Abstract
Spiny-rayed fishes, or acanthomorphs, comprise nearly one-third of all living vertebrates. Despite their dominant role in aquatic ecosystems, the evolutionary history and tempo of acanthomorph diversification is poorly understood. We investigate the pattern of lineage diversification in acanthomorphs by using a well-resolved time-calibrated phylogeny inferred from a nuclear gene supermatrix that includes 520 acanthomorph species and 37 fossil age constraints. This phylogeny provides resolution for what has been classically referred to as the “bush at the top” of the teleost tree, and indicates acanthomorphs originated in the Early Cretaceous. Paleontological evidence suggests acanthomorphs exhibit a pulse of morphological diversification following the end Cretaceous mass extinction; however, the role of this event on the accumulation of living acanthomorph diversity remains unclear. Lineage diversification rates through time exhibit no shifts associated with the end Cretaceous mass extinction, but there is a global decrease in lineage diversification rates 50 Ma that occurs during a period when morphological disparity among fossil acanthomorphs increases sharply. Analysis of clade-specific shifts in diversification rates reveal that the hyperdiversity of living acanthomorphs is highlighted by several rapidly radiating lineages including tunas, gobies, blennies, snailfishes, and Afro-American cichlids. These lineages with high diversification rates are not associated with a single habitat type, such as coral reefs, indicating there is no single explanation for the success of acanthomorphs, as exceptional bouts of diversification have occurred across a wide array of marine and freshwater habitats.
Keywords: Actinopterygii, Cichlidae, Percomorpha, Teleostei
With more than 18,000 species, acanthomorph fishes comprise almost one-third of living vertebrates (1–3). Acanthomorphs, or spiny-rayed fishes, are present in nearly all marine and freshwater habitats from tropical coral reefs, freezing waters around Antarctica, alpine mountain lakes, and hadal trenches in the deepest parts of the world’s oceans known to harbor animal life (2). Acanthomorphs exhibit substantial morphological disparity, with body plans as diverse as flatfishes, pufferfishes, swordfishes, seahorses, and flyingfishes (3), and adult body sizes that range from among the smallest of all vertebrates to the largest bony fishes (4, 5). In addition, acanthomorphs include some of the most economically and scientifically important fish species, such as cods, tunas, sticklebacks, and cichlids. Investigating the processes that have resulted in the success of acanthomorphs is key to understanding the origins of the rich biodiversity of living vertebrates.
Resolution of the phylogenetic relationships among acanthomorphs, particularly the species-rich Percomorpha, has ranked among the most important and vexing problems in vertebrate biology (3, 6–9). High species richness in particular has posed a challenge to investigating relationships of the group. For example, Percomorpha contains more than 17,000 species, has been shown to exhibit exceptional species richness relative to other lineages of jawed vertebrates (10, 11), and is described as “a bush at the top” of the teleost tree in reference to the historical lack of resolution of its basic phylogenetic intrarelationships (12). Studies using morphological and molecular datasets offer alternative perspectives on the evolutionary relationships of acanthomorphs (6, 8, 13–15). Molecular phylogenies support the monophyly of several traditionally recognized taxonomic groups of percomorphs as well as the resolution of several novel clades (13, 14, 16–19); however, a consistent and well-supported resolution of the phylogenetic relationships among these major groups has eluded the scientific community.
In addition to the uncertainty surrounding phylogenetic relationships, there are conflicting observations from molecular phylogenies and the fossil record regarding the temporal history of acanthomorph diversification (20). Recent analyses of lineage diversification using time-calibrated molecular phylogenies identified an unnamed subclade of percomorphs, with an estimated age of ∼110 Ma (9–11, 19), as exhibiting a shift to an exceptionally high lineage diversification rate (10, 11). In contrast, acanthomorph and percomorph species preserved in the fossil record show relatively little morphological disparity through the Late Cretaceous, but extensive expansion of morphospace from the Cretaceous–Paleogene (K-Pg) boundary to the early Eocene (7). The signature of increased morphological disparity in the fossil record of acanthomorphs was interpreted in the context of recovery from the K-Pg mass extinction event (7). These two results indicate that the timing of a shift to an increased lineage diversification rate in acanthomorphs is at least 45 My older than the expansion of morphological disparity. However, the taxon sampling in the molecular time tree analyses were designed to assess patterns of diversification among more inclusive vertebrate lineages, and did not target specific acanthomorph clades that may have diversified around the K-Pg (10, 11). Despite these important observations, it is not known how the K-Pg mass extinction event affected the accumulation of lineage diversity in acanthomorphs, or if there are global shifts in rates of lineage origination coincident with the increase of morphological disparity following the K-Pg.
We investigated the timing and tempo of acanthomorph diversification by using a time-calibrated molecular phylogeny inferred by using 10 nuclear genes sampled from 579 species that included 520 acanthomorphs, representing all major lineages and traditionally recognized taxonomic orders and suborders (2). The analyses of diversification rates through time in acanthomorphs are analyzed in the context of the K-Pg mass extinction event and the timing of morphological expansion detected in the fossil record. We also investigate clade-specific shifts in diversification rates, which identify several percomorph lineages that exhibit exceptionally high rates of lineage accumulation. Our results provide a phylogenetic and temporal perspective from which to investigate the evolutionary processes that resulted in the origin of one of the most species rich clades of living vertebrates.
Results and Discussion
The supermatrix of 10 combined nuclear genes is 84.5% complete, with the proportion of missing taxa among the 10 genes ranging from 3.6% for patched domain containing 4 (Ptr) to 36.5% for super conserved receptor 2 (sreb2). The aligned DNA sequences are available from the Dryad Digital Repository (DOI 10.5061/dryad.d3mb4). Maximum likelihood and Bayesian analyses resulted in well-resolved and consistent phylogenetic trees in which more than 78% of the nodes in the maximum likelihood tree are supported with bootstrap values (BS) ≥70% (Fig. 1A and Figs. S1 and S2). Similar to separate analyses of nine of the 10 individual nuclear genes (9), the acanthomorph molecular phylogeny resolves Paracanthopterygii as polyphyletic with a clade (BS = 91%) containing Polymixia and the trout-perches (Percopsiformes) as the earliest diverging lineage of acanthomorphs. A clade uniting Zeiformes (dories), Stylephorus (tube-eye), and Gadiformes (cods) is resolved with strong support (BS = 100%). Previous phylogenetic analyses had resolved Lampriformes (opahs) as the sister lineage of all other acanthomorphs (8, 15, 21, 22), but our molecular phylogeny places the clade in a more derived position (BS = 99%) as the sister lineage of Acanthopterygii or the clade containing dories, tube-eyes, and cods (Fig. 1B and Fig. S1). Beryciformes (squirrelfishes and pricklefishes) is monophyletic and resolved as the sister lineage of Percomorpha (BS = 79%), a result in agreement with assessments using whole mtDNA genomes and nuclear genes (9, 13), but divergent from morphological and molecular studies that support beryciform paraphyly (8, 19, 23).
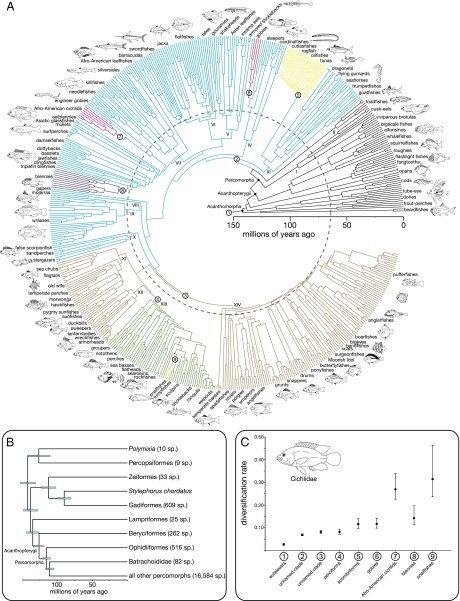
Fig. 1.
Time-calibrated phylogenetic tree of Acanthomorpha and results of lineage diversification rate analyses. (A) Bayesian inferred maximum clade credibility time tree of 520 acanthomorph species calibrated with 37 fossil-based age constraints. Branch colors and circled numbers identify the nine best-fitting lineage diversification regimes identified using MEDUSA. The dashed gray circle at 66 Ma denotes the K-Pg boundary. The 14 major percomorph clades delimited in this phylogeny are labeled with Roman numerals (I–XIV). Maximum likelihood bootstrap and Bayesian posterior clade support values are given in Figs. S1 and S2. (B) Time-calibrated phylogeny detailing the earliest divergences within Acanthomorpha. Bars at nodes represent 95% highest posterior density intervals of age estimates, with light blue bars showing Bayesian posterior clade support of 0.95 or greater. (C) Net diversification rate (birth rate minus death rate) estimates with 95% CIs of the nine identified lineage diversification regimes. Colors and circled numbers correspond to clades denoted in Fig. 1A. Fish line drawings are by A.D., K. L. Tang, and W.L.S.
Our phylogenetic analyses provide well-supported resolution of relationships among major lineages of the species-rich Percomorpha, which has remained the last frontier in the phylogeny of living vertebrates (2, 3). This phylogeny resolves all sampled percomorphs into 14 major lineages with BS >75%, some of which are incongruent with current taxonomic classifications (2, 19, 24). Percomorph lineages discovered in previous molecular analyses are supported by our phylogenies (6, 9, 13, 14, 19), including the early divergence of cusk-eels (Ophidiiformes) and toadfishes (Batrachoidiformes; Fig. 1B), a clade uniting anglerfishes and pufferfishes, as well as Ovalentaria, a clade containing cichlids, engineer gobies (Pholidichthys), silversides, livebearers, and ricefish (Atherinomorpha), surfperches (Embiotocidae), damselfishes (Pomacentridae), mullets (Mugilidae), clingfishes (Gobiesocidae), and blennies (6). Our results also support a clade uniting swamp eels (Synbranchiformes), armored sticklebacks (Indostomus), Asian leaffishes (Nandidae), gouramies and snakeheads (Anabantomorpha), flatfishes (Pleuronectiformes), jacks (Carangidae), sharksuckers (Echeneidae), billfishes (Xiphioidei), and barracudas (Sphyraena), as well as a clade containing temperate basses (Moronidae) and a large collection of marine percomorphs that include butterflyfishes (Chaetodontidae), grunts (Haemulidae), angelfishes (Pomacanthidae), anglerfishes (Lophiiformes), and pufferfishes (Tetraodontiformes; Fig. 1A and Figs. S1 and S2).
In addition to providing much-needed resolution for one of the most unyielding problems in vertebrate phylogenetics, calibrating the molecular phylogeny with the teleost fossil record provides important insights into the timing and tempo of acanthomorph diversification. A set of time-calibrated phylogenies were estimated using Bayesian methods and 37 well-justified fossil-based prior age constraints that included 33 acanthomorph taxa, of which 24 were percomorphs (SI Text), ranging from Early Cretaceous (∼134 Ma) to Miocene (∼12 Ma) in age. The estimated ages of Acanthomorpha, between 133 and 152 Ma, and Percomorpha, between 109 and 120 Ma (Table S1), are consistent with those implied by the fossil record and previous molecular clock analyses (7, 9–11, 25). Major nonpercomorph acanthomorph lineages, such as the Beryciformes, originate in the Cretaceous (Figs. 1 A and B and 2A). However, we find no clear signature of lineage origination centered on the K-Pg because many percomorph lineages predate or postdate the age of the mass extinction event (Figs. 1A and 2A and Table S1). For example, the species rich clades Gobiiformes, Syngnathiformes, and Ovalentaria all originate well before the K-Pg, between 91.4 and 88.1 Ma, whereas Perciformes sensu stricto, Centrarchiformes, and Scombriformes diversified after the K-Pg between 54.6 and 34.5 Ma (Fig. 2A and Table S1).
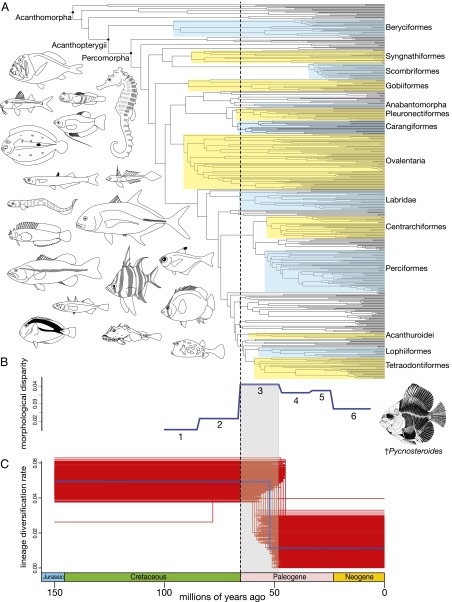
Fig. 2.
Temporal scale of acanthomorph diversification during the past 140 My. The dashed line at 66 Ma denotes the K-Pg boundary. (A) Time-calibrated phylogenetic tree of Acanthomorpha with select major clades highlighted with alternating blue and yellow boxes that correspond with group names. (B) Morphological disparity of acanthomorph fossil taxa measured in six numbered stratigraphic bins that extend from the Cretaceous through the Neogene. The drawing of the Cretaceous fossil taxon †Pycnosteroides is taken from ref. 7. (C) Maximum-likelihood BDS estimates of diversification rates. The blue line represents the diversification rate estimated from the maximum clade credibility time tree (Fig. 1A), and the red lines denote the diversification rate estimates from BDS analyses across 1,000 posterior time trees. Fish line drawings are by A.D. and K. L. Tang.
Similar to patterns of lineage origination, there is no correlation of the K-Pg with the ages of percomorph lineages that exhibit elevated rates of diversification. By using a diversity tree that contained species richness information for all unsampled lineages and a stepwise information theoretic approach to detect shifts in lineage diversification rates (10), we identify eight clades that exhibit higher rates of lineage diversification relative to the background rate of euteleost fishes (Fig. 1 A and C and Table S2). Two of the detected shifts occur at unnamed percomorph clades that predate the K-Pg, including one that was identified as exceptionally diverse in a previous analysis that sampled all major lineages of jawed vertebrates (10). Within these two unnamed percomorph lineages, there are five inclusive clades of percomorphs that exhibit exceptionally high rates of lineage diversification (Fig. 1A and Table S2). However, each is younger than the K-Pg, some by as much as 45 My (Fig. 1A and C). Among these five clades, gobies, blennies, and Afro-American cichlids have been proposed to contain unusually high species diversity (2, 26, 27); however, exceptional lineage diversification rates have never been detected in tunas (Scombriformes) and snailfishes (Liparidae).
We used a maximum likelihood-based birth-death-shift (BDS) model to explicitly test for global changes in the diversification of acanthomorph lineages through time (28). The BDS analysis does not detect a global shift in lineage diversification of acanthomorphs at the K-Pg, but instead supports a model of constant lineage origination from their origin, through the K-Pg, and into the Paleogene (Fig. 2C). The diversification rate of acanthomorphs declined globally at ∼50 Ma. In contrast, previous analyses show that morphological disparity of acanthomorph fossil taxa was low through the Cretaceous (Fig. 2B), but increased significantly in the time interval from the K-Pg to the early Eocene (∼48 Ma). This increase in disparity involved expansion into regions of morphospace previously occupied by nonacanthomorph teleost lineages, such as pachycormiforms, ichthyodectiforms, and enchodontids, which all make last appearances in the fossil record shortly before the K-Pg. (7, 29). However, the decline in diversification rates in the Eocene is coincident with the age of stratigraphic bin 3 (Fig. 2B), bounded by the K-Pg and the early Eocene, in which morphological disparity of fossil acanthomorphs increases significantly following the K-Pg and does not change appreciably over the younger bins (7) (Fig. 2 B and C). The temporal congruence between the increase in morphological disparity and decrease in lineage diversification rates may reflect the results of a rapid filling of vacant niches by acanthomorphs in the Paleogene, and eventual tapering off of lineage diversification in response to limited ecological opportunities following a period of rapid morphological and ecological expansion (30).
The lack of a detected shift in lineage diversification rates of acanthomorphs through the K-Pg boundary indicates that the patterns of species richness among one-third of all living vertebrates were not entirely a result of recovery from the K-Pg mass extinction event (Fig. 2C). The end Cretaceous mass extinction has been interpreted as a key event in the evolution of terrestrial vertebrates such as snakes, lizards, birds, and mammals (31–34); however, analyses of time-calibrated molecular phylogenies show that many major clades of birds and mammals predate the K-Pg and do not exhibit pulses of diversification around 66 Ma (35–37). These results, along with our estimates of Cretaceous origins for many species-rich percomorph clades and constant lineage diversification of acanthomorphs across the K-Pg, suggest that much of the living biodiversity of aquatic vertebrates was not generated solely in response to the K-Pg mass extinction event.
The phylogenetic resolution of all major acanthomorph and percomorph lineages presented in this study offers insight into the evolutionary relationships of nearly one-third of all vertebrate species, as well as revealing that the currently accepted classification for these fishes is dramatically discordant with the inferred phylogeny (2, 24). Previous explanations for the species richness of acanthomorphs have focused on a narrow set of marine habitats, namely coral reefs, as driving diversification (38, 39). Our temporal analyses reveal that the five percomorph lineages with the highest rates of diversification comprise more than 5,400 species (Fig. 1C and Table S2) and occupy diverse habitat types that include freshwater (cichlids), pelagic oceanic (tunas and allies), cold temperate seafloor (snailfishes), and coral reefs (blennies and gobies). Although specific habitat types may drive exceptional lineage diversification in teleosts (38, 40), our results suggest that acanthomorphs generally, and percomorphs specifically, have radiated into a wide range of freshwater and marine habitat types, many of which provided opportunities for exceptional diversification and species richness.
Materials and Methods
Phylogenetic Data and Analyses.
Standard phenol-chloroform extraction protocol or Qiagen DNeasy Blood and Tissue kits were used to isolate DNA from tissue biopsies sampled from 579 teleost species (Table S3). Previously published PCR primers (41, 42) were used to amplify and sequence a single exon from each of 10 unlinked nuclear encoded genes: ENC1, Glyt, myh6, plagl2, Ptr, rag1, SH3PX3, sreb2, tbr1, and zic1. These 10 protein coding gene regions were aligned by using the inferred amino acid sequences. No frame mutations or DNA substitutions that resulted in stop codons were observed in the aligned sequences. The combined 10-gene dataset contained 8,577 bp. Seven sampled ostariophysan species were used as outgroup taxa in all phylogenetic analyses (Table S3).
Thirty data partitions were designated that corresponded to the three separate codon positions for each of the 10 protein coding genes. A GTR+G substitution model was used in a partitioned maximum likelihood analysis using the computer program RAxML 7.2.6 (43), run with the –D option, and 1,000 maximum likelihood searches. Support for nodes in the RAxML tree was assessed with a thorough bootstrap analysis (option –f i) with 1,000 replicates.
Relaxed-Molecular Clock Analyses.
Divergence times of the sampled teleost species were estimated by using an uncorrelated lognormal (UCLN) model of molecular evolutionary rate heterogeneity implemented in the computer program BEAST, version 1.6.1 (44, 45). The 10-gene dataset was partitioned as in the maximum likelihood phylogenetic analysis described earlier, unlinking the nucleotide substitution models among the 30 codon-based partitions and UCLN clock model among the 10 genes. Thirty-seven calibration priors from the fossil record of teleost fishes were used in the UCLN analyses (SI Text).
The BEAST analyses were run four times with each run consisting of 1.0 × 109 generations, sampling every 10,000 generations. The resulting trees and log files from each of the four runs were combined by using the computer program LogCombiner, version 1.6.1 (http://beast.bio.ed.ac.uk/LogCombiner). Convergence of model parameter values and node-height estimates was assessed by plotting the marginal posterior probabilities versus the generation state in the computer program Tracer, version 1.5 (http://beast.bio.ed.ac.uk/Tracer). Effective sample size values were calculated for each parameter to ensure adequate mixing of the Markov chain Monte Carlo. All effective sample size values were greater than 200 (Table S4). The posterior probability density of the combined tree and log files was summarized as a maximum clade credibility tree by using TreeAnnotator, version 1.6.1 (http://beast.bio.ed.ac.uk/TreeAnnotator). The posterior probabilities of inferred clades and the mean and 95% highest posterior density interval of divergence time estimates were visualized by using the computer program FigTree, version 1.3.1 (http://beast.bio.ed.ac.uk/FigTree).
Assessing Patterns of Lineage Diversification.
To determine if any acanthomorph lineages exhibit shifts in their diversification rate relative to a background rate of euteleost lineage diversification, we used a stepwise information theoretic approach, MEDUSA, to incrementally fit increasingly complex models of lineage diversification to a diversity tree that contained information on species richness for all unsampled lineages (10). The diversity tree was constructed by collapsing lineages with missing taxa and assigning a species richness value from the Catalog of Fishes (1) to these resulting stem lineages (Table S5). We calculated the fit, based on the Akaike information criterion (AIC), of incrementally adding two-parameter birth-death or single-parameter Yule (pure birth) models of diversification with an associated shift point to the time-calibrated diversity tree. We repeated the model selection process using the stepwise function, retaining the more complex parameter rich model and comparing its fit to a model that includes an additional model of cladogenesis and shift point parameter, until the iterative model building process no longer offered an improvement in AIC score.
To estimate diversification rate and shifts over time among living acanthomorph lineages, we used the TreePar package (28) in R (46). The euteleost diversity tree constructed for the MEDUSA analysis described earlier was used to account for unsampled lineages (Table S5), but all nonacanthomorph lineages were pruned from this phylogeny. We calculated AIC values for models estimating zero to six shifts in the rate of lineage diversification over time at 1-My intervals ranging from 40 to 141.9 Ma (mean crown age estimate for acanthomorphs). The lowest AIC supported a model with a single rate shift at ∼50 Ma (ΔAIC for 0–1 shifts, 18.674). We also estimated the 95% highest posterior density in magnitude and timing of rate shifts by using a wrapper for TreePar to include 1,000 phylogenies sampled randomly from the posterior distribution of BEAST inferred time-calibrated acanthomorph phylogenies (Fig. 2C).
Morphometric Analysis and Measurement of Disparity.
Patterns of morphological diversification in fossil Acanthomorpha shown in Fig. 2B are taken from a previous publication (7). However, we include a brief description of analytical protocols. A series of 22 landmarks were placed on each of 1,336 images representing photographs or specimen drawings of whole-body acanthomorph fossils. Landmarks were digitized by using the software tpsDig (http://life.bio.sunysb.edu/morph/morphmet/tpsdig2w32.exe). Fourteen fixed landmarks were tied to specific anatomical features, such as the position of fin insertions or the lower jaw joint. Eight sliding semilandmarks were distributed evenly across the dorsal and ventral margins of the body to capture the curvature of overall shape. This landmark scheme corresponds closely to those applied in studies of shape-based variation in extant fishes (47). Taphonomically distorted specimens were avoided, but landmark constellations for individuals showing smooth curvature along the spine resulting from postmortem muscle contraction were retrodeformed by using the software package tpsUtil (http://life.bio.sunysb.edu/morph/morphmet/tpsutilw32.exe). The 1,336 images contained examples of 605 nominal species, so landmark coordinates for congeneric specimens were averaged. Landmark constellations were subject to Procrustes alignment and relative warps analysis by using the software package tpsRelw (http://life.bio.sunysb.edu/morph/morphmet/tpsrelww32.exe).
Only those relative warp axes explaining more than 5% of overall variance were retained for subsequent disparity analysis. Species were divided into six composite stratigraphic bins of more comparable duration than standard stage- or epoch-level chronostratigraphic units, and which allowed for the placement of taxa of somewhat imprecisely constrained ages. These bins are as follows: the early Late Cretaceous (Cenomanian–Santonian; duration, 16.9 My), the late Late Cretaceous (Campanian–Maastrichtian; duration, 17.6 My), the Paleocene–early Eocene (Danian–Ypresian; duration, 18.2 My), the middle-late Eocene (Lutetian–Priabonian; duration, 13.9 My), the Oligocene (Rupelian–Chattian; duration, 10.87 My), and the Miocene (Aquitanian–Messinian; duration, 17.697 My). Morphological variation, or “disparity,” within each of these time bins was calculated as the sum of variances on retained morphospace axes (multivariate variance). In contrast to other measures of disparity, multivariate variance has the desirable property of being relatively insensitive to variation in sample size (48).
Supplementary Material
Acknowledgments
We thank K.-T. Shao (Biodiversity Research Museum, Academia Sinica); J. Friel (Cornell University Museum of Vertebrates); P.A. Hastings and H.J. Walker (Scripps Institution of Oceanography); K.P. Maslenikov and T. W. Pietsch (Burke Museum of Natural History and Culture, University of Washington); L.M. Page and R. Robbins (Florida Museum of Natural History); C.D. Roberts and A. Stewart (Te Papa Museum of New Zealand); and A.C. Bentley and E.O. Wiley (Biodiversity Institute of the University of Kansas) for gifts of tissue specimens; T.M. Berra, C.D. Hulsey, M. Miya, and P.J. Unmack for additional specimens; J.W. Brown for making available the latest version of MEDUSA; T. Stadler for assistance with our use of TreePar; and G. Watkins-Colwell for assistance with museum collections. This work was supported by the Peabody Museum of Natural History; National Science Foundation Grants DEB-0444842, DEB-0716155, DEB-0717009, DEB-0732642, ANT-0839007, DEB-1060869, DEB-1061806, and DEB-1061981; and Natural Environment Research Council Grants NE/I005536/1 and NE/J022632/1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KF139346–KF141634).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304661110/-/DCSupplemental.
References
- 1. Eschmeyer WN, Fricke R, eds (2012) Catalog of Fishes, Electronic Version (12 Jan 2012) (California Academy of Sciences, San Francisco). Available at http://research.calacademy.org/research/ichthyology/catalog/fishcatmain.asp. Accessed February 20, 2012.
- 2.Nelson JS. Fishes of the World. 4th Ed. Hoboken, NJ: Wiley; 2006. [Google Scholar]
- 3. Stiassny MLJ, Wiley EO, Johnson GD, de Carvalho MR (2004) Gnathostome fishes. Assembling the Tree of Life, eds Cracraft J, Donoghue MJ (Oxford Univ Press, New York), pp 410–429.
- 4.Watson W, Walker HJ. The world’s smallest vertebrate, Schindleria brevipinguis, a new paedomorphic species in the family Schindleriidae (Perciformes: Gobioidei) Rec Aust Mus. 2004;56:139–142. [Google Scholar]
- 5.Helfman GS, Collette BB, Facey DE, Bowen BW. The Diversity of Fishes. Chichester, UK: Wiley; 2009. [Google Scholar]
- 6.Wainwright PC, et al. The evolution of pharyngognathy: A phylogenetic and functional appraisal of the pharyngeal jaw key innovation in labroid fishes and beyond. Syst Biol. 2012;61(6):1001–1027. doi: 10.1093/sysbio/sys060. [DOI] [PubMed] [Google Scholar]
- 7.Friedman M. Explosive morphological diversification of spiny-finned teleost fishes in the aftermath of the end-Cretaceous extinction. Proc Biol Sci. 2010;277(1688):1675–1683. doi: 10.1098/rspb.2009.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson GD, Patterson C. Percomorph phylogeny: A survey of acanthomorphs and a new proposal. Bull Mar Sci. 1993;52(1):554–626. [Google Scholar]
- 9.Near TJ, et al. Resolution of ray-finned fish phylogeny and timing of diversification. Proc Natl Acad Sci USA. 2012;109(34):13698–13703. doi: 10.1073/pnas.1206625109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alfaro ME, et al. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proc Natl Acad Sci USA. 2009;106(32):13410–13414. doi: 10.1073/pnas.0811087106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santini F, Harmon LJ, Carnevale G, Alfaro ME. Did genome duplication drive the origin of teleosts? A comparative study of diversification in ray-finned fishes. BMC Evol Biol. 2009;9:194. doi: 10.1186/1471-2148-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nelson G (1989) Phylogeny of major fish groups. The Hierarchy of Life: Molecules and Morphology in Phylogenetic Analysis, eds Fernholm B, Bremer K, Jôrnvall H (Elsevier, Amsterdam), pp 325–336.
- 13.Miya M, Satoh TR, Nishida M. The phylogenetic position of toadfishes (order Batrachoidiformes) in the higher ray-finned fish as inferred from partitioned Bayesian analysis of 102 whole mitochondrial genome sequences. Biol J Linn Soc Lond. 2005;85(3):289–306. [Google Scholar]
- 14.Li B, et al. RNF213, a new nuclear marker for acanthomorph phylogeny. Mol Phylogenet Evol. 2009;50(2):345–363. doi: 10.1016/j.ympev.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 15.Smith WL, Wheeler WC. Venom evolution widespread in fishes: A phylogenetic road map for the bioprospecting of piscine venoms. J Hered. 2006;97(3):206–217. doi: 10.1093/jhered/esj034. [DOI] [PubMed] [Google Scholar]
- 16.Kawahara R, et al. Interrelationships of the 11 gasterosteiform families (sticklebacks, pipefishes, and their relatives): A new perspective based on whole mitogenome sequences from 75 higher teleosts. Mol Phylogenet Evol. 2008;46(1):224–236. doi: 10.1016/j.ympev.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 17.Setiamarga DHE, et al. Interrelationships of Atherinomorpha (medakas, flyingfishes, killifishes, silversides, and their relatives): The first evidence based on whole mitogenome sequences. Mol Phylogenet Evol. 2008;49(2):598–605. doi: 10.1016/j.ympev.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 18.Lautredou A-C, et al. New nuclear markers and exploration of the relationships among Serraniformes (Acanthomorpha, Teleostei): The importance of working at multiple scales. Mol Phylogenet Evol. 2013;67(1):140–155. doi: 10.1016/j.ympev.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 19.Betancur-R R, et al. The tree of life and a new classification of bony fishes. PLoS Curr. 2013;5(Apr):18. doi: 10.1371/currents.tol.53ba26640df0ccaee75bb165c8c26288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alfaro M, Santini F. Evolutionary biology: A flourishing of fish forms. Nature. 2010;464(7290):840–842. doi: 10.1038/464840a. [DOI] [PubMed] [Google Scholar]
- 21.Wiley EO, David Johnson G, Wheaton Dimmick W. The interrelationships of Acanthomorph fishes: A total evidence approach using molecular and morphological data. Biochem Syst Ecol. 2000;28(4):319–350. doi: 10.1016/s0305-1978(99)00069-1. [DOI] [PubMed] [Google Scholar]
- 22.Stiassny MLJ, Moore JA. A review of the pelvic girdle of acanthomorph fishes, with comments on hypotheses of acanthomorph intrarelationships. Zool J Linn Soc. 1992;104:209–242. [Google Scholar]
- 23.Moore JA. Phylogeny of the Trachichthyiformes (Teleostei: Percomorpha) Bull Mar Sci. 1993;52(1):114–136. [Google Scholar]
- 24. Wiley EO, Johnson GD (2010) A teleost classification based on monophyletic groups. Origin and Phylogenetic Interrelationships of Teleosts, eds Nelson JS, Schultze H-P, Wilson MVH (Verlag Dr. Friedrich Pfeil, Munich), pp 123–182.
- 25.Patterson C. An overview of the early fossil record of acanthomorphs. Bull Mar Sci. 1993;52(1):29–59. [Google Scholar]
- 26.Day JJ, Cotton JA, Barraclough TG. Tempo and mode of diversification of lake Tanganyika cichlid fishes. PLoS ONE. 2008;3(3):e1730. doi: 10.1371/journal.pone.0001730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.López-Fernández H, Arbour JH, Winemiller KO, Honeycutt RL. Testing for ancient adaptive radiations in neotropical cichlid fishes. Evolution. 2013;67(5):1321–1337. doi: 10.1111/evo.12038. [DOI] [PubMed] [Google Scholar]
- 28.Stadler T. Mammalian phylogeny reveals recent diversification rate shifts. Proc Natl Acad Sci USA. 2011;108(15):6187–6192. doi: 10.1073/pnas.1016876108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman M. Ecomorphological selectivity among marine teleost fishes during the end-Cretaceous extinction. Proc Natl Acad Sci USA. 2009;106(13):5218–5223. doi: 10.1073/pnas.0808468106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harmon LJ, Schulte JA, 2nd, Larson A, Losos JB. Tempo and mode of evolutionary radiation in iguanian lizards. Science. 2003;301(5635):961–964. doi: 10.1126/science.1084786. [DOI] [PubMed] [Google Scholar]
- 31.Ericson PGP, et al. Diversification of Neoaves: Integration of molecular sequence data and fossils. Biol Lett. 2006;2(4):543–547. doi: 10.1098/rsbl.2006.0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wible JR, Rougier GW, Novacek MJ, Asher RJ. Cretaceous eutherians and Laurasian origin for placental mammals near the K/T boundary. Nature. 2007;447(7147):1003–1006. doi: 10.1038/nature05854. [DOI] [PubMed] [Google Scholar]
- 33.Longrich NR, Bhullar B-AS, Gauthier JA. Mass extinction of lizards and snakes at the Cretaceous-Paleogene boundary. Proc Natl Acad Sci USA. 2012;109(52):21396–21401. doi: 10.1073/pnas.1211526110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Longrich NR, Tokaryk T, Field DJ. Mass extinction of birds at the Cretaceous-Paleogene (K-Pg) boundary. Proc Natl Acad Sci USA. 2011;108(37):15253–15257. doi: 10.1073/pnas.1110395108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown JW, Rest JS, García-Moreno J, Sorenson MD, Mindell DP. Strong mitochondrial DNA support for a Cretaceous origin of modern avian lineages. BMC Biol. 2008;6:6. doi: 10.1186/1741-7007-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meredith RW, et al. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science. 2011;334(6055):521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- 37.Bininda-Emonds ORP, et al. The delayed rise of present-day mammals. Nature. 2007;446(7135):507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 38.Alfaro ME, Santini F, Brock CD. Do reefs drive diversification in marine teleosts? Evidence from the pufferfishes and their allies (Order Tetraodontiformes) Evolution. 2007;61(9):2104–2126. doi: 10.1111/j.1558-5646.2007.00182.x. [DOI] [PubMed] [Google Scholar]
- 39. Bellwood DR, Wainwright PC (2002) The history and biogeography of fishes on coral reefs. Coral Reef Fishes: Dynamics and Diversity in a Complex Ecosystem, ed Sale PF (Academic, Boston), pp 5–32.
- 40.Cowman PF, Bellwood DR. Coral reefs as drivers of cladogenesis: Expanding coral reefs, cryptic extinction events, and the development of biodiversity hotspots. J Evol Biol. 2011;24(12):2543–2562. doi: 10.1111/j.1420-9101.2011.02391.x. [DOI] [PubMed] [Google Scholar]
- 41.Li CH, Ortí G, Zhang G, Lu GQ. A practical approach to phylogenomics: The phylogeny of ray-finned fish (Actinopterygii) as a case study. BMC Evol Biol. 2007;7:44. doi: 10.1186/1471-2148-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopez JA, Chen WJ, Ortí G. Esociform phylogeny. Copeia. 2004;(3):449–464. [Google Scholar]
- 43.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 44.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4(5):e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2012. [Google Scholar]
- 47.Chakrabarty P. Testing conjectures about morphological diversity in cichlids of lakes Malawi and Tanganyika. Copeia. 2005;(2):359–373. [Google Scholar]
- 48.Ciampaglio CN, Kemp M, McShea DW. Detecting changes in morphospace occupation patterns in the fossil record: Characterization and analysis of measures of disparity. Paleobio. 2001;27(4):695–715. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.