proposal. In any event, it is clear that the conformation of the cNOR is not dependent on the ligation of E211 to FeB. This finding is consistent with the idea that the glutamate ligand may dissociate from FeB as part of the catalytic cycle (5, 30).
It should be noted that the cytochrome cbb3 oxygen reductase from P. stutzeri exhibits NOR activity with a kcat ∼ 2 s−1 (45), which is comparable to the P. stutzeri cNOR, kcat ∼ 2−7 s−1 (13). Although the C-family (cyt cbb3) oxygen reductases are related to the cNORs, the cytochrome cbb3 contains CuB instead of FeB at the active site and also lacks the equivalent of the glutamates present in most cNORs (E211, E215, and E280 in TtcNOR). Hence, these glutamates and FeB are not essential for catalytic NOR function. However, the KM values are very different for the NO reductase activity of the P. stutzeri cNOR (1–2 nM) (13) and the P. stutzeri cbb3 oxygen reductase (12–14 μM) (45), so the cNOR is clearly catalytically superior in the physiologically relevant range of substrate concentrations.
Pathways for Proton Delivery to the Active Site.
There is no question that the protons required for the chemistry catalyzed at the active site of the cNORs are all taken from the bulk solution on the periplasmic side of the membrane. This finding is opposite of the heme-copper oxygen reductases, in which all of the protons are taken from the cytoplasmic side of the membrane. In the case of the oxygen reductases, the proton delivery channels can be identified by patterns of conserved residues, unique to each family (A, B, or C) of enzymes, and confirmed by site-directed mutagenesis (7, 46, 47). The proton-input channels of the oxygen reductases facilitate proton diffusion from the cytoplasmic bulk water along hydrogen bonds formed by internal water molecules and protonatable amino acid side chains. Single point mutations of conserved residues in the proton-input channels in the oxygen reductases can result in the virtual elimination of catalytic function. In contrast, there is no pattern of conserved residues within the cNORs that can be used to define a pathway leading from the periplasm to the vicinity of the region of the propionates of heme b3 (Table S1). With the exception of the tyrosine that ligates the Ca2+, and the cytochrome c binding residues (including a CXXCH motif), no residues in the NorC subunit is anywhere near totally conserved (Table S1). However, the lack of conserved residues in NorC is accompanied by a large number of internal water molecules that are either resolved in the X-ray structure of the PacNOR (5) or postulated by computational methods (32). These internal water molecules are present both in those portions of the NorB and NorC subunits that separate the heme propionates from the bulk periplasmic water, and are important components in each of the three proposed proton input channels proposed for cNORs. It is very likely, therefore, that there is no proton input pathway that is universally shared by all cNORs, but that small structural differences might open or close different pathways in individual cNOR variants. Multiple proton input pathways might easily be used in each cNOR, so no residue would be absolutely required for this function. This result would be consistent with the mutagenesis data in the current work on the TtcNOR. The lack of conservation of residues in the proposed channels and the abundance of internal water molecules means that conclusions concerning proton pathways in the TtcNOR from mutagenesis may not apply generally to other cNORs. Based on these mutagenesis data (Table 2), channels 2 and 3 may be used in the T. thermophilus cNOR, whereas channel 1 seems less likely. It is interesting that molecular dynamics studies suggest that channel 2 is not likely to function to deliver protons in PacNOR because there is a region in the NorB subunit where the connection between internal water molecules is broken (32). If this is also the case for the T. thermophilus cNOR, then the major proton pathway would be that corresponding to channel 3 in PacNOR. Channel 3 was proposed for PacNOR based on molecular dynamics studies showing that small side-chain motions would open a pathway from the bulk solution to internal waters near the Ca2+ site and the heme propionates. The critical amino acids that anchor the internal water molecules in this region of the protein are also structurally important, most notably the two glutamates required for Ca2+ binding (E134 and E137 in TtcNOR). These two glutamates were postulated to be important for proton input before information about the structure of the PacNOR or their role in Ca2+ binding (6, 10, 41).
Several pathways have been proposed for protons to reach their final destination at the bimetallic active site from the region around the heme b3 propionates (22, 32). The most prominent suggestion is for protons to follow a pathway from propionate-D of heme b3→(water)→T330→S277→E280→E211 in TtcNorB (Fig. 3). Although these residues are highly conserved (Table S1), mutagenesis does not indicate that any one of these residues is absolutely essential for function, including the FeB ligand E211. Mutants do have significantly lower activity: E280Q (26%), E280M (0%), S277L (43%), and T330L (45%). Previously, it was reported that the E267A mutation in the cNOR from P. denitrificans (equivalent to E280 in TtcNOR) has 5% activity (6, 22). Hence, this pathway remains a possibility and is not excluded by mutagenesis data. On the other hand, the proximity of these residues to heme b3 and FeB make it equally likely that mutations might reduce activity independent of any effects on proton transfer.
Materials and Methods
The operon that was cloned for the expression of cNOR from T. thermophilus was obtained from genomic DNA from strain PRQ25. This was expressed in E. coli strain C43(DE3) using the pET22b vector. All details for the cloning, expression, mutagenesis, cell growth, protein expression, and purification are included in the SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank H. Nikaido (University of California, Berkeley) for the pRARE2-Km plasmid and J. Fee (deceased; Scripps Research Institute) for the HB27 strain; Jon Hosler (University of Mississippi Medical Center) for the TtcNOR metal analysis; and all the laboratory members, especially Paige Sheraden and Mariana Lencina, for their help and useful discussions. This work was supported by National Institutes of Health Grant HL16101 (to R.B.G.); Spanish Ministry of Economy and Competitivity Grant BIO2010-18875 (to J.B.); an institutional grant from Fundación Ramón Areces (to the Centro de Biología Molecular Severo Ochoa); and a Formacion de Personal Investigator fellowship (to C.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301731110/-/DCSupplemental.
References
- 1.Burney S, Tamir S, Gal A, Tannenbaum SR. A mechanistic analysis of nitric oxide-induced cellular toxicity. Nitric Oxide. 1997;1(2):130–144. doi: 10.1006/niox.1996.0114. [DOI] [PubMed] [Google Scholar]
- 2.Ravishankara AR, Daniel JS, Portmann RW. Nitrous oxide (N2O): The dominant ozone-depleting substance emitted in the 21st century. Science. 2009;326(5949):123–125. doi: 10.1126/science.1176985. [DOI] [PubMed] [Google Scholar]
- 3.Canfield DE, Glazer AN, Falkowski PG. The evolution and future of Earth’s nitrogen cycle. Science. 2010;330(6001):192–196. doi: 10.1126/science.1186120. [DOI] [PubMed] [Google Scholar]
- 4.Hedlund BP, et al. Potential role of Thermus thermophilus and T. oshimai in high rates of nitrous oxide (N2O) production in ∼80 °C hot springs in the US Great Basin. Geobiology. 2011;9(6):471–480. doi: 10.1111/j.1472-4669.2011.00295.x. [DOI] [PubMed] [Google Scholar]
- 5.Hino T, et al. Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science. 2010;330(6011):1666–1670. doi: 10.1126/science.1195591. [DOI] [PubMed] [Google Scholar]
- 6.Thorndycroft FH, Butland G, Richardson DJ, Watmough NJ. A new assay for nitric oxide reductase reveals two conserved glutamate residues form the entrance to a proton-conducting channel in the bacterial enzyme. Biochem J. 2007;401(1):111–119. doi: 10.1042/BJ20060856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hemp J, et al. Comparative genomics and site-directed mutagenesis support the existence of only one input channel for protons in the C-family (cbb3 oxidase) of heme-copper oxygen reductases. Biochemistry. 2007;46(35):9963–9972. doi: 10.1021/bi700659y. [DOI] [PubMed] [Google Scholar]
- 8.Buschmann S, et al. The structure of cbb3 cytochrome oxidase provides insights into proton pumping. Science. 2010;329(5989):327–330. doi: 10.1126/science.1187303. [DOI] [PubMed] [Google Scholar]
- 9.Matsumoto Y, et al. Crystal structure of quinol-dependent nitric oxide reductase from Geobacillus stearothermophilus. Nat Struct Mol Biol. 2012;19(2):238–245. doi: 10.1038/nsmb.2213. [DOI] [PubMed] [Google Scholar]
- 10.Reimann J, Flock U, Lepp H, Honigmann A, Adelroth P. A pathway for protons in nitric oxide reductase from Paracoccus denitrificans. Biochim Biophys Acta. 2007;1767(5):362–373. doi: 10.1016/j.bbabio.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Flock U, et al. Defining the proton entry point in the bacterial respiratory nitric-oxide reductase. J Biol Chem. 2008;283(7):3839–3845. doi: 10.1074/jbc.M704615200. [DOI] [PubMed] [Google Scholar]
- 12.Heiss B, Frunzke K, Zumft WG. Formation of the N-N bond from nitric oxide by a membrane-bound cytochrome bc complex of nitrate-respiring (denitrifying) Pseudomonas stutzeri. J Bacteriol. 1989;171(6):3288–3297. doi: 10.1128/jb.171.6.3288-3297.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kastrau DH, Heiss B, Kroneck PM, Zumft WG. Nitric oxide reductase from Pseudomonas stutzeri, a novel cytochrome bc complex. Phospholipid requirement, electron paramagnetic resonance and redox properties. Eur J Biochem. 1994;222(2):293–303. doi: 10.1111/j.1432-1033.1994.tb18868.x. [DOI] [PubMed] [Google Scholar]
- 14.Hoglen J, Hollocher TC. Purification and some characteristics of nitric oxide reductase-containing vesicles from Paracoccus denitrificans. J Biol Chem. 1989;264(13):7556–7563. [PubMed] [Google Scholar]
- 15.Carr GJ, Ferguson SJ. The nitric oxide reductase of Paracoccus denitrificans. Biochem J. 1990;269(2):423–429. doi: 10.1042/bj2690423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakurai N, Sakurai T. Genomic DNA cloning of the region encoding nitric oxide reductase in Paracoccus halodenitrificans and a structure model relevant to cytochrome oxidase. Biochem Biophys Res Commun. 1998;243(2):400–406. doi: 10.1006/bbrc.1998.8106. [DOI] [PubMed] [Google Scholar]
- 17.Sakurai T, Nakashima S, Kataoka K, Seo D, Sakurai N. Diverse NO reduction by Halomonas halodenitrificans nitric oxide reductase. Biochem Biophys Res Commun. 2005;333(2):483–487. doi: 10.1016/j.bbrc.2005.05.149. [DOI] [PubMed] [Google Scholar]
- 18.Matsuda Y, et al. Nitric oxide-reductase homologue that contains a copper atom and has cytochrome c-oxidase activity from an aerobic phototrophic bacterium Roseobacter denitrificans. J Biochem. 2002;131(6):791–800. doi: 10.1093/oxfordjournals.jbchem.a003167. [DOI] [PubMed] [Google Scholar]
- 19.Matsuda Y, Uchida T, Hori H, Kitagawa T, Arata H. Structural characterization of a binuclear center of a Cu-containing NO reductase homologue from Roseobacter denitrificans: EPR and resonance Raman studies. Biochim Biophys Acta. 2004;1656(1):37–45. doi: 10.1016/j.bbabio.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Timóteo CG, et al. Low-spin heme b(3) in the catalytic center of nitric oxide reductase from Pseudomonas nautica. Biochemistry. 2011;50(20):4251–4262. doi: 10.1021/bi101605p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumita H, et al. NO reduction by nitric-oxide reductase from denitrifying bacterium Pseudomonas aeruginosa: Characterization of reaction intermediates that appear in the single turnover cycle. J Biol Chem. 2004;279(53):55247–55254. doi: 10.1074/jbc.M409996200. [DOI] [PubMed] [Google Scholar]
- 22.Flock U, Lachmann P, Reimann J, Watmough NJ, Adelroth P. Exploring the terminal region of the proton pathway in the bacterial nitric oxide reductase. J Inorg Biochem. 2009;103(5):845–850. doi: 10.1016/j.jinorgbio.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 23.Koyama Y, Hoshino T, Tomizuka N, Furukawa K. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J Bacteriol. 1986;166(1):338–340. doi: 10.1128/jb.166.1.338-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cava F, Zafra O, da Costa MS, Berenguer J. The role of the nitrate respiration element of Thermus thermophilus in the control and activity of the denitrification apparatus. Environ Microbiol. 2008;10(2):522–533. doi: 10.1111/j.1462-2920.2007.01472.x. [DOI] [PubMed] [Google Scholar]
- 25.Alvarez L, Bricio C, Gómez MJ, Berenguer J. Lateral transfer of the denitrification pathway genes among Thermus thermophilus strains. Appl Environ Microbiol. 2011;77(4):1352–1358. doi: 10.1128/AEM.02048-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arslan E, Schulz H, Zufferey R, Künzler P, Thöny-Meyer L. Overproduction of the Bradyrhizobium japonicum c-type cytochrome subunits of the cbb3 oxidase in Escherichia coli. Biochem Biophys Res Commun. 1998;251(3):744–747. doi: 10.1006/bbrc.1998.9549. [DOI] [PubMed] [Google Scholar]
- 27.Kim HS, Nagore D, Nikaido H. Multidrug efflux pump MdtBC of Escherichia coli is active only as a B2C heterotrimer. J Bacteriol. 2010;192(5):1377–1386. doi: 10.1128/JB.01448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oshima T, Imahori K. Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a nonsporulating Thermophilic bacterium from a Japanese thermal spa. Int J Syst Bacteriol. 1974;24(1):102–112. [Google Scholar]
- 29.Keightley JA, Sanders D, Todaro TR, Pastuszyn A, Fee JA. Cloning and expression in Escherichia coli of the cytochrome c552 gene from Thermus thermophilus HB8. Evidence for genetic linkage to an ATP-binding cassette protein and initial characterization of the cycA gene products. J Biol Chem. 1998;273(20):12006–12016. doi: 10.1074/jbc.273.20.12006. [DOI] [PubMed] [Google Scholar]
- 30.Hino T, Nagano S, Sugimoto H, Tosha T, Shiro Y. Molecular structure and function of bacterial nitric oxide reductase. Biochim Biophys Acta. 2012;1817(4):680–687. doi: 10.1016/j.bbabio.2011.09.021. [DOI] [PubMed] [Google Scholar]
- 31.Grönberg KL, et al. A low-redox potential heme in the dinuclear center of bacterial nitric oxide reductase: implications for the evolution of energy-conserving heme-copper oxidases. Biochemistry. 1999;38(42):13780–13786. doi: 10.1021/bi9916426. [DOI] [PubMed] [Google Scholar]
- 32.Pisliakov AV, Hino T, Shiro Y, Sugita Y. Molecular dynamics simulations reveal proton transfer pathways in cytochrome C-dependent nitric oxide reductase. PLOS Comput Biol. 2012;8(8):e1002674. doi: 10.1371/journal.pcbi.1002674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fee JA, et al. Integrity of Thermus thermophilus cytochrome c552 synthesized by Escherichia coli cells expressing the host-specific cytochrome c maturation genes, ccmABCDEFGH: Biochemical, spectral, and structural characterization of the recombinant protein. Protein Sci. 2000;9(11):2074–2084. doi: 10.1110/ps.9.11.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ouyang H, et al. Functional importance of a pair of conserved glutamic acid residues and of Ca(2+) binding in the cbb(3)-type oxygen reductases from Rhodobacter sphaeroides and Vibrio cholerae. Biochemistry. 2012;51(37):7290–7296. doi: 10.1021/bi3006847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376(6542):660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 36.Tsukihara T, et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272(5265):1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 37.Soulimane T, et al. Structure and mechanism of the aberrant ba(3)-cytochrome c oxidase from Thermus thermophilus. EMBO J. 2000;19(8):1766–1776. doi: 10.1093/emboj/19.8.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abramson J, et al. The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site. Nat Struct Biol. 2000;7(10):910–917. doi: 10.1038/82824. [DOI] [PubMed] [Google Scholar]
- 39.Hemp J, Christian C, Barquera B, Gennis RB, Martínez TJ. Helix switching of a key active-site residue in the cytochrome cbb3 oxidases. Biochemistry. 2005;44(32):10766–10775. doi: 10.1021/bi050464f. [DOI] [PubMed] [Google Scholar]
- 40.Hemp J, et al. Evolutionary migration of a post-translationally modified active-site residue in the proton-pumping heme-copper oxygen reductases. Biochemistry. 2006;45(51):15405–15410. doi: 10.1021/bi062026u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butland G, Spiro S, Watmough NJ, Richardson DJ. Two conserved glutamates in the bacterial nitric oxide reductase are essential for activity but not assembly of the enzyme. J Bacteriol. 2001;183(1):189–199. doi: 10.1128/JB.183.1.189-199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blomberg MRA, Siegbahn PEM. Mechanism for N₂O generation in bacterial nitric oxide reductase: A quantum chemical study. Biochemistry. 2012;51(25):5173–5186. doi: 10.1021/bi300496e. [DOI] [PubMed] [Google Scholar]
- 43.Blomberg LM, Blomberg MR, Siegbahn PE. Reduction of nitric oxide in bacterial nitric oxide reductase—A theoretical model study. Biochim Biophys Acta. 2006;1757(4):240–252. doi: 10.1016/j.bbabio.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 44.Lin Y-W, et al. Roles of glutamates and metal ions in a rationally designed nitric oxide reductase based on myoglobin. Proc Natl Acad Sci USA. 2010;107(19):8581–8586. doi: 10.1073/pnas.1000526107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forte E, et al. The cytochrome cbb3 from Pseudomonas stutzeri displays nitric oxide reductase activity. Eur J Biochem. 2001;268(24):6486–6491. doi: 10.1046/j.0014-2956.2001.02597.x. [DOI] [PubMed] [Google Scholar]
- 46.Chang HY, Hemp J, Chen Y, Fee JA, Gennis RB. The cytochrome ba3 oxygen reductase from Thermus thermophilus uses a single input channel for proton delivery to the active site and for proton pumping. Proc Natl Acad Sci USA. 2009;106(38):16169–16173. doi: 10.1073/pnas.0905264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Konstantinov AA, Siletsky S, Mitchell D, Kaulen A, Gennis RB. The roles of the two proton input channels in cytochrome c oxidase from Rhodobacter sphaeroides probed by the effects of site-directed mutations on time-resolved electrogenic intraprotein proton transfer. Proc Natl Acad Sci USA. 1997;94(17):9085–9090. doi: 10.1073/pnas.94.17.9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.