Abstract
The nuclear antisense properties of a series of tricyclo (tc)-DNA oligonucleotide 9–15mers, targeted against the 3′ and 5′ splice sites of exon 4 of cyclophilin A (CyPA) pre-mRNA, were evaluated in HeLa cells and compared with those of corresponding LNA-oligonucleotides. While the 9mers showed no significant antisense effect, the 11–15mers induced exon 4 skipping and exon 3+4 double skipping to about an equal extent upon lipofectamine mediated transfection in a sequence- and dose-dependent manner, as revealed by a RT–PCR assay. The antisense efficacy of the tc-oligonucleotides was found to be superior to that of the LNA-oligonucleotides in all cases by a factor of at least 4–5. A tc-oligonucleotide 15mer completely abolished CyPA mRNA production at 0.2 µM concentration. The antisense effect was confirmed by western blot analysis which revealed a reduction in CyPA protein to 13% of its normal level. Fluorescence microscopic investigations with a fluorescein labeled tc-15mer revealed a strong propensity for homogeneous nuclear localization of this backbone type after lipofectamine mediated transfection, while the corresponding lna 15mer showed a less clear cellular distribution pattern. Transfection without lipid carrier showed no significant internalization of both tc- and LNA- oligonucleotides. The obtained results confirm the power of tc-DNA for nuclear antisense applications. Moreover, CyPA may become an interesting therapeutic target due to its important role in the early steps of the viral replication of HIV-1.
INTRODUCTION
Antisense oligonucleotides that are stable against nucleolytic degradation and that do not induce RNase H catalyzed hydrolysis of complementary mRNA were shown to be powerful modulators of pre-mRNA splicing in vitro and in vivo (1,2). Such oligonucleotides have successfully been used in the past to shift pre-mRNA splicing of CFTR (3), IL-5R (4), c-myc (5), tau (6), SMN-2 (7), bcl-x (8–10), β-globin (11,12) and dystrophin (13–15). Amongst the oligonucleotide analogues most intensively investigated for splice shifting are the anionic analogues 2′-OMe-RNA and 2′-methoxyethyl (MOE)-RNA, as well as the charge neutral analogues morpholino-DNA and PNA. A direct comparison of the four different backbone types has recently been performed in a in vitro assay in which the modified oligonucleotides blocked aberrant and restored correct splicing of modified enhanced green fluorescent protein (EGFP) pre-mRNA (16). It was found that free uptake and the antisense efficacy of neutral morpholino derivatives and of PNA carrying 4 C-terminal lysine residues were significantly higher than that of the negatively charged 2′-O-alkyl RNAs. Interestingly, the same assay in a transgenic mouse model showed inverted results with the 2′-O-MOE oligonucleotides showing much higher activity than the lysine modified PNA or morpholino-oligonucleotides.
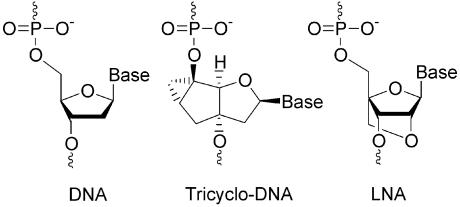
High potential in terms of antisense properties is given to the class of conformationally constrained oligonucleotide analogues such as for example LNA (17,18) or tricyclo (tc)-DNA (19) (Fig. 1), which show significantly enhanced target binding properties and enhanced biological stability. In the case of LNA, efficient antisense activity was reported for a G-protein coupled receptor in rat brain (20), HIV-1 TAR (21), VR1 (22,23) and human telomerase (24). Given the fact that both backbone systems do not mediate RNaseH activity, they seem particularly well suited for nuclear antisense applications. We recently investigated tc-DNA for correction of aberrant β-globin splicing in a cellular assay and found a 100-fold increased antisense activity relative to 2′OMe-PS-RNA (25). In order to compare the nuclear antisense properties of the two backbone systems we set out to study their performance side by side in a cyclophilin A (CyPA) splice assay.
Figure 1.
Molecular structures of tc-DNA and LNA in comparison to DNA.
We chose CyPA because of recent data, suggesting it to be a target for the inhibition of HIV-1 replication. CyPA is a proline cis–trans isomerase that was initially discovered as the cellular ligand of the immunosuppressive drug cyclosporin A (CsA) (26). Although CyPA has been implicated in diverse functions such as for example in the heat-shock response (27), in the nuclear import of some proteins (28), and in various signal transduction pathways (29,30), its exact function is still not yet fully understood. Surprisingly, it was shown that CyPA was specifically incorporated into HIV-1 virions and was required to maintain efficient viral replication (31–34). Virions lacking CyPA show normal morphologies, package normal levels of viral proteins and RNAs, and exhibit normal reverse transcriptase activity (31,35). However, the viral replication is blocked at some step after viral entry but before the reverse transcription starts (31). More recently, it was shown that CyPA might also function at the viral entry step of HIV-1 infection (36,37). Taken together, these observations imply that CyPA not only participates in post-entry events, but also contributes to the early events of HIV-1 replication (36–38).
We found that tc-oligonucleotides, 11–15 nt in length, targeted to the 3′-splice site of exon 4 of the CyPA pre-mRNA efficiently redirect splicing by skipping exon 3 and 4 simultaneously in a dose-dependent manner. Down-regulation of CyPA was confirmed by western blot analysis. In the given assay, tc-DNA outperformed LNA in antisense efficacy. Intracellular distribution of a 3′-fluorescein labeled tc-oligonucleotide 15mer in HeLa cells after lipofection revealed selective accumulation of the oligonucleotide in the nucleus which might be a reason for the observed strong nuclear antisense effect.
MATERIALS AND METHODS
Synthesis and purification of oligonucleotides
Tc-oligonucleotides (Table 1) were prepared on the 0.2 and 1.3 µmol scale on a PerSeptive Biosystems Expedite or on a Pharmacia Gene Assembler Special™ DNA-synthesizer using the modified phosphoramidite chemistry procedure as reported previously (19). The oligomers tc1 and tc2 were synthesized on universal solid support (GlenResearch). The oligomers tc3–tc5 were assembled on a universal solid support (CT-Gen, San Jose, CA). The 3′-fluorescein-labeled oligomer tc6 was synthesized on the 1.3 µmol scale using LC fluorescein CPG (Roche Diagnostics). All tc-oligomers were synthesized with an additional nucleotide unit at its 5′ end which is cleaved during deprotection, resulting in a 5′-terminal phosphate group (39). The coupling time was set to 10 min, and 5-(ethylthio)-1H-tetrazole (0.25 M in CH3CN) was used as activator. Coupling yields were >95% per step. Deprotection and cleavage from the solid support were performed under standard conditions (conc. NH3, 16–60 h, 55–65°C). The molecular compositions of all oligonucleotides were determined by ESI– mass spectrometry (Table 1). The experimental masses of tc1 and tc2 showed the presence of a 3′-terminal 2,5-dimethoxytetrahydrofuran-3,4-diol unit arising from the universal solid support. Oligonucleotide tc-NS had already been synthesized before and was used in the present study as a nonsense-sequence (25). LNA phosphoramidites with the bases adenine, thymine, guanine and 5-methylcytosine were purchased from Exiqon. The LNA oligonucleotides were prepared on the 1.3 µmol scale on a Pharmacia Gene Assembler Special™ DNA-synthesizer according to the manufacturer’s protocol on either universal solid support from CT-Gen (lna1–lna5) or on LC fluorescein CPG (lna6). Crude oligonucleotides were purified by DEAE ion exchange HPLC and/or reversed phase HPLC. Oligonucleotide tc6 was prepurified by gel filtration over Sephadex G-25 and then purified by reversed phase HPLC. Also in these cases the compositions of the oligonucleotides were confirmed by ESI-MS (Table 1).
Table 1. Oligonucleotides prepared and used in this study.
| Oligomer | Sequence 5′ → 3′a | Target site (exon 4) | Tm (°C)b | ESI-MSc, calc. | ESI-MSc, found |
|---|---|---|---|---|---|
| tc1 | pCTTAC′CACT* | 5′ss | 56.4 | 3281.4 | 3282.9 |
| tc2 | pCCTTAC′CACTC* | 5′ss | 69.3 | 3935.8 | 3937.0 |
| tc3 | pCCCTTAC′CACTCA | 5′ss | 77.4 | 4388.2 | 4389.0 |
| tc4 | pACCCTTAC′CACTCAG | 5′ss | 83.2 | 5106.7 | 5107.6 |
| tc5 | pACCCTTAA′CTCTCAG | 5′ss | 60.2 | 5121.7 | 5121.5 |
| tc6 | pACCACC′CTGTCAA-FAM | 3′ss | – | 5035.0 | 5034.8 |
| tcNS | pCATTATTGCCCTGAAAGa | – | – | 6210.2 | 6209.3 |
| lna1 | CTTAC′CACT | 5′ss | 82.6 | 2942.5 | 2941.0 |
| lna2 | CCTTAC′CACTC | 5′ss | ≈95 | 3605.3 | 3604.0 |
| lna3 | CCCTTAC′CACTCA | 5′ss | >95 | 4277.9 | 4276.2 |
| lna4 | ACCCTTAC′CACTCAG | 5′ss | >95 | 4976.3 | 4976.1 |
| lna5 | ACCCTTAA′CTCTCAG | 5′ss | 92.3 | 4977.3 | 4975.0 |
| lna6 | ACCACC′CTGTCAA-FAM | 3′ss | – | 4908.3 | 4907.0 |
aBold italic: mismatch position; C: 5-methylcytosine; a: 2′-deoxyadenosine; (′) denotes the splice position on the pre-mRNA; (*) denotes a 3′-terminal modification by a 2,5-dimethoxytetrahydrofuran-3,4-diol unit (see Materials and Methods).
bTm values (2 µM duplex in 10 mM NaH2PO4, 150 mM NaCl, pH 7.0) were measured with the RNA complement r(ACUGAGUGGUAAGGGUA).
cMolecular masses were recorded in the negative ion mode and are indicated as [M-H]–.
Cell culture, transfection and analysis
HeLa cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C in an incubator containing 5% CO2. For transfection, 2.5 × 105 HeLa cells were seeded in a six-well plate. After 12 h, 0.05, 0.1 or 0.2 µM of oligonucleotides (final concentration in 1 ml of transfection solution) were complexed with 1.5, 3 or 6 µl LipofectAMINE (Invitrogen/Life Technologies), respectively in 0.2 ml of OptiMEM (Invitrogen/Life Technologies) at room temperature for 30 min. Then the complex was diluted with OptiMEM to a final volume of 1 ml and added to the cells for further incubation. The medium was replaced by normal growth medium after 5 h.
TRI-Reagent Kit (MRC, Cincinnati, OH) was used for total RNA isolation, and the RNA was usually isolated 48 h after transfection. To detect CyPA mRNA, 250 ng of total RNA was analyzed by RT–PCR using rTth DNA polymerase (Applied Biosystems) in a final volume of 50 µl with forward and reverse primers corresponding to the sequences of exon 1 and exon 5, respectively. CyPA 5′-primer: (1671–1690, CyPA exon 1): 5′-CACCGTGTTCTTCGACATTG; 3′-primer: (6260–6279, CyPA exon 5), 5′-CCATGGCCTCCACAA TATTC. The reverse transcribed CyPA mRNA was amplified with 32P-labeled dCTP for 31 cycles (94°C for 1 min, 60°C for 1 min, 72°C for 10 min). The RT–PCR products were separated on 6% non-denaturing polyacrylamide gels. Dried gels were exposed to storage phosphor screens (Molecular Dynamics) and visualized with the AIDA program (version 2.31, Raytest Isotopenmessgeräte GmbH, Straubenhardt, Germany). Data were analyzed using ImageQuant software (version 3.3).
Western blot
HeLa cells were transfected with oligonucleotide tc4. After 72 h, the cells were lysed in SDS sample buffer and separated by 12% SDS–PAGE. The gel was transferred to a nitrocellulose membrane overnight. Polyclonal rabbit anti-human CyPA- (1:1000, gift from Dr Uta Vonschwedler, University of Utah) or anti-actin- (Sigma, 1:500) antibodies were used as the first antibodies, and anti-rabbit horseradish peroxidase-linked antibody (Promega, 1:2500) as the secondary antibody. Detection was carried out using the ECL chemiluminescence technique (Amersham Pharmacia) and the blot was exposed to an AGFA X-ray film. The protein level was quantified with ImageQuant software (version 3.3). The signals were corrected to the actin loading level.
Cellular uptake and microscopy
HeLa cells were cultured and transfected with 0.2 µM (final concentration) of fluorescein labeled oligonucleotide tc6 or lna6, complexed with 6 µl lipofectAMINE as described before, and analyzed after 24 h by fluorescence microscopy (Leica Microsystems, Leica TCS SP2; microscope: Leica DM IRE2). For transfection without lipofectamine, HeLa cells were cultured and treated with a 20 µM solution of tc6 or lna6 in DMEM+/+ medium at 37°C for 24 h. The cells were then washed twice with PBS solution, and fresh DMEM+/+ was added. The living cells were first analyzed by fluorescence microscopy and then counted with a fluorescence activated cell sorter (FACS).
RESULTS
Design of oligonucleotides and general splicing pattern
We decided to investigate the modification of CyPA pre-mRNA splicing patterns in HeLa cell cultures using oligonucleotides that were targeted to the 3′- or 5′-splice sites of exon 4 (Table 1).
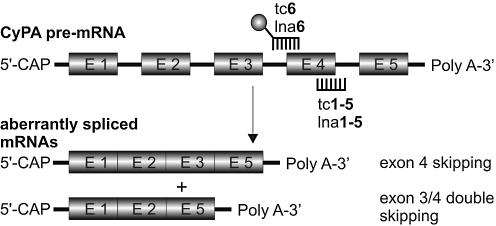
More precisely, sequences complementary to the 5′-splice site between exon 4 and intron 4 were used to compare the efficacy, minimal length requirements and mismatch sensitivity of tc- and LNA-oligonucleotides (tc1–5 and lna1–5). The 3′-splice site between intron 3 and exon 4 was the target of tc6 and lna6 which were 3′-fluorescein (FAM) labeled. This sequence was primarily used to follow cellular uptake and distribution. In all cases, we observed a general splicing pattern in which skipping of exon 4 and double skipping of exons 3+4 occurred on the mRNA level. A schematic representation of the redirection of CyPA pre-mRNA splicing in the presence of antisense oligonucleotides is given in Figure 2.
Figure 2.
Schematic representation of the splicing pattern of CyPA pre-mRNA. The ball at tc6 or lna6 represents the fluorescein label.
Thermal stability of tc- and LNA oligonucleotides with complementary RNA
Thermal stabilities of duplexes with the RNA 17mer representing the 5′-splice site of CyPA pre-mRNA were assessed by UV-melting curve analysis, and the corresponding Tms are listed in Table 1. The Tms of tc1–4 range from 56 to 83°C. The mismatched 15mer tc5 shows a decrease in thermal stability relative to the matched 15mer tc4 by 23°C. Its thermal stability is thus between that of the matched 9- and 11mer (tc1 and tc2). The corresponding oligonucleotides with the LNA backbone bind significantly more efficiently to the corresponding RNA target. While lna1 shows a Tm of 82.6°C, that of lna2 was already at 95°C. For lna3 and lna4, no sigmoidal melting curves could be observed indicating even higher Tms. The mismatched 15mer lna5 showed a Tm of 92°C which lies in between that of the 9mer lna1 and 11mer lna2, as in the case of tc-DNA. Thus LNA binds RNA with comparable selectivity but overall higher affinity by ca. 2.5–3.0°C/per mod. relative to tc-DNA. Whether the increase in affinity of LNA is entirely due to the backbone or arises in part by the base 5-methylcytosine used instead of cytosine remains open.
Antisense activity of tc-oligonucleotides
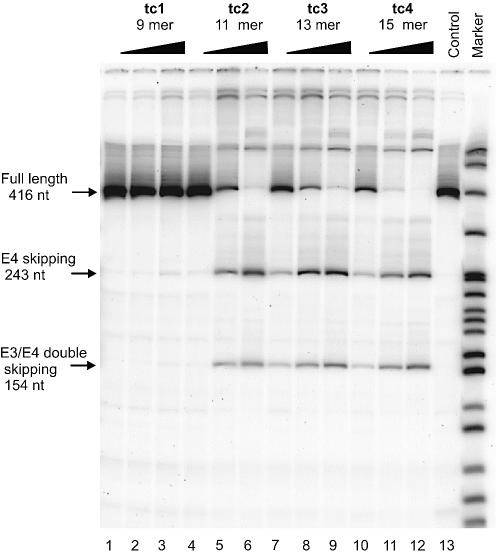
HeLa cells were transfected with three different concentrations (0.05, 0.1 and 0.2 µM) of oligonucleotides tc1–5. After 48 h total RNA was isolated, amplified by RT–PCR, and the products separated by PAGE (Fig. 3). To obtain quantitative data, we determined the residual full-length CyPA mRNA as a fraction of total RNA (full length + exon skipped RNA) by phosphor imaging (Fig. 4).
Figure 3.
RT–PCR analysis of CyPA pre-mRNA splicing in HeLa cells in the presence of varying concentrations (0.05, 0.1 and 0.2 µM) of antisense tc-oligonucleotides (lanes 1–12). Lane 13 (control): RNA from untreated cells.
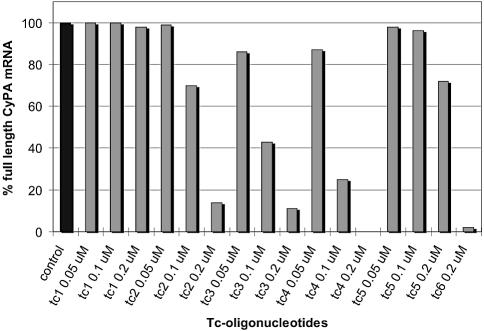
Figure 4.
Quantitative evaluation of full-length CyPA mRNA after treatment with tc-oligonucleotides. Values are normalized averages of two independent experiments. Control: RNA from untreated cells.
With the exception of tc1 (9mer), all oligonucleotides led to a significant amount of exon 4 skipping and exon 3+4 double skipping. The degree of splice shifting increased with increasing length and increasing dose of the antisense oligonucleotides. At the lowest (50 nM) concentration investigated, essentially no antisense effect was observed for tc2 (11mer) while a reduction in full length RNA by ca. 10% occurs for tc3 (13mer) and tc4 (15mer). At 0.1 µM concentration of tc2, tc3 and tc4, full length RNA is reduced to 70%, 43% and 25%, respectively, relative to the control without oligonucleotide. At 0.2 µM concentration, the level of full length RNA was below 20% for tc2 and 3, and was essentially undetectable in the case of tc4, indicating quantitative splice shifting in the corresponding pre-mRNA. In all cases, the bands corresponding to mRNA in which only exon 4 was skipped were slightly more intense than that of exon 3+4 double skipping.
Sequence specificity in the antisense response was investigated with oligonucleotide tc5, showing the same length as tc4 but two mismatches in the center of the sequence. In this case, we observed a reduction in full length RNA (Fig. 4) by 28% only at high (0.2 µM) concentration. Thus its antisense activity lies between those of the matched 9- and 11mers and is far inferior to that of the fully matched 15mer tc4. An additional control with the non-sequence related oligonucleotide tcNS showed no exon skipping at all by RT–PCR over the same concentration range (data not shown).
From these results, we conclude that the observed change in the splice pattern of CyPA pre-mRNA is due to a true antisense effect of the oligonucleotide analogues. Furthermore, a sequence length of 9 is insufficient for eliciting significant antisense effects in the submicromolar range while with 15, complete reorientation of splicing is achieved at 0.2 µM concentration.
The fluorescein labeled oligonucleotide tc5 targeted to the 3′ splice site displayed an antisense activity comparable to the oligonucleotides directed to the 5′-splice site, displaying the same single and double skipping pattern of exons 3 and 4. We found that tc6 reduced the full-length CyPA mRNA by 98% at 0.2 µM concentration (Fig. 4).
Antisense activity of LNA-oligonucleotides
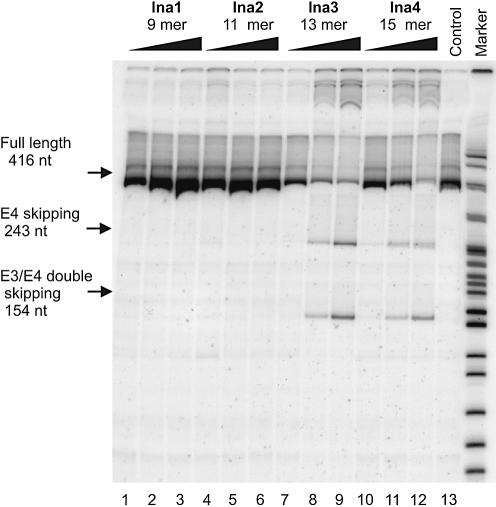
In order to compare the antisense efficacy of tc-DNA with that of LNA, the same experiments were repeated with the LNA-oligonucleotides lna1–4. A representative gel of the RT–PCR products is given in Figure 5, and the corresponding data for residual full-length CyPA mRNA were determined as before (Fig. 6).
Figure 5.
RT–PCR analysis of CyPA pre-mRNA splicing in HeLa cells in the presence of varying concentrations (0.05, 0.1 and 0.2 µM) of antisense lna-oligonucleotides (lanes 1–12). Lane 13 (control): RNA from untreated cells.
Figure 6.
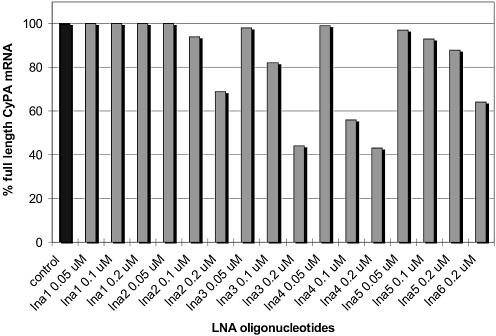
Quantitative evaluation of full-length CyPA mRNA after treatment with lna-oligonucleotides. All values are normalized averages with standard deviations from three independent experiments. Control: RNA from untreated cells.
As in the case of tc-DNA, exon 3 skipping and exon 3+4 double skipping to about equal ratios was observed with lna3 and lna4 while no effect occurred with lna1 (9mer). Also here nuclear antisense effects increased with increasing dose and length of the oligonucleotides. However, full-length RNA was only reduced to 69% with lna2 (11mer) at 0.2 µM concentration. This indicates roughly a 5-fold less efficient splice shifting compared to tc2. Interestingly, even with lna3 and lna4 at 0.2 µM, 44 and 43%, respectively, of full length RNA was still observed. This contrasts with the efficacy of the tricyclo-backbone which reduced the amount of full-length RNA to 11 and 0%, respectively. Furthermore, no increase in nuclear antisense activity was observed between the 13mer and 15mer LNA at 0.2 µM which indicated that the optimum length/activity relation is already met at the level of a 13mer.
As in the case of tc-DNA, the selectivity of the antisense response was investigated with the double mismatch sequence lna5. With this oligonucleotide a full-length RNA production of 88% relative to untreated controls was observed. Also here, the antisense activity is not only dose- but also sequence-dependent and reminiscent of a true antisense effect.
We also tested the fluorescently labeled oligonucleotide lna6 for antisense activity. As with tc6, lna6 also showed antisense activity with single and double exon skipping, but again with less efficiency (64% remaining full-length RNA) as compared to tc6. Thus in the cases investigated here, tricyclo-oligonucleotides show stronger nuclear antisense effects than corresponding lna-oligonucleotides by at least a factor of 4–5 although the overall affinity of tc-DNA to an RNA target is inferior to that of LNA.
Western blot
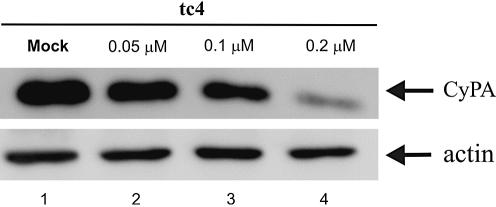
To ensure that exon skipping induced by tc-oligonucleotides also led to a reduced CyPA protein level, a western blot was performed. To this end, HeLa cells were transfected with tc4 (15mer) and left for 72 h before blotting. As can be seen from Figure 7, CyPA protein is down-regulated in a concentration-dependent manner. β-Actin was analyzed as a loading control. At 0.1 µM concentration of tc4, only 45% of the normal CyPA level was produced while at 0.2 µM, expression was reduced to 13%. This experiment lends further evidence for tc-DNA acting as specific and potent nuclear antisense oligonucleotides.
Figure 7.
Western blot, demonstrating the down-regulation of CyPA protein by tc4. CyPA protein was probed with polyclonal anti-CyPA antibodies. A polyclonal anti-actin antibody was used for loading control.
Cellular uptake and distribution
Given the surprisingly strong nuclear antisense effects, it was of interest to follow the cellular uptake and distribution of the tc- and lna-oligonucleotides. For this purpose we produced the fluorescently labeled oligonucleotide tc6 and lna6 (13mers) targeting the 3′-splice site upstream of exon 4. Free cellular uptake by HeLa cells was investigated by fluorescence microscopy via direct addition of a high concentration (20 µM) of tc6 or lna6 to the cell culture. For both backbone systems, we found no significant fluorescence accumulation in the cytosol or in the nucleus after 24 h of incubation. Only a high background fluorescence associated with the cell membrane was observed, indicating that no significant internalization occurred. A FACS analysis revealed a final free oligonucleotide uptake of only 3% in the case of tc6 and 0% for lna6.
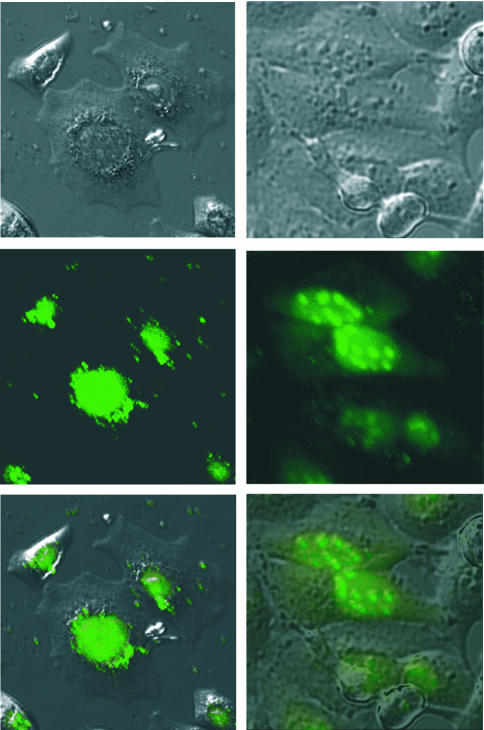
In a second experiment, we investigated the cellular distribution of tc6 and lna6 24 h after lipofection at a concentration of 0.2 µM (Fig. 8). We found some differences in the cellular distribution of the two oligonucleotide analogues. Tc-DNA (Fig. 8, left) is almost exclusively accumulated in the nucleus and virtually absent in the cytosol. Within the nucleus, tc-DNA seems to be homogeneously distributed with no obvious compartmentalization. The situation is somewhat different in the case of LNA (Fig. 8, right). Also here, most of the oligonucleotide is localized in the nucleus, although some fluorescence still remains in the cytosol. Within the nucleus, LNA is unevenly distributed and preferentially enriched in small bodies, most likely the nucleoli.
Figure 8.
Fluorescence microscopic images of the cellular distribution of tc6 (left) and lna6 (right) in HeLa cells (top: phase contrast image; center: UV image; and bottom: overlay of both images).
DISCUSSION
Tc-DNA and LNA both belong to the class of conformationally constrained DNA analogues. They share many common features, such as the anionic nature of the backbone, increased affinity for RNA targets, overall A-like conformation in duplexes with RNA, inability to activate RNaseH and increased biostability (18,19,25). There are, however, differences in the relative dimensions of some of these features that translate into different biological properties. For example, the average increase in Tm of a modified unit in RNA target recognition relative to a DNA unit is between 4 and 7°C in the case of LNA (17) and between 2 and 3°C in the case of tc-DNA (19). The higher RNA affinity of LNA relative to tc-DNA is corroborated by the observed differences in the thermal stabilities of LNA/RNA and tc-DNA/RNA duplexes, where Tm differences of ca. 2.5–3.0°C per nucleotide unit were found. Furthermore, tc-DNA possesses a sugar–phosphate backbone which is more hydrophobic due to the extended carbon framework of the sugar unit. This might affect cellular uptake and distribution. Moreover, there may also be differences as to the extent of biostability against nucleolytic activity of both backbone systems. In previous experiments with tc-DNA we found a remarkable stability of tc-DNA against snake venom phosphodiesterase (39) as well as in heat deactivated fetal calf serum (25).
Tc- and lna-oligonucleotides with a length of 15 nt, used in the submicromolar concentration range, can efficiently change RNA maturation and largely abolish the production of the associated protein in the cell. This goes along with no significant level of toxicity (measured in terms of cell number) or alteration of cell morphology upon transfection. Furthermore, we found that all antisense effects observed were sequence-dependent, and thus true antisense effects. This was demonstrated with the double mismatch oligonucleotides which showed largely reduced antisense efficacy, and with a nonsense 18mer tc-DNA which showed no antisense effect at all. Remarkably, the differential increase in full-length RNA production between tc4 and tc5 is higher (Δ = 70%) than in the pair lna4 and lna5 (Δ = 45%). This indicates a slightly higher selectivity of the tc-system in the antisense effect, compared to LNA. With a sequence length of 15, genome-wide statistical selectivity on the level of RNA is unlikely to be compromised. Thus, strong binding oligonucleotide analogues such as tc-DNA or LNA enable a reduction in the size of antisense oligonucleotides, which for typical applications are 20–25 nt in length. This could potentially reduce costs and material load and increase selectivity.
According to the RT–PCR assays, tc-DNA shows increased nuclear antisense effects by a factor of 4–5 or more, compared to LNA. This contrasts with RNA target affinity where LNA is superior to tc-DNA. Thus there is an inverse affinity/antisense activity relation between the two systems. Differences in the antisense response could in principle arise from different cellular distribution patterns of the oligonucleotide systems. LNA was previously found to occur in all compartments of HeLa and other cells (20,24). In our experiments, we observed some differences in the cellular distribution pattern of tc-DNA and LNA. tc-DNA showed a higher propensity for nuclear localization and a more homogeneous distribution within the nucleus as compared to LNA (Fig. 8). This could be one reason for the higher antisense efficacy of the former system. Along the same lines, the observed accumulation of LNA presumably in the nucleoli, the site of assembly of the ribosomes, may competitively reduce its binding to the spliceosome.
As a note of care, we add that the observed cellular distribution could also be a property of the lipofectamine/oligonucleotide complex and not a property of the oligonucleotidic system alone. Thus we cannot exclude that the distribution pattern changes when the nature of the cationic lipid is varied. It was shown before in HeLa cells that 2′-OMe and 2′-MOE-oligonucleotides have higher nuclear accumulation when transfected with lipofectAMINE compared to when scrape loaded (16). Also, we cannot exclude the possibility that other effects, such as higher nuclease stability of tc-DNA, also contribute to the observed higher efficacy of tc-DNA.
The fact that no significant uptake was found for tc-oligonucleotides applied at high doses but in the absence of transfecting agents is in line with observations on 2′-O-alkyl RNAs and also for LNA (24). However, we showed previously that small amounts of tc-DNA, sufficient to give weak antisense effects, may still enter the cell under these conditions (25). Nevertheless the increased hydrophobicity of the tc-DNA backbone does not significantly increase passive penetration into cells. Efficient membrane passage is therefore still a privilege of charge neutral DNA analogues such as morpholino-DNA or cationic PNAs (16).
Interestingly, not only exon 4 is skipped, but also exon 3+4 double skipping occurs in spite of the fact that the oligonucleotides were designed to bind exclusively to the splice sites flanking exon 4. This fact seems to be neither dependent on the oligonucleotide type used, as it occurs with both LNA- and tc-oligonucleotides, nor is it dependent on the length of the oligonucleotides. A sequence alignment revealed no further binding site with nine or more matched base pairs in the CyPA pre-mRNA (we note explicitly that neither of the matched 9mers lna1 or tc1 showed a significant antisense effect). Furthermore, there is no difference in the splicing pattern whether the 5′- or the 3′-splice site is targeted. Using antisense U7 snRNAs targeting either exon 4 or exon 3 of CyPA, the same phenomenon also occurs (D.Schümperli and co-workers, manuscript in preparation). Multiple exon skipping was already observed before with antisense oligonucleotides or U7 snRNAs targeting exon 23 splice sites of the mouse dystrophin gene (40,41). This implies that certain groups of exons may be recognized by the splicing machinery in a concerted fashion so that they tend to be coordinately included in or excluded from the mRNA. However, the mechanism for this is still unknown.
In summary, we have shown that tc-oligonucleotide 11– 15 mers, targeted to the 5′-splice site of exon 4 efficiently change the splice pattern of CyPA pre-mRNA. A direct comparison with the analogous LNA-oligonucleotides revealed a higher antisense efficacy of tc-DNA by at least a factor of 4–5. Thus, tc-DNA emerges as a very powerful nuclear antisense system. Whether tc-DNA is also useful for cytosolic antisense applications has still to be shown. Investigations in this direction with tc-DNA and tc-DNA–DNA/tcDNA gapmers are currently under way in our laboratories.
Acknowledgments
ACKNOWLEDGEMENTS
D.R. and C.J.L. gratefully acknowledge financial support from the Swiss National Science Foundation (grant No. 200020–100178). Work in the laboratory of D.S. was supported by the Swiss AIDS Research Program (grant no. 3339-63429).
REFERENCES
- 1.Vacek M., Sazani,P. and Kole,R. (2003) Antisense-mediated redirection of mRNA splicing. Cell. Mol. Life Sci., 60, 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurreck J. (2003) Antisense technologies. Improvement through novel chemical modifications. Eur. J. Biochem., 270, 1628–1644. [DOI] [PubMed] [Google Scholar]
- 3.Friedman K.J., Kole,J., Cohn,J.A., Knowles,M.R., Silverman,L.M. and Kole,R. (1999) Correction of aberrant splicing of the cystic fibrosis transmembrane conductance regulator (CFTR) gene by antisense oligonucleotides. J. Biol. Chem., 274, 36193–36199. [DOI] [PubMed] [Google Scholar]
- 4.Karras J.G., McKay,R.A., Dean,N.M. and Monia,B.P. (2000) Deletion of individual exons and induction of soluble murine interleukin-5 receptor-alpha chain expression through antisense oligonucleotide-mediated redirection of pre-mRNA splicing. Mol. Pharmacol., 58, 380–387. [DOI] [PubMed] [Google Scholar]
- 5.Giles R.V., Spiller,D.G., Clark,R.E. and Tidd,D.M. (1999) Antisense morpholino oligonucleotide analog induces missplicing of C-myc mRNA. Antisense Nucleic Acid Drug Dev., 9, 213–220. [DOI] [PubMed] [Google Scholar]
- 6.Kalbfuss B., Mabon,S.A. and Misteli,T. (2001) Correction of alternative splicing of tau in frontotemporal dementia and parkinsonism linked to chromosome 17. J. Biol. Chem., 276, 42986–42993. [DOI] [PubMed] [Google Scholar]
- 7.Lim S.R. and Hertel,K.J. (2001) Modulation of survival motor neuron pre-mRNA splicing by inhibition of alternative 3′ splice site pairing. J. Biol. Chem., 276, 45476–45483. [DOI] [PubMed] [Google Scholar]
- 8.Mercatante D.R., Mohler,J.L. and Kole,R. (2002) Cellular response to an antisense-mediated shift of Bcl-x pre-mRNA splicing and antineoplastic agents. J. Biol. Chem., 277, 49374–49382. [DOI] [PubMed] [Google Scholar]
- 9.Taylor J.K., Zhang,Q.Q., Wyatt,J.R. and Dean,N.M. (1999) Induction of endogenous Bcl-xS through the control of Bcl-x pre-mRNA splicing by antisense oligonucleotides. Nat. Biotechnol., 17, 1097–1100. [DOI] [PubMed] [Google Scholar]
- 10.Mercatante D.R., Bortner,C.D., Cidlowski,J.A. and Kole,R. (2001) Modification of alternative splicing of Bcl-x pre-mRNA in prostate and breast cancer cells. Analysis of apoptosis and cell death. J. Biol. Chem., 276, 16411–16417. [DOI] [PubMed] [Google Scholar]
- 11.Lacerra G., Sierakowska,H., Carestia,C., Fucharoen,S., Summerton,J., Weller,D. and Kole,R. (2000) Restoration of hemoglobin A synthesis in erythroid cells from peripheral blood of thalassemic patients. Proc. Natl Acad. Sci. USA, 97, 9591–9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suwanmanee T., Sierakowska,H., Lacerra,G., Svasti,S., Kirby,S., Walsh,C.E., Fucharoen,S. and Kole,R. (2002) Restoration of human beta-globin gene expression in murine and human IVS2-654 thalassemic erythroid cells by free uptake of antisense oligonucleotides. Mol. Pharmacol., 62, 545–553. [DOI] [PubMed] [Google Scholar]
- 13.Errington S.J., Mann,C.J., Fletcher,S. and Wilton,S.D. (2003) Target selection for antisense oligonucleotide induced exon skipping in the dystrophin gene. J. Gene Med., 5, 518–527. [DOI] [PubMed] [Google Scholar]
- 14.Pramono Z.A., Takeshima,Y., Alimsardjono,H., Ishii,A., Takeda,S. and Matsuo,M. (1996) Induction of exon skipping of the dystrophin transcript in lymphoblastoid cells by transfecting an antisense oligodeoxynucleotide complementary to an exon recognition sequence. Biochem. Biophys. Res. Commun., 226, 445–449. [DOI] [PubMed] [Google Scholar]
- 15.Gebski B.L., Mann,C.J., Fletcher,S. and Wilton,S.D. (2003) Morpholino antisense oligonucleotide induced dystrophin exon 23 skipping in mdx mouse muscle. Hum. Mol. Genet., 12, 1801–1811. [DOI] [PubMed] [Google Scholar]
- 16.Sazani P., Kang,S.H., Maier,M.A., Wei,C., Dillman,J., Summerton,J., Manoharan,M. and Kole,R. (2001) Nuclear antisense effects of neutral, anionic and cationic oligonucleotide analogs. Nucleic Acids Res., 29, 3965–3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imanishi T. and Obika,S. (2002) BNAs: novel nucleic acid analogs with a bridged sugar moiety. Chem. Commun., 1653–1659. [DOI] [PubMed] [Google Scholar]
- 18.Petersen M. and Wengel,J. (2003) LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol., 21, 74–81. [DOI] [PubMed] [Google Scholar]
- 19.Renneberg D. and Leumann,C.J. (2002) Watson–Crick base-pairing properties of tricyclo-DNA. J. Am. Chem. Soc., 124, 5993–6002. [DOI] [PubMed] [Google Scholar]
- 20.Wahlestedt C., Salmi,P., Good,L., Kela,J., Johnsson,T., Hökfelt,T., Broberger,C., Porreca,F., Lai,J., Ren,K. et al. (2000) Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl Acad. Sci. USA, 97, 5633–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arzumanov A., Walsh,A.P., Rajwanshi,V.K., Kumar,R., Wengel,J. and Gait,M.J. (2001) Inhibition of HIV-1 Tat-dependent trans activation by steric block chimeric 2′-O-methyl/LNA oligoribonucleotides. Biochemistry, 40, 14645–14654. [DOI] [PubMed] [Google Scholar]
- 22.Grunweller A., Wyszko,E., Bieber,B., Jahnel,R., Erdmann,V.A. and Kurreck,J. (2003) Comparison of different antisense strategies in mammalian cells using locked nucleic acids, 2′-O-methyl RNA, phosphorothioates and small interfering RNA. Nucleic Acids Res., 31, 3185–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurreck J., Wyszko,E., Gillen,C. and Erdmann,V.A. (2002) Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res., 30, 1911–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elayadi A.N., Braasch,D.A. and Corey,D.R. (2002) Implications of high-affinity hybridization by locked nucleic acid oligomers for inhibition of human telomerase. Biochemistry, 41, 9973–9981. [DOI] [PubMed] [Google Scholar]
- 25.Renneberg D., Bouliong,E., Reber,U., Schümperli,D. and Leumann,C.J. (2002) Biological and antisense properties of tricyclo-DNA. Nucleic Acids Res., 30, 2751–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Handschumacher R.E., Harding,M.W., Rice,J., Drugge,R.J. and Speicher,D.W. (1984) Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science, 226, 544–547. [DOI] [PubMed] [Google Scholar]
- 27.Sykes K., Gething,M.J. and Sambrook,J. (1993) Proline isomerases function during heat shock. Proc. Natl Acad. Sci. USA, 90, 5853–5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ansari H., Greco,G. and Luban,J. (2002) Cyclophilin A peptidyl-prolyl isomerase activity promotes ZPR1 nuclear export. Mol. Cell. Biol., 22, 6993–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brazin K.N., Mallis,R.J., Fulton,D.B. and Andreotti,A.H. (2002) Regulation of the tyrosine kinase Itk by the peptidyl-prolyl isomerase cyclophilin A. Proc. Natl Acad. Sci. USA, 99, 1899–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weisman R., Creanor,J. and Fantes,P. (1996) A multicopy suppressor of a cell cycle defect in S. pombe encodes a heat shock-inducible 40 kDa cyclophilin-like protein. EMBO J., 15, 447–456. [PMC free article] [PubMed] [Google Scholar]
- 31.Braaten D., Franke,E.K. and Luban,J. (1996) Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J. Virol., 70, 3551–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franke E.K., Yuan,H.E. and Luban,J. (1994) Specific incorporation of cyclophilin A into HIV-1 virions. Nature, 372, 359–362. [DOI] [PubMed] [Google Scholar]
- 33.Gamble T.R., Vajdos,F.F., Yoo,S., Worthylake,D.K., Houseweart,M., Sundquist,W.I. and Hill,C.P. (1996) Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell, 87, 1285–1294. [DOI] [PubMed] [Google Scholar]
- 34.Thali M., Bukovsky,A., Kondo,E., Rosenwirth,B., Walsh,C.T., Sodroski,J. and Gottlinger,H.G. (1994) Functional association of cyclophilin A with HIV-1 virions. Nature, 372, 363–365. [DOI] [PubMed] [Google Scholar]
- 35.Steinkasserer A., Harrison,R., Billich,A., Hammerschmid,F., Werner,G., Wolff,B., Peichl,P., Palfi,G., Schnitzel,W., Mlynar,E. et al. (1995) Mode of action of SDZ NIM 811, a nonimmunosuppressive cyclosporin A analog with activity against human immunodeficiency virus type 1 (HIV-1): interference with early and late events in HIV-1 replication. J. Virol., 69, 814–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saphire A.C., Bobardt,M.D. and Gallay,P.A. (2002) Cyclophilin a plays distinct roles in human immunodeficiency virus type 1 entry and postentry events, as revealed by spinoculation. J. Virol., 76, 4671–4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saphire A.C., Bobardt,M.D. and Gallay,P.A. (1999) Host cyclophilin A mediates HIV-1 attachment to target cells via heparans. EMBO J., 18, 6771–6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luban J. (1996) Absconding with the chaperone: essential cyclophilin-Gag interaction in HIV-1 virions. Cell, 87, 1157–1159. [DOI] [PubMed] [Google Scholar]
- 39.Steffens R. and Leumann,C.J. (1999) Synthesis and thermodynamic and biophysical properties of tricyclo-DNA. J. Am. Chem. Soc., 121, 3249–3255. [Google Scholar]
- 40.Dunckley M.G., Manoharan,M., Villiet,P., Eperon,I.C. and Dickson,G. (1998) Modification of splicing in the dystrophin gene in cultured Mdx muscle cells by antisense oligoribonucleotides. Hum. Mol. Genet., 7, 1083–1090. [DOI] [PubMed] [Google Scholar]
- 41.Brun C., Suter,D., Pauli,C., Dunant,P., Lochmüller,H., Burgunder,J.M., Schümperli,D. and Weis,J. (2003) U7 snRNAs induce correction of mutated dystrophin pre-mRNA by exon skipping. Cell. Mol. Life Sci., 60, 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]