Abstract
Sixty percent of the world ocean by area is contained in oligotrophic gyres [Longhurst A (1995) Prog Oceanog 36:77–16], the biomass of which is dominated by picophytoplankton, including cyanobacteria and picoeukaryotic algae, as well as picoheterotrophs. Despite their recognized importance in carbon cycling in the surface ocean, the role of small cells and their detrital remains in the transfer of particulate organic matter (POM) to the deep ocean remains disputed. Because oligotrophic marine conditions are projected to expand under current climate trends, a better understanding of the role of small particles in the global carbon cycle is a timely goal. Here we use the lipid profiles, radiocarbon, and stable carbon isotopic signatures of lipids from the North Pacific Subtropical Gyre to show that in the surface ocean, lipids from submicron POM (here called extra-small POM) are distinct from larger classes of suspended POM. Remarkably, this distinct extra-small POM signature dominates the total lipids collected at mesopelagic depth, suggesting that the lipid component of mesopelagic POM primarily contains the exported remains of small particles. Transfer of submicron material to mesopelagic depths in this location is consistent with model results that claim the biological origin of exported carbon should be proportional to the distribution of cell types in the surface community, irrespective of cell size [Richardson TL, Jackson GA (2007) Science 315:838–840]. Our data suggest that the submicron component of exported POM is an important contributor to the global biological pump, especially in oligotrophic waters.
Keywords: biogeochemistry, biomarkers, oceanography, carbon isotopes
Picoplankton are Bacteria, Archaea, and Eukarya smaller than 2–3 µm in diameter (1–3). Whereas picoplankton biomass constitutes a majority of the unicellular particulate organic matter (POM) in oligotrophic waters, its role in export is poorly known (4–8), because up to 40–70% of these cells are small enough to escape detection under the most common definition of suspended POM (9, 10). In the majority of carbon flux studies, suspended POM is defined operationally by using filters with a 0.7-µm or greater pore size. By excluding most submicron material [extra-small POM (X-POM)], such methods miss this component of the standing stock of POM as well as its contribution to the export flux (for further discussion, see ref. 11). X-POM has long been recognized as comprising >20% of total POM (12, 13), and accordingly many studies of POM bulk molecular classes have included particles as small as 0.1–0.2 µm in diameter (14–18). In contrast, neglecting submicron particles in flux studies often is considered to be insignificant (11), because picoplankton cells should not sink passively due to their small size. Consequently, picoplanktonic remains are generalized as contributing little to particle cycling and sequestration of CO2 in the deep ocean (8, 19). However, new understanding of aggregation–disaggregation processes (20) raises the prospect that submicron particulate biomass may enter the mesopelagic ocean more readily than expected. Such processes would be important additions to the recognized pathways for small particle export via fecal pellets or mesoplankton feeding structures, because self-aggregation minimizes the codependence of export on large cells (20, 21). Thus, both bacterivory and physical aggregation may be routes for the transfer of very small cells to the ocean’s interior.
Several recent studies quantifying the specific role of picoplankton in export have focused on autotrophs, either by tracing pigments through the water column (6) or by measuring the 15N content of taxonomically sorted cells to model the relative contributions of Cyanobacteria and small Eukarya (7). However, because X-POM also includes heterotrophic biomass and detritus, studies of autotrophs do not scale proportionally to total carbon export. Other studies have quantified the contribution of both heterotrophic and autotrophic bacterial biomass to mesopelagic POM by isolating specific biomarkers (d-amino acids) (15–18). In such work, the fraction of submicron POM at a depth that is specifically due to surface-derived export is difficult to quantify. Thus, more work is needed to establish a general picture of the sources of exported POM. Such a model would include surface and deep-sourced biomass, heterotrophic and autotrophic metabolisms, and prokaryotic and eukaryotic cells. Here we begin to tackle this problem by examining carbon isotopic signatures of fatty acid profiles of POM.
Capturing and Characterizing “X-POM”: Approach and Results
POM was collected from the oligotrophic North Pacific Subtropical Gyre (NPSG). The oceanographic environment of the NPSG is well characterized in association with the Hawaii Ocean Time-series (HOT) (22). Using sequential filtration of surface waters (21 m), we obtained a >0.5-µm (suspended plus sinking POM) size class and also isolated the very smallest fraction (0.2–0.5 µm or X-POM) (23). At 670 m we captured the total POM >0.2 µm, which includes both X-POM and typical suspended plus sinking POM. From all samples we characterized the fatty acid distributions of these size fractions, along with the compound-specific δ13C and Δ14C values of the fatty acids. Fatty acids are ubiquitous in Bacteria and Eukarya and thus derive from the majority of biological sources contributing to POM (excluding Archaea). Sampling and lipid extraction techniques, and data analysis, are described in ref. 23 and in SI Text 1 and Figs. S1 and S2.
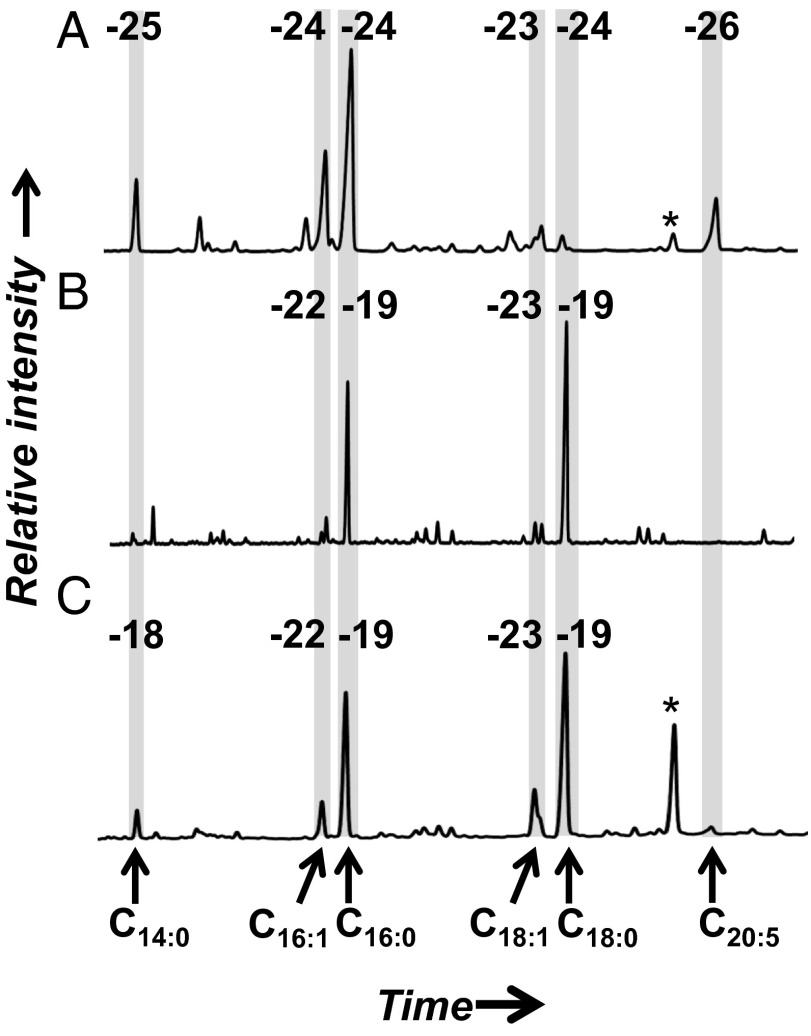
The profile of total fatty acids from the >0.5-µm size class at 21 m is typical of the NPSG mixed phytoplankton community captured by glass microfiber grade GF/F (0.7-μm nominal pore size) filtration (24): a dominance of C14:0, C16:0, C16:1, and C18-chain lengths with one to three unsaturations (Fig. 1A). The measured values of δ13C (−23‰ to −26‰) also are consistent with values reported elsewhere for marine planktonic lipids from the same size class of POM (25). In contrast, the fatty acid profile of 0.2- to 0.5-μm X-POM is markedly different: There is a prominent C18:0 peak, with slightly less C16:0, and all other compounds are significantly lower in abundance (Fig. 1B). All saturated, even-chain-length compounds in this size class are 3–4‰ enriched in 13C (−18‰ to −19‰), compared with the unsaturated compounds (−22‰ to −25‰). The mesopelagic sample (670 m depth; Fig. 1C), which includes total POM >0.2 μm, has a fatty acid and 13C profile remarkably similar to the surface-derived 0.2- to 0.5-μm X-POM size fraction. All compounds measured from all samples have natural 14C contents consistent with a carbon source deriving from surface waters (Δ14C > 0‰; Table 1, Table S1, and Figs. S1 and S2).
Fig. 1.
Gas chromatograms and δ13C values (‰) of fatty acids. (A) Surface (21 m) particulate organic matter >0.5 μm. (B) Surface (21 m) particulate organic matter, 0.2–0.5 μm. (C) Deep (670 m) particulate organic matter >0.2 μm. Each peak represents an individual compound; chromatograms are aligned according to the shaded boxes, compounds are identified at the bottom, and values of δ13C are identified at the top. The peak area is equivalent to relative abundance of each compound. Peaks marked with * are an added C19:0 internal standard.
Table 1.
Relative abundance and isotopic data for fatty acids from the NPSG
| Deep, >0.2 μm |
Surface, 0.2–0.5 μm |
Surface, >0.5 μm |
|||||||
| Compound | Fraction | δ13C ± 1σ | ∆14C* ± 1σ | Fraction | δ13C ± 1σ | ∆14C* ± 1σ | Fraction | δ13C ± 1σ | ∆14C* ± 1σ |
| 14:0 | 0.18 | −18.3 ± 0.2 | 114 ± 76 | 0.17 | —† | −201 ± 261‡ | 0.22 | −25.3 ± 0.2 | — |
| i-15:0 | — | — | — | — | — | — | 0.13 | −22.7 ± 0.3 | 48 ± 14 |
| 16:1 | 0.29 | −21.7 ± 0.2 | 66 ± 47 | 0.13 | −22.4 ± 0.2 | — | 0.51 | −23.8 ± 0.1 | — |
| 16:0 | 1.00 | −18.8 ± 0.2 | 57 ± 13 | 1.00 | −19.0 ± 0.2 | 86 ± 28 | 1.00 | −23.6 ± 0.2 | 51 ± 5 |
| 17:1 | 0.15 | −25.0 ± 0.2 | — | 0.12 | −22.8 ± 0.7 | — | — | — | — |
| 17:0 | 0.09 | −21.5 ± 0.2 | 57 ± 74 | 0.07 | — | 86 ± 112 | 0.03 | — | — |
| 18:1 | 0.52 | −22.7 ± 0.2 | 58 ± 25 | 0.11 | −23.3 ± 0.3 | −2 ± 58 | 0.14 | −22.8 ± 1.2 | 52 ± 10 |
| 18:0 | 1.48 | −18.5 ± 0.2 | 80 ± 11 | 1.78 | −18.8 ± 0.2 | — | 0.05 | −23.7 ± 1.5 | 93 ± 23 |
| 20:5 | — | — | — | — | — | — | 0.27 | −25.7 ± 0.9 | — |
| 22:0 | — | — | — | — | — | — | 0.03 | — | −201 ± 24‡ |
For details of isotopic measurements and background corrections, as well as AMS facility sample identifiers, see expanded Table S1, and Figs. S1 and S2.
Final value after all blank corrections (SI Text 1).
No value indicates insufficient abundance for measurement.
Samples in italics were eliminated due to small size and/or unexplained contaminants.
Additionally, the community captured on each filter was characterized by bacterial and archaeal cell counts and DNA community profiling (PhyloChip hybridization of DNA amplicons of 16S ribosomal RNA genes) (26). We confirmed the similarity of our samples to annual averages (27), using fluorescent catalyzed reporter deposition in situ hybridization (CARD-FISH) with probes EUB338 and ARC915, using methods from ref. 28 and the permeabilization method specific for archaeal cells from ref. 29. The proportion of bacterial cells in the surface was 85% and that in the mesopelagic was 57%—both values are within 1 SD of the annual average data from ref. 27. Our PhyloChip results further show that surface POM contained abundant Cyanobacteria, SAR-11, SAR-86, and other Alpha- and Gammaproteobacteria. The mesopelagic (670 m) sample was rich in MG-A, Epsilon- and Gammaproteobacteria, and Oceanospiralles (SI Text 2 and Fig. S3). Such patterns of community organization in the NPSG are well established (30).
A Lipid and Isotope Balance Model for Sources of POM to the Mesopelagic Ocean
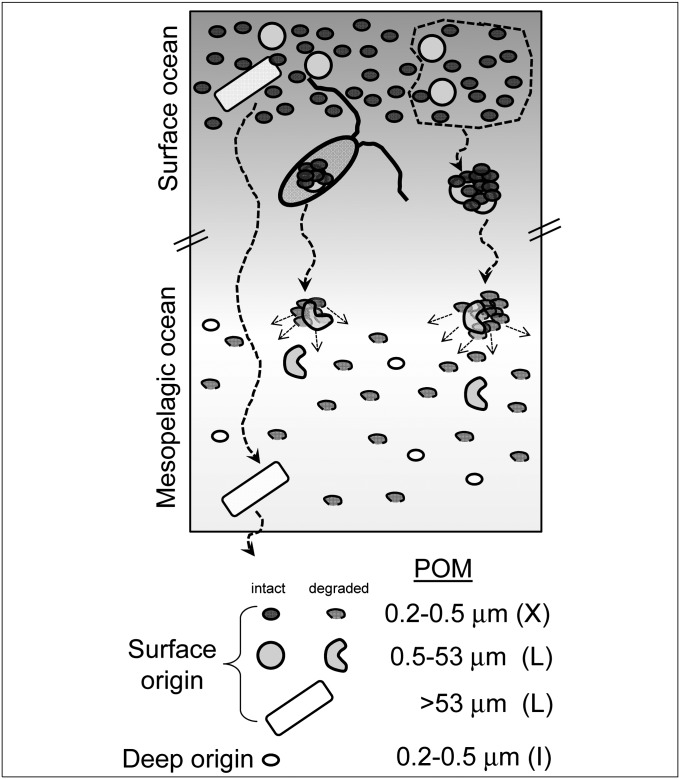
Because the lipids in the mesopelagic sample (670 m) have a surface-water 14C signature, this material could originate solely from the direct sinking of freshly synthesized POM from surface waters. A conventional interpretation involving export of large particles is problematic, however, because the total fatty acid profile at 670 m does not resemble the fatty acid profile of the larger particles obtained from 21 m (POM > 0.5 μm; Fig. 1A). Instead, the profile at 670 m qualitatively resembles the X-POM fraction at 21 m. An alternate interpretation is that aggregation, sinking, and disaggregation of the total pool of surface-derived POM transfers carbon to mesopelagic depths, regardless of the original particle size (4), and that much of this exported material was originally X-POM (Fig. 2).
Fig. 2.
Cartoon of hypothesized particle export in the NPSG, an environment in which X-POM is isotopically and compositionally distinct from other suspended POM. Total POM is packaged through aggregation and grazing, thereby contributing to sinking POM regardless of the original particle size. Disaggregation and lysis injects this material into the mesopelagic, where it provides metabolic substrates for the in situ prokaryotic community.
To specifically address the contribution of submicron X-POM to exported lipids, we model the mesopelagic—or deep (D)—lipid and isotopic content as a mixture of surface large POM (L-POM) (L, >0.5 μm), surface X-POM (X, 0.2–0.5 μm), and in situ mesopelagic biomass (I). We construct a mixing model based on the five major fatty acids present in these samples (C14:0, C16:1, C16:0, C18:1, and C18:0). The model is developed in three parts, two of which depend only on the data presented here and one of which incorporates these new data with our previous results (23, 31).
The model assumes that the relative proportions of fatty acids in POM are controlled by source inputs and not by differential degradation in the water column. It also assumes that the compound and isotopic distributions represent a steady-state signature of NPSG plankton. The latter assumption may be valid, because the magnitude of production in the NPSG is weakly seasonal, and our surface samples were collected within the depth zone of highest primary productivity (e.g., ref. 32). We thus consider that lipid signatures in our surface sample likely represent average lipids exported out of the euphotic zone. The first assumption, that planktonic fatty acid end members retain their characteristic profiles, is supported by work in lakes, estuaries, and the ocean (e.g., refs. 33–35). Here it also is substantiated by the specific finding that the proportion of C18:0 lipid increases dramatically with depth. Although unsaturated C18:1 and C18:2 as well as >C18-carbon fatty acids could degrade to yield C18:0, these potential precursors account for only ∼20% of fatty acids in the L fraction, suggesting it would be difficult to explain the relative increase in C18:0 via degradative transformation of surface L lipids. In addition, although unsaturated fatty acids are known to degrade more quickly than saturated forms (e.g., ref. 35), the overall loss rate constants for both forms are of the same order (36), making it difficult to greatly skew the C18:0 abundance solely through selective loss of other compounds.
1. The Minimum Lipid Contribution from in Situ Mesopelagic Bacteria (I): Lipid Profiles.
First, we calculate a best-fit mixture of the two surface fractions to predict the small:large particle export ratio for the case where mesopelagic POM would derive maximally from surface material (X + L) and minimally from I.; i.e., we test the ability of X + L to mimic the total mesopelagic profile, D. All possible mixing ratios between fatty acid profiles for surface large POM (L, >0.5 μm) and X-POM (X, 0.2–0.5 μm) size classes were calculated (0–100% of each end member, stepping by 0.2%). The relative abundance of each fatty acid (i) in the mixture M is
where
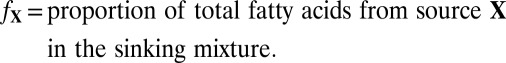 |
The modeled mixture (M) was optimized to mimic the actual deep profile (D) (Fig. 1C). The resulting best-fit estimate shows that if all mesopelagic fatty acids originated from the surface, 89% would be sourced from X and only 11% from L (for details, see SI Text 3 and Fig. S4).
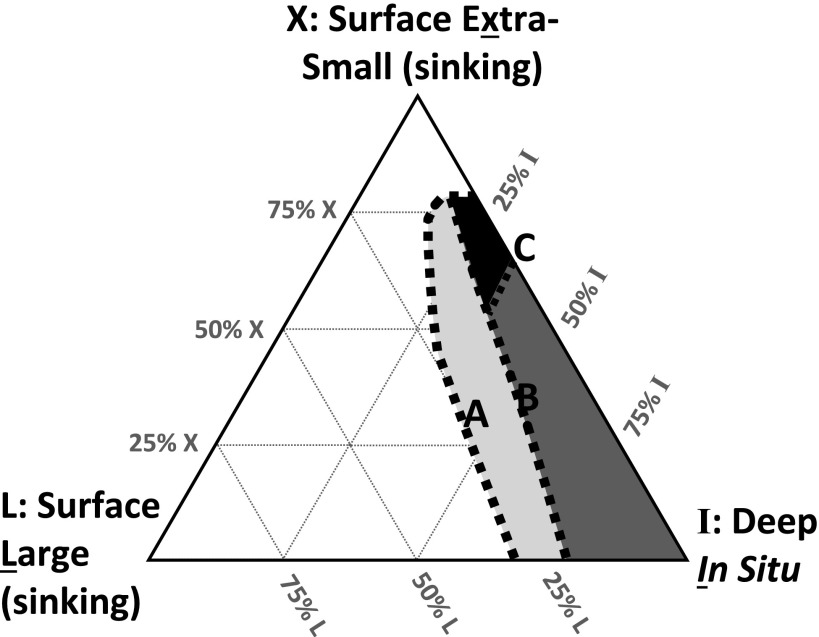
However, lipids from the mesopelagic sample (D) also must contain at least some material produced by the in situ bacterial community at 670 m (I). Using our mixing model, we calculated the minimum proportion of the deep fatty acid profile that remained unexplained; i.e., we computed the residuals between the model, X + L = M, and the observation, D. This calculation suggests that if (X + L) + I = D, the in situ component (I) must contribute at minimum 14% of the total fatty acids. Alternatively, the fatty acids contributed by I could mimic a surface-derived signature even if they did not represent truly “exported” carbon. This could occur if active mesopelagic cells synthesize their lipids de novo from fresh POM delivered from surface waters and coincidentally generate lipid and δ13C profiles that are similar to those of the surface-derived X end member. If so, up to 100% of the mesopelagic sample could represent in situ bacterial consumers living on modern carbon. Thus, the widest allowed boundaries on I are 14–100% (Fig. 3, line A).
Fig. 3.
Ternary diagram showing modeled origin of mesopelagic (670 m) fatty acids as a mixture of three end members: sinking POM from the submicron (0.2–0.5 μm) X-POM size class (X) in the surface ocean (21 m), sinking POM from the larger (>0.5 μm) size class (L) in the surface ocean (21 m), and POM produced in situ (I) in the mesopelagic ocean (670 m). The shaded regions represent the solution spaces determined by models based on lipid profiles and compound-specific natural 13C and 14C content. The area with light shading defines the solution space allowed by a mixing model based on fatty acid chromatograms only; the boundaries of this space (A) are equivalent to line A in Fig. S5. The region with dark shading includes a constraint on the upper limit for the contribution from large surface particles, based on the 13C content of fatty acids; the boundary of this upper limit (B) is equivalent to line B in Fig. S5. The solid area is constrained further by the 14C content of mesopelagic DNA (31) and fatty acids; the boundary of this space (C) is equivalent to line C in Fig. S5.
2. Boundaries on Lipid Export from Surface L-POM (L) vs. X-POM (X): 13C-Constraint.
To estimate the magnitude of the end-members X and L, we use a compound-specific, isotope mass-balance model based on 13C. The solution is governed primarily by the presence of 13C-enriched fatty acids in the X-POM fraction (Fig. 1B) and the repetition of this pattern in the mesopelagic sample (Fig. 1C). This distinctive 13C-enriched signature argues against the fatty acids of deep (670 m) POM being primarily derived from disaggregation and/or modification of exported larger particles (L) from the surface ocean. The fatty acid profile and 13C signature from the surface large size class lipids (Fig. 1A) are both different from those of the mesopelagic material (Fig. 1C).
To determine the boundaries for the contribution of L to the mesopelagic sample, we include isotope mass balance, where δ = δ13C value of fatty acid “i”:
Using Eqs. 1–3, we can solve for the proportion and δ13C value of each fatty acid from the in situ community (χI,i and δI,i, respectively) across the allowable range of M (via its components X and L) (SI Text 4 and Fig. S5). The solution field for each in situ component (χI,i δI,i) contains all values that permit isotopic mass balance with the deep sample (χD,i, δD,i) within the measurement errors of the data for δXi, δLi, and δDi. We further constrain the modeled values of δI,i to a maximum value of −16‰ or no more than 2.5‰ more positive than the highest measured value of δ in the entire system, based on the argument of limited trophic-level enrichment of 13C in bacterial heterotrophy (37, 38)—thus accounting for the potential effects of heterotrophy on the observed signatures, but placing these effects within reasonable limits. The result of this isotope constraint is that L contributes a maximum of 23% in the three-component mixture (X + L) + I = D (Fig. 3, line B).
3. Further Constraint on the Lipid Contribution from in Situ Mesopelagic Bacteria (I): Radiocarbon.
Next we calculate the expected 14C signature of in situ Bacteria and use this value to further constrain their maximum contribution (I) to total mesopelagic fatty acids (D). Radiocarbon measurements from DNA at this location indicate that the integrated mesopelagic community uses some “aged”, or subsurface, carbon (31); however, these DNA measurements include contributions from Archaea, which are not a source of fatty acids. We therefore used isotope mass balance to remove the archaeal component from the total mesopelagic ∆14CDNA values, leading to a predicted value of ∆14CI, i.e., the value of ∆14C that is specific to in situ Bacteria. The end-member ∆14C value for Archaea at this depth and location is −112‰ (23). Using the entire error-bounded range of mesopelagic total ∆14CDNA (−157‰ to −69‰) (31) and the Bacteria:Archaea cell counts we measured (and verified against ref. 27), we calculate ∆14CI = −191‰ to −37‰ for in situ mesopelagic Bacteria (Table S2 and SI Text 5 and SI Text 6). This conservative approach places a broad window on the Δ14CI value of the in situ bacterial community. The optimized mixing of fatty acid profiles (section 1) suggests that at least 14% of total fatty acids in the mesopelagic (D) must derive from the in situ mesopelagic component (I). Using the compound-specific Δ14C values (Table 1), we can calculate that such a contribution (≥14%) from I is compatible with our estimate for Δ14CI. The abundance-weighted Δ14C value for total fatty acids at 670 m is 68 ± 34‰ (aggregate value for D; Table 1). Biomass sinking from the surface (X + L) should carry the Δ14C value of surface dissolved inorganic carbon (DIC) (71 ± 3‰) (23). Although some of our surface Δ14C values are lower than 71‰, the error ranges reported for these values are large, and choosing 71‰ also yields the most conservative outcome (i.e., greatest potential for in situ contributions from I at 670 m). Regardless, it is difficult to solve an isotope mass balance between surface-derived material (X + L) and the in situ fraction (I) to yield the deep sample (D), because on average, the calculated proportion I is indistinguishable from zero. However, taking into consideration the full span of error ranges, the maximum allowed proportion of lipids from I could be up to 36%:
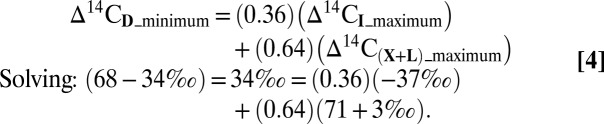 |
If the in situ contribution from Bacteria (I) contributes up to 36% of total deep fatty acids, then exported material (X + L) contributes ≥64% of the mesopelagic lipids. The boundary of this range is defined as line C in Fig. 3, and it constrains the earlier 14–100% range for I more narrowly to 14–36% (Fig. 3, Line C).
In sum, the boundaries on the sinking contribution from large particles (L < 23%) and on the in situ mesopelagic bacterial component (I ∼ 14–36%) suggest that X must be ≥50%. These calculations are largely independent—the first derives from fatty acid profiles and 13C mixing models, whereas the second derives from Δ14C measurements of bulk DNA, fatty acids, and archaeal lipids. Because of the large uncertainties in these calculations, we resist assigning consensus numbers. Instead, we suggest the data are consistent with contributions to mesopelagic fatty acids from surface extra-small particles > in situ mesopelagic sources ∼ surface larger particles, shown as the solid solution space defined in Fig. 3. All calculations remain consistent with the idea that exported X-POM (X) accounts for at least half of the total lipid recovered at 670 m (Fig. 2).
Model Validation: Comparison with Community Profiles and Estimates of Export Based on Cell Counts
As a check on the value of X, we estimate independently the proportion of mesopelagic lipids expected to derive specifically from exported Bacteria. By analyzing the same filter samples from the NPSG, Ingalls et al. (23) used natural 14C signatures to calculate that 14 ± 7% of archaeal lipids recovered at 670 m derive from export of surface-derived Archaea. If we assume that bacterial and archaeal cell debris aggregate (or are grazed) proportionally in the upper water column, an analogous value for the export of Bacteria can be calculated using Bacteria:Archaea cell ratios.
Cell ratios for the present calculation were taken from direct counts of our samples and from the literature (SI Text 5 and SI Text 6) (27). Archaeal cells in the mesopelagic NPSG average 2.26 × 104 cells/mL. Using the assumption that all Archaea have roughly the same cellular quota of lipid (39) and that the exported cells mainly are dead (i.e., their RNA is sufficiently degraded that they would not be counted by fluorescent in situ hybridization), the 14 ± 7% of surface-derived archaeal lipids are contributed without being counted as part of the deep in situ population. By this reasoning, the number of “cell equivalents” of archaeal lipids in the mesopelagic should be scaled up to 2.43–2.86 × 104 cells/mL, meaning the surface-derived contribution is equivalent to 1,700–6,000 cells/mL. Because there are 3.78 × 104 archaeal cells/mL in surface waters, this export represents 4.5–15.9% of the original concentration in the surface. Assuming that Bacteria are exported in similar proportions, we would expect that the equivalent also would be true for Bacteria: lipid equivalent to 4.5–15.9% of the surface bacterial cells would be exported to mesopelagic depths. In the NPSG, Bacteria average 3.07 × 105 cells/mL in surface waters, so lipids from 1.38 to 4.88 × 104 cells/mL would be expected to reach the mesopelagic. Compared with in situ mesopelagic bacterial cell counts of 3.01 × 104 cells/mL, this input of cell equivalents would constitute around half (31–62%) of the total recovered bacterial lipid at 670 m. Minor seasonal variations in cell numbers may place additional uncertainty on this range (e.g., ref. 32); but notably, our samples were not significantly different from the annual averages (SI Text 5) (27). The greater uncertainty comes from equating this 31–62% estimate to the parameter, X, because of the challenge of equating the particle size classes directly to cell counts (SI Text 7). Therefore, although this result is consistent with our other calculations, we do not use it to further constrain the final estimates shown in Fig. 3 and simply note that it is compatible with our model result of X ≥ 50%. Both the cell count approach and the radiocarbon portion of the mixing model (section 3), rest on empirical evidence that lipids and DNA are exported differently. To derive the Δ14C value of mesopelagic Bacteria in section 3, we use measurements of Δ14CDNA and assert that all nucleic acids represent local production; i.e., none survive within POM sinking from the surface ocean (SI Text 5). We thus imply that nucleic-acid–stained fluorescent cell counts will not detect surface-derived detrital cell debris, because this debris is a source of lipid but not of DNA. Our results from DNA community profiling by PhyloChip (26) as well as other data from the NPSG (27, 30) support this assertion. The DNA measured at 670 m is distinct from prokaryotic DNA in the surface ocean and thus likely represents primarily the in situ community (Fig. S3).
In addition, these sharp phylogenetic differences argue against an alternative hypothesis in which a community of particle-attached Bacteria produces uniform lipid and isotopic transformations throughout the surface and mesopelagic—the living communities in our samples are phylogenetically different. Alternatively, it remains possible that lipids from the living fractions of the surface and mesopelagic communities are coincidentally similar; this cannot be ruled out. However, we argue that the overall 13C and 14C similarity between surface X-POM lipids and mesopelagic lipids (Fig. 1 B and C) is evidence for a common, but largely detrital, source of lipids that can be exported to depth.
Discussion and Implications
Other data from the NPSG additionally support our conclusions. Roland et al. (40) observe that bulk POM >0.1 μm is ∼3.5‰ enriched in 13C relative to POM >0.7 μm, implying that it is specifically the X-POM fraction that carries a heavy 13C signature. Their results also indicate that the size-based isotopic distinction we observe in fatty acids is not lipid specific—it is reflected in the total POM as well as the lipids. Relatively light δ13C values for 0.1–60 µm POM previously were observed in the NPSG (14), but our data and those of Roland et al. (40) show that it is L-POM that contains the 13C-depleted fraction, whereas X-POM is rich in 13C. Additionally, within the POM >0.1 μm from 500 to 750 m depth in the NPSG, ≥75–80% of bacteria-specific d-amino acids and muramic acid are not associated with living bacterial cells, supporting the idea that the majority of mesopelagic POM is of (recent) detrital origin (18). Such results are consistent with other reports of abundant bacterial amino acids in X-POM (15–18). Inverse models of equatorial Pacific carbon cycling (4, 5) also indicate that significant prokaryotic biomass must be exported from the upper water column to mesopelagic depths. These findings are consistent with the hypothesis that much of the POM collected at depth is from detrital biomass of Bacteria, whether autotrophic or heterotrophic, and that surface export is a major source of this POM (Fig. 2).
Our data capture an export fingerprint and associated lipid signature of 13C-enriched carbon that may be missed by sediment trap measurements. Estimates of carbon export based on respiration (oxygen utilization) rates and nitrogen cycling (f-ratio) indicate that sediment traps undersample a significant source of exported organic matter to the pelagic ocean (11, 41). Additional evidence for a missing source of organic matter comes from the difference between particulate organic carbon (POC) concentrations obtained from sediment traps and those obtained by water column filtration (11, 42). POC flux models based on sediment trap data would predict a 90% decrease in POC concentration between surface waters and 650 m depth in the NPSG (43). In contrast, observations by Wakeham (24) for total filter-collected suspended material (≥0.7 µm) show there is only a 66–76% reduction in lipid and bulk POM concentration over this same depth. This highlights the challenge of comparing “sinking” vs. “suspended” carbon, and it could indicate that the different hydrodynamics of large and small particles may provide some explanation for the “missing” flux.
We suggest that continuous disaggregation and reaggregation (20) of POM during sinking could transfer X-POM to depth while subsequently veiling this material in the bulk properties of the sinking particle pool and/or obscuring its mass flux by sequestering it frequently in the disaggregated (suspended) state. The latter could be especially problematic if—due to its small (usually uncollected) size—the majority of the residence time of the transit was as small particles where this material would not be captured either by traps or by GF/F filtration (44–47) and therefore escape quantification altogether. Calculations of particle flux, disaggregation rates, and respiration rates may operate on different time constants, making it difficult to achieve accurate mass balance. Regardless, our data indicate that in the NPSG, larger particles are not the sources of most of the disaggregated material that exists in the mesopelagic as X-POM. Compound distributions and 13C and 14C isotopic signatures indicate that at mesopelagic depths, most of the standing stock of total POM >0.2 μm originated within the X-POM size class itself.
Operationally, most X-POM exists in a size-based “no-man’s land” (48): Typically it either is not collected or is sampled as part of “dissolved” organic matter (DOM). However, our observation that X-POM has a chemical signature distinct from typical suspended and sinking POM (0.7–53 µm) (24) and DOM (49) specifically suggests that it should be treated as a unique class of particulate matter. In particular, submicron cells should not be included in the DOM operational size class or assumed primarily to contribute recalcitrant (but dissolved) exudates (50), as X-POM should engage in particle surface reactions, and intact cells within X-POM are likely to be metabolically alive. To further elucidate carbon transformations in the water column it will be essential to sample a wide size range of organic material.
Our results support the idea that POM of picoplanktonic origin should be incorporated into global carbon flux models (4–6). Picoplanktonic X-POM likely is important fuel for mesopelagic metabolisms (44, 45). With picoplankton-dominated ecosystems projected to expand in a warming climate (2, 3), it will be vital to better understand the dynamics of extra-small POM in the water column.
Materials and Methods
Samples were obtained in 2005 at the Natural Energy Laboratory of Hawaii Authority (NELHA) seawater pipelines. The samples discussed here are different lipid fractions of the same filtered samples of biomass described previously (23). At that time, fractions containing lipids of Archaea were separated by SiO2-gel chromatography from fractions containing the fatty acids prepared as fatty acid methyl esters (FAMEs) (this work). Methods for the identification, separation, and isotopic analysis of these FAMEs are described in SI Text 1. Brief descriptions of our isotope mixing models are given in the main text, with detailed derivations and complete notation shown in SI Text S3–S7. PhyloChip (26) is described in SI Text 2.
Supplementary Material
Acknowledgments
We thank Tom Daniel, Barbara Lee, Jan War, and the staff of the Natural Energy Laboratory of Hawaii Authority for access to the sampling facility; Susan Carter and Katherine Goldfarb for laboratory assistance; Sheila Griffin for assistance with accelerator-mass spectrometry (AMS) preparation and analysis; Li Xu for assistance with preparative capillary gas chromatography; and John Southon, Tom Guilderson, Ann McNichol, and all of the staff members of the Keck Carbon Cycle AMS facility at the University of California, Irvine, Lawrence Livermore National Laboratories, and National Ocean Sciences Accelerator Mass Spectrometry accelerator facilities. Part of this work was performed at Lawrence Berkeley National Laboratory under the auspices of the University of California Department of Energy Contract DE-AC02-05CH11231. This work was supported by National Science Foundation Grants OCE-0241363 and OCE-0927290 (to A.P.) and OCE-0242160 (to L.I.A.) and by the National Aeronautics and Space Administration Astrobiology Institute and an ExxonMobil Geoscience grant (to H.G.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217514110/-/DCSupplemental.
References
- 1.Longhurst A. Seasonal cycles of pelagic production and consumption. Prog Oceanogr. 1995;36(2):77–167. [Google Scholar]
- 2.Karl D, Bidigare R, Letelier R. Long-term changes in plankton community structure and productivity in the North Pacific Subtropical Gyre: The domain shift hypothesis. Deep Sea Res Part II Top Stud Oceanogr. 2001;48:1449–1470. [Google Scholar]
- 3.Morán XAG, López-Urrutia Á, Calvo-Díaz A, Li WKW. Increasing importance of small phytoplankton in a warmer ocean. Glob Change Biol. 2010;16:1137–1144. doi: 10.1111/j.1365-2486.2009.01960.x. [DOI] [Google Scholar]
- 4.Richardson TL, Jackson GA. Small phytoplankton and carbon export from the surface ocean. Science. 2007;315(5813):838–840. doi: 10.1126/science.1133471. [DOI] [PubMed] [Google Scholar]
- 5.Stukel MR, Landry MR. Contribution of picophytoplankton to carbon export in the equatorial Pacific: A reassessment of food web flux inferences from inverse models. Limnol Oceanogr. 2010;55:2669–2685. doi: 10.4319/lo.2010.55.6.2669. [DOI] [Google Scholar]
- 6.Lomas MW, Moran SB. Evidence for aggregation and export of cyanobacteria and nano-eukaryotes from the Sargasso Sea euphotic zone. Biogeosciences. 2011;8:203–216. doi: 10.5194/bg-8-203-2011. [DOI] [Google Scholar]
- 7.Fawcett SE, Lomas MW, Casey JR, Ward BB, Sigman DM. Assimilation of upwelled nitrate by small eukaryotes in the Sargasso Sea. Nat Geosci. 2011;4(10):1–6. doi: 10.1038/ngeo1265. [DOI] [Google Scholar]
- 8.Lomas MW, et al. Increased ocean carbon export in the Sargasso Sea linked to climate variability is countered by its enhanced mesopelagic attenuation. Biogeosciences. 2010;7(1):57–70. [Google Scholar]
- 9.Altabet MA, Organic C. N, and stable isotopic composition of particulate matter collected on glass-fiber and aluminum oxide filters. Limnol Oceanogr. 1990;35:902–909. [Google Scholar]
- 10.Koike I, Shigemitsu H, Kazuki T, Kazuhiro K. Role of sub-micrometre particles in the ocean. Nature. 1990;345:242–244. [Google Scholar]
- 11.Burd AB, et al. Assessing the apparent imbalance between geochemical and biochemical indicators of meso- and bathypelagic biological activity: What the @$#! is wrong with present calculations of carbon budgets? Deep Sea Res Part II Top Stud Oceanogr. 2010;57:1557–1571. doi: 10.1016/j.dsr2.2010.02.022. [DOI] [Google Scholar]
- 12.Sharp JH. Size classes of organic carbon in seawater. Limnol Oceanogr. 1973;18:441–447. [Google Scholar]
- 13.Bishop JKB, Edmond JM. A new large volume filtration system for the sampling of oceanic particulate organic matter. J Mar Res. 1976;34(1):181–198. [Google Scholar]
- 14.Hernes PJ, Benner R. Transport and diagenesis of dissolved and particulate terrigenous organic matter in the North Pacific Ocean. Deep Sea Res Part I Oceanogr Res Pap. 2002;49:2119–2132. [Google Scholar]
- 15.Sannigrahi P, Ingall ED, Benner R. Cycling of dissolved and particulate organic matter at station Aloha: Insights from 13C NMR spectroscopy coupled with elemental, isotopic and molecular analyses. Deep Sea Res Part I Oceanogr Res Pap. 2005;52:1529–1544. [Google Scholar]
- 16.Kaiser K, Benner R. Major bacterial contribution to the ocean reservoir of detrital organic carbon and nitrogen. Limnol Oceanogr. 2008;53(1):99–112. [Google Scholar]
- 17.Kaiser K, Benner R. Biochemical composition and size distribution of organic matter at the Pacific and Atlantic time-series stations. Mar Chem. 2009;113(1):63–77. [Google Scholar]
- 18.Kawasaki N, Sohrin R, Ogawa H, Nagata T, Benner R. Bacterial carbon content and the living and detrital bacterial contributions to suspended particulate organic carbon in the North Pacific Ocean. Aquat Microb Ecol. 2011;62(2):165–176. doi: 10.3354/ame01462. [DOI] [Google Scholar]
- 19.Michaels AF, Silver MW. Primary production, sinking fluxes and the microbial food web. Deep-Sea Res. 1988;35:473–490. [Google Scholar]
- 20.Burd AB, Jackson GA. Particle aggregation. Annu Rev Mar Sci. 2009;1:65–90. doi: 10.1146/annurev.marine.010908.163904. [DOI] [PubMed] [Google Scholar]
- 21.Waite AM, Safi KA, Hall JA, Nodder SD. Mass sedimentation of picoplankton embedded in organic aggregates. Limnol Oceanogr. 2000;45(1):87–97. [Google Scholar]
- 22.Karl D, Lukas R. The Hawaii Ocean Time-series (HOT) program: Background, rationale and field implementation. Deep Sea Res Part II Top Stud Oceanogr. 1996;43:129–156. doi: 10.1016/0967-0645(96)00005-7. [DOI] [Google Scholar]
- 23.Ingalls AE, et al. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Natl Acad Sci USA. 2006;103(17):6442–6447. doi: 10.1073/pnas.0510157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wakeham S. Lipid biomarkers for heterotrophic alteration of suspended particulate organic matter in oxygenated and anoxic water columns of the ocean. Deep Sea Res Part I: Oceanogr Res Pap. 1995;42:1749–1771. [Google Scholar]
- 25.Tolosa I, et al. Distribution of pigments and fatty acid biomarkers in particulate matter from the frontal structure of the Alboran Sea (SW Mediterranean Sea) Mar Chem. 2004;88(3):103–125. doi: 10.1016/j.marchem.2004.03.005. [DOI] [Google Scholar]
- 26.DeSantis TZ, et al. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb Ecol. 2007;53(3):371–383. doi: 10.1007/s00248-006-9134-9. [DOI] [PubMed] [Google Scholar]
- 27.Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409(6819):507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- 28.Pernthaler A, Pernthaler J, Amann R. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl Environ Microbiol. 2002;68(6):3094–3101. doi: 10.1128/AEM.68.6.3094-3101.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teira E, Reinthaler T, Pernthaler A, Pernthaler J, Herndl GJ. Combining catalyzed reporter deposition-fluorescence in situ hybridization and microautoradiography to detect substrate utilization by bacteria and Archaea in the deep ocean. Appl Environ Microbiol. 2004;70(7):4411–4414. doi: 10.1128/AEM.70.7.4411-4414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeLong EF, et al. Community genomics among stratified microbial assemblages in the ocean’s interior. Science. 2006;311(5760):496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 31.Hansman RL, et al. The radiocarbon signature of microorganisms in the mesopelagic ocean. Proc Natl Acad Sci USA. 2009;106(16):6513–6518. doi: 10.1073/pnas.0810871106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Letelier RM, Dore JE, Winn CD, Karl DM. Seasonal and interannual variations in photosynthetic carbon assimilation at Station ALOHA. Deep Sea Res Part II Top Stud Oceanogr. 1996;43:467–490. [Google Scholar]
- 33.Meyers PA, Eadie BJ. Sources, degradation and recycling of organic matter associated with sinking particles in Lake Michigan. Org Geochem. 1993;20(1):47–56. [Google Scholar]
- 34.Canuel EA. Relations between river flow, primary production and fatty acid composition of particulate organic matter in San Francisco and Chesapeake Bays: A multivariate approach. Org Geochem. 2001;32:563–583. [Google Scholar]
- 35.Wakeham SG, et al. Compositions and transport of lipid biomarkers through the water column and surficial sediments of the equatorial Pacific Ocean. Deep Sea Res Part II Top Stud Oceanogr. 1997;44:2131–2162. [Google Scholar]
- 36.Niggemann J, Schubert CJ. Fatty biogeochemistry of sediments from the Chilean coastal upwelling region: Sources and diagenetic changes. Org Geochem. 2006;37:626–647. [Google Scholar]
- 37.DeNiro M, Epstein S. Influence of diet on the distribution of carbon isotopes in animals. Geochim Cosmochim Acta. 1978;42:495–506. [Google Scholar]
- 38.Blair N, et al. Carbon isotopic fractionation in heterotrophic microbial metabolism. Appl Environ Microbiol. 1985;50(4):996–1001. doi: 10.1128/aem.50.4.996-1001.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sinninghe Damsté JS, et al. Distribution of membrane lipids of planktonic Crenarchaeota in the Arabian Sea. Appl Environ Microbiol. 2002;68(6):2997–3002. doi: 10.1128/AEM.68.6.2997-3002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roland LA, McCarthy MD, Peterson TD, Walker BD. A large-volume microfiltration system for isolating suspended particulate organic matter: Fabrication and assessment versus GFF filters in central North Pacific. Limnol Oceanogr Methods. 2008;6:64–80. [Google Scholar]
- 41.Steinberg DK, et al. Bacterial vs. zooplankton control of sinking particle flux in the ocean’s twilight zone. Limnol Oceanogr. 2008;53:1327–1338. [Google Scholar]
- 42.Buesseler KO, et al. Revisiting carbon flux through the ocean’s twilight zone. Science. 2007;316(5824):567–570. doi: 10.1126/science.1137959. [DOI] [PubMed] [Google Scholar]
- 43.Lamborg C, et al. The flux of bio- and lithogenic material associated with sinking particles in the mesopelagic “twilight zone” of the northwest and North Central Pacific Ocean. Deep Sea Res Part II Top Stud Oceanogr. 2008;55:1540–1563. doi: 10.1016/j.dsr2.2008.04.011. [DOI] [Google Scholar]
- 44.Cho B, Azam F. Major role of bacteria in biogeochemical fluxes in the ocean’s interior. Nature. 1988;332:441–443. [Google Scholar]
- 45.Arístegui J, Gasol JM, Duarte CM, Herndl G. Microbial oceanography of the dark ocean’s pelagic realm. Limnol Oceanogr. 2009;54:1501–1529. [Google Scholar]
- 46.Benitez-Nelson C, Buesseler KO, Karl DM, Andrews J. A time-series study of particulate matter export in the North Pacific Subtropical Gyre based on Th-234:U-238 disequilibrium. Deep Sea Res Part I Oceanogr Res Pap. 2001;48:2595–2611. [Google Scholar]
- 47.Buesseler KO, Boyd PW. Shedding light on processes that control particle export and flux attenuation in the twilight zone of the open ocean. Limnol Oceanogr. 2009;54:1210–1232. [Google Scholar]
- 48.Fuhrman JA. Bacterioplankton roles in cycling of organic matter: The microbial food web. In: Falkowski PG, Woodhead AD, editors. Primary Productivity and Biogeochemical Cycles in the Sea. New York: Plenum; 1992. pp. 361–382. [Google Scholar]
- 49.Loh AN, Canuel EA, Bauer JE. Potential source and diagenetic signatures of oceanic dissolved and particulate organic matter as distinguished by lipid biomarker distributions. Mar Chem. 2008;112(3):189–202. [Google Scholar]
- 50.Jiao N, et al. Microbial production of recalcitrant dissolved organic matter: Long-term carbon storage in the global ocean. Nat Rev Microbiol. 2010;8(8):593–599. doi: 10.1038/nrmicro2386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.