Abstract
Homogentisate 1,2-dioxygenase (HGDO) uses a mononuclear nonheme Fe2+ to catalyze the oxidative ring cleavage in the degradation of Tyr and Phe by producing maleylacetoacetate from homogentisate (2,5-dihydroxyphenylacetate). Here, we report three crystal structures of HGDO, revealing five different steps in its reaction cycle at 1.7–1.98 Å resolution. The resting state structure displays an octahedral coordination for Fe2+ with two histidine residues (His331 and His367), a bidentate carboxylate ligand (Glu337), and two water molecules. Homogentisate binds as a monodentate ligand to Fe2+, and its interaction with Tyr346 invokes the folding of a loop over the active site, effectively shielding it from solvent. Binding of homogentisate is driven by enthalpy and is entropically disfavored as shown by anoxic isothermal titration calorimetry. Three different reaction cycle intermediates have been trapped in different HGDO subunits of a single crystal showing the influence of crystal packing interactions on the course of enzymatic reactions. The observed superoxo:semiquinone-, alkylperoxo-, and product-bound intermediates have been resolved in a crystal grown anoxically with homogentisate, which was subsequently incubated with dioxygen. We demonstrate that, despite different folds, active site architectures, and Fe2+ coordination, extradiol dioxygenases can proceed through the same principal reaction intermediates to catalyze the O2-dependent cleavage of aromatic rings. Thus, convergent evolution of nonhomologous enzymes using the 2-His-1-carboxylate facial triad motif developed different solutions to stabilize closely related intermediates in unlike environments.
Keywords: dioxygen activation, non-heme iron, amino acid degradation, Pseudomonas putida, alkaptonuria
Homogentisate (2,5-dihydroxyphenylacetate, HG) is the central metabolite in the degradation pathways of phenylalanine and tyrosine in aerobic organisms ranging from soil bacteria like Pseudomonas putida to man (1, 2). Oxidative ring cleavage is catalyzed by homogentisate 1,2-dioxygenase (HGDO), which incorporates the atoms of molecular oxygen into HG to produce maleylacetoactate (1, 3–6). A deficiency of HGDO is known to cause the autosomal recessive disorder alkaptonuria in humans, the first genetic defect to be recognized as such (6–8). The crystal structure of HGDO from man (HGDOHs) revealed that the enzyme belongs to the cupin fold type of nonheme iron-dependent dioxygenases, forming a homohexamer consisting of a dimer of trimers (6). The active site of HGDOHs employs the 2-His-1-carboxylate facial triad to bind an essential Fe2+ ion with a distorted square-pyramidal arrangement (6). The facial triad motif is found in various nonhomologous types of nonheme Fe2+-dependent enzymes and commonly serves to control the reactivity of the Fe2+ site and to activate an aromatic or aliphatic substrate together with dioxygen (9) and is also featured in extradiol-type dioxygenases such as 2,3-dihydroxybiphenyl 1,2-dioxygenase (BphC) and homoprotocatechuate 2,3-dioxygenase (HPCD) (9, 10). Structures of BphC and HPCD in complex with their substrates show that the ortho hydroxyl groups of these catecholic substrates bind as an asymmetric bidentate ligand of Fe2+ preparing the metal to activate O2 (11–13). HGDO may be grouped into a class of extradiol dioxygenases containing the cupin fold and differs from BphC and HPCD by cleaving an aromatic ring with hydroxyl groups in para position, implying a way of Fe2+-substrate interaction distinct from the interaction of HPCD and BphC with their Fe2+-chelating substrates.
Despite the long history of research on HGDO, we are still lacking an understanding of how the enzyme achieves substrate activation and its O2-dependent cleavage. In crystallo turnover of a substrate has successfully been used in the past to elucidate the structure of reaction intermediates of nonheme iron-containing enzymes (12, 14–16) and has also been exploited in this study. Here, we present five different states along the reaction pathway of HGDO revealing closely related reaction mechanisms among nonhomologous extradiol-type dioxygenases.
Results and Discussion
Characterization of HGDOPp.
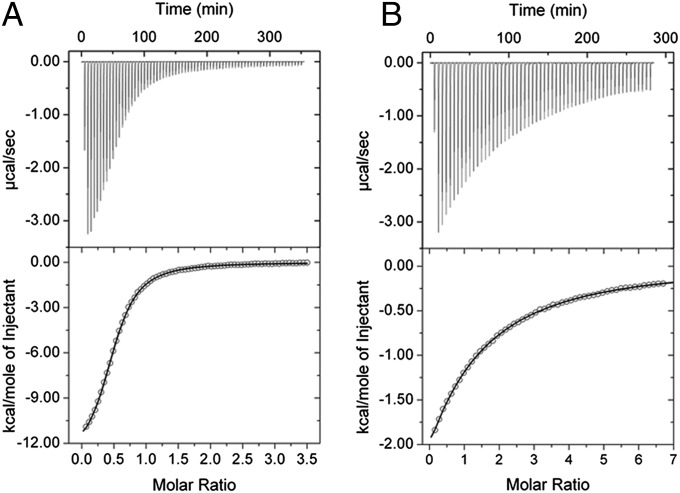
In the present study, homogentisate 1,2-dioxygenase from P. putida (HGDOPp) was heterologously produced and purified to homogeneity with a ferrous ion content of 0.56 ± 0.04 mol of iron per mol of protein. The Vmax of HGDOPp at 25 °C was 99.4 ± 0.6 µmol⋅min−1⋅mg−1 of protein (apparent kcat = 79.5 s−1) with an apparent Km of 53.2 ± 4.2 µM for homogentisate. Isothermal titration calorimetry (ITC) under strictly anoxic conditions was used to follow the binding of HG to the mononuclear ferrous iron site of HGDOPp in the absence of dioxygen (Fig. 1A). HG bound to the ferrous ion of HGDOPp with a Kd of 15.8 ± 1.4 µM. The ratio of bound HG to HGDOPp (N) was 0.51 ± 0.03, which agrees well with the iron content. Binding of HG was entropically disfavored (ΔSbind = −22.6 cal⋅mol−1⋅deg−1) and enthalpically driven (ΔHbind = −13.3 kcal⋅mol−1), resulting in a free enthalpy of binding of ΔGbind = −12.8 kcal⋅mol−1 at room temperature.
Fig. 1.
ITC data for the titration of HG to wild-type (A) and Y346F (B) HGDOPp. (Upper) The binding isotherms with exothermic heat change. (Lower) The fits of the integrated enthalpic changes to a one-site binding model. Dissociation constants (Kd) were determined to be 15.8 and 909 µM for wild-type and Y346F variant of HGDOPp, respectively.
Structure of HGDOPp.
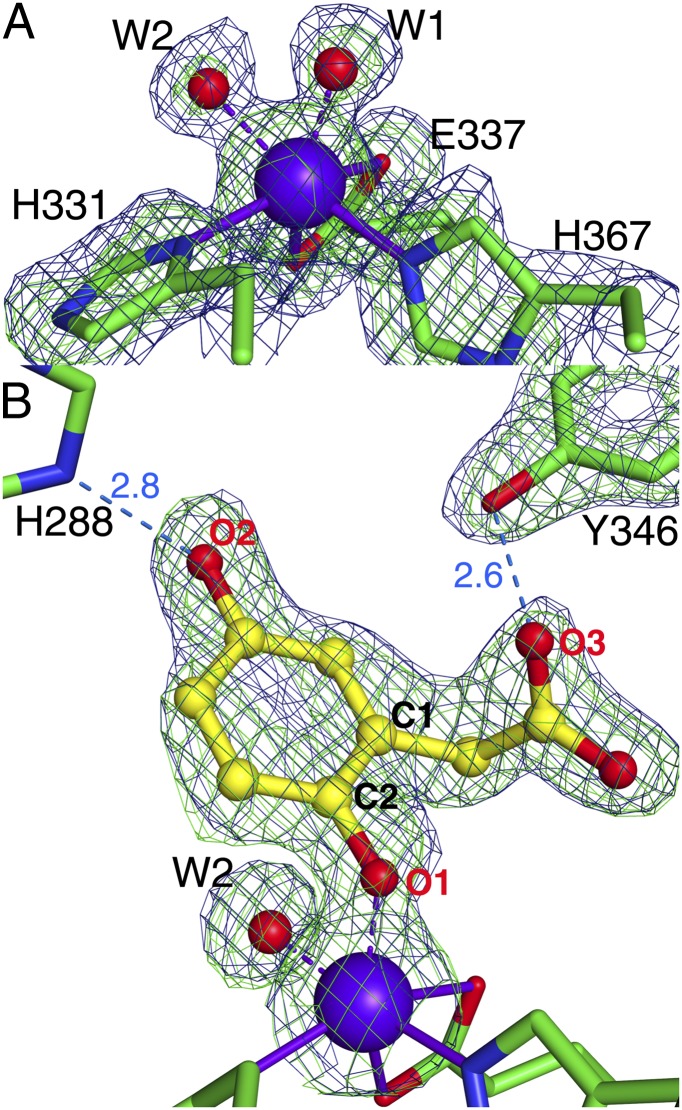
The crystal structure of resting state HGDOPp was determined at 1.95 Å resolution (“Fe2+-only” in SI Appendix, Table S1). The twelve molecules in the asymmetric unit (a.u.) create two complete hexamers each built up by a dimer of trimers. All residues except for G344-E351 are defined in the electron density. HGDOHs and HGDOPp share a sequence identity of 49%, and consistently the overall structure of HGDOPp is very similar to HGDOHs (6) (PDB ID code 1EYZ), with an rmsd of 1.8 Å for Cα atoms (SI Appendix, Fig. S1A). The active site pocket with its Fe2+ ion is freely accessible from the outside through a wide opening (SI Appendix, Fig. S1C). All twelve subunits in the a.u. contain fully occupied Fe sites, indicating that only Fe-containing HGDOPp has crystallized. The active site of human HGDOHs after soaking with ferrous iron shows Fe2+ coordinated by the 2-His-1-carboxylate facial triad (H335-Nδ1, H371-Nε2, and E341-Oε2), resulting in a distorted square pyramidal arrangement whose overall geometry remained ill-defined as no solvent molecules were observed (SI Appendix, Fig. S1B) (6). The structure of the resting state of HGDOPp reveals the ferrous ion to be coordinated by H331-Nδ1 and H367-Nε2, the carboxylate of E337 (E337-Oε1 and -Oε2), and two water molecules completing the observed octahedral coordination, as the carboxylate acts as a bidentate ligand (Fig. 2A). The observed bond lengths of the Fe2+ ligands are in the range typically found for Fe–N and Fe–O bonds as reported for other dioxygenases (SI Appendix, Fig. S2A and Table S2) (11–13).
Fig. 2.
Active site structures of resting state HGDOPp (A) and its complex with the substrate HG (B). Electron density maps in the vicinity of the active site are shown as blue mesh for the 2Fobs − Fcalc map and as green mesh for the Fobs − Fcalc simulated-annealing omit map. The weighted 2Fobs − Fcalc maps of A and B are contoured at 1.2 σ. The Fobs − Fcalc omit maps in A and B are contoured at 3.5 σ and were generated after removing modeled atoms from the active site structure. Possible hydrogen–bonding interactions are shown as blue dashed lines with distances given in angstroms. Iron and solvent molecules are shown as purple and red spheres, respectively.
Cocrystallization of HGDOPp with HG was achieved under anoxic conditions. A complete dataset was recorded from a cocrystal to a resolution of 1.98 Å (“Fe2+:HG” in SI Appendix, Table S1). In contrast to the substrate-free structure, the structure of HGDOPp in complex with HG allowed the facile tracing of residues G344-E351 in all twelve copies in the a.u. Residues G344-E351 form a loop, which closes the substrate channel by acting as a lid for the active site pocket shielding it from solvent once HG is bound (SI Appendix, Fig. S1D). A large patch of positive difference density was observed in the active site above the Fe2+ ion (Fig. 2B). Incorporating HG in the model explained this difference density, and HG refined with an average temperature factor of 18.3 Å2 and 89% occupancy in the 12 molecules. HG replaced one of the Fe2+-coordinating water molecules found in the resting state (W1 in Fig. 2A) and binds to the Fe2+ ion with its phenolate group (O1HG – Fe = 1.98 Å) (SI Appendix, Table S2), preserving the octahedral arrangement of the iron ligands. No deviation from planarity was observed for the ring atoms of HG. Additionally, the second hydroxyl group of HG is in H-bonding distance to H288 (O2HG – H288-Nε2 = 2.8 Å) and its 1-acetate group to the hydroxyl group of Y346 (O3HG – Y346-OH = 2.6 Å) (Fig. 2B and SI Appendix, Fig. S2B). Y346 is part of the active site lid (G344-E351), and its interaction with HG is most likely responsible for the closure of the active site lid once HG binds. Replacement of Y346 by phenylalanine in Y346F-HGDOPp decreased the affinity for HG more than 60-fold (Fig. 1B; Kd = 897 ± 5.7 µM) and reduced the apparent kcat 20-fold to 4.2 ± 0.03 s−1, resulting in a decrease of the specificity constant (kcat/Kd) by three orders of magnitude (wild-type HGDOPp: kcat/Kd = 5,038 mM−1⋅s−1; Y346F-HGDOPp: kcat/Kd = 4.6 mM−1⋅s−1) agreeing with a central role of Y346 in substrate binding and turnover. Noticeable change in turnover of HG was observed when H288 was substituted with glutamine, resulting in an ∼75-fold reduction in kcat (1.03 ± 0.06 s−1), and no signal could be recorded when H288Q-HGDOPp was titrated with HG in the ITC. The lower turnover may indicate that H288 plays a crucial role as a base catalyst to abstract a proton from the p-hydroxyl group of HG.
Structures of HGDO During Turnover.
When a cocrystal of HGDOPp grown anoxically with HG was exposed to air, it slowly turned light yellowish, due to the formation of the product maleylacetoacetate that absorbs light in the region 300–400 nm (4). Thus, HGDOPp is catalytically active in the crystal. To gain further insights into the reaction mechanism of HGDOPp, we allowed crystals grown anoxically in the presence of HG to react under oxic conditions in the presence of additional HG and stopped the reaction by rapid cooling of the crystals in liquid nitrogen. The structure of a “reacting” crystal was determined at 1.70 Å resolution (“Fe2+:HG-O2 structure” in SI Appendix, Table S1).
Four (A, C, F, and L) of the twelve subunits present in the a.u. displayed previously unobserved positive densities near the Fe2+ ion after refinement of HGDOPp with HG (Fig. 3 and SI Appendix, Figs. S3 and S4) whereas the other eight subunits showed electron densities indistinguishable from the untreated HGDOPp–HG complex. Stepwise interpretation and modeling of the electron densities in the active sites of the four subunits was performed as explained in SI Appendix, and the resulting structures are described next.
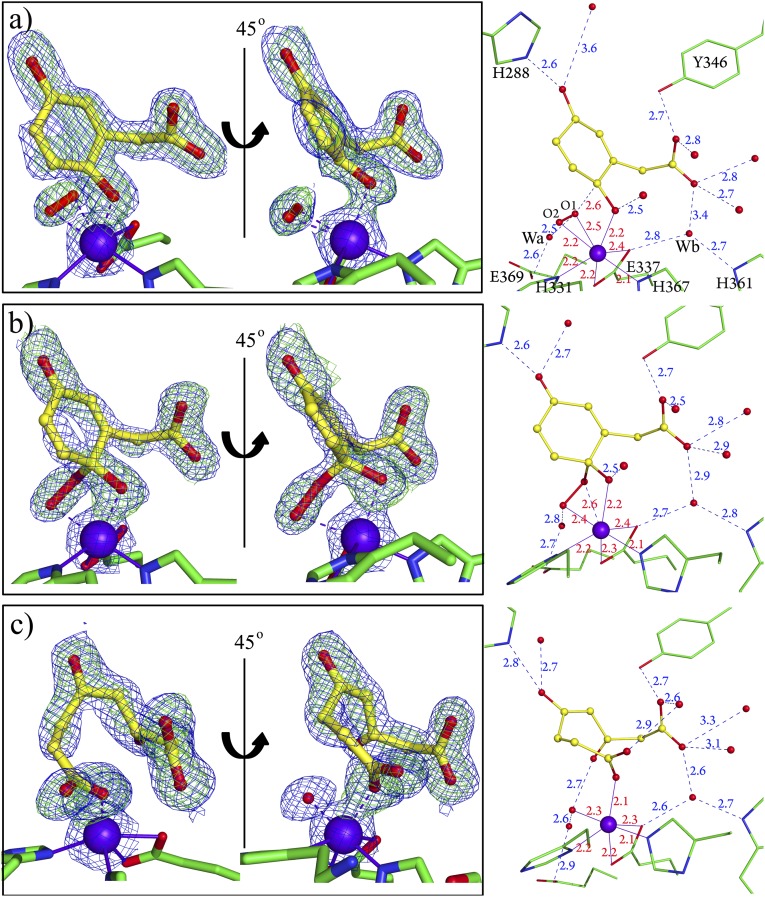
Fig. 3.
Three different reaction intermediates found in the asymmetric unit of an HG-containing HGDOPp crystal after incubation with air. (A) Semiquinone state of HG and side-on binding of dioxygen in subunit C. (B) Alkylperoxo intermediate in subunit F. (C) Ring-open product in subunit A. The weighted 2Fobs − Fcalc maps (blue mesh) are contoured at 1.2 σ (A–C). The Fobs − Fcalc omit maps (green mesh) generated after removing modeled atoms from the active site structure are contoured at 3.2 σ (A–C). (Right) Interactions of the observed species with second coordination sphere atoms are shown, and distances are given in angstroms. Color code is as in Fig. 2.
Subunit C shows a positive peak at the Fe2+ ion (Fig. 3A), which could not be modeled as a single OHx ligand but clearly indicated a diatomic species bound to Fe2+ in trans to H367 (W2 position in Fig. 2A). Modeling with dioxygen satisfied the observed difference density and resulted after refinement into an asymmetrically side-on bound O2 ligand (O1–O2 = 1.35 Å, O2–Fe(II) = 2.2 Å and O1–Fe(II) = 2.5 Å). The geometry of bound HG clearly deviates from the planar ring structure observed in the absence of dioxygen and shows a pronounced puckering of the ring around the C2 atom. We attribute the puckering to the presence of a semiquinone radical state of HG (Fig. 3A). The distance of 2.6 Å between the O1 atom of dioxygen and C2HG is consistent with a slight overlap of the van der Waals radii of both atoms. The side-on bound O2 ligand shows no further interactions with the protein, and only a water molecule (Wa) is in short hydrogen-bonding distance (2.5 Å) to the O1 atom of dioxygen (Fig. 3A and SI Appendix, Fig. S5A).
As the substrate-binding cavity is shielded from the bulk solvent by folding of the flexible loop, binding of HG likely blocks direct access of dioxygen from bulk solvent (SI Appendix, Fig. S1D). However, a cavity placed around the threefold axis of HGDOPp hexamer connects the surface of the protein (SI Appendix, Fig. S6A) with an interior cavity (SI Appendix, Fig. S6B). This interior cavity reaches to the active site of each subunit through a short channel that ends at the observed binding site of O2 at Fe2+ in trans to H367 (Fig. 3A and SI Appendix, S6C) and thus may act as a channel for O2.
The structure of subunits F and L show similar continuous electron densities between O2 and HG, and the two molecules refine to a short distance between O1O2 and C2HG (Fig. 3B). The observed connection between O1O2 and C2HG (Fig. 3B) indicates the presence of an alkylperoxo intermediate, and the structure refined to a bond length of 1.44 Å for C2HG–O1O2 with 129° between atoms C2HG–O1O2–O2O2 and a tetrahedral geometry of C2HG agreeing with an sp3-hybridized C atom. The bond length for O1O2–O2O2 refined to 1.53 Å. The distal oxygen atom interacts with a water molecule, Wa (SI Appendix, Fig. S5B).
Subunit A finally shows a disconnected electron density for the ring atoms and a distance of 3.5 Å between carbon atoms C1 and C2 of HG, showing that the product maleylacetoacetate has been formed (Fig. 3C). The oxygen atom of the downward-pointing carbonyl group at C1HG is stabilized by a solvent molecule in the former position of dioxygen at Fe2+, and the carboxyl group formed at C2HG acts as a monodentate ligand of the Fe2+ ion (Fig. 3C). A superposition of the active site structures containing the different reaction intermediates highlights three ligand anchors in the active site (SI Appendix, Fig. S7). The first anchor is the Fe2+ ion to which the phenolate group and the activated dioxo-species are coordinated. His288, which interacts with the para hydroxyl group provides the second anchor whereas the third is Tyr346 recruited by the binding of the substrate.
It is striking to observe these different structures in the individual subunits, as they must be distinguished in their reactivities for some reason. We don’t see obvious structural differences in the twelve subunits, and the overall flexibility of the chains as indicated by the average B-factors for the chains are very similar. Differences in reactivity and the accumulation of different reaction intermediates in the independent subunits of the a.u. have been reported for other enzymes, notably including nonheme oxygenases (12, 15), and these differences were related to intermolecular contacts in the crystal lattice. In the crystal structure of HGDOPp, the four reacting subunits interact with each other (SI Appendix, Fig. S8), and these interactions include the active site lids in the subunits where HG has reacted. It is thus feasible that the decreased flexibility of the active site lid and its interaction with the substrate via Y346 favored the accumulation of the observed intermediates.
Mechanism Proposed from the Structures.
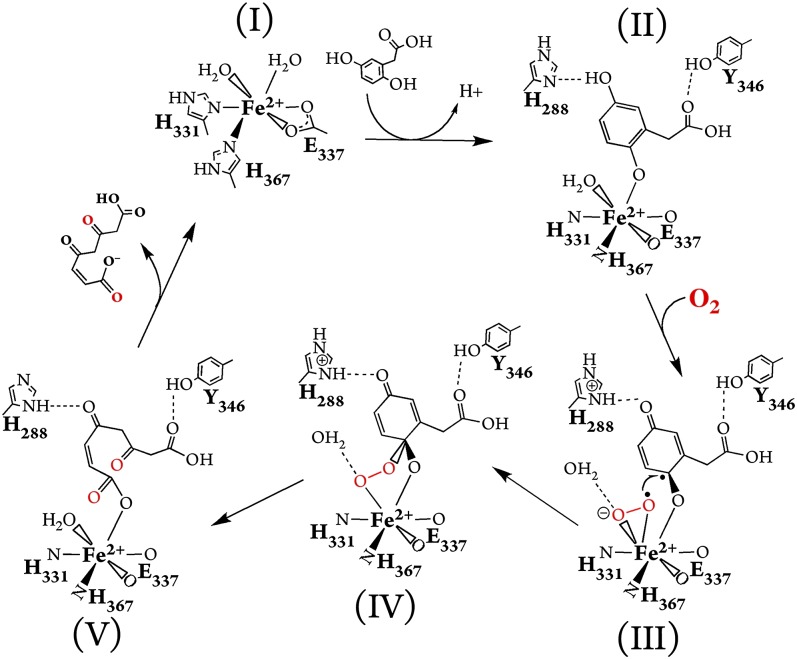
Based on the structures identified in this study and those of HPCD (12, 17), we propose a mechanism for the oxidative ring-cleavage reaction catalyzed by HGDO (Fig. 4). (I) In the absence of HG, the Fe2+ ion of HGDO has an octahedral coordination including two water ligands. (II) Binding of HG to the Fe2+ ion replaces one water molecule and likely leads to the deprotonation of the C2 hydroxyl group with a water molecule in suitable distance to act as proton acceptor. HG further interacts with H288 and Y346; the latter closes the active site lid protecting the active site from bulk solvent. The coordination site trans to H367 remains occupied by a water ligand, and the strength of the Fe–OH2 interaction seems not to be affected by HG binding as we don’t observe a change in the Fe–OH2 bond length (SI Appendix, Table S2). (III) Dioxygen can diffuse through a tunnel ending at the vacant coordination site of Fe2+, where it is activated by inducing an electron transfer from HG to O2, resulting in the formation of two juxtaposed radical species, a side-on bound superoxo-radical and a semiquinone-radical on HG. H288 acts as a base and accepts a proton from HG to form the neutral semiquinone radical. (IV) Recombination of the two radicals generates the alkylperoxo intermediate, which is stabilized by H-bonds with water molecules. Attack of the superoxo ligand on HG at the hydroxylated C2 carbon is additionally favored by the proximity of both reactants with a distance of 2.6 Å between C2 and the nearest oxygen atom whereas the adjacent carbon atoms (C1 and C3) of HG show shortest O–C distances of more than 3 Å to the superoxo ligand. (V) The alkylperoxo intermediate may react under O–O bond fission via a gem-diol and a seven-member lactone intermediate (SI Appendix, Fig. S9), both unobserved in this study, before the ring opens and forms maleylacetoacetate that we captured in the crystal. O–O bond cleavage is likely facilitated by protonation of the oxygen atom bound to Fe2+ in which the water molecule denoted as Wa can act as proton donor. The linearized product maleylacetoacetate is released after protonation from H288 and keto/enol tautomerization, which is assisted by an opening movement of the active site lid.
Fig. 4.
Reaction mechanism for the oxidative aromatic ring-cleavage of HG by HGDO based on the five active site structures found in this study. (I) Fe2+ site before substrate binding. Water molecules occupy two coordination sites completing the octahedral arrangement of the iron ligands. (II) HG acts as a monodentate ligand. Bound HG is further stabilized by hydrogen bonds to H288 and Y346. (III) Oxidized HG (semiquinone state) is formed by reduction of dioxygen to the superoxo state. (IV) Alkylperoxo intermediate. (V) Ring-fission product. States IV and V are possibly connected by formation of gem-diol and lactone intermediates.
Recently, the mechanism of HGDO was investigated using hybrid density functional theory (18). In the absence of a crystal structure of HGDO with bound substrate, HG had been modeled to define the starting geometry, and it was assumed to act as a bidentate ligand of the ferrous ion (18). With this assumption, an arrangement leaving the coordination site trans to E337-Oε2 (HGDOPp numbering) vacant was found to be the lowest-energy configuration, and accordingly dioxygen was proposed to bind end-on in trans to E337-Oε2 (18). Attack of the Fe-bound dioxygen was supposed to occur at C1, the acetate chain carrying ring carbon atom and reactions with the ring carbon C2 or C3 were disfavored (18). Our crystal structures deviate in most aspects from the modeled HG:HGDO complex and the proposed reaction mechanism: (i) HG acts as mono- not a bidentate ligand; (ii) dioxygen binds side-on in trans to H367 not end-on in trans to E337-Oε2; (iii) the reaction between superoxide and HG proceeds by reaction at the OH-group carrying C2 atom of HG, not the C1 atom, to form the alkylperoxo intermediate at C2HG, thus making the formation of the gem-diol intermediate the likely next step, which was disfavored in the computational study (18).
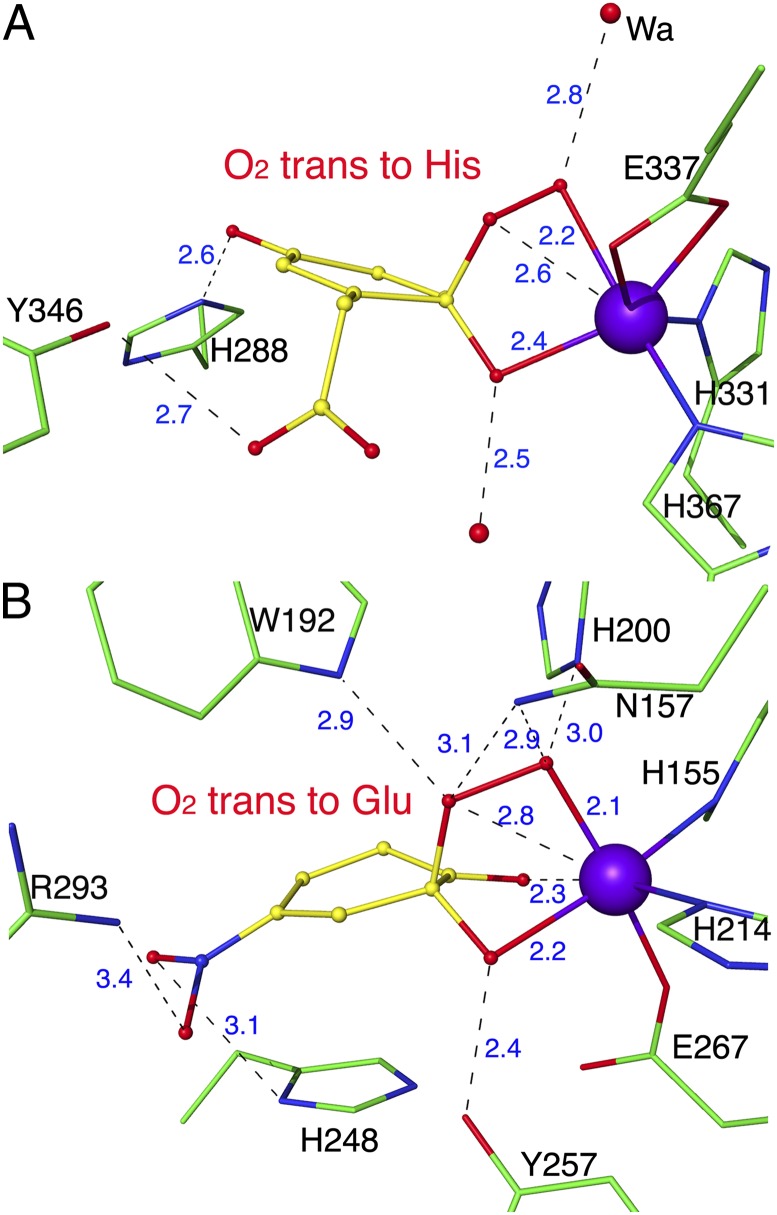
Our results bear parallels to the recent structural investigations of the extradiol dioxygenase HPCD with the substrate analog 4-nitrocatechol (12). HGDO and HPCD share no detectable sequence similarity, display different folds, and are thus nonhomologous enzymes. Whereas HPCD is a prototypical type I extradiol dioxygenase acting on catechol-type substrates, HGDO reacts with a substrate containing hydroxyl groups in para position, which acts as a monodentate ligand when bound to the active site Fe2+ ion. Nevertheless, the reaction mechanisms of HPCD and HGDO show surprising parallels in the formation of closely related intermediates to catalyze the cleavage of dihydroxylated aromatic rings (12, 17). However, the similar intermediates develop in different active site environments (Fig. 5). Whereas, in HPCD, the Fe2+-bound superoxo ligand is stabilized by a network of hydrogen bonds originating from surrounding amino acids, no interaction between the protein and the superoxo ligand was observed in HGDOPp, and the only candidates for stabilization of the superoxo ligand by hydrogen-bonding interactions are water molecules (Fig. 5 and SI Appendix, Fig. S5). The situation is similar for the alkylperoxo intermediate, which is stabilized by five possible hydrogen bonds with amino acid side chains in HPCD whereas, in HGDOPp, this intermediate only interacts with water molecules. It is therefore not surprising that several residues in the second coordination sphere of the active site Fe2+ of HPCD have been shown to be critical for dioxygen activation and catalysis (17). H200 of HPCD stabilizes Fe2+-bound O2, favors the formation of the alkylperoxo intermediate, and may also facilitate its breakdown by protonating it to form the gem-diol intermediate (19). Y257 in HPCD is hydrogen-bonded to the C2-O− group of the bound substrate but also forces the ring atoms of the substrate to adopt a strained conformation by pushing against the C2 atom, both factors favoring the formation of the semiquinone radical intermediate (20, 21). No amino acids corresponding to these two critical residues of HPCD can be found in HGDOPp, where most interactions with the intermediates are mediated by water molecule, and it is not obvious how strained conformations of the substrate and intermediates could be produced (Fig. 5 and SI Appendix, Fig. S5). These structural differences may serve as precedence that even nonhomologous Fe2+-dependent dioxygenases have evolved to react through the same principal intermediates to catalyze cleavage of aromatic rings, and that this is likely an intrinsic property of the vicinal facial triad coordinating Fe2+. Like proteases and the different classes of carboanhydrases, nonheme iron containing extradiol dioxygenases are thus a class of enzymes where convergent evolution resulted in the development of nonhomologous but nearly iso-mechanistic enzymes.
Fig. 5.
Comparison of two alkylperoxo intermediates. (A) Alkylperoxo intermediate structure of HGDOPp. (B) Alkylperoxo structure of HPCD with the nonphysiological substrate 4-nitrocatechol (subunit D from PDB ID code 2IGA) (12). Distances are given in angstroms.
Materials and Methods
Protein Preparation and Activity Assay.
The gene encoding homogentisate 1,2-dioxygenase (HGDOPp) was cloned from the genomic DNA of P. putida. Escherichia coli BL21(DE3) was used for protein expression in LB media at 37 °C. Purification of HGDO was achieved by two classical chromatography steps, ion exchange using Source 30Q and hydroxyapatite columns. The whole-plasmid PCR method was applied to generate the Y346F variant. Expression and purification of the Y346F-HGDOPp variant were achieved as described for the wild-type protein. Determination of the nonheme iron content of HGDOPp was carried out by the method of Fish (22) using Ferene S as a specific ferrous chelator. HGDOPp activity measurements were performed spectrophotometrically by measuring accumulation of the product maleylacetoacetate at 318 nm (ε318 = 11,900 M−1⋅cm−1) (4). Steady-state kinetic parameters Km and Vmax were determined by varying homogentisate concentrations (0–500 µM) in three independent measurements. The unit of activity is defined as micromoles of maleylacetoacetate produced per minute. See further details in SI Appendix, Materials and Methods.
Isothermal Titration Calorimetry.
Isothermal titration calorimetry (ITC) measurements were performed using a MicroCal VP-ITC at 25 °C installed in a glove box (Labstar workstation; MBRAUN) with an atmosphere containing less than 0.5 ppm of molecular oxygen. Concentrations of protein and ligand were determined by UV absorption, ε280 = 61,880 M−1⋅cm−1 and ε291 = 3,500 M−1⋅cm−1 (4), respectively. Titration experiments were conducted using 2.5 mM HG for the wild type (0.15 mM) and 10 mM homogentisate for the Y346F (0.3 mM) and H288Q (0.16 mM) HGDOPp variants. Heats of dilution in titrations of wild type and Y346F HGDOPp were corrected. To determine thermodynamic parameters (ΔHbind and ΔGbind = −RTlnKa) together with the dissociation constant (Kd), the corrected data were approximated assuming a one-site binding model as supplied from the manufacturer (Origin7). The entropy change (ΔS) was calculated using the thermodynamic law, ΔGbind = ΔHbind − TΔSbind.
Crystallization and Crystal Treatment.
HGDOPp at a concentration of 15 mg/mL was mixed with an equal volume of a reservoir solution containing 16–17% PEG 3350, 0.02 M Na,K phosphate, and 0.1 M Bis-Tris propane, pH 8.0, in the presence or absence of 2 mM homogentisate using the vapor diffusion method under anoxic conditions inside a glovebox (model B; COY Laboratory Products) under an atmosphere of 95% N2/5% H2. Crystals were flash-cooled in the crystallization solution supplemented with 20% (vol/vol) glycerol as cryoprotectant.
For snap freezing under turnover conditions, HGDOPp crystals grown under anoxic conditions and in the presence of HG for typically 1 wk were transferred to a cryo-solution containing 2 mM HG and 1 mM TCEP (Tris-2-carboxyethylphosphine). The reaction was started by removing the crystal tray from the anoxic glovebox and thus exposing the crystals to air. After 10–60 min, crystals turned yellowish and were flash-cooled in liquid N2 to stop the reaction.
Data Collection, Structure Determination, and Model Refinements.
Diffraction data were collected on BL14.2 operated by the Joint Berlin MX-Laboratory at the BESSY II electron storage ring (Berlin-Adlershof, Germany) (23). Data sets were integrated and scaled using XDS (24). The structure of HGDOPp was solved by Patterson search techniques using the program Phaser (25), in which the crystal structure of human HGDO (6) (PDB ID code 1EYB) served as homologous search model. The solvent content of the HGDOPp crystal was calculated as ∼38%. Model building was carried out with COOT (26). Iterative refinement cycles were performed with PHENIX (27) and BUSTER (28). Although noncrystallographic symmetry (NCS) restraints were applied for the twelve molecules in the asymmetric unit during the early stages of the structure refinements, the reported final coordinates were refined without NCS restraints. The structure of HG was modeled into the observed Fobs − Fcalc omit map density in the Fe2+:HG structure (Fig. 2). Initial refinement of the Fe2+:HG-O2 structure was performed using the Fe2+-bound HG model. Although eight chains of the Fe2+:HG-O2 structure showed electron density as found in the Fe2+:HG structure, clear differences in 2Fobs − Fcalc and Fobs − Fcalc maps were observed in the other four chains (chains A, C, F, and L) of the Fe2+:HG-O2 structure. Further adaptations of the model were based on interpretation of the observed Fobs − Fcalc difference maps after the initial refinements (SI Appendix, Figs. S3 and S4). In chain C, additional positive peaks in the Fobs − Fcalc difference maps were interpreted as the semiquinone state of HG and a side-on binding of dioxygen molecule based on the observation of puckered electron density at C2 atom of HG and an elongated positive peak, which could not be fit with a single water ligand (Fig. 3A and SI Appendix, Fig. S3). In chain F and L, additional electron density below the C2 atom of HG and further toward the Fe2+ ion was modeled as an alkylperoxo moiety, which fully explained the calculated Fourier maps (Fig. 3B and SI Appendix, Fig. S4). Further details on ligand modeling and refinement are described in SI Appendix, Materials and Methods. The ring-open product state was modeled and refined based on the observation of a break in the electron density between the C1 and C2 atoms of HG in the Fobs − Fcalc omit map (Fig. 3C). Modeling of the carbonyl oxygen atom at C1HG as pointing upwards compared with the model in Fig. 3C deviates from the observed density and clashes with the oxygen atom of the acetate group of HG. Restraints for all ligands were obtained using PHENIX (27) based on the model described in Fig. 4. Distances between the Fe2+ ion and its ligands were not restrained during refinement. Data collection and refinement statistics of the reported structures including occupancies and B-factors of ligands are shown in SI Appendix, Table S1.
Detailed descriptions of the materials and experimental procedures are included in SI Appendix.
Supplementary Material
Acknowledgments
For help with data collection, we thank scientists at beamline BL14.2, operated by the Joint Berlin MX-Laboratory at the Berliner Elektronenspeicherring- Gesellschaft für Synchrotronstrahlung II electron storage ring (Berlin-Adlershof, Germany) (23), and we thank members of the structural biology/biochemistry group of the Humboldt-Universität zu Berlin for discussions. This study was supported by Deutsche Forschungsgemeinschaft Grant DO 785/2-3.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4AQ2 (Fe2+-only state), 4AQ6 (Fe2+:HG state), and 3ZDS (Fe2+:HG-O2 state)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302144110/-/DCSupplemental.
References
- 1.Arias-Barrau E, et al. The homogentisate pathway: A central catabolic pathway involved in the degradation of L-phenylalanine, L-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J Bacteriol. 2004;186(15):5062–5077. doi: 10.1128/JB.186.15.5062-5077.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim YH, et al. Analysis of aromatic catabolic pathways in Pseudomonas putida KT 2440 using a combined proteomic approach: 2-DE/MS and cleavable isotope-coded affinity tag analysis. Proteomics. 2006;6(4):1301–1318. doi: 10.1002/pmic.200500329. [DOI] [PubMed] [Google Scholar]
- 3.Veldhuizen EJ, et al. Steady-state kinetics and inhibition of anaerobically purified human homogentisate 1,2-dioxygenase. Biochem J. 2005;386(Pt 2):305–314. doi: 10.1042/BJ20041370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amaya AA, Brzezinski KT, Farrington N, Moran GR. Kinetic analysis of human homogentisate 1,2-dioxygenase. Arch Biochem Biophys. 2004;421(1):135–142. doi: 10.1016/j.abb.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 5.Fernández-Cañón JM, Peñalva MA. Spectrophotometric determination of homogentisate using Aspergillus nidulans homogentisate dioxygenase. Anal Biochem. 1997;245(2):218–221. doi: 10.1006/abio.1996.9957. [DOI] [PubMed] [Google Scholar]
- 6.Titus GP, et al. Crystal structure of human homogentisate dioxygenase. Nat Struct Biol. 2000;7(7):542–546. doi: 10.1038/76756. [DOI] [PubMed] [Google Scholar]
- 7.Rodríguez JM, et al. Structural and functional analysis of mutations in alkaptonuria. Hum Mol Genet. 2000;9(15):2341–2350. doi: 10.1093/oxfordjournals.hmg.a018927. [DOI] [PubMed] [Google Scholar]
- 8.Zatkova A. An update on molecular genetics of Alkaptonuria (AKU) J Inherit Metab Dis. 2011;34(6):1127–1136. doi: 10.1007/s10545-011-9363-z. [DOI] [PubMed] [Google Scholar]
- 9.Koehntop KD, Emerson JP, Que L., Jr The 2-His-1-carboxylate facial triad: A versatile platform for dioxygen activation by mononuclear non-heme iron(II) enzymes. J Biol Inorg Chem. 2005;10(2):87–93. doi: 10.1007/s00775-005-0624-x. [DOI] [PubMed] [Google Scholar]
- 10.Vaillancourt FH, Bolin JT, Eltis LD. The ins and outs of ring-cleaving dioxygenases. Crit Rev Biochem Mol Biol. 2006;41(4):241–267. doi: 10.1080/10409230600817422. [DOI] [PubMed] [Google Scholar]
- 11.Sato N, et al. Crystal structures of the reaction intermediate and its homologue of an extradiol-cleaving catecholic dioxygenase. J Mol Biol. 2002;321(4):621–636. doi: 10.1016/s0022-2836(02)00673-3. [DOI] [PubMed] [Google Scholar]
- 12.Kovaleva EG, Lipscomb JD. Crystal structures of Fe2+ dioxygenase superoxo, alkylperoxo, and bound product intermediates. Science. 2007;316(5823):453–457. doi: 10.1126/science.1134697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han S, Eltis LD, Timmis KN, Muchmore SW, Bolin JT. Crystal structure of the biphenyl-cleaving extradiol dioxygenase from a PCB-degrading pseudomonad. Science. 1995;270(5238):976–980. doi: 10.1126/science.270.5238.976. [DOI] [PubMed] [Google Scholar]
- 14.Yi C, et al. Iron-catalysed oxidation intermediates captured in a DNA repair dioxygenase. Nature. 2010;468(7321):330–333. doi: 10.1038/nature09497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovaleva EG, Lipscomb JD. Intermediate in the O-O bond cleavage reaction of an extradiol dioxygenase. Biochemistry. 2008;47(43):11168–11170. doi: 10.1021/bi801459q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burzlaff NI, et al. The reaction cycle of isopenicillin N synthase observed by X-ray diffraction. Nature. 1999;401(6754):721–724. doi: 10.1038/44400. [DOI] [PubMed] [Google Scholar]
- 17.Borowski T, Georgiev V, Siegbahn PE. Catalytic reaction mechanism of homogentisate dioxygenase: A hybrid DFT study. J Am Chem Soc. 2005;127(49):17303–17314. doi: 10.1021/ja054433j. [DOI] [PubMed] [Google Scholar]
- 18.Kovaleva EG, Lipscomb JD. Versatility of biological non-heme Fe(II) centers in oxygen activation reactions. Nat Chem Biol. 2008;4(3):186–193. doi: 10.1038/nchembio.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groce SL, Lipscomb JD. Aromatic ring cleavage by homoprotocatechuate 2,3-dioxygenase: Role of His200 in the kinetics of interconversion of reaction cycle intermediates. Biochemistry. 2005;44(19):7175–7188. doi: 10.1021/bi050180v. [DOI] [PubMed] [Google Scholar]
- 20.Kovaleva EG, Lipscomb JD. Structural basis for the role of tyrosine 257 of homoprotocatechuate 2,3-dioxygenase in substrate and oxygen activation. Biochemistry. 2012;51(44):8755–8763. doi: 10.1021/bi301115c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mbughuni MM, Meier KK, Münck E, Lipscomb JD. Substrate-mediated oxygen activation by homoprotocatechuate 2,3-dioxygenase: Intermediates formed by a tyrosine 257 variant. Biochemistry. 2012;51(44):8743–8754. doi: 10.1021/bi301114x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fish WW. Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol. 1988;158:357–364. doi: 10.1016/0076-6879(88)58067-9. [DOI] [PubMed] [Google Scholar]
- 23.Mueller U, et al. Facilities for macromolecular crystallography at the Helmholtz-Zentrum Berlin. J Synchrotron Radiat. 2012;19(Pt 3):442–449. doi: 10.1107/S0909049512006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst. 1993;26:795–800. [Google Scholar]
- 25.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bricogne GBE, et al. BUSTER. Cambridge, United Kingdom: Global Phasing Ltd; 2011. Version 2.10.0. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.