Abstract
The maintenance of cooperation in populations where public goods are equally accessible to all but inflict a fitness cost on individual producers is a long-standing puzzle of evolutionary biology. An example of such a scenario is the secretion of siderophores by bacteria into their environment to fetch soluble iron. In a planktonic culture, these molecules diffuse rapidly, such that the same concentration is experienced by all bacteria. However, on solid substrates, bacteria form dense and packed colonies that may alter the diffusion dynamics through cell–cell contact interactions. In Pseudomonas aeruginosa microcolonies growing on solid substrate, we found that the concentration of pyoverdine, a secreted iron chelator, is heterogeneous, with a maximum at the center of the colony. We quantitatively explain the formation of this gradient by local exchange between contacting cells rather than by global diffusion of pyoverdine. In addition, we show that this local trafficking modulates the growth rate of individual cells. Taken together, these data provide a physical basis that explains the stability of public goods production in packed colonies.
Keywords: biofilm, evolution, noise, variability, ecology
Iron is required for many enzymatic processes, and is therefore an essential nutrient. To overcome the low abundance of free iron in aerobic environments, bacteria, as well as other microorganisms, synthesize and secrete iron-chelating molecules called siderophores (1). Once released in the extracellular environment, siderophores diffuse and complex with iron. Because other bacteria can import them, siderophores can be regarded as public goods (2) and siderophore secretion is often cited as a model system for studying the stability of cooperation (3, 4). In this context, nonproducing mutants, which can enjoy the benefit of siderophores without bearing the cost of their production, have a selective advantage over the WT-producing strain and could, in principle, displace them on evolutionary time scales.
This argument usually assumes that public good molecules are readily available to all, as is the case in planktonic cultures, where molecules diffuse freely and rapidly between cells. In liquid conditions, mutants that do not produce siderophores have, in fact, been shown to outcompete WT strains (5–8). However, the limited spatial dispersal of public goods can challenge this picture and has been proposed as a general mechanism for explaining the maintenance of cooperation (8–12). When dispersal is limited, public good molecules tend to stay in the vicinity of the producing subpopulations, allowing them to benefit preferentially from their own production, and thus to balance the advantage of opportunistic nonproducing strains. In many ecological situations, bacteria form complex, tightly packed, and spatially structured colonies, such as biofilms, where public good dispersal may be severely reduced compared with planktonic cultures. This raises the questions of how public good molecules circulate between cells in these natural conditions and how cooperation is affected by the population structure (8, 13–15). Experimental realizations of limited public good dispersal were obtained by tuning the viscosity (16), the distance between colonies of producers and nonproducers (17), or the amount of cell attachment (18). Although the dispersal of public goods has been shown to influence the outcome of ecological competitions, direct measurements of public goods diffusion in structured population are still lacking.
Here, we address this question by taking advantage of the natural fluorescence of pyoverdine (Pvd), a siderophore produced by the human opportunistic pathogenic bacterium Pseudomonas aeruginosa (19). We monitored under a microscope the diffusion of Pvd in bacterial microcolonies forming monolayers. This minimal model for a spatially structured population of packed cells enables us to measure the distribution and spatiotemporal dynamics of siderophore concentrations precisely. Based on our observations at the single-cell level, we propose a model of local Pvd exchange and test its predictions on global, colony-wide observables. In particular, the model quantitatively explains the formation of a concentration gradient measured across the colony. We further show that the traffic of Pvd between cells has an impact on the growth rate at an individual level and discuss the ecological implications through observation-driven computer simulations of competition between producing and nonproducing strains.
Results
Dynamics of Pvd Distribution Inside Clonal Microcolonies.
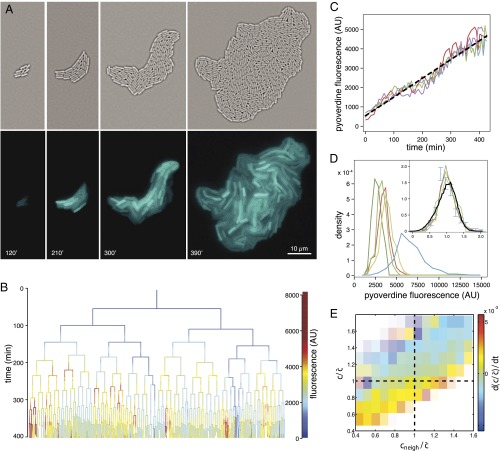
WT P. aeruginosa bacteria (PAO1) were inoculated at a low density on top of a nutritional gel sealed with a coverslip. We monitored colony growth by video microscopy thanks to a newly developed correlation imaging technique based on bright-field images taken at various focal positions (SI Appendix, Fig. S2). Every 3 min over the course of 7 h, we measured the endogenous fluorescence of free Pvd in each cell of a growing microcolony. We observed a strong variability in Pvd concentration across bacteria, as well as within the same bacteria at different times (Fig. 1A and SI Appendix, Movie S1). We reconstructed the genealogical trees of 10 microcolonies (Fig. 1B). Along each lineage, the fluorescence per pixel in cells fluctuated strongly in time (by about 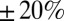 ), with a correlation time of ∼93 min (i.e., longer than the averaged division time of ∼40 min). Although the average fluorescence per pixel in the colony increased linearly with time over the course of nine cell divisions (Fig. 1C), the distributions of cell fluorescence normalized by the average fluorescence of the colony remained invariant over time and across different colonies (Fig. 1D and SI Appendix, Fig. S3). The analysis of Pvd dynamics in individual cells reveals that the concentrations in both the cell and its neighbors appear to contribute to free Pvd dynamics (Fig. 1E and SI Appendix). Additionally, a gradient of Pvd is established between the border and the center of the colony, suggesting that Pvd dynamics are local.
), with a correlation time of ∼93 min (i.e., longer than the averaged division time of ∼40 min). Although the average fluorescence per pixel in the colony increased linearly with time over the course of nine cell divisions (Fig. 1C), the distributions of cell fluorescence normalized by the average fluorescence of the colony remained invariant over time and across different colonies (Fig. 1D and SI Appendix, Fig. S3). The analysis of Pvd dynamics in individual cells reveals that the concentrations in both the cell and its neighbors appear to contribute to free Pvd dynamics (Fig. 1E and SI Appendix). Additionally, a gradient of Pvd is established between the border and the center of the colony, suggesting that Pvd dynamics are local.
Fig. 1.
Dynamics and variability of Pvd concentrations in P. aeruginosa (PAO1) microcolonies. (A) Time-lapse fluorescence images of a growing microcolony, with correlation images (Upper) and Pvd intrinsic fluorescence (Lower). (B) Genealogical tree of the colony. The level of Pvd in each cell c is color-coded along each lineage (for clarity, only a subset of the tree is displayed). (C) Dynamics of the mean level of Pvd 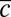 in four different colonies. AU, arbitrary unit. (D) Distributions of Pvd concentrations in cells at various time points in a given microcolony (green), in a different microcolony (red), and in a medium supplemented with human transferrin (blue). (Inset) Distributions are normalized by the mean in the microcolony and the predicted distribution of our model (black). Error bars represent the SEM. (E) Dependence on the relative Pvd concentration in the cell
in four different colonies. AU, arbitrary unit. (D) Distributions of Pvd concentrations in cells at various time points in a given microcolony (green), in a different microcolony (red), and in a medium supplemented with human transferrin (blue). (Inset) Distributions are normalized by the mean in the microcolony and the predicted distribution of our model (black). Error bars represent the SEM. (E) Dependence on the relative Pvd concentration in the cell 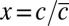 and its neighbors
and its neighbors 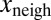 of the variation
of the variation 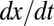 . Color transparency indicates the uncertainty of measurements (light color means uncertain). The correlation between
. Color transparency indicates the uncertainty of measurements (light color means uncertain). The correlation between 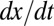 and
and 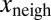 is assessed with a P value less than
is assessed with a P value less than 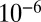 .
.
Model of Pvd Dynamics in Individual Cells.
The overall dynamics of Pvd are a product of a complex pathway involving synthesis, export, and import of the molecule. Pvd biosynthesis starts in the cytoplasm and ends in the periplasm (SI Appendix, Fig. S1). It involves four nonribosomal peptide synthetases and probably seven different other enzymes (20–22). Newly synthesized Pvd is stored in the bacterial periplasm (21) before secretion into the extracellular medium by the efflux system PvdRT-OpmQ (23). After iron chelation in the extracellular medium, ferri-Pvd is imported across the outer membrane by the ferri-Pvd outer membrane transporter FpvA (24) and iron is released from the siderophore in the periplasm (25). Free Pvd is then recycled into the extracellular medium by the efflux pump PvdRT-OpmQ (26, 27). Pvd biosynthesis is under positive feedback, regulated by a σ-factor PvdS and its anti–σ-factor FpvR, triggered when bacteria import ferri-Pvd (28, 29). Free Pvd is naturally fluorescent, whereas ferri-Pvd is not.
To model the spatial dynamics of Pvd inside microcolonies, we reasoned that the net effect of production, export, import, dilution (from cell division rate v), and degradation depends on the periplasmic concentration (dilution, export), on the concentration in the immediate vicinity of a bacterium (import), or on both (through feedbacks). Keeping only linear terms in the concentrations leads to a dynamical equation (SI Appendix):
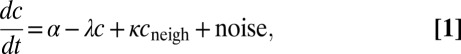 |
where c and 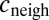 stand for the concentration in the cell and its neighbors; α is the basal production rate; and
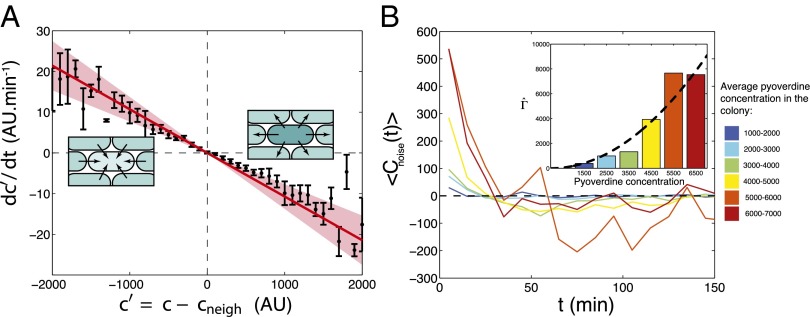
stand for the concentration in the cell and its neighbors; α is the basal production rate; and 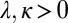 are phenomenological parameters determined from the data (see below), reflecting the effective efflux of Pvd out of a cell and the influx from its neighbors. Note that because of the negative contribution from feedback and import, λ is expected to be smaller than v (Materials and Methods). The observed linear increase of the average fluorescence in the colony indicates a balance between efflux and influx,
are phenomenological parameters determined from the data (see below), reflecting the effective efflux of Pvd out of a cell and the influx from its neighbors. Note that because of the negative contribution from feedback and import, λ is expected to be smaller than v (Materials and Methods). The observed linear increase of the average fluorescence in the colony indicates a balance between efflux and influx, 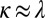 . This balance defines an exchange mechanism formally equivalent to an effective diffusion process on the lattice formed by adjacent neighbors (SI Appendix). Within the mean-field approximation (SI Appendix), the above model predicts that local fluctuations in Pvd,
. This balance defines an exchange mechanism formally equivalent to an effective diffusion process on the lattice formed by adjacent neighbors (SI Appendix). Within the mean-field approximation (SI Appendix), the above model predicts that local fluctuations in Pvd, 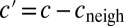 , obey an Ornstein–Uhlenbeck process,
, obey an Ornstein–Uhlenbeck process, 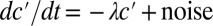 , meaning that the local fluctuations are damped proportionally to their amplitude with a measured rate of
, meaning that the local fluctuations are damped proportionally to their amplitude with a measured rate of 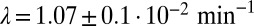 (Fig. 2A), which is independent of the average concentration in the colony (SI Appendix, Fig. S4). Consistent with our model, these local fluctuations are uniform within the colony (SI Appendix, Fig. S5) and spatially uncorrelated (SI Appendix, Fig. S7). The noise spectrum was observed to be white, and its intensity scaled quadratically with the average Pvd concentration in the colony:
(Fig. 2A), which is independent of the average concentration in the colony (SI Appendix, Fig. S4). Consistent with our model, these local fluctuations are uniform within the colony (SI Appendix, Fig. S5) and spatially uncorrelated (SI Appendix, Fig. S7). The noise spectrum was observed to be white, and its intensity scaled quadratically with the average Pvd concentration in the colony: 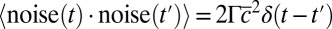 , with
, with 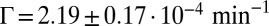 (Fig. 2B and SI Appendix, Figs. S8 and S9). With λ and
(Fig. 2B and SI Appendix, Figs. S8 and S9). With λ and 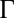 measured on data collected from 10 growing microcolonies of WT bacteria, we solved our model to predict the spatiotemporal dynamics of Pvd and its variability in packed microcolonies.
measured on data collected from 10 growing microcolonies of WT bacteria, we solved our model to predict the spatiotemporal dynamics of Pvd and its variability in packed microcolonies.
Fig. 2.
Measurement of the model parameters. (A) Average change in relative Pvd concentration in a given cell is plotted (points with error bars) vs. its average relative concentration (relative to its nearest neighbors). (Insets) Observed linear correlation is explained by a simple model: Bacteria exchange Pvd with their neighbors in proportion to the difference of their concentrations. The proportionality coefficient λ is estimated by a linear fit to the data (continuous line). Error bars represent the SEM. (B) Temporal autocorrelation function of the noise for various values of the mean Pvd concentration in the colony, 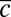 . This function can be approximated by
. This function can be approximated by 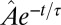 , which we fit for each Pvd level. Because τ is small, the noise autocorrelation function can be approximated by a δ-function:
, which we fit for each Pvd level. Because τ is small, the noise autocorrelation function can be approximated by a δ-function: 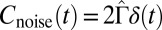 , with
, with 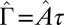 . (Inset)
. (Inset) 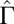 increases quadratically with the concentration:
increases quadratically with the concentration: 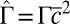 (dashed line: variation of
(dashed line: variation of 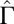 as predicted from the estimation of
as predicted from the estimation of 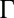 deduced from SI Appendix, Fig. S9).
deduced from SI Appendix, Fig. S9).
Predictions of the Model for WT Microcolonies.
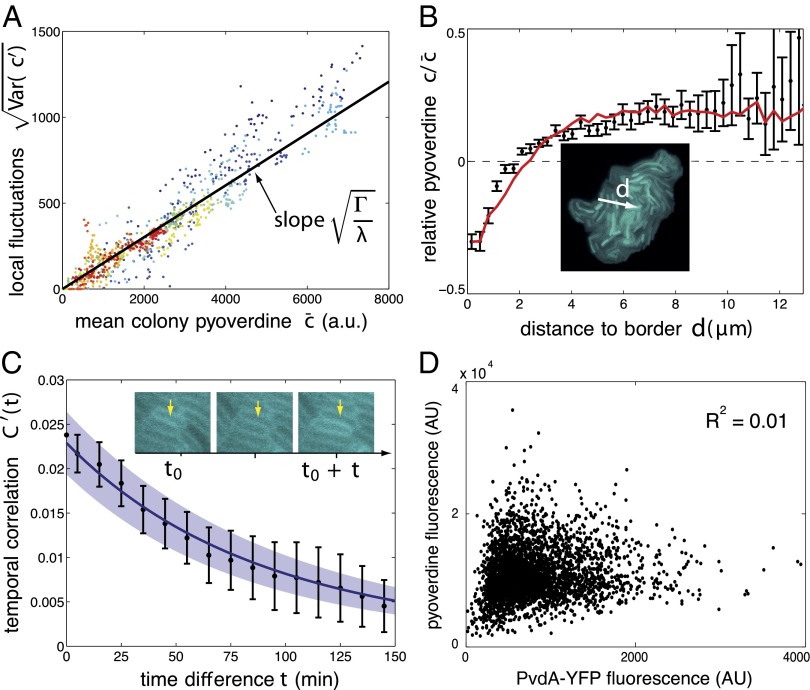
First, the experimental data confirmed the prediction of the model on the variability in the population. The variability is shown to scale with the average fluorescence in the colony, with a proportionality constant given by 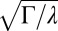 (Fig. 3A and SI Appendix, Fig. S3). This linear relation, which also holds in a well-mixed condition (SI Appendix, Fig. S17), is unusual. In most descriptions of gene regulation, noise stems from fluctuations in the small number of molecules (30). The magnitude of these fluctuations is expected to increase not linearly but as the square root of the mean number of molecules. A linear scaling similar to ours has been reported for proteins produced in high copy numbers (31, 32), however, and has been attributed to sources of extrinsic noise. In the Pvd system, this high level of noise may be due to fluctuations in the number of efflux pumps and transporters.
(Fig. 3A and SI Appendix, Fig. S3). This linear relation, which also holds in a well-mixed condition (SI Appendix, Fig. S17), is unusual. In most descriptions of gene regulation, noise stems from fluctuations in the small number of molecules (30). The magnitude of these fluctuations is expected to increase not linearly but as the square root of the mean number of molecules. A linear scaling similar to ours has been reported for proteins produced in high copy numbers (31, 32), however, and has been attributed to sources of extrinsic noise. In the Pvd system, this high level of noise may be due to fluctuations in the number of efflux pumps and transporters.
Fig. 3.
Model predictions for the WT strain PAO1. (A) SD of local fluctuations in Pvd is proportional to the mean fluorescence in the colony, as predicted by the model, with coefficient 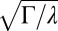 (black line). Colors code for different colonies
(black line). Colors code for different colonies 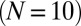 . Each point is calculated from the distribution of fluorescence in a colony measured at a given time. (B) Pvd gradient is established from the edge to the center of the colony. The black points are obtained by pooling the data from 10 microcolonies in discrete bins according to their distances to the edge (white arrow). The red curve is the model prediction. Error bars represent the SEM. (C) Comparison between the measured (black) and the predicted (blue) temporal autocorrelation functions. The shaded areas show the uncertainty in parameter estimation. Error bars represent SEM (N = 10). The yellow arrow points to the same cell at different time points. Note that in A–C, the predicted curves are not the result of a fit. (D) Level of Pvd in individual bacteria is only weakly related to its production rate assessed by the reporter strain PvdA-YFP (AU).
. Each point is calculated from the distribution of fluorescence in a colony measured at a given time. (B) Pvd gradient is established from the edge to the center of the colony. The black points are obtained by pooling the data from 10 microcolonies in discrete bins according to their distances to the edge (white arrow). The red curve is the model prediction. Error bars represent the SEM. (C) Comparison between the measured (black) and the predicted (blue) temporal autocorrelation functions. The shaded areas show the uncertainty in parameter estimation. Error bars represent SEM (N = 10). The yellow arrow points to the same cell at different time points. Note that in A–C, the predicted curves are not the result of a fit. (D) Level of Pvd in individual bacteria is only weakly related to its production rate assessed by the reporter strain PvdA-YFP (AU).
Second, our model of local exchange explains the gradient of Pvd concentration observed away from the center of the colony (Fig. 3B). Cells on the edge of the colony have fewer neighbors to import from, and thus have lower Pvd levels. In turn, these cells contribute less Pvd to their direct neighbors. This effect is propagated toward the center, resulting in a gradient of Pvd across the colony. This gradient was calculated by solving Eq. 1 over the whole population (SI Appendix) and is in close agreement with the observed experimental gradient (Fig. 3B).
Third, adding the calculated spatial heterogeneities to the local fluctuations, we have been able to explain and predict the shape of the normalized Pvd distribution (Fig. 1D, Inset). In this framework, the variability is governed mainly by local exchanges rather than by fluctuations in production activity. Consistent with this interpretation, we found that the variability in production rate accounts for only 1% of the observed variance of the concentration of Pvd in cells expressing a PvdA fusion, the gene coding for the first enzyme involved in Pvd synthesis (Fig. 3D).
Finally, using Eq. 1, we computed the temporal correlations of spatial fluctuations (SI Appendix). This correlation decays exponentially with a characteristic time, 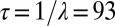 min, in agreement with our measurements (Fig. 3C).
min, in agreement with our measurements (Fig. 3C).
Predictions of the Model in Other Contexts.
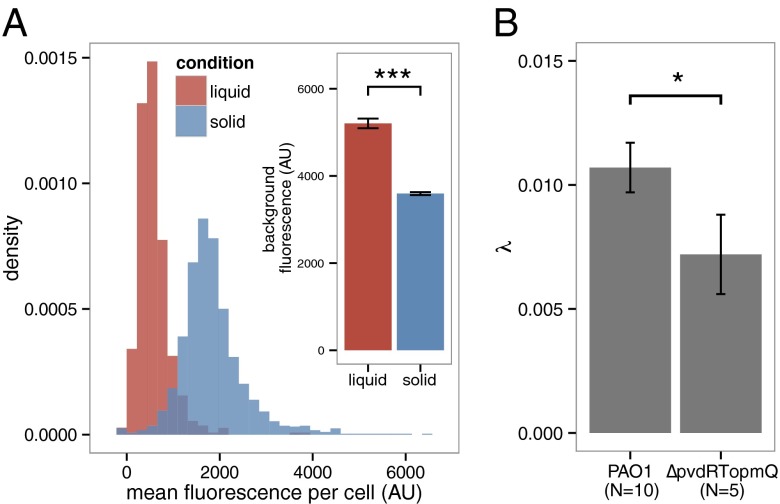
An important consequence of these local exchanges is that Pvd should be lost in the environment if bacteria do not interact physically by cell–cell contact. Accordingly, we observed that when bacteria are grown in stirred liquid conditions, Pvd is mainly found in the environment and the concentration in isolated bacteria is much lower than in packed bacteria (Fig. 4A and SI Appendix, Fig. S10). In contrast to passive diffusion, which leads to the fast release of Pvd in the extracellular environment in stirred conditions, the local exchanges between cells on solid substrate actively slow down the leak of Pvd out of the colony. We further challenged the predictions of our model by examining the effects of mutations in the Pvd pathway (SI Appendix, Fig. S1). In 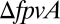 microcolonies, bacteria are not able to import Pvd from their neighbors and local trafficking of Pvd is prevented. In this strain, microcolonies cannot accumulate Pvd and no gradient is formed (SI Appendix, Movie S3). For a strain partially deficient in Pvd export, the model predicts that local fluctuations should be damped with a characteristic time longer than for a WT strain (SI Appendix). Consistent with this prediction, we measured a lower λ in the export-deficient
microcolonies, bacteria are not able to import Pvd from their neighbors and local trafficking of Pvd is prevented. In this strain, microcolonies cannot accumulate Pvd and no gradient is formed (SI Appendix, Movie S3). For a strain partially deficient in Pvd export, the model predicts that local fluctuations should be damped with a characteristic time longer than for a WT strain (SI Appendix). Consistent with this prediction, we measured a lower λ in the export-deficient 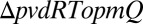 strain than in WT (Fig. 4B).
strain than in WT (Fig. 4B).
Fig. 4.
Model predictions for a planktonic culture and for a mutant. (A) Model prediction for the internal Pvd concentration for well-mixed planktonic cultures vs. 2D growth on an agar plate. The distribution of Pvd in cells grown in liquid and solid conditions shows that cells from liquid cultures retain less Pvd in their periplasm, whereas the concentration in the environment is higher (Inset). This suggests higher leakage when cells are not in contact. Error bars represent the SEM (n = 10). (B) As expected by the model, a partial mutant of Pvd export (ΔpvdRTopmQ) exhibits a smaller Pvd exchange rate λ (evaluated as in Fig. 2A) than for the WT strain (PAO1). Error bars represent the SEM, and the number is indicated for each condition on the bar plot. The two samples are different with a P value of 0.04. The number of asterisks indicates the significance level (0.01<*<0.05; ***<0.001).
Local Exchange Affects Individual Fitness.
The dynamics described above imply that iron uptake is mediated by local trafficking of Pvd when bacteria grow in packed colonies. To test whether this mechanism could enhance individual fitness, we measured the dependence of the individual growth rate on the history, 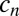 , of Pvd concentration in the neighboring cells (SI Appendix) through linear fits (SI Appendix, Fig. S11). On the addition of a competing iron chelator [human transferrin (Tsf)], growth became limited by iron. Individual growth rates then showed a positive dependence on the history of Pvd concentration in neighboring cells (Fig. 5A) and a moderate negative dependence on both the distance to the edge of the colony and the internal concentration (SI Appendix, Fig. S11). No dependence was observed in absence of Tsf (SI Appendix, Fig. S12). Interestingly, we observed that in iron-depleted conditions [succinate minimal medium (SMM) + Tsf], WT cells formed double layers at significantly earlier stages of microcolony development (SI Appendix, Fig. S12) than in control conditions (SMM), extending the exchange mechanism to the vertical dimension at the onset of biofilm formation, and thus limiting Pvd leaks from the colony.
, of Pvd concentration in the neighboring cells (SI Appendix) through linear fits (SI Appendix, Fig. S11). On the addition of a competing iron chelator [human transferrin (Tsf)], growth became limited by iron. Individual growth rates then showed a positive dependence on the history of Pvd concentration in neighboring cells (Fig. 5A) and a moderate negative dependence on both the distance to the edge of the colony and the internal concentration (SI Appendix, Fig. S11). No dependence was observed in absence of Tsf (SI Appendix, Fig. S12). Interestingly, we observed that in iron-depleted conditions [succinate minimal medium (SMM) + Tsf], WT cells formed double layers at significantly earlier stages of microcolony development (SI Appendix, Fig. S12) than in control conditions (SMM), extending the exchange mechanism to the vertical dimension at the onset of biofilm formation, and thus limiting Pvd leaks from the colony.
Fig. 5.
Local trafficking affects the individual fitness and stabilizes cooperation. (A) Mean relative growth rate of cells (black points) is plotted against the distance to the colony edge (normalized by the mean radius of the colony 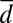 ) and the recent history of Pvd concentration in the cell’s neighborhood
) and the recent history of Pvd concentration in the cell’s neighborhood 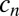 (normalized by
(normalized by 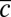 ). The colored planes are bivariate linear fits to the data. No significant dependence is found when the level of iron is low (SMM) (SI Appendix, Fig. S11). By contrast, when iron is depleted (SMM + Tsf), the individual growth rates depend on both the position inside the colony and the Pvd history in the nearest neighbors. (B) Phase diagram of the sustainability of cooperation in an in silico competition experiment between defectors and cooperators. The result of the simulation after 4,000 generations is shown as a function of the local exchange rate of Pvd λ and the cost of production. Cooperators are found to dominate in a wide range of values around the measured exchange rate
). The colored planes are bivariate linear fits to the data. No significant dependence is found when the level of iron is low (SMM) (SI Appendix, Fig. S11). By contrast, when iron is depleted (SMM + Tsf), the individual growth rates depend on both the position inside the colony and the Pvd history in the nearest neighbors. (B) Phase diagram of the sustainability of cooperation in an in silico competition experiment between defectors and cooperators. The result of the simulation after 4,000 generations is shown as a function of the local exchange rate of Pvd λ and the cost of production. Cooperators are found to dominate in a wide range of values around the measured exchange rate 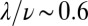 . The white line depicts the initial ratio. The black line depicts the measured value of λ, and the dashed lines depict the confidence interval. The time evolution of the proportion of nonproducers at the points (I, II and III) marked in B is shown in SI Appendix, Fig. S14.
. The white line depicts the initial ratio. The black line depicts the measured value of λ, and the dashed lines depict the confidence interval. The time evolution of the proportion of nonproducers at the points (I, II and III) marked in B is shown in SI Appendix, Fig. S14.
In Silico Competition of Producers vs. Nonproducers.
Spatially structured environments are known to promote cooperation (8–10) and to maintain phenotypic diversity (33–35). To investigate the stability of producers in the context of mixed, packed populations on evolutionary time scales, we implemented numerical simulations using our model for Pvd dynamics. Producers and nonproducers were allowed to compete on a square lattice with a constant population size (SI Appendix and SI Appendix, Fig. S13). We varied the range of effective diffusion through exchanges and the cost of production (Fig. 5B and SI Appendix, Movie S4). When the rate of exchange, 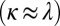 , is faster than the growth rate, v, producers always go extinct (SI Appendix, Fig. S14). In contrast, cooperation was found to be dominant if
, is faster than the growth rate, v, producers always go extinct (SI Appendix, Fig. S14). In contrast, cooperation was found to be dominant if 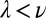 . In the observed range of experimental costs (5, 6) (SI Appendix) and at the value of λ we measured, the population of producers was found to be stable. Hence, the effective slowing down of diffusion by active exchange between contacting cells limits the dispersal of the public goods to the producers and their kin, minimizing exchanges with nonproducer mutants.
. In the observed range of experimental costs (5, 6) (SI Appendix) and at the value of λ we measured, the population of producers was found to be stable. Hence, the effective slowing down of diffusion by active exchange between contacting cells limits the dispersal of the public goods to the producers and their kin, minimizing exchanges with nonproducer mutants.
Discussion
In aquatic ecosystems, Pvd diffuses freely and is homogenized well by the agitation of the medium, such that the secreted molecules are shared by all. However, P. aeruginosa is also known to colonize many solid habitats. In rhizospheres, plants, and human lungs, colonies mature into biofilm in which Pvd diffusion is expected to be dominated by local interactions rather than by free diffusion. Our data show that competition for Pvd is governed by local exchange between close neighbors, which actively confines its diffusion within the microcolony. It challenges the general assumption that secreted molecules diffuse freely when bacteria are packed. In particular, if the bacteria exchange this public good at a rate that is slower than their growth rate, Pvd exchange benefits mostly the clonal patches of producers, ensuring the maintenance of cooperation. From an evolutionary perspective, our results emphasize how the spatial structure of the colony might constrain its evolution. On solid substrate, offspring remain physically close to each other after division, causing cells to interact more with cells of their own kin than with other cells. When exchanges are local, nonproducer cells may only benefit from the public good if they are in contact with producers, which only occurs at the boundary between patches of producing and nonproducing cells.
Although small molecules  diffuse at high rates in water, it is possible for microscale chemical gradients to form within biofilms. At the microscopic scale, our data provide a complete description of how local exchange can generate gradients when bacteria are physically in contact with each other. At the cellular level, differential responses to these local environments [e.g., as gradients of gene expression (36) or position-dependent cell specialization (37)] may increase phenotypic variability and promote bacterial adaptation during infection (38). The gradients formed inside packed colonies may also be used by bacteria as a cue for positioning during biofilm development (39). In the case of P. aeruginosa, Pvd has been identified as a virulence factor in the development of infectious biofilms (28, 40–42) and confocal imaging has revealed that the Pvd pathway is not activated uniformly within biofilms (43, 44). Thus, elucidating the formation of these gradients at early stages of microcolony development may help us to understand better the mechanisms responsible for physiological heterogeneities in biofilms and their role in pathogenicity.
diffuse at high rates in water, it is possible for microscale chemical gradients to form within biofilms. At the microscopic scale, our data provide a complete description of how local exchange can generate gradients when bacteria are physically in contact with each other. At the cellular level, differential responses to these local environments [e.g., as gradients of gene expression (36) or position-dependent cell specialization (37)] may increase phenotypic variability and promote bacterial adaptation during infection (38). The gradients formed inside packed colonies may also be used by bacteria as a cue for positioning during biofilm development (39). In the case of P. aeruginosa, Pvd has been identified as a virulence factor in the development of infectious biofilms (28, 40–42) and confocal imaging has revealed that the Pvd pathway is not activated uniformly within biofilms (43, 44). Thus, elucidating the formation of these gradients at early stages of microcolony development may help us to understand better the mechanisms responsible for physiological heterogeneities in biofilms and their role in pathogenicity.
Contact-dependent interactions between bacteria are known to operate via tube-like secretion systems that bridge the periplasms of two neighboring bacteria (45–47). In contrast, Pvd must diffuse in the environment to chelate iron. Like Pvd, diffusible molecules have been shown to function over short distances (48). Restricted diffusion by cell compaction enhances access to public goods, which has been shown to provide a growth advantage (18, 49). However, the molecular mechanisms responsible for their confinement remain to be elucidated. Several hypotheses can be considered. If the secreted molecules have an affinity for the LPSs or other components of the bacterial cell wall, they would preferentially diffuse in the vicinity of the bacteria. Similarly, if a polymeric network fills the space between bacteria in packed colonies, the diffusible molecules could be jammed inside its mesh.
Beyond the social aspects of public goods diffusion in packed colonies, the local exchange between adjacent bacteria is reminiscent of paracrine signaling in eukaryotic tissues. Altogether, these aspects emphasize the growing recognition that bacterial colonies might share many more similarities with multicellular organisms than previously assumed.
Materials and Methods
Culture Conditions.
Strains are described in SI Appendix, Table S1. They were inoculated in LB from glycerol stocks and grown overnight at 37 °C. The next day, the culture was washed and resuspended at a 100-fold dilution in fresh SMM for an overnight growth at 28 °C. SMM is a medium with a low iron concentration that activates Pvd synthesis (composition: 6 g⋅L−1 K2HPO4, 3 g⋅L−1 KH2PO4, 1 g⋅L−1 [NH4]2SO4, 0.2 g⋅L−1 MgSO4 7H2O, and 4 g⋅L−1 sodium succinate, with the pH adjusted to 7.0 by adding NaOH). On the third day, bacteria were diluted 104-fold and plated on a gel pad (1% agarose in SMM). The preparation was sealed with double-sided tape (Gene Frame; Fisher Scientific) to a glass coverslip. A duct was cut at the center of the pad to let oxygen diffuse into the gel. Microcolonies grew on the pad at 30 °C using a custom-made temperature controller.
To create iron-depleted conditions, 5 μM of human Tsf (Sigma) supplemented with 20 mM NaHCO3 was added to SMM and agarose gel. At this concentration and after 24 h of growth, the total yield of a ΔpvdA strain was reduced by a factor of 10 (SI Appendix, Fig. S15).
Image Acquisition and Analysis.
The dynamics of Pvd concentration in each cell of a microcolony were monitored by time-lapse microscopy and epifluorescence. Colonies were observed with bright-field illumination using a magnification of 100× and 1.35-N.A. objective (Apo-ph1; Olympus) on an automated inverted microscope (IX81; Olympus) equipped with an Orca-R2 CCD camera (Hamamatsu). The microscope, the camera, and the stage were actuated with a LabView interface (National Instruments). Correlation images were obtained by acquiring z-stacks of bright-field images. For each pixel, we computed the correlation of the intensity profile along the vertical direction with a reference kernel (SI Appendix, Fig. S2). Fluorescence excitation was achieved with a mercury vapor light source (EXFO X-Cite 120Q). Pvd was imaged with a 390(40)/475(64)-nm filter set using a dichroic beam-splitter at 405 nm (Semrock). Cell segmentation was performed on correlation images, and cell lineage was computed using a MATLAB (MathWorks) code developed in the laboratory of Michael Elowitz (California Institute of Technology) (51, 52). To compute the concentration of Pvd c in a cell, we subtracted the average fluorescence per pixel of the background from the average intensity per pixel in the given cell. In this study, an average intensity of 1,000 arbitrary units for a cell corresponds to ∼500 molecules of Pvd.
Significance of the Dependence of 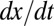 on
on 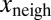 .
.
To test whether 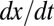 also depended on
also depended on 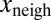 and not only on x (Fig. 1E), we performed a multivariate fit of
and not only on x (Fig. 1E), we performed a multivariate fit of 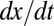 with respect to x and
with respect to x and 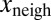 . The linear coefficient of
. The linear coefficient of 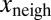 was found to be positive. We tested the significance of this positive correlation using a permutation test. We divided the whole dataset into 400 bins according to the value of x. Inside each of these bins, the values of
was found to be positive. We tested the significance of this positive correlation using a permutation test. We divided the whole dataset into 400 bins according to the value of x. Inside each of these bins, the values of 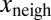 were randomly permuted, and the same multivariate regression was performed on the permuted data points. Repeated 1 million times, this procedure never yielded a linear coefficient for xneigh as high as the one observed in the dataset that had not undergone any permutations, thus implying a P-value <10−6.
were randomly permuted, and the same multivariate regression was performed on the permuted data points. Repeated 1 million times, this procedure never yielded a linear coefficient for xneigh as high as the one observed in the dataset that had not undergone any permutations, thus implying a P-value <10−6.
Percentage of the Variability Explained by Production.
To assess the contribution of production in the variability of siderophore concentration, we measured the level of Pvd in single cells of a colony grown on agar pads and the level of PvdA (using the fusion reporter strain pvdA-YFP).We estimated the percentage of the total unexplained variability by subtracting the linear contribution of production measured by fitting Pvd against PvdA-YFP and computing the remaining variability (Fig. 3D). The fraction of unexplained variability (1 − R2 = 99%) is the ratio of this remaining variability to the total variability. We also measured the correlation between the level of Pvd and the level of production in liquid conditions to avoid the variability due to cell–cell contacts (SI Appendix, Fig. S16).
Exchange Model.
The exchange model described by Eq. 1 can be rationalized in two ways. First, as shown in Fig. 1E, the time derivative of Pvd concentration correlates positively with the concentration in the cell’s immediate neighborhood and negatively with the cell’s own concentration. Eq. 1 is the simplest model one can write that accounts for this observation.
Alternatively, one can write a model accounting for all the events that may affect Pvd concentration and expand it to the lowest (linear) order. The evolution of Pvd concentration in a single cell is governed by production (21), export (23, 27), import (53), dilution from cell division and possible degradation, and positive feedback (19, 20, 28):
where 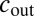 is the concentration outside the cell; α is the production rate;
is the concentration outside the cell; α is the production rate; 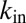 and
and 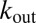 are the import and export constants, respectively; v accounts for dilution due to growth rate (and possible degradation); and f accounts for positive feedback depending on Pvd uptake [consistent with the observation of positive feedback loops involving the import channel (28)]. Because cells are tightly packed, it is not possible to estimate
are the import and export constants, respectively; v accounts for dilution due to growth rate (and possible degradation); and f accounts for positive feedback depending on Pvd uptake [consistent with the observation of positive feedback loops involving the import channel (28)]. Because cells are tightly packed, it is not possible to estimate 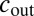 directly from microscopy images. Instead, we assume it to be approximated by a linear combination of the concentration inside the cell c and the average concentration in its nearest neighbors (defined as a corona of one cell’s width around the cell):
directly from microscopy images. Instead, we assume it to be approximated by a linear combination of the concentration inside the cell c and the average concentration in its nearest neighbors (defined as a corona of one cell’s width around the cell): 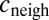 :
: 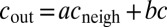 . Regrouping terms, we obtain Eq. 1, with
. Regrouping terms, we obtain Eq. 1, with 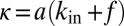 and
and 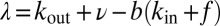 .
.
Comparison Between Liquid and Solid Cultures.
To assess the importance of cell–cell contact, cells were inoculated at the same bacterial density in parallel on agar pads and in liquid. They were then allowed to grow for 9 h. Bacteria from the liquid culture were plated directly without dilution 5 min before fluorescence measurements (SI Appendix, Fig. S11).
Simulation of Spatial Competition for Mixed Population.
We performed simulations of bacterial evolution on a 30 × 30 square lattice with periodic boundary conditions. Each point was occupied by a bacterium, which was either a producer or a nonproducer. At each point, we simulated the evolution of Pvd, external iron, and internal iron using Euler’s integration method with 10 time steps per generation. In cells, Pvd evolution is given by Eq. 1, where 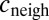 was taken as the average in the four nearest neighbors.
was taken as the average in the four nearest neighbors.
Supplementary Material
Acknowledgments
We thank Michael Elowitz, Rob Phillips, Minus Van Baalen, Gordon Hamilton, Clement Nizak, Olivier Rivoire, Giuseppe Lia, Jean-Baptiste Boulé, and Julien Mozziconacci for their comments on the manuscript. We thank Michael Elowitz for providing us with Schnitzcell, which was used to retrieve the lineage of the microcolonies. We also thank Antoine Decrule, Marie-Cécilia Duvernoy, and Maxime Ardré for their contributions to this work. This work was supported by the Centre National de la Recherche Scientifique Grant “Prise de Risque” (to N.D.), and ANR Grants ANR-08-BLAN-0309-02 (to I.J.S.) and ANR-2011-JSV5-005-01 (to N.D).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301428110/-/DCSupplemental.
References
- 1.Hider RC, Kong X. Chemistry and biology of siderophores. Nat Prod Rep. 2010;27(5):637–657. doi: 10.1039/b906679a. [DOI] [PubMed] [Google Scholar]
- 2.Buckling A, et al. Siderophore-mediated cooperation and virulence in Pseudomonas aeruginosa. FEMS Microbiol Ecol. 2007;62(2):135–141. doi: 10.1111/j.1574-6941.2007.00388.x. [DOI] [PubMed] [Google Scholar]
- 3.West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4(8):597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 4.West SA, Griffin AS, Gardner A. Evolutionary explanations for cooperation. Curr Biol. 2007;17(16):R661–R672. doi: 10.1016/j.cub.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Griffin AS, West SA, Buckling A. Cooperation and competition in pathogenic bacteria. Nature. 2004;430(7003):1024–1027. doi: 10.1038/nature02744. [DOI] [PubMed] [Google Scholar]
- 6.Ross-Gillespie A, Gardner A, West SA, Griffin AS. Frequency dependence and cooperation: Theory and a test with bacteria. Am Nat. 2007;170(3):331–342. doi: 10.1086/519860. [DOI] [PubMed] [Google Scholar]
- 7.Kümmerli R, Gardner A, West SA, Griffin AS. Limited dispersal, budding dispersal, and cooperation: An experimental study. Evolution. 2009;63(4):939–949. doi: 10.1111/j.1558-5646.2008.00548.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhang XX, Rainey PB. Exploring the sociobiology of pyoverdin-producing Pseudomonas. Evolution. 2013 doi: 10.1111/evo.12183. [DOI] [PubMed] [Google Scholar]
- 9.Nowak MA, May RM. Evolutionary games and spatial chaos. Nature. 1992;359:826–829. [Google Scholar]
- 10.Doebeli M, Knowlton N. The evolution of interspecific mutualisms. Proc Natl Acad Sci USA. 1998;95(15):8676–8680. doi: 10.1073/pnas.95.15.8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hauert C, Doebeli M. Spatial structure often inhibits the evolution of cooperation in the snowdrift game. Nature. 2004;428(6983):643–646. doi: 10.1038/nature02360. [DOI] [PubMed] [Google Scholar]
- 12.Wakano JY, Nowak MA, Hauert C. Spatial dynamics of ecological public goods. Proc Natl Acad Sci USA. 2009;106(19):7910–7914. doi: 10.1073/pnas.0812644106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xavier JB, Foster KR. Cooperation and conflict in microbial biofilms. Proc Natl Acad Sci USA. 2007;104(3):876–881. doi: 10.1073/pnas.0607651104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xavier JB. Social interaction in synthetic and natural microbial communities. Mol Syst Biol. 2011;7:483. doi: 10.1038/msb.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitri S, Xavier JB, Foster KR. Social evolution in multispecies biofilms. Proc Natl Acad Sci USA. 2011;108(Suppl 2):10839–10846. doi: 10.1073/pnas.1100292108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kümmerli R, Griffin AS, West SA, Buckling A, Harrison F. Viscous medium promotes cooperation in the pathogenic bacterium Pseudomonas aeruginosa. Proc Biol Sci. 2009;276(1672):3531–3538. doi: 10.1098/rspb.2009.0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross-Gillespie A, Gardner A, Buckling A, West SA, Griffin AS. Density dependence and cooperation: Theory and a test with bacteria. Evolution. 2009;63(9):2315–2325. doi: 10.1111/j.1558-5646.2009.00723.x. [DOI] [PubMed] [Google Scholar]
- 18.Koschwanez JK, Foster KR, Murray AW. Sucrose utilization in budding yeast as a model for the origin of undifferentiated multicellularity. PLoS Biol. 2011;9(8):e1001122. doi: 10.1371/journal.pbio.1001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schalk IJ. Metal trafficking via siderophores in gram-negative bacteria: Specificities and characteristics of the pyoverdine pathway. J Inorg Biochem. 2008;102(5-6):1159–1169. doi: 10.1016/j.jinorgbio.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 20.Visca P, Imperi F, Lamont IL. Pyoverdine siderophores: From biogenesis to biosignificance. Trends Microbiol. 2007;15(1):22–30. doi: 10.1016/j.tim.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Yeterian E, et al. Synthesis of the siderophore pyoverdine in Pseudomonas aeruginosa involves a periplasmic maturation. Amino Acids. 2010;38(5):1447–1459. doi: 10.1007/s00726-009-0358-0. [DOI] [PubMed] [Google Scholar]
- 22.Ravel J, Cornelis P. Genomics of pyoverdine-mediated iron uptake in pseudomonads. Trends Microbiol. 2003;11(5):195–200. doi: 10.1016/s0966-842x(03)00076-3. [DOI] [PubMed] [Google Scholar]
- 23.Hannauer M, Yeterian E, Martin LW, Lamont IL, Schalk IJ. An efflux pump is involved in secretion of newly synthesized siderophore by Pseudomonas aeruginosa. FEBS Lett. 2010;584(23):4751–4755. doi: 10.1016/j.febslet.2010.10.051. [DOI] [PubMed] [Google Scholar]
- 24.Schalk IJ, Lamont IL, Cobessi D. Structure-function relationships in the bifunctional ferrisiderophore FpvA receptor from pseudomonas aeruginosa. Biometals. 2009;22(4):671–678. doi: 10.1007/s10534-008-9203-2. [DOI] [PubMed] [Google Scholar]
- 25.Greenwald J, et al. Real time fluorescent resonance energy transfer visualization of ferric pyoverdine uptake in Pseudomonas aeruginosa. A role for ferrous iron. J Biol Chem. 2007;282(5):2987–2995. doi: 10.1074/jbc.M609238200. [DOI] [PubMed] [Google Scholar]
- 26.Yeterian E, Martin LW, Lamont IL, Schalk IJ. An efflux pump is required for siderophore recycling by Pseudomonas aeruginosa. Environ Microbiol Reps. 2009;2(3):412–418. doi: 10.1111/j.1758-2229.2009.00115.x. [DOI] [PubMed] [Google Scholar]
- 27.Imperi F, Tiburzi F, Visca P. Molecular basis of pyoverdine siderophore recycling in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2009;106(48):20440–20445. doi: 10.1073/pnas.0908760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamont IL, Beare PA, Ochsner U, Vasil AI, Vasil ML. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2002;99(10):7072–7077. doi: 10.1073/pnas.092016999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Visca P, Leoni L, Wilson MJ, Lamont IL. Iron transport and regulation, cell signalling and genomics: Lessons from Escherichia coli and pseudomonas. Mol Microbiol. 2002;45(5):1177–1190. doi: 10.1046/j.1365-2958.2002.03088.x. [DOI] [PubMed] [Google Scholar]
- 30.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 31.Taniguchi Y, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329(5991):533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salman H, et al. Universal protein fluctuations in populations of microorganisms. Phys Rev Lett. 2012;108(23):238105. doi: 10.1103/PhysRevLett.108.238105. [DOI] [PubMed] [Google Scholar]
- 33.Kerr B, Riley MA, Feldman MW, Bohannan BJM. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418(6894):171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- 34.Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci USA. 2008;105(47):18188–18193. doi: 10.1073/pnas.0807935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ackermann M, et al. Self-destructive cooperation mediated by phenotypic noise. Nature. 2008;454(7207):987–990. doi: 10.1038/nature07067. [DOI] [PubMed] [Google Scholar]
- 36.Shank EA, Kolter R. Extracellular signaling and multicellularity in Bacillus subtilis. Curr Opin Microbiol. 2011;14(6):741–747. doi: 10.1016/j.mib.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flores E, Herrero A. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat Rev Microbiol. 2010;8(1):39–50. doi: 10.1038/nrmicro2242. [DOI] [PubMed] [Google Scholar]
- 38.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6(3):199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 39.Shank EA, et al. Interspecies interactions that result in Bacillus subtilis forming biofilms are mediated mainly by members of its own genus. Proc Natl Acad Sci USA. 2011;108(48):E1236–E1243. doi: 10.1073/pnas.1103630108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischbach MA, Lin H, Liu DR, Walsh CT. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat Chem Biol. 2006;2(3):132–138. doi: 10.1038/nchembio771. [DOI] [PubMed] [Google Scholar]
- 41.Schaible UE, Kaufmann SHE. Iron and microbial infection. Nat Rev Microbiol. 2004;2(12):946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 42.George AM, Jones PM, Middleton PG. Cystic fibrosis infections: Treatment strategies and prospects. FEMS Microbiol Lett. 2009;300(2):153–164. doi: 10.1111/j.1574-6968.2009.01704.x. [DOI] [PubMed] [Google Scholar]
- 43.Banin E, Vasil ML, Greenberg EP. Iron and Pseudomonas aeruginosa biofilm formation. Proc Natl Acad Sci USA. 2005;102(31):11076–11081. doi: 10.1073/pnas.0504266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang L, Nilsson M, Gjermansen M, Givskov M, Tolker-Nielsen T. Pyoverdine and PQS mediated subpopulation interactions involved in Pseudomonas aeruginosa biofilm formation. Mol Microbiol. 2009;74(6):1380–1392. doi: 10.1111/j.1365-2958.2009.06934.x. [DOI] [PubMed] [Google Scholar]
- 45.Aoki SK, et al. Contact-dependent inhibition of growth in Escherichia coli. Science. 2005;309(5738):1245–1248. doi: 10.1126/science.1115109. [DOI] [PubMed] [Google Scholar]
- 46.LeRoux M, et al. Quantitative single-cell characterization of bacterial interactions reveals type VI secretion is a double-edged sword. Proc Natl Acad Sci USA. 2012;109(48):19804–19809. doi: 10.1073/pnas.1213963109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dubey GP, Ben-Yehuda S. Intercellular nanotubes mediate bacterial communication. Cell. 2011;144(4):590–600. doi: 10.1016/j.cell.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 48.Egland PG, Palmer RJ, Kolenbrander PE. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: Signaling in flow conditions requires juxtaposition. Proc Natl Acad Sci USA. 2004;101(48):16917–16922. doi: 10.1073/pnas.0407457101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gore J, Youk H, van Oudenaarden A. Snowdrift game dynamics and facultative cheating in yeast. Nature. 2009;459(7244):253–256. doi: 10.1038/nature07921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meyer JM, Abdallah MA. The fluorescent pigment of pseudomonas fluorescens: Biosynthesis, purification and physicochemical properties. J Gen Microbiol. 1978;107:319–328. [Google Scholar]
- 51.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307(5717):1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 52.Locke JCW, Elowitz MB. Using movies to analyse gene circuit dynamics in single cells. Nat Rev Microbiol. 2009;7(5):383–392. doi: 10.1038/nrmicro2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schalk IJ, Abdallah MA, Pattus F. Recycling of pyoverdin on the FpvA receptor after ferric pyoverdin uptake and dissociation in Pseudomonas aeruginosa. Biochemistry. 2002;41(5):1663–1671. doi: 10.1021/bi0157767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.